Abstract
Shotgun proteomics using mass spectrometry is a powerful method for protein identification but suffers limited sensitivity in complex samples. Integrating peptide identifications from multiple database search engines is a promising strategy to increase the number of peptide identifications and reduce the volume of unassigned tandem mass spectra. Existing methods pool statistical significance scores such as p-values or posterior probabilities of peptide-spectrum matches (PSMs) from multiple search engines after high scoring peptides have been assigned to spectra, but these methods lack reliable control of identification error rates as data are integrated from different search engines. We developed a statistically coherent method for integrative analysis, termed MSblender. MSblender converts raw search scores from search engines into a probability score for all possible PSMs and properly accounts for the correlation between search scores. The method reliably estimates false discovery rates and identifies more PSMs than any single search engine at the same false discovery rate. Increased identifications increment spectral counts for all detected proteins and allow quantification of proteins that would not have been quantified by individual search engines. We also demonstrate that enhanced quantification contributes to improve sensitivity in differential expression analyses.
Keywords: integrative analysis, database search, peptide identification
Introduction
Analyses of mass spectrometry-based shotgun proteomics data rely heavily upon computational algorithms for automating peptide identification via database searching. Database search engines assign each tandem mass spectrum to the best-scoring peptide sequence in the database based on scoring functions using spectral features1–7. Several different search engines are available today, and peptides identified with high confidence often show good consensus across different engines8. Nevertheless, many high-quality MS/MS spectra remain unassigned to peptide sequences or have scores below chosen confidence thresholds9. Moreover, some spectra may be assigned to different peptides by different search engines, which vary in their scoring schemes4, 10. Provided that these issues are properly addressed, pooling peptide identifications from multiple search engines is expected to improve peptide identifications and to leave fewer mass spectra without assignment to peptide sequences.
To date, a few computational approaches have been proposed for integrating database search results. Alves et al. proposed a calibration of p-values from multiple search engines into a meta-analytic p-value for each peptide8. Searle et al. proposed a Bayes approach to adjust probability scores computed in individual search engines based on the agreement between search engines, in which the largest adjusted probability is taken as the final score for each peptide11. Although these methods allow for more efficient use of available data, the integration of search results still has room for further development. First, the number of peptide-spectrum matches (PSMs) identified in some but not all search engines grows at combinatorial rates as more search engines are considered for integration, and the scores must be properly calibrated for the PSMs identified by individual search engines to control the overall identification error rates in a unified manner. Second, since some search engines only report the best matching peptide sequence for each spectrum, potential matches to lower-ranking peptides are ignored in the report even if individual scores for those secondary matches are nearly as good as the best match score and thus are likely true hits. If data are integrated from different search engines, one must include lower-ranking PSMs from every search engine and recalibrate the scores into a unified score as was done in Searle et al11. The strategy of integrating data after the selection of high confidence PSMs (i.e., leaving out lower-ranking scores) may lead to inaccurate estimation of integrative probability scores unless search engines are sufficiently homogeneous12–14.
To address these issues, we developed a unified probabilistic approach for the integrative analysis of unique PSMs, termed MSblender (Figure 1). We use probability mixture models for distinguishing correct and incorrect identifications. The score distributions across search engines are jointly modeled using multivariate distributions up to the number of observed dimensions to accommodate the correlation in raw search scores. Using this model, MSblender computes a unified posterior probability of correct identification for all PSMs identified by search engines. The conversion into posterior probabilities automatically calibrates PSM scores reported by individual search engines in two ways: (1) the likelihood is marginalized to the search engines identifying individual PSMs, and (2) prior probability is adjusted for different combinations of search engines. More importantly, MSblender pools raw search scores for every possible PSM and directly models the distribution for all listed scores from the beginning, so it is not necessary to revisit lower-ranking PSMs to account for the PSMs not agreed upon by all search engines.
Figure 1.
Schematic view of MSblender. Each spectrum is listed with all unique peptide assignments across the database search engines considered for integration. MSblender considers all different cases. PSMs may be found by some search engines, but not by others (peptides 1, 2). Some spectra may be matched to different peptides by different search engines (peptides 3 – 9), where each PSM is treated differently.
We evaluate the performance of MSblender with respect to peptide identification and protein quantification by spectral counting using three independent datasets. First, we use a yeast dataset (Yeast YPD hereafter) to assess the sensitivity and specificity profile for bona fide identifications, where high-confidence identifications reproducibly reported in multiple published datasets can be used as a benchmark set. Next, we include a (Sigma) UPS2 dataset featuring a simple mixture of 48 human proteins, where concentrations are known for all proteins and thus the accuracy in both identification and quantification can be evaluated. Lastly, we use a dataset (iPRG09) from an Association of Biomolecular Resource Facilities (ABRF) proteome informatics research group (iPRG) 2009 study consisting of two biological samples, in which proteins present in only one sample are known and thus the influence of improved identifications can be evaluated by differential expression analysis. Through these examples, we show that integrative analysis by MSblender increases the number of identifications substantially with accurate estimation of low false discovery rate (FDR), and it improves quantitative analysis of protein concentrations.
Materials and Methods
Yeast YPD dataset
Yeast YPD is a yeast dataset from Ramakrishnan et al.15. Briefly, cell lysates were harvested from S. cerevisiae BY4741 grown in rich medium (YPD) in log phase, digested with trypsin and prepared for LC/LC-MS/MS analysis. We performed eight replicate LC-MS/MS using four salt steps on an SCX column (ammonium chloride solutions of varying molarity, namely 0, 15, 60, 900 mM or 0, 20, 100, 900 mM in a 5% acetonitrile, 0.1% formic acid background), followed by reverse-phase chromatography on a C18 column and MS/MS analysis on an LTQ-Orbitrap Classic (Thermo). 32 files were analyzed using S. cerevisiae sequences from EnsEMBL version 50 and randomly shuffled sequences as decoy. The raw dataset is available at http://www.marcottelab.org/users/MSdata/Data_02/.
UPS2 dataset
The dataset comprises 48 human proteins mixed in concentrations covering six orders of magnitude, from 0.5 fmol to 50,000 fmol (Sigma Aldrich). The sample was prepared as described before15 including cysteine alkylation, trypsin digestion and cleanup of the resulting peptides. The sample was re-suspended in 50 μl of buffer (95% H2O, 5% acetonitrile, 0.1% formic acid) and ten samples of different dilutions were used for LC-MS/MS analysis on an LTQ-Orbitrap Classic (Thermo) mass spectrometer in a 5 to 90% acetonitrile gradient over four hours. Dilutions ranged from none to 1:30, with 10μl injected per run. We used a sequence file downloaded from Sigma Aldrich website as the target database and a decoy database derived from their randomly shuffled protein sequences. The raw data are deposited at http://www.marcottelab.org/users/MSdata/Data_13/.
iPRG09 dataset
We used the ABRF iPRG 2009 study data downloaded from Tranche Proteome Commons. The data consist of two 1D gel separations of identical Escherichia coli cellular lysates (called the ‘yellow’ and ‘red’ samples). In each sample, one segment of the separation gel was cut out and discarded. The two discarded segments (‘green’ and ‘blue’) did not overlap in their position in the two samples, thus the proteins in these segments would be identified as differentially expressed proteins relative to the other sample. For each of the two samples, five LC-MS/MS data files were available. To compare our results with the original study, we used the same E. coli sequences as available from the ABRF website http://www.abrf.org/index.cfm/group.show/ProteomicsInformaticsResearchGroup.53.htm including reversed sequences as decoy instead of randomly shuffled sequences.
Data processing
We used the same database with target and decoy sequences for all individual runs with four different search engines: SEQUEST, X!Tandem with k-score, InsPecT and MyriMatch. We used default parameters in each search engine wherever possible, assuming that parameters had already been optimized for each scoring matrix. We allowed for up to 2 missed tryptic cleavages, and set static cysteine alkylation. More detailed information, such as software version and modified parameters, is reported in Table 1. In the search results, individual spectra may be reported multiple times in different PSMs, mainly because of different charge state assignments. For example, MyriMatch reported two PSMs for every spectrum with different charge estimates (+2 and +3 in our default setup). To guarantee consistency across search engines, we selected the best scoring PSM per spectrum based on the scores listed in Table 1. However, this is not a requirement and additional lower ranking PSMs might also be allowed if the total number of PSMs from each search engine is not significantly different.
Table 1.
Summary of search engine parameters. Parameters not reported in this table were not changed from default values used by the search engine.
| Name | Source | Version | Scores used in MSblender | Parameters |
|---|---|---|---|---|
| SEQUEST | Thermo Electron | Bioworks 3.3.1 SP1 |
Xcorr | Mass type: monoisotopic precursor and fragments Peptide tolerance: 25.0 ppm Fragment ion tolerance: 1.0 amu |
| X!Tandem (k-score) | REFERENCE | 2009.10.01.1 | E-value | Fragment monoisotopic mass error: 0.7 Parent monoisotopic mass error: 100 ppm Minimum peaks: 15 Minimum fragment m/z: 150 |
| InsPecT | REFERENCE | 20100331 | MQscore | TagCount: 50 PMTolerance: 2.5 |
| MyriMatch | REFERENCE | 1.6.62 (2009-12-4) | Mvh | NumChargeStates: 3 UseAvgMassOfSequences: false |
Statistical model
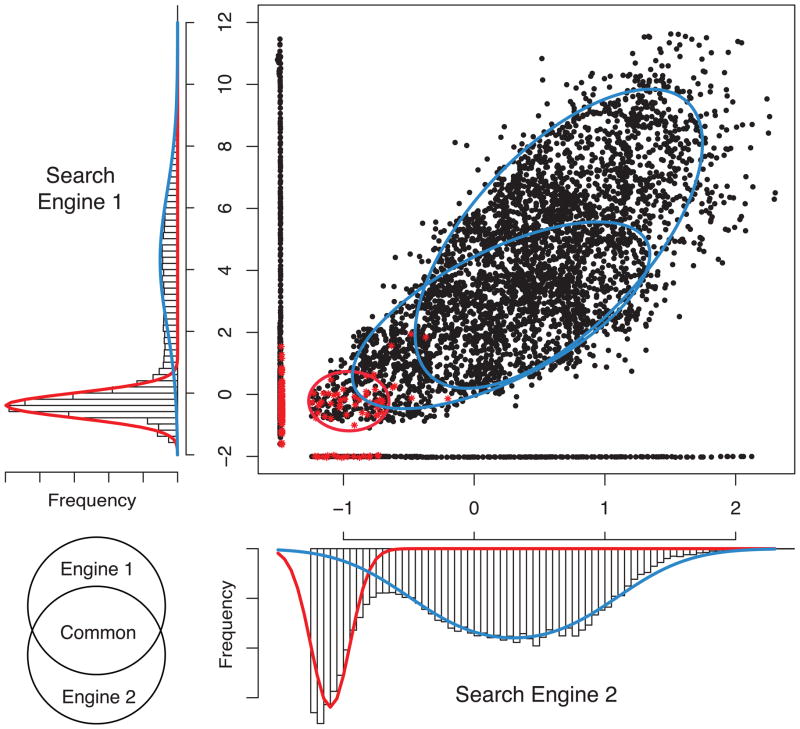
The statistical approach in MSblender is a probability mixture model for score distributions of correct and incorrect identifications, which has been widely used for scoring PSMs16. A novel feature of MSblender is that the mixture component distributions are modeled as multivariate distributions that appropriately account for the correlation between database search scores (Figure 2).
Figure 2.
An example of the statistical model for integrating scores from two database search engines. The scatter plot shows three groups of data: PSMs with scores reported from both engines (dots following a diagonal line), and PSMs identified uniquely by either of the two search engines (dots in lines parallel to each axis). Red stars indicate the PSMs assigned to decoy sequences. The elliptical contours in the scatter plot and the curves in the histograms are the estimated distributions (blue: correct identification, red: incorrect identification).
Suppose that the database search was repeatedly performed using K independent search engines, and M spectra were matched to peptide sequences by at least one search engine. Let Si=(Si1, Si2,…,SiK) denote the raw search score for spectrum i, where i=1, 2, …, M. We assume that the raw scores are median centered and scaled by setting unit standard deviation in all search engines. Note that some Sij can be missing if the j-th search engine does not report the same PSM. The joint probability density of search scores can be written as
where π is the proportion of spectra with correct peptide assignment in the data. g0 and g1 are the score distributions for PSMs in correct and incorrect identifications, respectively. We refer to them as negative and positive mixture component distributions from here on. To estimate these distributions, a sufficient number of PSMs must have scores across all database search algorithms included. However, not every spectrum is assigned to the same peptide sequence across all database search engines (Figure 1). Especially for decoy sequences, a spectrum is rarely assigned to the same decoy peptide by two or more search engines due to the random nature of incorrect peptide matches. It follows that the negative component g0 cannot be specified as a fully multivariate distribution due to the lack of usable data for estimation, and thus we assume g0(Si) = n=1,…,K g0n(Sin), i.e. scores from different search engines are conditionally independent for incorrect identifications. Furthermore, it is natural to assume different prior weights for true and false identifications when PSMs are identified in more search engines than others, i.e. frequent identification implies high prior belief of correct identification. Hence we vary the weight parameter π by each combination of search engines, as many as 2K-1.
To provide flexibility for accommodating variable shapes of score distributions, we allowed the mixture components g0 and g1 to be expressed as mixtures of multivariate Gaussian distributions themselves (g0 with a diagonal covariance matrix), where the number of subcomponents must be pre-specified by the user. Specifically,
where c is the number of subcomponents for the positive component distribution g1, MVNk stands for K-dimensional multivariate normal distribution, and Mc and Vc are the mean vector and the covariance matrix for the subcomponent distribution c with a respective mixing proportion λc (such that Σc=1,…,C λc = 1). In a typical run, we specified two subcomponents by default (c=2).
In the case of g0, the marginal negative component distribution g0n of an individual search engine n is expressed as a mixture of univariate Gaussian distributions to allow for the same flexibility as in the positive component, i.e.
where N denotes univariate normal distribution. Mean and variance parameters (mcn, Vcn) as well as the mixing proportion(s) π are estimated using the EM algorithm17, where the spectra assigned to decoy peptides are treated as known incorrect PSMs, rendering the mixture model semi-supervised18. Once we estimate the positive and negative distributions in the score distribution, we compute the posterior probability of correct identification for each PSM by Bayes’ rule:
where g(Si) = (1-π) g0(Si) + π g1(Si) for every spectrum i, and π varies by search engine combinations. Recall that Si is a fully K-dimensional vector without missing data only if PSM i is observed in all search engines. For a spectrum with scores from fewer than K database search engines, we compute the probability using the marginal distributions of observed scores only. After computing probabilities, the FDR at a probability threshold p* can be estimated by ΣiεS* (1 - pi) / |S*|, where S* is the set of PSMs such that pi ≥ p* and |A| is the size of a set A.
Benchmark data
In the Yeast YPD dataset, we compared the MSblender results to the proteins observed in previously published large-scale data under the same condition (the entire cellular lysate during logarithmic growth in rich medium). This list of proteins was prepared from 4 MS-based proteomics datasets and 3 non-MS-based datasets (see Ramakrishnan et al.15 and http://www.marcottelab.org/MSdata/gold_yeast.html). We used the list of 4,265 proteins observed in either two or more MS-datasets or any of non-MS-datasets as benchmark list.
Differential expression analysis by QSPEC
We applied a statistical method for selecting differentially expressed proteins based on spectral counts, termed QSPEC19, to the iPRG09 dataset analyzed by individual search engines and MSblender. QSPEC computes the odds (Bayes factors) of differential expression for individual proteins and reports log scaled odds multiplied by the sign determined by the direction of changes as the summary statistic. These quantities are used to estimate local fdr and FDR using nonparametric empirical Bayes methods20. We evaluated the sensitivity profile at various thresholds. We constructed receiver operating characteristic (ROC)-like curves using the benchmark set provided by the ABRF iPRG 2009 study committee. The ‘blue’ and ‘green’ segments contain positive sets of enriched proteins in the ‘red’ and ‘yellow’ data respectively. In the ROC plot, the horizontal coordinate corresponds to the number of proteins not included in the positive set, representing the false positive hits. Likewise, the vertical coordinate corresponds to the number of proteins included in the positive set, representing the sensitivity of detection.
Software Availability
The source for running MSblender can be downloaded from the URL http://www.marcottelab.org/index.php/MSblender.
Results and Discussion
Estimation of FDR
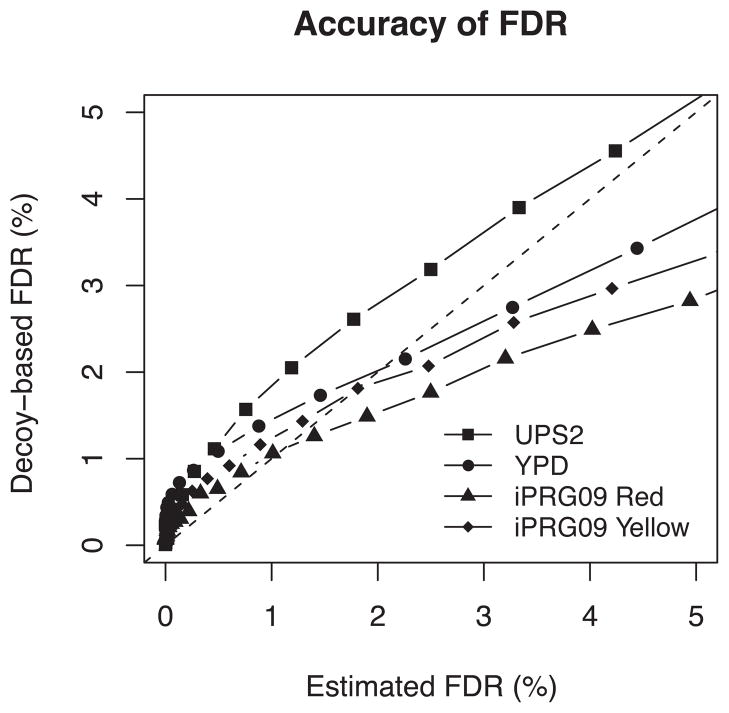
The fundamental challenge in data integration is accurate estimation of error rates such as FDR. This is particularly difficult when search engines are heterogeneous, i.e. conflicting PSMs occur frequently between search engines, and search scores are not in good agreement. Figure 3 plots the FDR estimated by MSblender (see Materials and Methods) against decoy-based FDR estimates in all three datasets. We estimated decoy-based FDR by labeling one half of the decoy PSMs as non-decoy PSMs and measuring their recurrence in the MSblender results (with proper scaling). Overall, the two estimates show good agreement in critical regions, i.e. where the error rate is low, in all datasets, with a trend of underestimation of FDR against decoys in UPS2 and Yeast YPD datasets. There was no evidence of such underestimation against decoys in iPRG09 datasets, particularly in the low error rate area. A possible explanation for the underestimation is that more consistent decoy PSMs were identified by multiple search engines in UPS2 and YPD datasets than in iPRG09 dataset. Since MSblender assigns higher prior weights for PSMs identified in more search engines than for PSMs identified in only one engine, many multi-engine (borderline scoring) decoy PSMs were assigned high probability.
Figure 3.
FDR estimated from posterior probabilities (Estimated FDR) against FDR estimated from decoy identifications (Decoy FDR). Estimated FDR is calculated by averaging PSM errors with a posterior probability threshold as described in Materials and Methods. Decoy-based FDR is calculated by recovery rate of decoy PSMs after labeling a half of decoy PSMs as target PSMs before running MSblender. Provided that decoys are truly random hits, the diagonal line indicates accurate FDR estimates.
To see this from a comparative angle, we first examined the union method, which selects PSMs in individual search engines at fixed FDRs and merging them. In Table 2 (FDR 0.5%), rows for MSblender and union show that the latter approach consistently includes more decoy PSMs than the former, leading to underestimation of error rates (actual error rate by decoy count is higher than the target 0.5%). A more sophisticated union approach with probability adjustments based on the search score agreement by Searle et al11 improves the accuracy of FDR estimates in the first two datasets but not in iPRG09 datasets (Supplement Figure 2; see below). In addition to estimated FDR, Table 3 shows that MSblender achieves a good trade-off between recovering more true targets (gold standard reference identification in Yeast YPD dataset; see Materials and Methods) and identifying false-positive proteins at FDR 1%, whereas union would have incurred higher error rates than MSblender to achieve similar sensitivity. However, there was no significant improvement at FDR 0.5%; union and MSblender reported nearly identical results, implying that at extremely low error cutoffs, a sophisticated statistical model, such as is used in MSblender, is not necessarily helpful.
Table 2.
Summary table of identification results by individual search engines and MSblender combining all search engine results. The entries were obtained at FDR 0.5%. In the PSM table, the rows referred to as ‘k engines’ indicate the number of PSMs identified with k search engines. In the same table, the numbers in the parentheses are the number of decoy PSMs identified at the same FDR (0.5%).
| PSM | UPS2 | Yeast YPD | iPRG09(Red) | iPRG09(Yellow) |
|---|---|---|---|---|
| Total MS/MS spectra observed | 74,602 | 240,781 | 69,416 | 70,970 |
| SEQUEST | 32,651 (87) | 57,955 (268) | 9,524 (98) | 9,492 (83) |
| X!Tandem | 27,264 (210) | 74,244 (332) | 15,147 (117) | 15,366 (112) |
| MyriMatch | 26,262 (79) | 41,179 (106) | 9,706 (88) | 9,134 (46) |
| InsPecT | 25,618 (64) | 69,341 (414) | 12,691 (202) | 13,295 (216) |
| Union | 40,829 (434) | 95,315 (1053) | 21,764 (505) | 21,684 (455) |
| MSblender | 39,273 (336) | 99,814 (1011) | 23,580 (153) | 23,717 (177) |
| 1 engine | 4,043 (190) | 10,441 (100) | 2,138 (38) | 2,073 (52) |
| 2 engines | 7,389 (89) | 16,861 (546) | 3,768 (76) | 3,878 (74) |
| 3 engines | 5,560 (35) | 32,111 (203) | 6,820 (24) | 6,816 (21) |
| 4 engines | 22,202 (24) | 38,257 (18) | 10,830 (3) | 10,826 (4) |
| Protein | UPS2 | Yeast YPD | iPRG09(Red) | iPRG09(Yellow) |
| Total proteins | 48 | 6,698 | 4,417 | 4,417 |
| SEQUEST | 38 | 1,391 | 757 | 749 |
| X!Tandem | 38 | 1,459 | 870 | 847 |
| MyriMatch | 36 | 1,241 | 722 | 657 |
| InsPecT | 29 | 1,527 | 877 | 902 |
| Union | 44 | 1,873 | 999 | 1,024 |
| MSblender | 42 | 1,911 | 1,185 | 1,147 |
Table 3.
Recovery of gold standard proteins in Yeast YPD dataset.
| FDR 0.5% | All proteins | False proteins | True proteins | False/Total |
|---|---|---|---|---|
| Total | 6698 | 2433 | 4265 | |
| SEQUEST | 1387 | 69 | 1318 | 4.97% |
| X!Tandem | 1453 | 62 | 1268 | 4.26% |
| MyriMatch | 1238 | 43 | 1195 | 3.47% |
| InsPecT | 1519 | 99 | 1420 | 6.51% |
| Union | 1899 | 161 | 1738 | 8.47% |
| MSblender | 1864 | 153 | 1711 | 8.20% |
| FDR 1% | All Proteins | False proteins | True proteins | False/Total |
|---|---|---|---|---|
| Total | 6698 | 2433 | 4265 | |
| SEQUEST | 1500 | 96 | 1404 | 6.40% |
| X!Tandem | 1628 | 98 | 1530 | 6.01% |
| MyriMatch | 1307 | 53 | 1254 | 4.05% |
| InsPecT | 1662 | 134 | 1528 | 8.06% |
| Union | 2218 | 252 | 1966 | 11.36% |
| MSblender | 2038 | 203 | 1835 | 9.96% |
The findings above show that choosing thresholds for each search engine before forming the union is non-trivial as merging data filtered at a fixed FDR in individual search engines results in the post-integration FDR being higher than the target FDR. In addition, there may not exist a unique solution to control the composite identification error rate since the combined error is not a simple function of the individual error rates. In this situation, FDR control by MSblender based on multivariate mixture model can be helpful despite its underestimation of error in extremely high confidence regions.
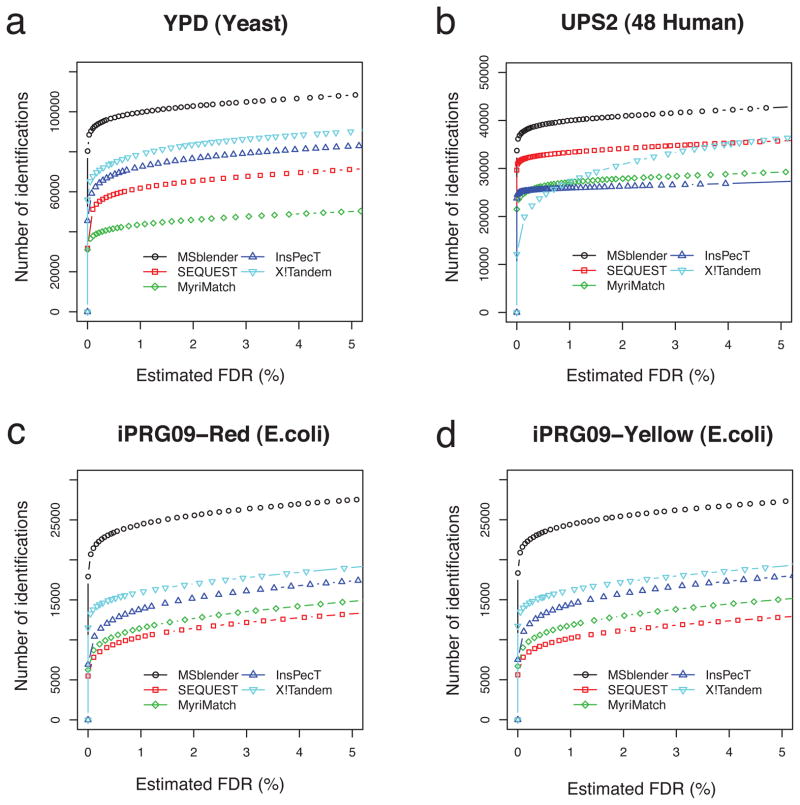
Sensitivity of identification
In addition to the accuracy of FDR estimates in high confidence selections, we evaluated the performance of MSblender in comparison to the individual search engines. Specifically, we examined the number of identifications for both PSMs and proteins at fixed FDRs (0.5% and 1%), and summarized the result in Figure 4. In all three datasets, MSblender clearly increases the number of identifications over individual search engines. For example, X!Tandem yielded the highest number of PSMs amongst other search engines at FDR 0.5% in the Yeast YPD dataset (Figure 4a). Table 2 shows that the dataset contains 240,781 PSMs, and X!Tandem identified 74,244 PSMs among these (30%). In comparison, MSblender identified 99,814 PSMs (41%) at the same FDR, increasing the number of identified PSMs by 11% of total spectra. From these additional PSMs, 452 new proteins were identified by MSblender in comparison to X!Tandem in this dataset. In the UPS2 dataset, the number of PSMs increased by ~20% in MSblender compared to the best performing search engine SEQUEST (Figure 4b), but the number of proteins increased only marginally (by 4 proteins) due to the low sample complexity (see Table 2). However, the additional PSMs helped to improve quantification by spectral counting (see below). In iPRG09 datasets (Figures 4c–4d), MSblender identified a substantial number of additional PSMs, e.g. roughly doubling the number of identified PSMs over SEQUEST, and many new proteins.
Figure 4.
Comparison of identification results across different database search engines and MSblender integrating all search engines. The plots show the number of PSMs assigned to non-decoy sequences against estimated FDR. Although the performance of individual search engine varies depending on the dataset, it is clear that integrated MSblender can identify substantially more PSMs at the same FDR. More figures with different combination of search engine results are available in the Supplementary Figure 1.
In general, MSblender consistently increased the number of identifications compared to individual search engines both when integrating all search engines and when integrating only a subset of the engines (Supplementary Figure 1). This result holds for both small and highly complex protein samples with thousands of proteins. Interestingly, MSblender was as good with some combinations of three search engines as with all four search engines, indicating that there exists saturation effect in terms of additional improvements by including additional search engines.
Comparison to agreement score adjustment
To compare MSblender to existing integrative methods, we implemented in-house code to carry out the procedure described in Searle et al11, which we term agreement score adjustment (personal communication with the author). For each PSM, the agreement score was calculated following Searle et al.’s description, the probability scores from individual search engines were adjusted reflecting this agreement score, and the largest probability from all available search engines was taken as the final score for each PSM. If a search engine does not report the PSM, we considered it as a zero probability event. Supplementary Figure 2 shows that agreement score adjustment not only produces as many PSMs as MSblender at fixed FDRs in UPS2 dataset, but also provides more accurate FDR estimates. The performance for the two methods was also similar in Yeast YPD dataset. However, MSblender offers much more accurate FDR estimates and more PSMs in both iPRG09 datasets. This is partly attributable to the fact that MSblender performs very well with highly heterogeneous search engine results, i.e. search engines that largely disagree on PSM identifications, as is the case in iPRG09 but not in UPS2 and Yeast YPD. Supplementary Table 1 shows that both decoy and non-decoy PSMs common in two or more search engines appeared more frequently in Yeast YPD and UPS2 datasets than in iPRG09 datasets. In the iPRG09 data, the method of agreement score adjustment operates close to the union method as it takes the largest probability, but probability adjustment occurs less frequently in iPRG09 than in other datasets (Supplementary Figure 2 panels 3b–4b).
Impact on label-free quantification by spectral counting
Enhanced PSM and protein identification by integrative analysis is expected to have a positive impact on spectral counting based protein quantification, in particular if additional PSMs contribute to total spectral counts per protein and thus refine the resolution of quantification. For instance, the difference between 1 spectrum and 2 spectra is less discernible than the difference between 10 spectra and 20 spectra. The increase in spectral counts is particularly important for low-abundance proteins, as they are often identified by one or a few PSMs only using a single search engine.
We first examined the number of proteins identified by individual search engines and MSblender across the range of spectral counts. In spectral counting, if a spectrum is matched to n different peptides at a given significance level, then we considered the PSM as 1/n count for each and every peptide (split spectral counts). We also investigated not using this correction and adding one count to each matched peptide, but this procedure did not perform differently in the low FDR range (not shown). Supplementary Figures 3a–3d illustrate the results, for the case of split spectral counts. When we plotted the number of identified proteins against the spectral counts normalized by their length (in log10 scale), we found that not only the spectral count per protein increased in MSblender compared to individual searches, but also additional low-abundance proteins were identified. This observation implies that MSblender not only increased spectral counts for proteins identified by individual search engines, but also identified additional proteins in low spectral counts that individual search engines missed.
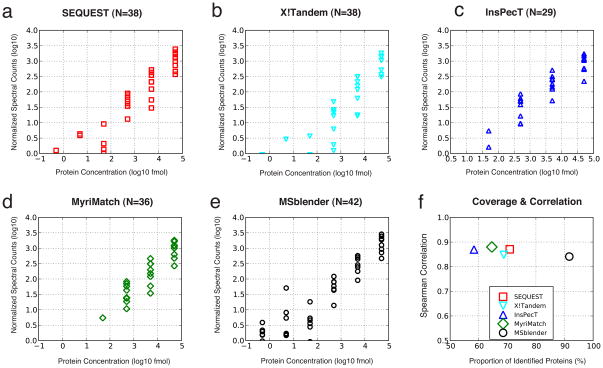
To assess the accuracy of quantification, we examined the correlation between spectral counts of 48 proteins of known concentrations (UPS2 dataset). Figure 5 shows the results for the individual search engines and MSblender combining all of them at FDR 0.5%. As in Yeast YPD data, spectral counts were normalized by protein length, and spectral counts and known concentrations were rescaled to log10. Figures 5a–5e show that MSblender allows for quantification of the largest number of proteins (42 proteins out of 48) while providing a similar correlation between observed and expected protein concentrations as individual searches, measured by rank correlation coefficients (Figure 5f). Even though tens of thousands of additional PSMs have been used by MSblender for quantification, length-normalized spectral counts were linearly correlated with the known concentrations. Based on the evenly distributed increase in identifications, we expect that this trend should hold in samples of high complexity. When we relaxed the error criterion (FDR 1%), the number of quantified proteins also increased in individual search engines, but the correlation decreased. FDR cutoffs less stringent than 1% admitted PSMs with probability score as low as 0.5 or 0.6, allowing noisy PSMs to compromise quantification accuracy.
Figure 5.
Length normalized spectral counts against known protein concentrations in the UPS2 dataset (FDR 0.5%). Spectral counts and concentrations were rescaled to log10. a–e: Individual search engines and MSblender. The number in subtitle parentheses reports the number of proteins identified. Across all protein concentrations, normalized spectral counts increase in MSblender, showing that MSblender improves the dynamic range of spectral counts. f: Spearman rank correlation coefficients (RS) against percentage of proteins identified, defined as the number of identified proteins divided by the total number of proteins known to exist (48). MSblender improves the protein identification substantially (x-axis), while maintaining a correlation between observed and known protein concentrations similar to single search engine results.
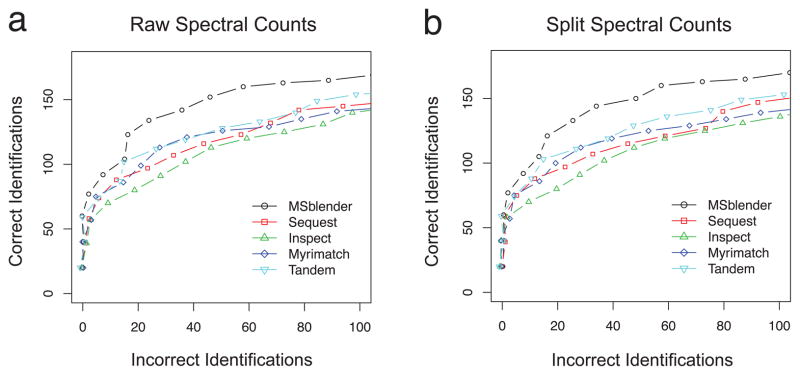
To further demonstrate that increased PSM identification improves quantification, we used iPRG09 datasets to examine the sensitivity/specificity profile during differential expression analysis. To evaluate the performance consistently, we applied the differential expression analysis tool QSPEC (see Materials and Methods) to spectral count data reported by individual search engines and MSblender. Figure 6 shows the comparative performance in terms of the receiver-operating characteristic (ROC) for split and raw spectral count data at FDR 0.5%. MSblender detected differentially expressed proteins with the highest sensitivity at all fixed error rates for both types of spectral count data.
Figure 6.
Comparison of QSPEC analyses using the search results from individual search engines and MSblender integrating all search engines (FDR 0.5%). Correct and incorrect identifications were determined from proteins removed from each sample provided by the ABRF iPRG 2009 committee. See Materials and Methods for details in each samples and segments.
Conclusion
This work presents an efficient probabilistic approach that substantially improves identification of PSMs at low error rates, reducing the volume of unassigned spectra in mass spectrometry-based shotgun proteomics experiments. Because scores are computed for all PSMs from the start, PSMs subject to search engine discrepancy are automatically considered for all possible peptide sequences and thus there is no need to trace back lower-ranking PSMs in individual searches. The final probability of correct identification does not have to rely on the probability from individual search engines which was calculated ignoring statistical dependence of raw search scores. The method can be applied to any number and combination of database search engines. Since the score is directly calculated from raw scores, the underlying statistical model allows a unified control of identification error rates.
Integration of unique PSMs in MSblender provides justifiable grounds for more coherent quantification than the post-assignment integration methods, particularly if spectral counting is employed. Interestingly, we observed that the identification and quantification improved not only for low abundance proteins with MSblender, but also for high abundance proteins. This observation implies that MSblender substantially expanded the dynamic range of detectable protein concentrations without compromising quantification accuracy.
For practical applications, we remind the readers that the fundamental challenge of integrative analysis is accurate estimation of identification errors, and it is important to understand the benefits and risks for error estimation when integrating different search engines of varying heterogeneity. MSblender delivers a tool to estimate correct integrated error rates even across heterogeneous datasets. In homogeneous datasets, i.e. where many search engines share PSMs including decoy PSMs, the performance of MSblender can be improved by modeling the negative component distribution in multivariate form as the positive component distribution. In practice this is not feasible because the proportion of decoy PSMs occurring in two or more search engines is too small. We leave further improvements in statistical modeling to future work.
Supplementary Material
Comparison of identification results across different database search engines and MSblender integrating 2, 3, and 4 search engines. The counts of PSMs assigned to non-decoy sequences are plotted versus the estimated FDR. The figure shows that MSblender integration using combinations of three search engines performs comparable to MSblender with all four search engines.
Comparison of MSblender and the agreement score method of Searle et al. with respect to the number of identifications (1a–4a) and the consistency between the estimated FDR and the decoy-based FDR (1b–4b). MSblender identifies more PSMs than the agreement score method except for the UPS2 dataset. The agreement method controls errors more accurately (closer to the diagonal) in the YPD and UPS2 datasets; MSblender estimates errors more accurately for the iPRG09 dataset.
The number of identified proteins as a function of MS/MS spectral counts. Spectral counts per protein were normalized by protein length and rescaled to log10. The improvement to protein identification by MSblender is similar for proteins from all abundance ranges, and does not show strong bias e.g. towards high abundance proteins.
Heterogeneity of PSM agreement across search engines before filtering.
Acknowledgments
We thank Dr. Brian Searle for generously providing his code excerpt from Scaffold. This work was supported grants from the NIH, NSF, the Welch (F1515) & Packard Foundations (to E.M.M.), and NIH grants R01-GM094231 and R01-CA126239 (to A.I.N.).
Abbreviations
- FDR
false discovery rate
- LC
liquid chromatography
- MS
mass spectrometry
- PSM
peptide-spectrum match
- ROC
receiver operating characteristic
Footnotes
Supporting Information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20(9):1466–7. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 2.Eng JK, McCormack AL, Yates Iii JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 3.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3(5):958–64. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 4.Kapp EA, Schutz F, Connolly LM, Chakel JA, Meza JE, Miller CA, Fenyo D, Eng JK, Adkins JN, Omenn GS, Simpson RJ. An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics. 2005;5(13):3475–90. doi: 10.1002/pmic.200500126. [DOI] [PubMed] [Google Scholar]
- 5.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 6.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6(2):654–61. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem. 2005;77(14):4626–39. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- 8.Alves G, Wu WW, Wang G, Shen RF, Yu YK. Enhancing peptide identification confidence by combining search methods. J Proteome Res. 2008;7(8):3102–13. doi: 10.1021/pr700798h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ning K, Fermin D, Nesvizhskii AI. Computational analysis of unassigned high-quality MS/MS spectra in proteomic data sets. Proteomics. 2010;10(14):2712–8. doi: 10.1002/pmic.200900473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sultana T, Jordan R, Lyons-Weiler J. Optimization of the Use of Consensus Methods for the Detection and Putative Identification of Peptides via Mass Spectrometry Using Protein Standard Mixtures. J Proteomics Bioinform. 2009;2(6):262–273. doi: 10.4172/jpb.1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Searle BC, Turner M, Nesvizhskii AI. Improving sensitivity by probabilistically combining results from multiple MS/MS search methodologies. J Proteome Res. 2008;7(1):245–53. doi: 10.1021/pr070540w. [DOI] [PubMed] [Google Scholar]
- 12.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25(1):117–24. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 13.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol Cell Proteomics. 2005;4(10):1487–502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Zybailov B, Mosley AL, Sardiu ME, Coleman MK, Florens L, Washburn MP. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. J Proteome Res. 2006;5(9):2339–47. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
- 15.Ramakrishnan SR, Vogel C, Prince JT, Li Z, Penalva LO, Myers M, Marcotte EM, Miranker DP, Wang R. Integrating shotgun proteomics and mRNA expression data to improve protein identification. Bioinformatics. 2009;25(11):1397–403. doi: 10.1093/bioinformatics/btp168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 17.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J Royal Statist Soc. 1977;39(1):1–38. [Google Scholar]
- 18.Choi H, Nesvizhskii AI. Semisupervised model-based validation of peptide identifications in mass spectrometry-based proteomics. J Proteome Res. 2008;7(1):254–65. doi: 10.1021/pr070542g. [DOI] [PubMed] [Google Scholar]
- 19.Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Mol Cell Proteomics. 2008;7(12):2373–85. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efron B, Tibshirani R, Storey JD, Tusher T. Empirical Bayes analysis of a microarray experiment. J Am Statist Assoc. 2001;96:1151–1160. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of identification results across different database search engines and MSblender integrating 2, 3, and 4 search engines. The counts of PSMs assigned to non-decoy sequences are plotted versus the estimated FDR. The figure shows that MSblender integration using combinations of three search engines performs comparable to MSblender with all four search engines.
Comparison of MSblender and the agreement score method of Searle et al. with respect to the number of identifications (1a–4a) and the consistency between the estimated FDR and the decoy-based FDR (1b–4b). MSblender identifies more PSMs than the agreement score method except for the UPS2 dataset. The agreement method controls errors more accurately (closer to the diagonal) in the YPD and UPS2 datasets; MSblender estimates errors more accurately for the iPRG09 dataset.
The number of identified proteins as a function of MS/MS spectral counts. Spectral counts per protein were normalized by protein length and rescaled to log10. The improvement to protein identification by MSblender is similar for proteins from all abundance ranges, and does not show strong bias e.g. towards high abundance proteins.
Heterogeneity of PSM agreement across search engines before filtering.