Abstract
Proteomics approach was used to elucidate the molecular interactions taking place at the stem cell wall level when tomato species were inoculated with Ralstonia solanacearum, a causative agent of bacterial wilt. Cell wall proteins from both resistant and susceptible plants before and after the bacterial inoculation were extracted from purified cell wall with salt buffers and separated with 2-D IEF/SDS–PAGE and with 3-D IEF/SDS/SDS–PAGE for basic proteins. The gels stained with colloidal Coomassie revealed varied abundance of protein spots between two species (eight proteins in higher abundance in resistant and six other in susceptible). Moreover, proteins were regulated differentially in response to bacterial inoculation in resistant (seven proteins increased and eight other decreased) as well as in susceptible plants (five proteins elevated and eight other suppressed). Combination of MALDI-TOF/TOF MS and LC-ESI-IonTrap MS/MS lead to the identification of those proteins. Plants responded to pathogen inoculation by elevating the expression of pathogenesis related, other defense related and glycolytic proteins in both species. However, cell wall metabolic proteins in susceptible, and antioxidant, stress related as well as energy metabolism proteins in resistant lines were suppressed. Most of the proteins of the comparative analysis and other randomly picked spots were predicted to have secretion signals except some classical cytosolic proteins.
Keywords: Defense and metabolic proteins, R. solanacearum, Secretory proteins, Stem cell wall proteome, Tomato species, 3-D PAGE
Introduction
The cell wall of the plant is one of the distinguishing features and a dynamic structure that consists predominantly of polysaccharides. Primary cell wall of dicotyledon plants possess less than 10% proteins of the total cell wall mass relative to 90% polysaccharides but still constitute several hundred in number (Jamet et al. 2008a). The primary cell wall is formed during cell growth and elongation where as the secondary cell wall is deposited after the cessation of cell growth inwards to the primary wall. The secondary walls of xylem fibers, tracheids and sclereids are further strengthened by the incorporation of lignin (Cassab and Varner 1988). A common cell wall model for the structure and architecture of the primary cell wall describes the existence of interwoven networks of polysaccharides and proteins (Cosgrove 2005).
Terrestrial plants are subjected to many biotic and abiotic stresses during their lifetime and correspondingly evolved a wide range of defence mechanisms to protect themselves. The plant defense is provided both at the constitutive as well as at induced level when they come into contact with invading pathogen. The interactions between plants and microbes lead either to disease resistance or plant disease depending on the type of interactions, however, the latter case could cause a huge economic loss. Bacterial wilt caused by Ralstonia solanacearum is one of the most devastating, systemic vascular wilt diseases causing up to 75–100% tomato yield loss in the lowland and highland tropics and subtropics (Denny 2006). Due to the wide host range and the variability of the pathogen, control measures based on the use of resistant cultivars remain the most effective, economical and environment friendly method (Denny 2006). The bacterial wilt resistance in tomato is a polygenic trait and was reported to be present in the mid-stem when plants were root inoculated with R. solanacearum (Dahal et al. 2009). Our previous analysis of the tomato stem proteome revealed the regulation of pathogenesis, stress related and metabolic proteins in susceptible species but not in resistant plants (Dahal et al. 2009). To further elucidate the interactions, sub-cellular fractions such as the stem cell wall were analysed to increase the sensitivity of the performed analysis. The cell wall is deeply involved in both constitutive and inducible defences through structural changes and modification during the interactions with pathogens (Carpita and McCann 2000; Jamet et al. 2008a). Histochemical analysis showed various differences on the constitutive level and changes after pathogen interaction on the level of pectic polysaccharide, though the differences could not conclusively explain the entire background of the resistance reactions (Wydra and Beri 2006). Since proteins perform the enzymatic, regulatory and structural functions in a biological system, the role of proteins secreted into the plant cell wall by the plants and pathogen during the host-pathogen interactions can play vital roles in establishing the outcome of plant–microbe interactions. The roles of plant cell wall proteins as structural, antimicrobial and enzyme molecules has been established, however, cell wall proteins associated both with susceptible and resistance tomato lines in the interaction with R. solanacearum were not characterized. Proteomics approach was therefore, undertaken to simultaneously analyze the broad spectrum of the cell wall protein (CWP) profiles that could provide further insight to understand the defense responses of the plant and also the outcome of the host-pathogen interaction. The sub-cellular proteome analysis should also present additional information and increase the usefulness of our earlier results from whole stem analysis. Extraction of wide range of cell wall proteins from mature tomato stems to a substantial purity is a challenging task due to the structural complexities of the cell wall and the nature of CWP (Jamet et al. 2008b). Therefore, disruptive and two steps salt extraction method was used to enrich cell wall proteins after purification of stem cell walls by rigorous washing with both aqueous and organic buffers to remove cytoplasmic contaminants (Watson et al. 2004).
Materials and methods
Plant material and inoculum preparation
Tomato plants were grown from highly resistant Hawaii7996 (Solanum lycopersicum) and susceptible WVa700 (Solanum pimpinellifolium) tomato lines against bacterial wilt. The seeds from both species were obtained from Asian Vegetables Research and Development Centre (AVRDC, Taiwan). The inoculum from the highly virulent R. solanacearum strain ToUdk2, race 1, biovar 3 (kindly provided by N. Thaveechai, Kasetsart University, Bangkok) was prepared by adjusting the bacterial cell concentration to about 7.8 × 107 colony-forming units per millilitre (cfu/mL) as described by Dahal et al. 2009.
Plant growth conditions and inoculation
Tomato plants were grown in the greenhouse for 4–6 weeks (20°C, 14 h photoperiod per day, 30 K lux and 70% relative humidity, RH). Each plant was then transferred to an individual pot with approximately 330 g of soil (Fruhstorfer Erde, type P: 150 mg/L N, 150 mg/L P2O5, and 250 mg/L K2O) and grown in climate chamber (30/28°C day/night temperature, 14 h photoperiod, 30 K lux and 85% RH). Some plants were root inoculated after transplantation by pouring 25 mL of the prepared inoculum per pot, to reach a final concentration of approximately 107 cfu/g of soil and the soil was watered up to soil field capacity.
Bacterial quantification
The number of bacteria residing in the stem was determined at 5 days post inoculation (dpi) considering the time the bacteria need to reach and multiply in the stem. The pathogen quantification was done as explained by Dahal et al. 2009. The pathogen developed as an elevated fluidal colony with red centre after 48 h/30°C incubation on triphenyl tetrazolium chloride (TTC) medium were quantified as colony-forming units per gram of fresh weight of plant stem (cfu/g).
Disease symptoms evaluation
Ten pathogen inoculated plants from each susceptible and resistant species were observed for symptoms assessment. The symptoms were evaluated over the period of 4 weeks in six disease severity classes from 0 to 5 (Dahal et al. 2009). The wilt incidence (WI) and disease severity (DS) were calculated as the percentage of dead plants (class 5) to the total number of plants at the evaluation date and as the mean of disease scores at the evaluation date, respectively.
Protein extraction from cell walls of tomato stems
The stem cell wall proteins from both Hawaii7996 and WVa700 species were extracted before and after pathogen inoculation at 5 dpi. Individual samples were prepared by combining mid stems of two to three individual plants to approximately 8 g. The samples were stored at −80°C until the extraction was performed from purified cell walls with salt solutions (Watson et al. 2004).
Approximately 8 g of mature tomato stem was powdered in liquid nitrogen followed by several washes and filtration through 47 μm2 nylon mesh membrane (SEFAR Nitex, Germany) to purify cell wall material for the protein extraction. The washing started with 100 mL of grinding buffer (50 mM sodium acetate pH 5.5, 50 mM NaCl, and 30 mM ascorbic acid) followed successively by 50 mL wash buffer 1 (100 mM NaCl), 100 mL bidest, 250 mL ice-cold 100% acetone and finally with 50 mL wash buffer 2 (10 mM sodium acetate pH 5.5). The extraction of protein from the purified cell wall was performed in two sequential steps: first, by shaking the debris with 8 mL of extraction buffer 1 (200 mM CaCl2 and 50 mM sodium acetate pH 5.5) twice each for 1 h, and then with 15 mL of extraction buffer 2 (3 M LiCl and 50 mM sodium acetate pH 5.5) overnight. The total volume of protein extract recovered from each sample was adjusted to 30 mL owing to some losses during filtration. The protein solution was concentrated in a centrifugal concentrator with a molecular mass cut off at 5 kDa (Vivaspin 6, Vivascience, Germany) at 5,080 g × 4°C × 2–3 h until a final volume of 100 μL was obtained. The concentrate was washed with double volume of bidest water and precipitated with a commercial 2-D clean up kit (Bio-Rad, Germany).
Protein separation with 2-D and 3-D SDS–PAGE
The complexity of the proteome extracted from the stem cell wall was resolved by two- dimensional sodium dodecyl sulphate–polyacrylamide gel electrophoresis (2-D IEF/SDS–PAGE) and for basic proteins by 3-D IEF/SDS/SDS–PAGE. In case of 2-D gels, three replicate gels were prepared for each species and treatment from separate biological samples where as only 2 replicates were prepared for 3-D gels.
Approximately 700 μg of total protein quantified by Bradford assay (Coomassie protein assay reagent, Fluka biochemical) was dissolved in 350 μL of “rehydration buffer” [8 M urea, 2% w/v CHAPS, 0.5% v/v carrier ampholyte mixture (IPG buffer 3–11 non linear (NL), GE Healthcare, Germany), 30 mM DTT, and 0.002% w/v (7 μg/350 μL) Bromophenol Blue] with the addition of 4.2 μL of DeStreak reagent (GE healthcare, Munich, Germany) to facilitate the basic proteins separation. The protein was then separated in first dimension by IEF using 18 cm NL IPG strip pH 3–11 (GE HealthCare, Munich, Germany) and in second dimension by vertical 12% SDS–PAGE (20 cm × 20 cm × 1 mm) using Protean II electrophoresis unit (Biorad, Hercules, CA, USA) as described by Werhahn and Braun 2002 and Schagger and Von Jagow 1987, respectively.
2-D IEF/SDS–PAGE of the cell wall proteome of all samples resulted in repetitive vertical streaking in very basic side of the gel and making 2-D gels inappropriate for comparative analysis of the basic protein spots. Therefore, a three-dimensional PAGE system was established, which is based on the transfer of the very basic gel region of a 2-D PAGE gel, before fixing and staining them, horizontally onto 3rd SDS–PAGE. For this approach, extra new samples were first required to be resolved and visualised by 2-D PAGE to check the exact position of the streaking on the gel so that these regions, without fixing and visualising, can be transferred correctly onto the 3rd gel.
Protein staining, gel scanning and image analysis
After completion of SDS–PAGE, proteins were first fixed and stained with colloidal Commassie solution (Neuhoff et al. 1985, 1990). The stained 2-D gels were washed with bidest water and 3-D gels additionally with 20% methanol to clean background staining. The gels were scanned with a UMAX Power Look III Scanner (UMAX Technologies, Fremont, USA) and analysed by using the ImageMaster™ 2D platinum software 6.0 (GE Healthcare, Germany) in order to accomplish the spots detection and calculate the quantitative values of the differentially regulated spots. The abundance of each protein spot was estimated by the percentage volume (% vol), which represents the contribution of each protein spot (volume) to the total protein spots (volume) on the whole gel, and thus minimizes inter-gel loading differences. Master gels of each treatment and species (each comprising at least three biological replications) were created to allow statistical comparison and only those spots present in all triplicate gels were included in the statistical analysis. Only statistically significant difference in the abundance of protein spot with Student’s t test P ≤ 0.05 (probability value) was considered. Moreover, quantitative differences of ±30% variance in a spot ratio (>1.3 or <0.7), were considered as biologically significant based on previous study (Asirvatham et al. 2002).
Tryptic digestion
All except few big and intense gel spots were handpicked from all three gel replicates to increase the protein amount for digestion. Proteins were destained by gently shaking in 20 mM NH4HCO3, 50% acetonitrile (ACN) for 30 min at 37°C. This step was repeated until spots became clear. The gel was first dehydrated in 100% ACN for 5 min followed by drying in a speed vac system (Eppendorf, Germany). The dried gel piece was directly treated with trypsin (4 ng/μL) in 20 mM NH4HCO3/10% ACN on ice with 1 h incubation because proteins were incubated with DTT followed by iodoacetamide during the equilibration steps before SDS–PAGE. The remaining trypsin solution was removed and digestion was carried out at 37°C over night after adding 20 mM NH4HCO3/10% ACN to cover gel pieces. The supernatant containing peptides was collected and the gel pieces were re-extracted using 0.2% TFA, containing increasing amounts of ACN (10–50%). All peptide containing solutions of each spots were combined, dried in a speed vac and stored at 4°C until further analysis.
Protein analysis by MALDI-TOF/TOF MS
Peptides were dissolved in 10 μL of 5% ACN containing 0.2% TFA. A saturated solution of alpha-cyano-4-hydroxycinnamic acid, CHCA (4 mg/mL) in 50% ACN and 0.2% TFA was diluted 1:10 with ethanol and 0.8 μL of the CHCA matrix was spotted on each spot of a matrix assisted laser desorption and ionization (MALDI) Anchor Chip 800/384 target plate (Bruker Daltonic GmbH). After mixing 0.5 μL of each sample with the applied matrix, air-dried samples were recrystallized with 0.2 μL of ethanol containing 0.1% TFA. For MS calibration 0.5 μL of peptide calibration standard (Bruker Daltonik GmbH) were spotted on the target with 0.8 μL of CHCA matrix and recrystallized too. Samples were analyzed in a MALDI-time of flight/time of flight (TOF/TOF) mass spectrometer (Ultraflex, Bruker Daltonics) in reflectron mode. Peptides with a signal to noise ratio above 100 were MS/MS analyzed by using the LIFT technology that is embedded in the Ultraflex MS; on average ten MS/MS spectra were measured for each protein digest leading to 2–10 identified peptides. Data processing was done using the flex analysis and biotools software packages (Bruker Daltonik).
Protein analysis by electrospray (ESI) ion trap MS
Peptides were dissolved in 5% ACN, 0.1% formic acid and were applied to reversed phase chromatography high performance liquid chromatography (HPLC) system (Agilent Technologies, Germany) that was directly mounted to the ion source of the ion trap MS. The HPLC system consists of an auto sampler and a gradient pump. The sample was dissolved in eluent A (5% ACN and 0.1% formic acid) and an aliquot was injected onto a C18 column (Zorbax SB-C18, particle size 5 μm, 300 A, 0.5 mm inner diameter, length 150 mm) at a flow rate of 5 μL/min. After loading, the column was washed for 15 min with buffer A and the peptides were eluted using a gradient of eluent A and eluent B (70% ACN in 0.1% formic acid) from 0 to 53.9% eluent B in 60 min. Then, buffer B was increased to 100% for 10 min and subsequently, the column was equilibrated with buffer A for 20 min. The HPLC outlet was directly connected to the ion source of the ion trap MS. The spray was stabilized by N2 as nebulizer gas (5 l/min). Ionization voltage was set to 4,500 V and dry gas was applied at 5 psi and 250°C. Spectra were collected with an Esquire3000+ ion trap mass spectrometer (Bruker Daltonik) at a scan speed of 13,000 m/z per second. Using ESI in positive mode, mass spectra were acquired from m/z 50 to 1,600 in scanning mode and data dependent switching between MS and MS/MS analysis. After each MS scan, two precursor ions from one spectrum were selected for subsequent MS/MS analysis and active exclusion was set to 2 min to exclude precursor ions that had already been measured. Data processing was performed with the Data Analysis software (V. 3.0, Bruker Daltonik GmbH, Germany).
MS data analysis
Data analysis was performed using BioTools 3.0 software and the MASCOT search engine (Matrix Sciences, UK). Searches were performed using the following parameters: mass tolerance was set to 100 ppm (MALDI-TOF/TOF) and 200 ppm (ESI-ion trap) for precursor ions and 0.7 Da for fragment ions. Trypsin was set as proteolytic enzyme with 2 allowed missed cleavages. Charge state 1+, was used for MALDI-TOF/TOF and charge states 1+, 2+ and 3+ for ESI–MS. Carbamidomethylation of cysteine residues and oxidation of methionine residues was set as variable modification.
MSDB, swissprot and NCBInr databases were used to identify tomato proteins either with an in house MASCOT server or online available MASCOT server (Matrix Science, UK). Proteins were termed identified, if at least two peptides were identified with a MASCOT peptide ion score higher than 25. The threshold of 25 for the peptide ion score was obtained searching a reverse data base (MSDB) with all generated MS/MS spectra. All hits obtained from this reverse database search showed a peptide ion score significantly below 25. Additionally, if high peptide ion scores (>46) were obtained for single peptides and a y-ion series of nearly all ions was obtained, the corresponding proteins were termed identified even if only one peptide was found for the protein. The databases used in house were released in August 2006 (MSDB), in October 2007 (NCBInr) and May 2009 (Uniprot/Swissprot, release 15.2). The same database releases were used at Matrix Science.
Results
Symptom development and bacterial populations in stems
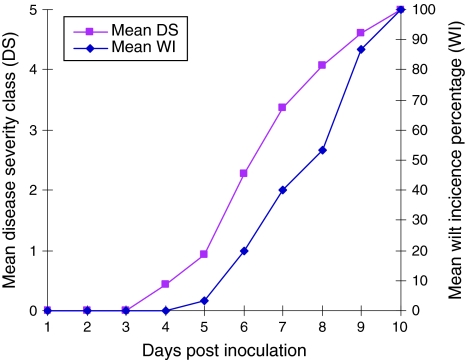
Plants of the susceptible species WVa700 started showing symptoms at 4 dpi and both the WI and DS gradually increased until all plants had died at 10 dpi. The mean WI and DS were calculated from three biological replications (Fig. 1). The resistant species Hawaii7996, on the other hand, did not show any level of symptoms until 30 dpi.
Fig. 1.
Mean disease severity (DS) and wilt incidence (WI) in susceptible tomato plants (WVa700) on the days following R. solanacearum strain To-udk2 inoculation. The mean value was calculated from three independent biological replications. Both DS and WI increased continuously to maximum by 10 dpi
Since each biological protein sample was prepared by combining two to three individual plants, the number of bacteria present in each sample was calculated by taking the mean of the bacterial population from the individual plants that were combined to make that sample. The pathogen population in three biological samples at 5 dpi was calculated as 25.6 × 107, 16.6 × 107 and 26.1 × 107 cfu/g of stem in susceptible species and 45.8 × 106, 33.3 × 106 and 22.2 × 106 cfu/g of stem in resistant species. All the susceptible plants considered for the sample preparation showed clear wilt symptoms of 1–3 degree of disease severity.
Cell wall protein analysis
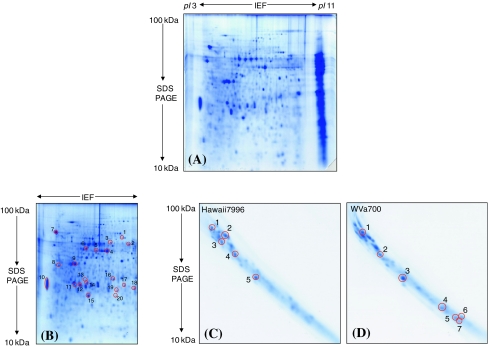
About 600–800 μg protein was obtained from 8 g of mid-stem sample. The resolution of approximately 700 μg protein loaded on each gel displayed an average of 370–470 protein spots in addition to several “poorly separated” spots on basic pI region of the gel (Fig. 2A, B). These unresolved spots upon further separation by 3-D SDS–PAGE revealed 25–35 spots detected with Image master (Fig. 2C, D). The comparison of the 2-D gels was performed within species and treatments. The differentially expressed spots were first analysed by MALDI-TOF tandem mass spectrometer (MS/MS), which gave 70% (29 out of 42 spots) successful identifications. The remaining 13 unidentified spots were unambiguously identified with liquid chromatography (LC)-ESI-Ion trap MS/MS which in one case (Spot 3, Table 4), correlated to two proteins.
Fig. 2.
Overview of the cell wall proteome analyzed from the mature tomato stem and separated in two as well as in three gel dimensions. (A) The resolution of proteome with 2-D IEF/SDS–PAGE in pI 3–11 non linear and 100–10 kDa molecular mass. The poorly resolved proteins on the basic pI range were seen as vertical streaking. (B) A representative 2-D IEF/SDS gel resolved in the same pI 3–11 non linear and 100–10 kDa molecular mass range but stained after cutting out the gel region with the poorly resolved vertical streak. The poorly resolved basic proteins without fixing and staining served as the starting material for the 3rd SDS–PAGE. Twenty spots were randomly picked out to check the presence of secretion signals of the cell wall proteins extracted with the method applied. The identity of each encircled spots and the information of secretion signals are given in Table 5. (C) and (D) 3rd dimension SDS gels showing the resolution of the unresolved vertical streak. The streaked gel piece was cut out before fixing as well as staining and separated again by SDS–PAGE. The encircled spots in (C) and (D) were differentially expressed in resistant (Hawaii7996) and susceptible (WVa700) species after pathogen inoculation, however in two replicates. The identity of all proteins was given in Table 4 with the spot numbers corresponds to each other
Table 4.
List of major basic proteins that were poorly resolved in 2-D IEF/SDS–PAGE but well separated in 3rd dimension SDS–PAGE
| Spota | Identityb | Accessionc | Organismd | Scoree | Mr/pIf | Peptidesg MS/MS |
Coverageh MS/MS |
SiP-SePi |
|---|---|---|---|---|---|---|---|---|
| Hawaii7996 (resistant genotype) | ||||||||
| 1 | Peroxidase | Q94IQ1 | Nicotiana tabacum | 480 | 39.06/5.99 | 8 | 27 | Y |
| 2 | Peroxidase prx14 | Q9M4Z3 | Spinacia oleracea | 139 | 37.22/9.29 | 2 | 7.7 | Y |
| 3* | Peroxidase | Q07446 | Solanum lycopersicum | 299 | 35.99/7.52 | 5 | 17 | Y |
| 3* | Pectinesterase | Q43143 | Solanum lycopersicum | 280 | 64.10/8.97 | 6 | 9 | Y |
| 4 | Glucan endo-1,3- β-glucosidase B | Q01413 | Solanum lycopersicum | 244 | 39.71/7.84 | 5 | 17 | Y |
| 5 | Osmotin-like protein precursor | Q41350 | Solanum lycopersicum | 404 | 27.26/8.15 | 6 | 29 | Y |
| WVa700 (susceptible genotype) | ||||||||
| 1* | Peroxidase | Q94IQ1 | Nicotiana tabacum | 391 | 39.06/5.99 | 6 | 14 | Y |
| 2 | Glucan endo-1,3-β-glucosidase B | Q01413 | Solanum lycopersicum | 172 | 39.71/7.84 | 2 | 6.7 | Y |
| 3 | Osmotin-like protein | Q41350 | Solanum lycopersicum | 284 | 27.26/8.15 | 2 | 10 | Y |
| 4 | Photosystem I reaction center subunit IV B | Q41229 | Nicotiana sylvestris | 131 | 15.22/9.74 | 2 | 19 | 0.82 |
| 5 | Oxygen-evolving complex protein 3 | Q672Q6 | Solanum lycopersicum | 201 | 24.57/9.64 | 3 | 11 | 0.65 |
| 6 | Oxygen-evolving complex protein 3 | Q672Q6 | Solanum lycopersicum | 73 | 24.57/9.64 | 1 | 4.3 | 0.65 |
| 7 | Photosystem I reaction center subunit II | P12372 | Solanum lycopersicum | 125 | 22.91/9.71 | 3 | 12 | 0.80 |
aAssigned spot number corresponding to the number used in the respective figures
b Identity of the proteins annotated by MALDI-TOF MS/MS
cProtein database accession number (UniProt)
dPlant species from which the protein was annotated
eMASCOT score
fTheoretical molecular mass and isoelectric point computed from ExPASy Mr/pI calculation tool
gNumber of matched peptides of MSMS data with the corresponding protein in MSDB, SwissProt and NCBInr databases
hPercentage of peptide sequences coverage for the identified protein by MS/MS
iThe results of SignalP (SiP) and SecretomP (SeP) analysis. Y presence of signal peptide evaluated from SignalP. The numbers are Secretom NN score calculated from SecretomP, in case no signal peptides were identified, and the NN score >0.5 was considered as secretory protein as suggested by the author
* Identity of the proteins revealed by LC-ESI-Ion Trap MS/MS
Protein regulation in resistant species
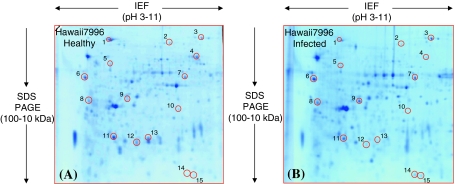
The proteomic reactions of resistant plants to pathogen invasion were evaluated by comparing triplicate gels developed before and after pathogen inoculation which revealed 15 spots of differential abundance (Fig. 3A, B). Seven spots identified as subtilase, peroxidase, hypothetical protein, luminal binding protein (BIP), fructokinase-2, nucleoside diphosphate kinase (NDPK) and PII like protein (spots 3, 6–9, 14 and 15 respectively) were increased. Eight other spots annotated as BIP, stress induced protein, catalase, enolase (2-phospho-D-glycerate hydrolase), vacuolar H+ ATPase (V-ATPase), oxygen evolving protein (OEE) 2, eukaryotic translation initiation factor 5A (eTIF 5A) -3 and eTIF 5A-4 (spots 1, 2, 4, 5, and 10–13, respectively) were decreased (Table 1).
Fig. 3.
“Zoom-in” view of the only area of the 2-D gels that shows differentially accumulated protein spots when the healthy plant of the resistant species Hawaii7996 (panel A) was inoculated with the pathogen R. solanacearum (panel B). The spot number is in accordance with the number used in Table 1. The tomato stem cell wall proteome was separated between pH 3–11 (non linear IPG stripes) and in the molecular mass range between 100 and 10 kDa. However, the basic pI side of the 2-D gel containing poorly separated proteins was removed before fixing and staining. All the shown spots regulation was consistently reproduced in three biological replications and is statistically significant (Student’s t test, P ≤ 0.05)
Table 1.
List of stem cell walls proteins which are differentially regulated in tomato genotypes Hawaii7996 (resistant) after R. solanacearum challenge. The listed proteins are those consistently reproduced in three biological replications and are statistically significant (Student’s t test, P ≤ 0.05)
| Spota | Identityb | Accessionc | Organismd | Scoree | Mr/pIf | Peptidesg | Coverageh | Regulationi | SiP-SePj |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Luminal-binding protein | P49118 | Solanum lycopersicum | 260 | 73.23/5.10 | 3 | 6 | 0.30 | Y |
| 2* | Stress induced protein | Q6H660 | Oryza sativa | 185 | 64.19/6.03 | 7 | 8.5 | 0.44 | 0.55 |
| 3* | Subtilase | O82777 | Solanum lycopersicum | 136 | 82.22/8.22 | 5 | 3 | 1.67 | Y |
| 4 | Catalase | P30265 | Solanum lycopersicum | 493 | 56.50/6.57 | 9 | 22 | 0.38 | 0.41 |
| 5* | Enolase | P26300 | Solanum lycopersicum | 92 | 47.79/5.68 | 3 | 6 | 0.54 | 0.52 |
| 6 | Peroxidase | Q9LWA2 | Solanum lycopersicum | 144 | 34.94/4.56 | 2 | 5.5 | 1.88 | Y |
| 7* | Hypothetical protein | Q9C6U3 | Arabidopsis thaliana | 88 | 34.70/7.17 | 5 | 3 | 1.62 | 0.61 |
| 8* | Luminal-binding protein | P49118 | Solanum lycopersicum | 219 | 73.23/5.10 | 7 | 9.9 | 1.72 | Y |
| 9* | Fructokinase-2 | Q42896 | Solanum lycopersicum | 510 | 34.76/5.76 | 9 | 33 | 1.91 | 0.68 |
| 10 | Vacuolar proton ATPase subunit E | Q9LKG0 | Solanum lycopersicum | 46 | 27.13/6.63 | 1 | 4.2 | 0.51 | 0.27 |
| 11 | Oxygen-evolving complex protein 2 | P29795 | Solanum lycopersicum | 362 | 27.79/8. 27 | 5 | 27 | 0.54 | 0.8 |
| 12 | Translation initiation factor 5A-3 | Q9AXQ4 | Solanum lycopersicum | 93 | 17.37/5.47 | 1 | 6.3 | 0.65 | 0.24 |
| 13 | Translation initiation factor 5A-4 | Q9AXQ3 | Solanum lycopersicum | 255 | 17.51/5.6 | 3 | 28 | 0.23 | 0.23 |
| 14 | Nucleoside diphosphate kinase | P47921 | Solanum lycopersicum | 165 | 15.67/7.04 | 2 | 12 | 1.75 | 0.40 |
| 15* | PII like protein | Q6T2D2 | Solanum lycopersicum | 201 | 21.73/9.33 | 5 | 22 | 1.75 | 0.88 |
aAssigned spot number corresponding to the number used in the respective figures
b Identity of the proteins annotated by MALDI-TOF MS/MS
cProtein database accession number (UniProt)
dPlant species from which the protein was annotated
eMASCOT score
fTheoretical molecular mass and isoelectric point computed from ExPASy Mr/pI calculation tool
gNumber of matched peptides of MSMS data with the corresponding protein in MSDB, SwissProt and NCBInr databases
hPercentage of peptide sequences coverage for the identified protein by MS/MS
iRegulation: The fold increase or decrese in % spot volume of each spot after the R. solanacearum inoculation. Variation: The ratio in the abundance of spot % spot volume between the species
jThe results of SignalP (SiP) and SecretomP (SeP) analysis. Y presence of signal peptide evaluated from SignalP. The numbers are Secretom NN score calculated from SecretomP, in case no signal peptides were identified, and the NN score >0.5 was considered as secretory protein as suggested by the author
* Identity of the proteins revealed by LC-ESI-Ion Trap MS/MS
Protein regulation in susceptible species
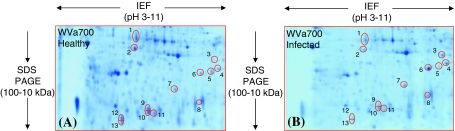
The proteomic reactions of the susceptible plants were also investigated in response to pathogen inoculation where 13 spots turned out to be differential abundance (Fig. 4A, B). Among them, five proteins (spot 3, 4 and 6–8) were increased while eight proteins (spot 1, 2, 5 and 9–13) were decreased upon pathogen inoculation. The identity of the increased spots was correspondingly shown as peroxidase, peroxidase cevi16, basic 30 kDa endochitinase, triose phosphate isomerise (TPI) and PR-5 like protein and of decreased as α-galactosidase (α-D-galactoside galactohydrolase), disulphide isomerase like protein (PDIs), xyloglucan endotransglucosylase-hydrolase7 (XTH7), two eTIF 5A-4, one eTIF 5A-1 and two glycine rich proteins (GRP) (Table 2).
Fig. 4.
“Zoom-in” view of the only area of the 2-D gels that shows differentially accumulated protein spots when the healthy plant of the susceptible species WVa700 (panel A) was inoculated with the pathogen R. solanacearum (panel B). The spot number is in accordance with the number used in Table 2. The tomato stem cell wall proteome was separated between pH 3–11 (non linear IPG stripes) and in the molecular mass range between 100 and 10 kDa. However, the basic pI side of the 2-D gel containing poorly separated proteins was removed before fixing and staining. All the shown spots regulation was consistently reproduced in three biological replications and is statistically significant (Student’s t test, P ≤ 0.05)
Table 2.
List of stem cell walls proteins which are differentially regulated in tomato genotypes WVa700 (susceptible) after R. solanacearum challenge. The listed proteins are those consistently reproduced in three biological replications and are statistically significant (Student’s t test, P ≤ 0.05)
| Spota | Identityb | Accessionc | Organismd | Scoree | Mr/pIf | Peptidesg | Coverageh | Regulationi | SiP-SePj |
|---|---|---|---|---|---|---|---|---|---|
| 1 | α-galactosidase, putative | Q9FWV8 | Oryza sativa | 137 | 44.66/5.47 | 2 | 5.6 | 0.58 | Y |
| 2 | Disulfide isomerase like protein | Q38JJ2 | Solanum tuberosum | 153 | 39.49/5.62 | 2 | 8.1 | 0.61 | Y |
| 3* | Peroxidase | Q07446 | Solanum lycopersicum | 273 | 35.99/7.52 | 5 | 17 | 2.99 | Y |
| 4 | Peroxidase cevi16 | Q4A3Y6 | Solanum lycopersicum | 170 | 31.74/7.71 | 3 | 15 | 1.86 | 0.44 |
| 5 | Xyloglucan endotransglucosylase-hydrolase XTH7 | Q6RHX8 | Solanum lycopersicum | 184 | 33.46/7.57 | 2 | 11 | 0.59 | Y |
| 6 | Basic 30 kDa endochitinase | Q05538 | Solanum lycopersicum | 266 | 34.34/6.19 | 4 | 18 | 3.56 | Y |
| 7 | Triose phosphate isomerase | Q6T379 | Solanum chacoense | 411 | 27.04/5.73 | 8 | 36 | 1.3 | 0.67 |
| 8 | PR 5-like protein | Q7Y1P9 | Solanum lycopersicum | 144 | 27.52/5.76 | 2 | 14 | 2.88 | Y |
| 9 | Translation initiation factor 5A-4 | Q9AXQ3 | Solanum lycopersicum | 255 | 17.51/5.60 | 3 | 28 | 0.3 | 0.23 |
| 10 | Translation initiation factor 5A-4 | Q9AXQ3 | Solanum lycopersicum | 255 | 17.51/5.60 | 3 | 28 | 0.37 | 0.23 |
| 11 | Translation initiation factor 5A-1 | Q9AXQ6 | Solanum lycopersicum | 139 | 17.30/5.71 | 2 | 18 | 0.66 | 0.23 |
| 12 | Glycine-rich protein | Q04130 | Solanum lycopersicum | 272 | 73.31/9.98 | 4 | 43 | 0.52 | 0.47 |
| 13 | Glycine-rich protein | Q04130 | Solanum lycopersicum | 210 | 73.31/9.98 | 4 | 61 | 0.63 | 0.47 |
aAssigned spot number corresponding to the number used in the respective figures
b Identity of the proteins annotated by MALDI-TOF MS/MS
cProtein database accession number (UniProt)
dPlant species from which the protein was annotated
eMASCOT score
fTheoretical molecular mass and isoelectric point computed from ExPASy Mr/pI calculation tool
gNumber of matched peptides of MSMS data with the corresponding protein in MSDB, SwissProt and NCBInr databases
hPercentage of peptide sequences coverage for the identified protein by MS/MS
iRegulation: The fold increase or decrese in % spot volume of each spot after the R. solanacearum inoculation. Variation: The ratio in the abundance of spot % spot volume between the species
jThe results of SignalP (SiP) and SecretomP (SeP) analysis. Y presence of signal peptide evaluated from SignalP. The numbers are Secretom NN score calculated from SecretomP, in case no signal peptides were identified, and the NN score >0.5 was considered as secretory protein as suggested by the author
* Identity of the proteins revealed by LC-ESI-Ion Trap MS/MS
Protein variation in genotypic comparison
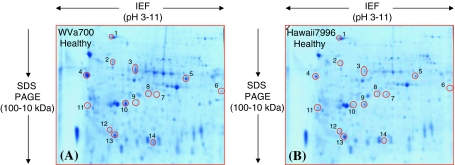
Fourteen genotypic differences were found comparing protein profiles between susceptible and resistant plants (Fig. 5A, B). Eight spots which were identified as BIP, three enolase, a hypothetical protein, fructokinase-2, nascent polypeptide-associated complex (NAC)-α-like protein 3 and OEE2 (spots 1, 2, 9, 12, 7, 8, 11, and 13, respectively) were of higher abundance in resistant species. Similarly, six further spots namely α-galactosidase, peroxidase, a hypothetical protein, ferredoxin-NADP reductase (FNR), OEE1, and eIF-5A-1 (spot 3, 4, 5, 6, 10 and 14, respectively) occurred in higher level in susceptible species (Table 3).
Fig. 5.
“Zoom-in” view of the only area of the 2-D gels that shows differentially accumulated protein spots with differential abundance between WVa700 (panel A) and Hawaii7996 (panel B) healthy plants. The spot number is in accordance with the number used in Table 3. The tomato stem cell wall proteome was separated between pH 3–11 (non linear IPG stripes) and in the molecular mass range between 100 and 10 kDa. However, the basic pI side of the 2-D gel containing poorly separated proteins was removed before fixing and staining. All the shown spots regulation was consistently reproduced in three biological replications and is statistically significant (Student’s t test, P ≤ 0.05)
Table 3.
List of differential proteins in between healthy tomato plants of genotypes Hawaii7996 (resistant) and WVa700 (susceptible). The listed proteins were those consistently reproduced in three biological replications and are statistically significant (Student’s t test, P ≤ 0.05)
| Spota | Identityb | Accessionc | Organismd | Scoree | Mr/pIf | Peptidesg | Coverageh | Variationi | SiP-SePj |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Luminal-binding protein | P49118 | Solanum lycopersicum | 260 | 73.23/5.10 | 3 | 6 | 0.53 | Y |
| 2* | Enolase | P26300 | Solanum lycopersicum | 92 | 47.79/5.68 | 3 | 6 | 0.60 | 0.52 |
| 3 | α-galactosidase, putative | Q9FWV8 | Oryza sativa | 137 | 44.66/5.47 | 2 | 5.6 | 1.79 | Y |
| 4 | Peroxidase | Q9LWA2 | Solanum lycopersicum | 144 | 34.94/4.56 | 2 | 5.5 | 2.92 | Y |
| 5* | Hypothetical protein T8G24.2 | Q9C6U3 | Arabidopsis thaliana | 88 | 34.70/7.17 | 5 | 3 | 1.69 | 0.61 |
| 6 | Ferredoxin-NADP reductase | O04397 | Nicotiana tabacum | 92 | 41.95/8.67 | 2 | 5.9 | 1.38 | 0.73 |
| 7 | Hypothetical protein | O04428 | Citrus paradisi | 92.8 | 32.64/5.46 | 1 | 4.1 | 0.38 | 0.50 |
| 8 | Fructokinase-2 | Q42896 | Solanum lycopersicum | 208 | 34.76/5.76 | 3 | 11 | 0.36 | 0.68 |
| 9* | Enolase | P26300 | Solanum lycopersicum | 291 | 47.79/5.68 | 5 | 14 | 0.50 | 0.52 |
| 10* | Oxygen-evolving enhancer protein 1 | P23322 | Solanum lycopersicum | 423 | 34.98/5.91 | 12 | 34 | 2.00 | 0.45 |
| 11 | NAC-alpha-like protein 3 | Q6ICZ8 | Arabidopsis thaliana | 151 | 22.10/4.41 | 2 | 16 | 0.44 | 0.6 |
| 12* | Enolase | P26300 | Solanum lycopersicum | 181 | 47.79/5.68 | 4 | 15 | 0.41 | 0.52 |
| 13 | Oxygen-evolving complex protein 2 | P29795 | Solanum lycopersicum | 362 | 27.79/8.27 | 5 | 27 | 0.69 | 0.8 |
| 14 | Translation initiation factor 5A-1 | Q9AXQ6 | Solanum lycopersicum | 139 | 17.30/5.71 | 2 | 18 | 4.12 | 0.23 |
aAssigned spot number corresponding to the number used in the respective figures
b Identity of the proteins annotated by MALDI-TOF MS/MS
cProtein database accession number (UniProt)
dPlant species from which the protein was annotated
eMASCOT score
fTheoretical molecular mass and isoelectric point computed from ExPASy Mr/pI calculation tool
gNumber of matched peptides of MSMS data with the corresponding protein in MSDB, SwissProt and NCBInr databases
hPercentage of peptide sequences coverage for the identified protein by MS/MS
iRegulation: The fold increase or decrese in % spot volume of each spot after the R. solanacearum inoculation. Variation: The ratio in the abundance of spot % spot volume between the species
jThe results of SignalP (SiP) and SecretomP (SeP) analysis. Y presence of signal peptide evaluated from SignalP. The numbers are Secretom NN score calculated from SecretomP, in case no signal peptides were identified, and the NN score >0.5 was considered as secretory protein as suggested by the author
* Identity of the proteins revealed by LC-ESI-Ion Trap MS/MS
Resolution of cell wall proteins at basic pI range
2-D IEF/SDS–PAGE of cell wall proteins resulted in repetitive vertical streaking and poor focussing/resolution in the basic pI region even though the DeStreak reagent (GE Healthcare, Germany) was used to facilitate basic protein separation (Fig. 2A). The problem of poor resolution was improved considerably by separating them further with SDS–PAGE in the 3rd dimension (Fig. 2C, D). Two 3-D gels were prepared from all species and treatments in order to check the reproducibility. This simple method provided reproducible gels for comparative proteomic analysis. The visual comparison of the proteome of both resistant and susceptible species in response to bacterial invasion revealed five and seven protein spots of differential abundance (Fig. 2C and D). They were identified by MS analysis as peroxidase [pathogenesis related (PR)-9], β-1, 3-endoglucanase (PR-2) and osmotin like protein (PR-5) among others (Table 4). All of these PR proteins constitute important components of the disease resistance response. However, they were not considered for discussion due to only two biological replications.
Prediction of secretion signals
Since the secretory pathway is involved in the biosynthesis of cell wall proteins and their transport to the cell wall, the presence of secretion signals in the above identified proteins was evaluated with SignalP (V. 3.0, www.cbs.dtu.dk/services/SignalP) and SecretomeP (V. 2.0, www.cbs.dtu.dk/services/SecretomeP) programs. Most of the proteins were predicted to have secretion signals (Tables 1, 2, 3, 4). In order to check the extracellular nature of cell wall proteins extracted by this particular method, 20 additional spots were picked randomly from the gel (Fig. 2B) and 90% of them showed signal peptides (Table 5). However, some glycolytic and other metabolic proteins that are conventionally not considered as extracellular proteins were also found in the cell wall.
Table 5.
Overview of stem cell wall proteins randomly picked from the 2-D gels
| Spota | Identityb | Accessionc | Organismd | Scoree | Mr/pIf | Peptidesg MS/MS |
Coverageh MS/MS |
SiP-SePi |
|---|---|---|---|---|---|---|---|---|
| 1 | Catalase isozyme | P30264 | Solanum lycopersicum | 211 | 56.50/6.60 | 2 | 5.3 | 0.34 |
| 2 | Peroxidase | Q07446 | Solanum lycopersicum | 149 | 35.99/7.52 | 5 | 16 | Y |
| 3 | NADH-glutamate dehydrogenase | Q8W1X4 | Solanum lycopersicum | 257 | 44.68/6.28 | 3 | 12 | 0.56 |
| 4* | Peroxidase | Q42964 | Nicotiana tabacum | 124 | 34.52/4.65 | 4 | 4 | Y |
| 5 | Glyceraldehyde 3-phosphate dehydrogenase | O04891 | Solanum lycopersicum | 42 | 31.94/5.93 | 1 | 4.8 | 0.39 |
| 6 | Hypothetical protein | O24329 | Ricinus communis | 80 | 40.00/7.56 | 1 | 4.4 | Y |
| 7 | Calreticulin | Q40401 | Nicotiana plumbaginifolia | 167 | 47.48/4.45 | 4 | 9.9 | Y |
| 8 | Ripening regulated protein | Q9FR30 | Solanum lycopersicum | 152 | 22.20/4.72 | 2 | 15 | 0.73 |
| 9 | Oxygen-evolving complex protein 1 | P23322 | Solanum lycopersicum | 205 | 34.94/5.91 | 2 | 8.2 | 0.44 |
| 10* | Calmodulin | P84339 | Agaricus bisporus | 277 | 16.78/4.15 | 6 | 18 | 0.70 |
| 11 | ATP synthase D chain | Q6L460 | Solanum demissum | 191 | 19.80/5.34 | 2 | 8.9 | 0.61 |
| 12 | Oxygen-evolving complex protein 2 | P29795 | Solanum lycopersicum | 292 | 27.79/8.28 | 4 | 19 | 0.80 |
| 13 | Soluble inorganic pyrophosphatase | Q43187 | Solanum tuberosum | 70 | 24.26/5.59 | 2 | 12 | 0.79 |
| 14 | Temperature-induced lipocalin | Q38JE1 | Solanum lycopersicum | 171 | 21.25/5.96 | 3 | 17 | 0.51 |
| 15 | Superoxide dismutase (Cu–Zn) 1 | P14830 | Solanum lycopersicum | 67 | 15.30/5.83 | 1 | 8.6 | 0.68 |
| 16 | Dehydroascorbate reductase | Q4VDN8 | Solanum lycopersicum | 200 | 23.53/6.32 | 2 | 13 | 0.36 |
| 17 | Hypothetical protein | Q5XEP2 | Arabidopsis thaliana | 67 | 64.52/5.85 | 3 | 7.5 | 0.58 |
| 18 | Tomato invertase inhibitor | O82001 | Solanum lycopersicum | 62 | 18.76/8.30 | 2 | 15 | Y |
| 19 | Translation initiation factor 5A-4 | Q9AXQ3 | Solanum lycopersicum | 102 | 17.51/5.60 | 1 | 6.3 | 0.23 |
| 20 | Nucleoside diphosphate kinase | Q2KK37 | Thlaspi caerulescens | 121 | 25.79/9.34 | 1 | 5 | 0.89 |
aAssigned spot number corresponding to the number used in the respective figures
b Identity of the proteins annotated by MALDI-TOF MS/MS
cProtein database accession number (UniProt)
dPlant species from which the protein was annotated
eMASCOT score
fTheoretical molecular mass and isoelectric point computed from ExPASy Mr/pI calculation tool
gNumber of matched peptides of MSMS data with the corresponding protein in MSDB, SwissProt and NCBInr databases
hPercentage of peptide sequences coverage for the identified protein by MS/MS
iThe results of SignalP (SiP) and SecretomP (SeP) analysis. Y presence of signal peptide evaluated from SignalP. The numbers are Secretom NN score calculated from SecretomP, in case no signal peptides were identified, and the NN score >0.5 was considered as secretory protein as suggested by the author
* Identity of the proteins revealed by LC-ESI-Ion Trap MS/MS
Discussion
Various roles of CWPs, particularly their involvement in the regulation of growth and development, defence against biotic or abiotic stresses, and contribution to wall architecture are increasingly studied (Jamet et al. 2008a). Regulation of plant cell wall proteins following pathogen invasion have sparingly been reported (Bradley et al. 1992; Brisson et al. 1994). However, the comprehensive analysis of a broad range of CWPs expressed in response to bacterial inoculation is lacking. Therefore, the current study was initiated firstly to find out the differences at the proteome level in mid-stem cell wall between susceptible and resistant species against bacterial wilt and followed by the simultaneous characterization of the cell wall protein profiles that are regulated in susceptible and resistant tomato plants due to R. solanacearum ingress. Due to the reported expression of bacterial wilt resistance in the mid-stem of tomato and the time needed by R. solanacearum to reach and grow extensively in the stem after soil inoculation, the cell wall proteome was analyzed from the mid-stem and at 5 dpi (Dahal et al. 2009). The specificity of proteins to one of the species and subsequent up or down regulation of the identified protein profiles after pathogen inoculation further elucidates the resistant and susceptible reactions of the species.
Expression of plant defense mechanisms
We observed the up regulation of several defense related proteins both in susceptible and resistant species at 5 dpi after pathogen inoculation. The susceptible plants responded by increasing the expression of endochitinase (PR-3) and PR-5 family proteins whereas the resistant plants showed the up regulation of NDPK and subtilase. The abundance of peroxidase (PR-9) was elevated in both species. When plants are challenged with pathogens, the general defense responses are generally inducted by the synthesis of PR proteins and fortification of plant of cell walls among others responses.
PR proteins can be constitutively present in different plant species at low level, however, they are increased dramatically upon challenge with pathogens and abiotic stresses. Plant chitinase, the majority of which are of the endo type, are believed to mediate defence responses because of their potential to degrade fungal cell walls. Many endochitinases also displayed a lysozyme activity enabling the hydrolysis of bacterial cell walls (Brunner et al. 1998). PR-5 family proteins contain several unique proteins with diverse functions including antifungal (Ibeas et al. 2000) as well as β-glucanase activity (Grenier et al. 1999). Many PR-5 protein isoforms (PR-5a to PR-5d) were accumulated in the extracellular space of tobacco plant cells (Koiwa et al. 1994). PR protein induction is considered a marker of the activation of the basal defense, thus increased also in susceptible hosts.
NDPK not only performs a house keeping functions of regulating nucleotide pools but also involved in signal transduction in plants. NDPK1 expression was reported to enhance multiple stress tolerance in transgenic plants by activating the MAPK cascade (Tang et al. 2008). Subtilisin-like serine proteases or subtilases are endoproteases secreted into the extracellular space of the plant. They have been associated with cellular defense and stress responses by mediating a restructuring and reinforcement of plant cell walls in order to arrest pathogen spread (Dixon and Lamb 1990).
Plant peroxidases are class III secretory peroxidases with a large number of isoforms performing a wide range of functions. In many plant species, increase in peroxidases expression was correlated with resistance due to their involvement in the production of reactive oxygen species (ROS), the fortification of the cell wall structure, and synthesis of secondary metabolites, and consequently controlling the penetration and cellular spread of the pathogen (Passardi et al. 2005).
The up regulation of these PR and other defense related proteins in plant stem cell wall in response to bacterial invasion could support their so far known physiological roles in plant defense. Both the tolerant and susceptible plants showed the elevation of the defense related proteins, however, with different functional roles suggesting their involvements in resistance response to R. solanacearum.
Interestingly, three other defense and/or stress related proteins namely BIP, stress induced protein1 and catalase were decreased in resistant plants after pathogen invasion. In fact, BIP was observed in two spots (spot 1 and 8) and accumulated in opposite ways. BIP is a member of the hsp70 family protein and functions as a molecular chaperone. They have also been implicated in disease resistance but still unclear how they can be related to the defense response. Stress induced protein STI1 may play a role in mediating the heat shock response of some hsp70 genes (Nicolet and Craig 1989). Catalase is a primary antioxidant and has been demonstrated to be present in the plant cell wall. The down regulation can be expected following pathogen inoculation if the elevation of H2O2 and ROS level serves as second messengers to further trigger the downstream defense responses (Foyer and Noctor 2005). Furthermore, increased ROS level, however, under a threshold not triggering a cell death, can inhibit pathogen growth. An increase in H2O2 with consequent reduction of catalase and superoxide dismutase activities was observed in jute under water stress (Chowdhuri and Chowdhuri 1985).
Both up and down regulation of these defense related proteins indicate the reactions of the plants to pathogen infection by regulating the expression of their resistance proteins. However, the outcome of susceptibility or resistance to the given pathogen could be determined by the temporal and spatial regulation of these responses and hence their efficacy.
Change in cell wall metabolism
The analysis of the cell wall proteome revealed cell wall metabolism proteins such as XTH7, α-galactosidase and GRP in susceptible plants, all of which were decreased in response to pathogen invasion. Both XTH and α-galactosidase are carbohydrate modifying proteins and belong to the glycoside hydrolases (GHs) family which are generally involved in reorganization/reconstruction of cell wall polysaccharides during active development, defense, signaling and mobilization of storage reserves (Minic 2008). XTHs can exhibit both endo-glycanase and endo-transglycosylase activities and have a potential to modify plant cell wall during synthesis and also disease responsive fortification processes (Fry 2004). The enzyme α-galactosidase, generally acidic forms, is common in plants and is suggested to have transglycosylase actions (Soh et al. 2006). Down-regulating α-galactosidase was shown to enhance freezing tolerance in transgenic Petunia at the whole plant level (Pennycooke et al. 2003). GRPs are plant cell wall structural proteins characterized with high content of glycine (20–70%) and are localized in lignified cell walls. Members of a group of GRPs have a signal peptide and were suggested to play a role in cell wall reinforcement or in signal transduction in addition to their roles in the development as well as repair of vascular tissues and wound healing (Park et al. 2001; Ryser et al. 1997). Down regulation of GRP due to water stress was reported with the argument that remodeling of the cell wall as part of the plant defense response not only requires accumulation of pathogen restricting proteins but also the reduction of some proteins that are more suitable for cell wall function during normal conditions (Harrak et al. 1999). Additionally, oxidative cross linking of GRP occurring during pathogen invasion can lead to their suppression (Bradley et al. 1992). The presence of GRP in two different spots and reduction of both isoforms indicates the significance of a possible post translation modification (PTM) which could be crucial in the outcome of reactions.
The lowered abundance of both cell wall hydrolases and GRP due to bacterial inoculation in our study may reflect the decline of cell wall polysaccharide metabolism and mechanical stability in susceptible plants during disease expression.
Metabolic activities alteration
Proteins that are known to have active roles in plant metabolism were identified in the cell wall fraction of our analysis, though more specific experiments such as immuno-cytochemistry has not been performed to confirm their presence. The occurrence of cytosolic proteins in the extracellular space of the plants was reported in several previous studies. The possibility of moonlighting functions of these intracellular proteins was also increasingly revealed. Therefore, the association of the following metabolic proteins with defense responses of the plants are discussed in more generalized concept rather than within the boundary of extracellular proteins.
Variation in primary metabolism
Proteins associated with glycolysis such as fructokinase, TPI and enolase were differentially accumulated in the two species. Fructokinase and TPI were correspondingly increased in resistant and susceptible lines while enolase was decreased in resistant plants. The increased abundance of fructokinase and TPI during bacterial infection can be assumed as consequences of increased metabolism to compensate for the cost of defences (Curto et al. 2006). PII like protein is considered to participate in metabolic regulatory mechanism and was increased in resistant varieties (Hsieh et al. 1998). On the other hand, the expression of enolase was suppressed in resistant plants. Enolase is known to catalyze the penultimate reversible reaction of glycolysis but also identified as a major glucan-associated cell wall protein in Arabidopsis and Medicago cell walls (Lee et al. 2004; Watson et al. 2004). Suppression of enolase in resistant plants could be correlated to inhibition of the pathogen by limiting the supply of sugars.
In overall, regulation of these metabolic proteins in both species after pathogen challenge supports the observation that the plant defense responses are associated with active metabolic changes in host plants.
Inhibition of energy metabolism
Both OEE 2 and V-ATPases sub unit E were decreased in resistant reactions. OEE is associated with the photosystem II complex and are believed to be important for photosynthesis and overall PSII stability (Raymond and Blankenship 2004). Therefore, any damage or inhibition of PSII due to stresses could lead to suppression of OEE proteins. Down regulation of OEE proteins was reported earlier due to water stress and tobacco mosaic virus infections (Echevarría-Zomeño et al. 2009; Lehto et al. 2003). V-ATPases are multimeric enzymes and can be associated with various membranes of the secretory system to promote cell growth or expansion and secretion of cell wall components (Cipriano et al. 2008).
Reduction of energy metabolism proteins in our analysis is in line with previous reports (Castillejo et al. 2004). It is assumed that root inoculation of tomato plant with R. solanacearum could inhibit the root carbohydrate oxidation pathways leading to the decrease in the overall energy production.
Variation in other proteins
The abundance of PDIs was reduced after pathogen invasion in susceptible plants. PDIs are involved in the folding, assembling and sorting of plant secretory or plasma membrane proteins, which are essential for the stability and activity of extracellular proteins (Wilkinson and Gilbert 2004). Four spots identified as eukaryotic translation initiation factor 5A (eIF-5A) were repressed in both susceptible (eIF-5A-1 and eIF-5A-4) and resistant (eIF-5A-3 and eIF-5A-4) interactions. eIF-5A is a nucleo-cytoplasmic shuttle protein (Jao and Chen 2005) and were also known to regulate cell division, cell wall expansion and cell death (Thompson et al. 2004). In Arabidopsis, plant eIF5A was involved in the development of disease symptoms induced by bacterial phytopathogen (Hopkins et al. 2008). Both isoforms of eIF-5A were repressed in both species suggesting the possible importance of their PTMs during host-pathogen interaction.
Constitutive differences in tomato species
BIP, enolase, fructokinase-2, OEE2, and NAC-α-like protein 3 occurred in higher abundance in resistant plants while α-galactosidase, peroxidase, FNR, OEE1, and eIF-5A-1 in susceptible plants. Again, the identification of these cytosolic proteins in our cell wall fraction does not validate their localization in the apoplast without additional experiments but their roles in the make-up of the defense reactions of the plant can be discussed.
The resistant species displayed the higher abundance of glycolytic proteins such as enolase observed in three different spots (spot 2, 9 and 12) and fructokinase-2 than the susceptible one. It was suggested that the maintenance of metabolic proteins such as glycolytic enzymes is important to fulfill the increased carbohydrate “fuel” demand required during pathogen invasion and for the recovery too. Moreover, resistant plant showed the elevation of NAC-α-like protein 3 which could assist in the prevention of disordered metabolism (Yan et al. 2005). Defense and stress related proteins also showed constitutive differences among the species. BIP was in higher abundance in resistant variety where as multifunctional peroxidase and α-galactosidase in susceptible plants. The susceptible species also differed from the resistant one in the expression of FNR, which is considered as detoxifying agents due to its free radical scavenging ability. Both species showed the abundance of OEE but were differed in the variation only in their isoforms. The increased abundance of OEE proteins is considered to improve the photosynthetic efficiency which could contribute to ward off the pathogen infection upon challenge. It should also be noted that an increase or decrease in the abundance of protein spots can also occur by selective decay of either protein in addition to the synthesis or suppression of the protein.
Several proteins identified in multiple spots such as enolase, OEE, BIP, GRP as well as eIF-5A and their isoforms were regulated in the same way (Tables 1, 2 and 3). However, the differences in the protein isoforms may indicate that the protein modifications could be a crucial phenomenon in determining the genetic status of the plants.
Based on the above finding, it can be argued that the metabolic proteins in addition to defense and stress related proteins, play crucial roles in determining the resistance or susceptibility of the plants. They are abundant and soluble proteins and hence often appeared in 2-D gels.
Nature of cell wall proteins
The presence of secretion signals that are characteristic to extracellular proteins were predicted with SignalP and SecretomP. Among 42 differentially expressed proteins, 29 were identified to have signal peptides and ten more were predicted to posses non-classical secretion signals (Tables 1, 2 and 3). Further 20 proteins from 2-D gels and 12 proteins from the comparative 3-D gels were tested for their secretory nature and 30 of them contain secretion signals. The overall results showed that most of the extracted cell wall proteins contain signals for extracellular localization. However, some proteins generally predicted to have other than extracellular locations, such as glycolytic proteins, V-ATPase, OEE, eIF-5A, BIP, enolase, catalase and others were detected in the cell wall fraction. Growing evidences suggest the possibility of them being “moonlighting” proteins due to the evidence of their localization in the extracellular space. These so called “moonlighting” proteins perform more than one function in the cell as a consequence of changing their cellular localization, oligomeric state or ligand concentration (Copley 2003). For example, enolase is also supposed to have additional non-glycolytic functions such as mitochondrial targeting of tRNA (Entelis et al. 2006). Therefore, further validations with immunolocalization or other methods would be required to confirm their presence in cell wall.
Resolution of basic proteins
We showed the use of simple 3rd dimension SDS–PAGE to separate the basic pI range proteins, that are often components of cell wall proteins, in a resolution enough for the comparative gel based proteomic analysis. The 3-D gels from both species and treatments consistently resolved the basic range of cell wall proteins. The identification of 12 major spots from 3-D gels showed the presence of proteins with theoretical pI higher than 7 (Table 4).
Conclusion
To conclude, the current study provides for the first time the broad spectrum analysis of the stem cell wall proteome of tomato species followed by their specific regulation after bacterial challenge. It unveiled constitutive proteomic differences between tomato species differing in resistance to bacterial wilt, and the differential regulation of their respective protein profiles in response to R. solanacearum invasion, which further extends the understanding of the molecular basis of the host-pathogen interactions. The selective differential expression of defense/stress related and metabolic proteins in both resistant and susceptible species triggered by the pathogen support their pivotal roles in the make-up of defense mechanism. Though, clear statements on the role of the proteins can only be made after their functional analysis and the demonstration of their role in susceptible and resistant species. The work further supports the hypothesis that the resistance mechanism is a complex interplay of several proteins where their activation kinetics might play more pivotal role than their number and type in the outcome of the host-pathogen interaction. Further experiments determining the physiological roles of each of the regulated proteins will help elucidating the resistance or susceptibility mechanisms.
Acknowledgments
This study was funded by the Federal Ministry for Economic Cooperation and Development (BMZ), Germany, in the framework of the collaborative project number 03.7860.4–001.00 with the Asian Vegetable Research and Development Center (AVRDC), Taiwan.
Abbreviations
- 2-, 3-D IEF/SDS–PAGE
2 or 3 dimensional isoelectric focusing/sodium dodecyl sulphate–protein gel electrophoresis
- MALDI-TOF/TOF MS
Matrix assisted laser desorption and ionization-time of flight tandem mass spectrometry
- LC-ESI-IonTrap MS/MS
Liquid chromatography-electrospray ionization-ion trap tandem mass spectrometry
- PR protein
Pathogenesis related proteins
References
- Asirvatham VS, Watson BS, Summer LW. Analytical and biological variance associated with proteomic studies of Medicago truncatula by two-dimensional polyacrylamide gel electrophoresis. Proteomics. 2002;2:960–968. doi: 10.1002/1615-9861(200208)2:8<960::AID-PROT960>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ. Elicitor-and wound-induced oxidative cross-linking of a proline-rich plant cell wall structural protein: a novel, rapid plant defense response. Cell. 1992;70:21–30. doi: 10.1016/0092-8674(92)90530-P. [DOI] [PubMed] [Google Scholar]
- Brisson LF, Tenhaken R, Lamb C. Function of oxidative cross-linking of cell wall structural proteins in plant disease resistance. Plant Cell. 1994;6:1703–1712. doi: 10.1105/tpc.6.12.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner F, Stintzi A, Fritig B, Legrand M. Substrate specificities of tobacco chitinases. Plant J. 1998;14:225–234. doi: 10.1046/j.1365-313X.1998.00116.x. [DOI] [PubMed] [Google Scholar]
- Carpita N, McCann M. The cell wall. In: Buchanan BB, Wilhelm G, Jones RL, editors. Biochemistry and molecular biology of plants. Rockville: American Society of Plant Physiologists; 2000. pp. 52–108. [Google Scholar]
- Cassab GI, Varner JE. Cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:321–353. doi: 10.1146/annurev.pp.39.060188.001541. [DOI] [PubMed] [Google Scholar]
- Castillejo MÁ, Amiour N, Dumas-Gaudot E, Rubiales D, Jorrín JV. A proteomic approach to studying plant response to crenate broomrape (Orobanche crenata) in pea (Pisum sativum) Phytochemistry. 2004;65:1817–1828. doi: 10.1016/j.phytochem.2004.03.029. [DOI] [PubMed] [Google Scholar]
- Chowdhuri RS, Chowdhuri MA. Hydrogen peroxide metabolism as an index of water stress tolerance in jute. Physiol Plant. 1985;65:503–507. doi: 10.1111/j.1399-3054.1985.tb08681.x. [DOI] [Google Scholar]
- Cipriano DJ, Wang Y, Bond S, Hinton A, Jefferies KC, Qi J, Forgac M. Structure and regulation of the vacuolar ATPases. Biochim Biophys Acta. 2008;1777:599–604. doi: 10.1016/j.bbabio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley SD. Enzymes with extra talents: moonlighting functions and catalytic propmiscuity. Curr Opin Chem Biol. 2003;7:265–272. doi: 10.1016/S1367-5931(03)00032-2. [DOI] [PubMed] [Google Scholar]
- Cosgrove DJ. Growth of the plant cell wall. Nat Rev Mol Cell Biol. 2005;6:850–861. doi: 10.1038/nrm1746. [DOI] [PubMed] [Google Scholar]
- Curto M, Camafeita LE, López JA, Maldonado AM, Rubiales D, Jorrín J. A proteomic approach to study pea (Pisum sativum) responses to powdery mildew (Erysiphe pisi) Proteomics. 2006;6:163–174. doi: 10.1002/pmic.200500396. [DOI] [PubMed] [Google Scholar]
- Dahal D, Heinz D, Dorsselaer AV, Braun HP, Wydra K. Pathogenesis and stress related, as well as metabolic proteins are regulated in tomato stems infected with Ralstonia solanacearum. Plant Physiol Biochem. 2009;47:838–846. doi: 10.1016/j.plaphy.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Denny TP. Plant pathogenic Ralstonia species. In: Gnanamanickam SS, editor. Plant-associated bacteria. Dordrecht: Springer; 2006. pp. 573–644. [Google Scholar]
- Dixon RA, Lamb CJ. Molecular communication in interactions between plants and microbial pathogens. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:339–367. doi: 10.1146/annurev.pp.41.060190.002011. [DOI] [Google Scholar]
- Echevarría-Zomeño S, David A, Inmaculada J, Christof L, Antonio DC, Jesús VJ, Rafael MN. Changes in the protein profile of Quercus ilex leaves in response to drought stress and recovery. J Plant Physiol. 2009;166:233–245. doi: 10.1016/j.jplph.2008.05.008. [DOI] [PubMed] [Google Scholar]
- Entelis N, Irina B, Piotr K, Igor AK, Robert PM, Ivan T. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 2006;20:1609–1620. doi: 10.1101/gad.385706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Oxidant and antioxidant signaling in plants: a re-evaluation of the concept of oxidative stress in a physiological context. Plant Cell Environ. 2005;28:1056–1071. doi: 10.1111/j.1365-3040.2005.01327.x. [DOI] [Google Scholar]
- Fry SC. Primary cell wall metabolism: tracking the careers of wall polymers in living plant cells. New Phytol. 2004;161:641–675. doi: 10.1111/j.1469-8137.2004.00980.x. [DOI] [PubMed] [Google Scholar]
- Grenier J, Potvin C, Trudel J, Asselin A. Some thaumatin-like proteins hydrolyse polymeric beta-1, 3-glucans. Plant J. 1999;19:473–480. doi: 10.1046/j.1365-313X.1999.00551.x. [DOI] [PubMed] [Google Scholar]
- Harrak H, Chamberland H, Plante M, Bellemare G, Lafontaine JG, Tabaeizadeh Z. A proline-, threonine-, and glycine-rich protein down-regulated by drought is localized in the cell wall of xylem elements. Plant Physiol. 1999;121:557–564. doi: 10.1104/pp.121.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M, Lampi Y, Wang TW, Liu Z, Thompson JE. Eukaryotic translation initiation factor 5A is involved in pathogen-induced cell death and development of disease symptoms in Arabidopsis. Plant Physiol. 2008;148:479–489. doi: 10.1104/pp.108.118869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MH, Lam HM, Loo FJ, Coruzzi G. A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc Natl Acad Sci USA. 1998;95:13965–13970. doi: 10.1073/pnas.95.23.13965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibeas JI, Lee H, Damsz B, Prasad DT, Pardo JM, Hasegawa PM, Bressan RA, Narasimhan ML. Fungal cell wall phosphomannans facilitate the toxic activity of a plant PR-5 protein. Plant J. 2000;23:375–383. doi: 10.1046/j.1365-313x.2000.00792.x. [DOI] [PubMed] [Google Scholar]
- Jamet E, Albenne C, Boudart G, Irshad M, Canut H, Pont-Lezica R. Recent advances in plant cell wall proteomics. Proteomics. 2008;8:893–908. doi: 10.1002/pmic.200700938. [DOI] [PubMed] [Google Scholar]
- Jamet E, Boudart G, Borderies G, Charmont S, Lafitte C, Rossignol M, Canut H, Pont-Lezica R. Isolation of plant cell wall proteins. Methods Mol Biol. 2008;425:187–201. doi: 10.1007/978-1-60327-210-0_17. [DOI] [PubMed] [Google Scholar]
- Jao DL, Chen KY. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J Cell Biochem. 2005;97:583–598. doi: 10.1002/jcb.20658. [DOI] [PubMed] [Google Scholar]
- Koiwa H, Sato F, Yamada Y. Characterization of accumulation of PR-5 proteins by IEF-immunoblot analysis. Plant Cell Physiol. 1994;35:821–827. doi: 10.1093/oxfordjournals.pcp.a078663. [DOI] [PubMed] [Google Scholar]
- Lee SJ, Saravanan RS, Damasceno CM, Yamane H, Kim BD, Rose JK. Digging deeper into the plant cell wall proteome. Plant Physiol Biochem. 2004;42:979–988. doi: 10.1016/j.plaphy.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Lehto K, Tikkanen M, Hiriart JB, Paakkarinen V, Aro EM. Depletion of the photosystem II complex in mature tobacco leaves infected by the Flavum strain of Tobacco mosaic virus. Mol Plant Microbe Interact. 2003;16:1135–1144. doi: 10.1094/MPMI.2003.16.12.1135. [DOI] [PubMed] [Google Scholar]
- Minic Z. Physiological roles of plant glycoside hydrolases. Planta. 2008;227:723–740. doi: 10.1007/s00425-007-0668-y. [DOI] [PubMed] [Google Scholar]
- Neuhoff V, Stamm R, Eibl H. Clear background and highly sensitive protein staining with Coomassie Blue dyes in polyacrylamide gels: a systematic analysis. Electrophoresis. 1985;6:427–448. doi: 10.1002/elps.1150060905. [DOI] [Google Scholar]
- Neuhoff V, Stamm R, Pardowitz I, Arold N, Ehrhardt W, Taube D. Essential problems in quantification of proteins following colloidal staining with Coomassie Brilliant Blue dyes in polyacrylamide gels and their solution. Electrophoresis. 1990;11:101–117. doi: 10.1002/elps.1150110202. [DOI] [PubMed] [Google Scholar]
- Nicolet CM, Craig EA. Isolation and characterization of STI1, a stress-inducible gene from Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3638–3646. doi: 10.1128/mcb.9.9.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park AR, Cho SK, Yun UJ, Jin MY, Lee SH, Sachetto-Martins G, Park OK. Interaction of the Arabidopsisreceptor protein kinase Wak1 with a glycine-rich protein, AtGRP-3. J Biol Chem. 2001;276:26688–26693. doi: 10.1074/jbc.M101283200. [DOI] [PubMed] [Google Scholar]
- Passardi F, Cosio C, Penel C, Dunand C. Peroxidases have more functions than a Swiss army knife. Plant Cell Rep. 2005;24:255–265. doi: 10.1007/s00299-005-0972-6. [DOI] [PubMed] [Google Scholar]
- Pennycooke JC, Jones ML, Stushnoff C. Down-regulating (alpha)-galactosidase enhances freezing tolerance in transgenic petunia1. Plant Physiol. 2003;133:901–909. doi: 10.1104/pp.103.024554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond J, Blankenship RE. The evolutionary development of the protein complement of photosystem. Biochim Biophys Acta. 2004;1655:133–139. doi: 10.1016/j.bbabio.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Ryser U, Schorderet M, Zhao GF, Studer D, Ruel K, Hauf G, Keller B. Structural cell-wall proteins in protoxylem development: evidence for a repair process mediated by a glycine-rich protein. Plant J. 1997;12:97–111. doi: 10.1046/j.1365-313X.1997.12010097.x. [DOI] [PubMed] [Google Scholar]
- Schagger H, Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Soh CP, Ali ZM, Lazan H. Characterisation of an alpha-galactosidase with potential relevance to ripening related texture changes. Phytochemistry. 2006;67:242–254. doi: 10.1016/j.phytochem.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Tang L, Kim MD, Yang KS, Kwon SY, Kim SH, Kim JS, Yun DJ, Kwak SS, Lee HS. Enhanced tolerance of transgenic potato plants overexpressing nucleoside diphosphate kinase 2 against multiple environmental stresses. Transgenic Res. 2008;17:705–715. doi: 10.1007/s11248-007-9155-2. [DOI] [PubMed] [Google Scholar]
- Thompson JE, Hopkins MT, Taylor C, Wang TW. Regulation of senescence by eukaryotic translation initiation factor 5A: implications for plant growth and development. Trends Plant Sci. 2004;9:174–179. doi: 10.1016/j.tplants.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Watson BS, Lei Z, Dixon RA, Sumner LW. Proteomics of Medicago sativa cell walls. Phytochemistry. 2004;65:1709–1720. doi: 10.1016/j.phytochem.2004.04.026. [DOI] [PubMed] [Google Scholar]
- Werhahn WH, Braun HP. Biochemical dissection of the mitochondrial proteome of Arabidopsis thaliana by three-dimensional gel electrophoresis. Electrophoresis. 2002;23:640–646. doi: 10.1002/1522-2683(200202)23:4<640::AID-ELPS640>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Wydra K, Beri H. Structural changes of homogalacturonan, rhamnogalacturonan I and arabinogalactan protein in xylem cell walls of tomato species in reaction to Ralstonia solanacearum. Physiol Mol Plant Pathol. 2006;68:41–50. doi: 10.1016/j.pmpp.2006.06.001. [DOI] [Google Scholar]
- Yan S, Tang Z, Su W, Sun W. Proteomic analysis of salt stress-responsive proteins in rice root. Proteomics. 2005;5:235–244. doi: 10.1002/pmic.200400853. [DOI] [PubMed] [Google Scholar]