Abstract
2-Nitropropane (2-NP), an important industrial solvent and a component of cigarette smoke, is mutagenic in bacteria and carcinogenic in rats. 8-Amino-2′-deoxyguanosine (8-amino-dG) is one of the types of DNA damage found in liver, the target organ in 2-NP-treated rats. To investigate the thermodynamic properties of 8-amino-dG opposite each of the four DNA bases, we have synthesized an 11mer, d(CCATCG*CTACC), in which G* represents the modified base. By annealing a complementary DNA strand to this modified 11mer, four sets of duplexes were generated each containing one of the four DNA bases opposite the lesion. Circular dichroism studies indicated that 8-amino-dG did not alter the global helical properties of natural right-handed B-DNA. The thermal stability of each duplex was examined by UV melting measurements and compared with its unmodified counterpart. For the unmodified 11mer, the relative stability of the complementary DNA bases opposite G was in the order C > T > G > A, as determined from their –ΔG° values. The free energy change of each modified duplex was lower than its unmodified counterpart, except for the G*:G pair that exhibited a higher melting transition and a larger –ΔG° than the G:G duplex. Nevertheless, the stability of the modified 11mer duplex also followed the order C > T > G > A when placed opposite 8-amino-dG. To explore if 8-amino-dG opposite another 8-amino-dG has any advantage in base pairing, a G*:G* duplex was evaluated, which showed that the stability of this duplex was similar to the G*:G duplex. Mutagenesis of 8-amino-dG in this sequence context was studied in Escherichia coli, which showed that the lesion is weakly mutagenic (mutation frequency ∼10–3) but still can induce a variety of targeted and semi-targeted mutations.
INTRODUCTION
2-Nitropropane (2-NP) has been widely used as an industrial solvent and as a component of paints, inks and varnishes (1,2). It is also present in cigarette smoke (3). 2-NP is mutagenic in Salmonella typhimurium (4) and a potent hepatocarcinogen in rats (5,6). Analysis of 2-NP-treated rat liver DNA showed 8-oxo-2′-deoxyguanosine (8-oxo-dG) and 8-amino-2′-deoxyguanosine (8-amino-dG) as two major DNA modifications (7). It was proposed that 2-NP is metabolized to hydroxylamine-O-sulfonate or acetate that is capable of forming the reactive nitrenium ion, NH4+, which in turn aminates the 8 position of dG (7). The well known oxidative DNA damage, 8-oxo-dG, is a mutagenic lesion (8–10). Extensive thermodynamic and structural studies on 8-oxo-dG have been carried out in an attempt to relate its conformation and base pairing properties with its mutagenic specificity and repair (11–13).
8-Amino-dG has not been studied as extensively as 8-oxo-dG. Early studies on 8-amino-G riboside explored its base pairing modes and ability to form triple helical DNA and tetrameric structures (14–16). In a more recent biological study, incorporation of 8-amino-dGTP by mammalian DNA polymerases on a template DNA was measured (17). 8-Amino-dG was incorporated more efficiently than 8-oxo-dG by both viral reverse transcriptases and mammalian DNA polymerases (17). In the only in vivo study in which the mutagenic effects of 8-amino-dG were investigated, this lesion was found to be mutagenic inducing 2–3% transversions (primarily G→T base substitutions) in simian kidney cells (18). Under comparable conditions mutation frequency of 8-oxo-dG was estimated to be 2–4-fold higher, but the mutational specificity of the two lesions appeared to be similar (18).
In the current work, we have evaluated the thermodynamic stability of 8-amino-dG opposite all four bases in DNA using an 11mer, 5′-d(CCATCG*CTACC)-3′, in which G* represents the modified base. We also determined the stability of 8-amino-dG located opposite 8-amino-dG in the complementary strand, because the mode of self-pairing was the subject of much speculation in early studies. Finally, we have investigated the mutagenicity of 8-amino-dG in Escherichia coli using the same sequence context as used for the thermodynamic studies.
MATERIALS AND METHODS
Materials
All chemicals and solvents required for the synthesis of 8-amino-dG phosphoramidite monomer for the DNA synthesis were purchased from Aldrich Chemical Co. (Milwaukee, WI). [γ-32P]ATP was from Du Pont New England Nuclear (Boston, MA). T4 DNA ligase and T4 polynucleotide kinase were obtained from New England Biolabs (Beverly, MA). Escherichia coli strains DL7 and GW5100 are available in our laboratory.
Synthesis of oligonucleotides
The synthesis of 8-amino-dG phosphoramidite and its incorporation into an oligonucleotide has been reported (19). Since 8-amino-dG is susceptible to air oxidation, deprotection and subsequent manipulations were carried out in the presence of 2-mercaptoethanol (19). The modified oligonucleotides were purified by reverse phase HPLC followed by denaturing polyacrylamide gel electrophoresis. The homogeneity of the purified oligonucleotides was analyzed on a polyacrylamide gel after radiolabeling with [γ-32P]ATP in the presence of T4 polynucleotide kinase, and the presence of the modified base was confirmed by mass spectrometry using electrospray ionization. The unmodified oligonucleotides were purchased from commercial sources and purified by reverse phase HPLC.
Preparation of oligonucleotide duplexes
The concentration of the oligonucleotides was estimated from the extinction coefficients according to Borer (20). Since in guanosine the effect of introducing the 8-amino substituent on the magnitude of ɛ260 is negligible (14,15), we used the same ɛ values for both the unmodified and modified oligonucleotides. The stock solution of each oligonucleotide was prepared in a buffer containing 10 mM NaH2PO4 pH 7.0, 1 mM EDTA, 0.2 M NaCl and 0.7 M 2-mercaptoethanol and mixing experiments were performed to verify that the 1:1 mixture showed the greatest hyperchromicity.
Optical melting studies
Temperature-dependent changes in absorption (at 260 nm) of the oligonucleotide duplexes were measured in a Hewlett-Packard HP 8452A diode-array spectrophotometer equipped with a Peltier temperature controller. Oligonucleotides were dissolved in 10 mM NaH2PO4 pH 7.0, 1 mM EDTA, 0.2 M NaCl and 0.7 M 2-mercaptoethanol. The solution was heated to 70°C and slowly cooled to 4°C before temperature-dependent absorption measurements were carried out. The heating rate for these studies was 0.5°C/min and absorption was taken at 260 nm. Thermodynamic parameters were calculated from van’t Hoff plots as described in detail by Marky and Breslauer (21). At least eight different experiments were performed for each set of duplexes. The standard deviations were calculated as described by Persmerk and Guengerich (22).
Circular dichroism (CD)
CD spectra were measured in 10 mM NaH2PO4 pH 7.0, 1 mM EDTA, 0.2 M NaCl and 0.7 mM 2-mercaptoethanol using a Jasco Model J-710 spetropolarimeter equipped with a refrigerated circulating waterbath.
Construction of M13 genome containing a single 8-amino-dG and replication in E.coli
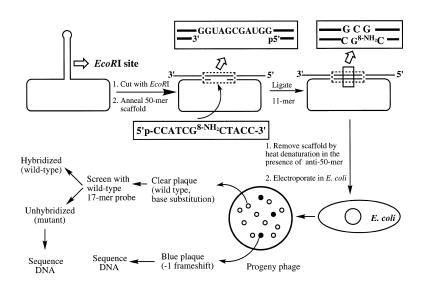
Construction of the modified and control M13 genome involved digestion of M13mp7L2 single-stranded DNA with EcoRI, annealing a 50mer scaffold and ligation of the 8-amino-dG-containing and control 11mer, which followed the protocol described earlier in detail (Scheme 1) 1 (23,24). Briefly, 400 µg bacteriophage M13mp7L2 DNA was digested with 3200 U of EcoRI for 2 h at 25°C in 1 ml 100 mM Tris–HCl pH 7.5, 5 mM MgCl2, 50 mM NaCl. An equimolar ratio of a scaffold 50mer was annealed to the linear single-stranded DNA at a concentration of 100 ng/ml by heating at 75°C for 15 min followed by slow cooling to room temperature over a period of 3–4 h. A 10-fold molar excess of the modified or unmodified 5′-phosphorylated 11mer was ligated into the gap of this annealed DNA in the presence of 800 U of T4 DNA ligase in 40 mM Tris–HCl buffer pH 7.8, 8 mM MgCl2, 16 mM dithiothreitol and 1 mM ATP at 16°C for 48 h. After ethanol precipitation, an additional round of EcoRI (5 U/µg DNA) digestion was carried out for 4 h to linearize any uncut or religated DNA. The efficiency of ligation was ∼34% for both the control and modified 11mer. To remove the 50mer scaffold from the M13 DNA, each DNA solution was heated at 100°C for 45 s and rapidly cooled to 0°C. Prior to heating, a 10-fold molar excess of an ‘anti-scaffold’ 50mer that contained the DNA sequence complementary to the scaffold oligomer was added to prevent the scaffold from reannealing on the M13 DNA.
Scheme 1. Construction of M13 genome containing a single 8-amino-dG and subsequent steps for mutational analysis.
Repair-competent E.coli (DL7) cells were grown in 100 ml cultures in Luria broth to 1 × 108 cells/ml and were made electrocompetent with and without SOS induction using UV light (254 nm; 20 J/m2) as described (23,24). For each transformation, 60 µl of the cell suspension was mixed with 60 ng M13 construct and transferred to the bottom of an ice-cold Bio-Rad Gene-Pulser cuvette (0.1 cm electrode gap). Electroporation of cells was carried out in a Bio-Rad Gene-Pulser apparatus at 25 µF and 1.8 kV with the pulse controller set at 200 Ω. Following a 1 h recovery at 37°C the cells were centrifuged to isolate the phage-containing supernatant. Analysis of progeny phage was carried out by oligonucleotide hybridization (23,24).
RESULTS
Thermodynamic stability of 8-amino-dG
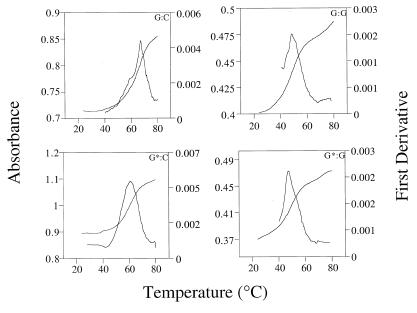
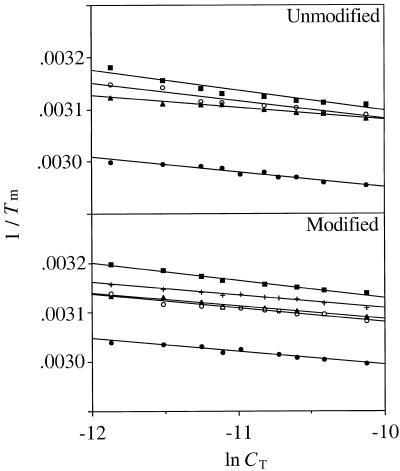
Thermal melting profile of the 8-amino-dG-containing and control 11mers showed cooperative transitions at all concentrations studied. Representative melting profiles are shown in Figure 1. The thermodynamic parameters were determined by van’t Hoff analysis of concentration-dependence of the optically monitored duplex melting transition as shown in the 1/Tm versus ln CT plots in Figure 2. For the unmodified duplex, C opposite G generated the most stable pair with a –ΔG° of ∼15 kcal/mol at 25°C followed by T, G and A, as expected (Table 1). For the duplexes containing 8-amino-dG, the trend appeared to be similar with a small decrease in stability of the modified pair compared to its unmodified counterpart, except for the G*:G duplex that was found to be more stable than the unmodified G:G pair. This suggests that amination of the 8 position of guanine destabilizes the helix but does not cause severe perturbations and that G*:G pair may have some additional favorable interactions relative to G:G mismatched pair. Since in the G:G mispair one of the Gs adopts a syn (or syn-like) structure while the other G remains in normal anti conformation (25), it is likely that the 8-amino group may allow for a more facile syn rotation. In the 1970s 8-amino-G was the subject of several studies that explored self-structure and triple helix formation, although these studies employed the ribonucleic acid polymers (14–16). We were therefore interested to determine the pairing mode of 8-amino-dG with another 8-amino-dG in the complementary strand. We found that the –ΔG° value of G*:G* in this sequence was very similar to that of G*:G. Even though G*:G* was more stable than the G:G pair, the difference is small and it does not appear to stabilize the duplex by some novel ordered structure.
Figure 1.
Representative thermal denaturation and first derivative curves for d(CCATCG*CTACC) and its unmodified analog annealed to d(GGTAGCGATGG) (left panels) or d(GGTAGGGATGG) (right panels).
Figure 2.
van’t Hoff plots of 1/Tm versus ln CT for helix-to-coil transitions of d(CCATCG*CTACC) and its unmodified analog annealed to an 11mer d(GGTAGNGATGG) where N denotes C (filled circle), T (triangle), G (open circle), A (square) or G* (+).
Table 1. Melting temperature and thermodynamic parameters for DNA duplexes containing dG or 8-amino-dG.
| Duplex |
Tm (°C)a |
–ΔG° (kcal/mol)b |
–ΔH° (kcal/mol)b |
–ΔS° (cal/mol.K)b |
| G:C |
63.4 |
15.0 ± 0.4 |
71.4 ± 6.0 |
189.3 ± 16.1 |
| G*:C |
59.4 |
14.7 ± 0.6 |
76.9 ± 6.0 |
208.7 ± 16.5 |
| G:T |
50.2 |
13.7 ± 0.5 |
87.9 ± 6.4 |
249.1 ± 18.3 |
| G*:T |
49.4 |
12.4 ± 0.5 |
75.1 ± 6.3 |
210.3 ± 17.9 |
| G:G |
49.2 |
11.2 ± 0.5 |
59.1 ± 6.6 |
160.6 ± 18.3 |
| G*:G |
50.2 |
12.3 ± 0.5 |
71.7 ± 6.1 |
134.4 ± 16.8 |
| G:A |
47.8 |
10.3 ± 0.4 |
50.4 ± 6.1 |
134.4 ± 16.8 |
| G*:A |
44.4 |
10.2 ± 0.2 |
56.0 ± 3.4 |
153.8 ± 9.5 |
| G*:G* | 46.8 | 12.0 ± 0.3 | 76.3 ± 3.9 | 215.7 ± 11.3 |
aConditions: 10 mM NaH2PO4 pH 7.0, 1 mM EDTA, 0.2 M NaCl, 0.7 mM 2-mercaptoethanol, 25 µM DNA. The precision of each determination was 0.5°C.
bObtained by plotting 1/Tm versus ln CT. At least eight concentrations were used in each case.
CD
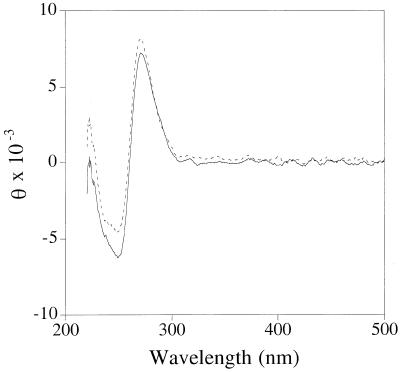
As shown in Figure 3, the CD spectra of unmodified and 8-amino-dG-containing 11mer duplexes opposite C were virtually identical suggesting that 8-amino-dG does not alter the global helical properties of natural right-handed B-DNA.
Figure 3.
CD spectra of modified (solid line) and unmodified 11mer (broken line) annealed to d(GGTAGCGATGG). For details see Materials and Methods.
Biological effects of 8-amino-dG
The construction of 8-amino-dG containing M13 genome, its replication in repair-competent E.coli cells, and analysis of the progeny phage were accomplished according to Scheme 1. Replication of the modified and control M13 genome in E.coli showed that the presence of 8-amino-dG did not affect viability, suggesting that the lesion does not interfere with translesion synthesis (Table 2). For screening mutant progeny, we used the following approach. The M13 construct formed by ligation of the 11mer is a +1 derivative of M13mp7, which gave clear plaques in the presence of IPTG and X-gal. A –1 deletion should restore the reading frame, resulting in the formation of blue plaques, which could be confirmed by DNA sequencing. Using this strategy we detected four one-base deletion mutants from a total of 8403 plaques screened. Only one of these contained a targeted G deletion and the rest contained semi-targeted one-base deletions (Table 3). To identify base substitution and other frame shift mutations, we used oligonucleotide hybridization using a 17mer probe complementary to the region where the 11mer was inserted. The probe was designed to bind to non-mutant plaques, and any plaques that did not hybridize or hybridized weakly were subjected to DNA sequencing (23,24). As shown in Table 3, of the 8403 clear plaques screened, only five base substitution and one one-base addition mutants were identified. None of these point mutations occurred at the lesion site. It is noteworthy that the induction of SOS did not have a significant impact on mutagenesis by 8-amino-dG. The combined mutation frequency was therefore 10 out of 8403. For the control construct, no mutants were detected after screening >9000 plaques. It is intriguing that the frequency of 8-amino-dG induced mutations is at least an order of magnitude higher in simian kidney cells (18) than in E.coli.
Table 2. Viability of 8-amino-dG containing M13 in E.colia.
| Experiment no.b | Relative survival | |||
| |
Control (%) |
|
8-amino-dG (%) |
|
| |
–SOS |
+SOSc |
–SOS |
+SOSc |
| 1 |
100 |
100 |
71 |
93 |
| 2 |
100 |
100 |
135 |
130 |
| 3 |
100 |
100 |
93 |
72 |
| Average | 100 | 100 | 100 | 98 |
aViability (%) was determined by comparing the number of plaques from the 8-amino-dG containing DNA with that of the control genome (assumed to have 100% viability).
bEach experiment refers to a separate construction of the control and the adducted genomes.
cSOS was induced by UV irradiation at 20 J/m2 as described by Bacolod et al. (24).
Table 3. Analysis of progeny phage.
| Experiment no. | Control | 8-amino-dG | ||
| |
Plaques screened |
Mutants |
Plaques screened |
Mutants |
| |
–SOS |
|
–SOS |
|
| 1 |
1680 |
0 |
907 |
1a |
| 2 |
1114 |
0 |
1151 |
0 |
| 3 |
980 |
0 |
1026 |
1b |
| Total |
3774 |
0 |
1026 |
2 |
| |
+SOS |
|
+SOS |
|
| 1 |
1480 |
0 |
1178 |
3c |
| 2 |
2166 |
0 |
2737 |
3d |
| 3 |
1934 |
0 |
1404 |
2e |
| Total | 5580 | 0 | 5319 | 8 |
Mutations found include:
aDeletion of C7 from 5′-C1C2A3T4C5G6C7T8A9C10C11-3′.
bBase substitution from C7→G.
cBase substitution from A3→T, T4→C, C5→G.
dDeletion of G6, addition of C, 3′ to G6, base substitution from A9→G.
eDeletion of A3, deletion of C5.
DISCUSSION
2-NP forms two lesions at the 8 position of 2′-deoxyguanosine to give 8-oxo-dG and 8-amino-dG. Although 8-amino-dG has not been studied as extensively as 8-oxo-dG, several notable differences between these two small lesions can be discerned. The 6,8-diketo form of 8-oxo-dG, which predominates under normal cellular condition (13), alters the hybridization state of N7 of dG and 8-oxo-dG exists in an equilibrium of anti and syn orientation. 8-Oxo-dG is mutagenic in bacterial and mammalian cells inducing primarily G→T substitutions (8,9), but it can also induce A→C transversions by misincorporation of 8-oxo-dGTP opposite dA (10). Spectroscopic and calorimetric studies of 13mer duplexes containing an 8-oxo-dG in a CG*C sequence context showed that free energy changes of the thermally induced order to disorder transition of the modified duplexes are lower than control duplexes (11). What is more interesting, however, is that whereas a thermodynamic comparison of the normal G:C pair with a mismatched G:A pair is characterized by a large free energy difference, which is the result of an unfavorable difference in transition enthalpy, the change from G8-oxo:C→G8-oxo:A pair is thermodynamically benign (11). Multiple DNA repair systems have been identified in bacterial and mammalian cells, which repair 8-oxo-dG (26,27). Escherichia coli utilizes a two-base excision repair glycosylase, Fpg (MutM) and MutY for 8-oxo-dG repair. Another enzyme, MutT, catalyzes the hydrolysis of 8-oxo-dGTP to 8-oxo-dGMP, thus preventing its incorporation into DNA. Functional homologs of some of these proteins in humans have been identified (27). By contrast, it is unknown if a DNA repair system that excises 8-amino-dG exists, even though it can no longer be characterized as a benign lesion as evidenced by its mutagenicity in simian kidney cells (18) and E.coli. In a study in vitro by Loeb and coworkers (17), incorporation efficiencies of 8-oxo-dGTP and 8-amino-dGTP by several reverse transcriptases and mammalian DNA polymerases were compared. HIV-1 reverse transcriptase, murine leukemia virus reverse transcriptase and pol α exhibit a strong bias against the incorporation of 8-oxo-dG, whereas facile utilization of 8-amino-dGTP was observed with all the enzymes. The repair polymerase pol β incorporates both analogs of dGTP efficiently. These data, taken together, may suggest that even though both 8-oxo-dG and 8-amino-dG can be classified as small non-distorting lesions, only the latter is capable of interacting with the DNA polymerases in a manner similar to undamaged dG or its triphosphate. The results of the current study are consistent with this hypothesis. The thermodynamic studies showed that 8-amino-dG destabilizes the DNA helix, albeit not very significantly. With the exception of G*:G and G*:G* pair, which showed favorable –ΔG° and –ΔH° values compared to the G:G mispair, each modified pair exhibited a slightly lower –ΔG° value than the unmodified duplex. A comparison of the extent of destabilization by 8-amino-dG and 8-oxo-dG can be made in the following manner. Relative to the G:C pair, G8-oxo:C destabilizes a 13mer duplex by 2.0 ± 0.7 kcal/mol (11), whereas 0.3 ± 1.0 kcal/mol destabilization was observed for the G8-amino:C 11mer duplex compared to its unmodified counterpart. Also noteworthy is the finding that relative to the G:A mismatch, 8-oxo-dG is stabilizing opposite dA, but no such stabilizing effect was observed for 8-amino-dG. Indeed, unlike 8-oxo-dG, which sometimes adopts a syn orientation, the thermodynamic data for 8-amino-dG do not suggest a significant departure from the pairing modes of dG. The results of the biological studies provide additional evidence that 8-amino-dG behaves like dG. Viability of 8-amino-dG-containing genome is similar to the control, and mutagenesis occurred at a frequency of 10–3. Nevertheless, various base substitution and frame shift mutations at or near the 8-amino-dG site were detected. Since 8-amino-dG does not stall DNA replication, it is conceivable that the high level of this damage may still contribute to significant mutagenic effects. In view of the potential link of 8-amino-dG with 2-NP carcinogenesis, further studies on its repair, persistence in DNA and mutagenesis should be carried out.
Acknowledgments
ACKNOWLEDGEMENTS
This study was supported by NIEHS grant ES09127 (to A.K.B.) and NCI grant CA47995 (to F.J.). A.K.B. is a recipient of a Research Career Development Award from the NIEHS (grant 1 K02 ES00318).
References
- 1. IARC (1982) IARC Monographs on the Evaluation of Carcinogenic Risk of Chemicals to Man. Some Industrial Chemicals and Dyestuffs. IARC, Lyon, Vol. 29, pp. 331–343.
- 2. National Toxicology Program (1998) Eighth Report on Carcinogens. US Department of Health and Human Services, Washington, DC, pp. 782–783.
- 3.Hoffmann D.H. and Rathkamp,G. (1968) Chemical studies on tobacco smoke. III. Primary and secondary nitroalkanes in cigarette smoke. Beitr. Tabakforsch., 4, 124–134. [Google Scholar]
- 4.Speck W.T., Meyer,L.W., Zeiger,E. and Rosenkranz,H.S. (1982) Mutagenicity and DNA modifying activity of 2-nitropropane. Mutat. Res., 104, 49–54. [DOI] [PubMed] [Google Scholar]
- 5.Lewis T.R., Ulrich,C.E. and Bushey,W.M. (1979) Subchronic inhalation toxicity of nitromethane and 2-nitropropane. J. Environ. Pathol. Toxicol., 2, 233–249. [PubMed] [Google Scholar]
- 6.Fiala E.S., Czerniak,R., Castonguay,A., Conaway,C.C. and Rivenson,A. (1987) Assay of 1-nitropropane, 2-nitropropane, 1-azoxypropane, and 2-azoxypropane for carcingenicity by gavage in Sprague-Dawley rats. Carcinogenesis, 8, 1947–1949. [DOI] [PubMed] [Google Scholar]
- 7.Sodum R.S., Nie,G. and Fiala,E.S. (1993) 8-Aminoguanine: a base modification produced in rat liver nucleic acids by the hepatocarcinogen 2-nitropropane. Chem. Res. Toxicol., 6, 269–276. [DOI] [PubMed] [Google Scholar]
- 8.Wood M.L., Dizdaroglu,M., Gajewski,E. and Essigmann,J.M. (1990) Mechanistic studies of ionizing radiation and oxidative mutagenesis: genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry, 29, 7024–7032. [DOI] [PubMed] [Google Scholar]
- 9.Moriya M. (1993) Single-stranded shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxoguanine in DNA induces targeted G.C → T.A transversions in simian kidney cells. Proc. Natl Acad. Sci. USA, 90, 1122–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng K.C., Cahill,D.S., Kasai,H., Nishimura,S. and Loeb,L.A. (1992) 8-Hydroxyguanine, and abundant form of oxidative DNA damage, causes G → T and A → C substitutions. J. Biol. Chem., 267, 166–172. [PubMed] [Google Scholar]
- 11.Plum G.E., Grollman,A.P., Johnson,F. and Breslauer,K.J. (1995) Influence of the oxidatively damaged adduct 8-oxodeoxyguanosine on the conformation, energetics, and thermodynamic stability of a DNA duplex. Biochemistry, 34, 16148–16160. [DOI] [PubMed] [Google Scholar]
- 12.Kouchakdjian M., Bodepudi,V., Shibutani,S., Eisenberg,M., Johnson,F., Grollman,A.P. and Patel,D.J. (1991) NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyguanosine in a DNA duplex. 9-oxo-7H-dG (syn).dA (anti) alignment at lesion site. Biochemistry, 30, 1403–1420. [DOI] [PubMed] [Google Scholar]
- 13.Cho B.P., Kadlubar,F.F., Culp,S.J. and Evans,F.E. (1990) 15N nuclear magnetic resonance studies on the tautomerism of 8-hydroxy-2′-deoxyguanosine, 8-hydroxyguanosine, and other C8-substituted guanine nucleosides. Chem. Res. Toxicol., 3, 445–452. [DOI] [PubMed] [Google Scholar]
- 14.Hattori M., Frazier,J. and Miles,H.T. (1975) Poly(8-aminoguanylic acid): formation of ordered self-structures and interaction with poly(cytidylic acid). Biochemistry, 14, 5033–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hattori M., Frazier,J. and Miles,H.T. (1975) Ordered forms of 5′-8-aminoguanylic acid. Biopolymers, 14, 2095–2106. [DOI] [PubMed] [Google Scholar]
- 16.Hattori M., Frazier,J. and Miles,H.T. (1976) The structure of triple-stranded G.2C polynucleotide helices. Biopolymers, 15, 523–531. [DOI] [PubMed] [Google Scholar]
- 17.Kamath-Loeb A.S., Hizi,A., Kasai,H. and Loeb,L.A. (1997) Incorporation of the guanosine triphosphate analogs 8-oxo-dGTP and 8-NH2-dGTP by reverse transcriptases and mammalian DNA polymerases. J. Biol. Chem., 28, 5892–5898. [DOI] [PubMed] [Google Scholar]
- 18.Tan X., Suzuki,N., Johnson,F., Grollman,A.P. and Shibutani,S. (1999) Mutagenic properties of the 8-amino-2′-deoxyguanosine DNA adduct in mammalian cells. Nucleic Acids Res., 27, 2310–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rieger R.A., Iden,C.R., Gonikberg,E. and Johnson,F. (1999) 8-Amino-2′-deoxyguanosine incorporation into oligomeric DNA. Nucl. Nucl., 18, 75–88. [DOI] [PubMed] [Google Scholar]
- 20.Borer P.N. (1975) In Fasman,G.D. (ed.), Handbook of Biochemistry and Molecular Biology, 3rd edn. CRC Press, Cleveland, OH, pp. 589.
- 21.Marky L.A. and Breslauer,K.J. (1987) Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers, 26, 1601–1620. [DOI] [PubMed] [Google Scholar]
- 22.Persmark M. and Guengerich,F.P. (1994) Spectroscopic and thermodynamic characterization of the interaction of N7-guanyl thioester derivative of d(TGCTG*CAAG) with potential complements. Biochemistry, 33, 8662–8672. [DOI] [PubMed] [Google Scholar]
- 23.Ramos L.A., Lipman,R., Tomasz,M. and Basu,A.K. (1998) The major mitomycin C-DNA monoadduct is cytotoxic but not mutagenic in Escherichia coli. Chem. Res. Toxicol., 11, 64–69. [DOI] [PubMed] [Google Scholar]
- 24.Bacolod M.D., Krishnasamy,R. and Basu,A.K. (2000) Mutagenicity of the 1-nitropyrene-DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene in Escherichia coli located in a nonrepetitive CGC sequence. Chem. Res. Toxicol., 13, 523–528. [DOI] [PubMed] [Google Scholar]
- 25.Cognet J.A.H., Gabarro-Arpa,J., Le Bret,M., van der Marel,G.A., van Boom,J.H. and Fazakerley,G.V. (1991) Solution conformation of an oligonucleotide containing a G.G mismatch determined by nuclear magnetic resonance and molecular mechanics. Nucleic Acids Res., 19, 6771–6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bessho T., Roy,R., Yamamoto,K., Kasai,H., Nishimura,S., Tano,K. and Mitra,S. (1993) Repair of 8-hydroxyguanine in DNA by mammalian N-methypurine-DNA glycosylase. Proc. Natl Acad. Sci. USA, 90, 8901–8904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.David S.S. and Williams,S.D. (1998) Chemistry of glycosylases and endonucleases involved in base-excision repair. Chem. Rev., 98, 1221–1261. [DOI] [PubMed] [Google Scholar]