Abstract
During our search for Streptomyces spp. as new producers of bioactive secondary metabolites, the ethyl acetate extract of the new terrestrial Streptomyces isolate TN262 delivered eight antimicrobially active compounds. They were identified as 1-acetyl-β-carboline (1), tryptophol (2), cineromycin B (3), 2,3-dihydrocineromycin B (4), cyclo-(tyrosylprolyl) (5), 3-(hydroxyacetyl)-indole (6), brevianamide F (7), and cis-cyclo-(l-prolyl-l-leucyl) (8). Three further metabolites were detected in the unpolar fractions using GC–MS and tentatively assigned as benzophenone (9), N-butyl-benzenesulfonamide (10), and hexanedioic acid-bis-(2-ethylhexyl) ester (11). This last compound is known as plasticizer derivatives, but it has never been described from natural sources. In this article, we describe the identification of the new Streptomyces sp. isolate TN262 using its cultural characteristics, the nucleotide sequence of the corresponding 16S rRNA gene and the phylogenetic analysis, followed by optimization, large-scale fermentation, isolation of the bioactive constituents, and determination of their structures. The biological activity of compounds (2), (3), (4), and those of the unpolar fractions was addressed as well.
Keywords: New Streptomyces sp.TN262, Identification, Antimicrobial activities, Macrolides, Natural hexanedioic acid-bis-(2-ethylhexyl) ester
Introduction
The dramatic rise of antibiotic resistance in human pathogenic microorganisms gave good reasons for the continuous development of novel antibiotics. Filamentous soil bacteria belonging to the genus Streptomyces are still rich sources of bioactive natural products with a great functional diversity [1], which are extensively used as pharmaceuticals and agrochemicals [1–4].
During our search program for bioactive compounds from actinomycetes, a new terrestrial bacterium was isolated, selected, and identified as Streptomyces sp. TN262 strain, followed by studies of the influences of different nutritional compounds on the biosynthesis of its bioactive molecules. The strain exhibited a potent activity against Micrococcus luteus LB 14110, Escherichia coli ATCC 8739, Staphylococcus aureus ATCC 6538, Salmonella enterica ATCC43972, and Fusarium sp. Chemical screening of the extract using TLC displayed the existence of four blue UV fluorescent (366 nm) or absorbing (254 nm) bands, respectively, besides a major one, which turned dark violet after spraying with anisaldehyde/sulfuric acid and showed only weak UV absorbance. Upscaling of the strain at optimum conditions followed by chromatographic separation afforded eight compounds 1–8. Further, components were tentatively assigned by GC–MS analysis of the less polar fractions I and II as benzophenone (9), N-butyl-benzenesulfonamide (10), and as the naturally hexanedioic acid-bis(2-ethylhexyl)ester (11).
Materials and Methods
Spectroscopic Measurements
The NMR spectra were measured on a Bruker AMX 300 (300.135 MHz), a Varian Unity 300 (300.145 MHz), and a Varian Inova 600 (150.820 MHz) spectrometer. ESI MS was recorded on a Finnigan LCQ with quaternary pump Rheos 4000 (Flux Instrument). EI mass spectra were recorded on a Finnigan MAT 95 spectrometer (70 eV) with perfluorkerosine as reference substance for EI HRMS. GC–MS was measured on a Trace GC–MS Thermo Finnigan instument (ionization mode EI, 70 eV), equipped with a capillary column CP-Sil 8 CB for amines (length = 30 m; inside diameter = 0.25 mm; outside diameter = 0.35 mm; film thickness = 0.25 µm). The analysis was carried out at a programmed temperature: initial temperature 40 °C (kept for 1 min), then increasing at a rate of 10 °C/min to a final temperature of 280 °C (kept for 10 min). In the EI mode, the injector and detector temperature was set to 250 °C; he was used as carrier gas at flow rate of 1 ml/min, total run time 27 min, injection volume 0.2 µl. Flash chromatography was carried out on silica gel (230–400 mesh). Rf values were measured on Polygram SIL G/UV254 (Macherey-Nagel & Co.). Size exclusion chromatography was done on Sephadex LH-20 (Pharmacia). Optical rotations were measured on a Perkin–Elmer polarimeter (model 343).
Microorganisms and Plasmids
The chromosomal DNA of the terrestrial Streptomyces sp. isolate TN262 was used to amplify the 16S rRNA gene. E. coli TOP10 (Invitrogen), F-mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG, and E. coli DH5α [1] were used as host strains. Bacterial strains M. luteus LB 14110, S. aureus ATCC 6538, E. coli ATCC 8739, and S. enterica ATCC43972 were used as indicator microorganisms for the antibacterial activity assays. Antifungal activity was determined against a Fusarium sp.
pIJ2925 derivative of pUC18 and pCR-Blunt vector (Invitrogen) Col E1 origin (pUC-derived) KnR were used as the cloning vectors. pLE1 is a derivative of pCR-Blunt vector carrying a 1.5-kb fragment corresponding to the whole 16S rRNA gene of the TN262 strain (this work). pLE2 and pLE3 are derivatives of pIJ2925 carrying, respectively, the 0.9- and 0.6-kb EcoRI–EcoRI DNA fragments of the 16S RNA gene of the TN262 strain (this work).
Culture Conditions
E. coli DH5α was grown on Luria–Bertani (LB) plates supplemented with ampicillin (50 μg/ml) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (40 μg/ml) when appropriate [1]. Transformation of E. coli DH5α with pIJ2925 derivatives was carried out according to Hanahan [5]. Growth and transformation of TOP10 E. coli strain with the pCR-Blunt vector derivative were carried out according to the manufacturer’s instructions (Invitrogen). For the isolation of actinomycete strains, soil and water samples collected from different Tunisian regions were spread on solid boiled bran barley medium [6]: 0.2% yeast extract and 2% agar were added to a supernatant of 4% boiled bran barley. The pH was adjusted to 7. After incubation at 30 or 40 °C for several days, colonies showing sporulation and filamentous morphology were picked and propagated on the same solid medium. For determination of antibacterial activities, indicator microorganisms were grown overnight in LB medium, M. luteus LB 14110 and S. enterica ATCC43972 at 30 °C and E. coli ATCC 8739 and S. aureus ATCC 6538 at 37 °C, then diluted 1:100 in LB medium and incubated for 5 h under constant agitation (200 rpm) at the appropriate temperature. For antifungal testing, Fusarium sp. was grown in potato dextrose agar for 7 days at 30 °C. Spores were collected in sterile distilled water and then concentrated to produce a suspension with approximately 104 spores/ml.
The TN262 strain was grown in tryptic soy broth (TSB) at 30 g/l for the preparation of genomic DNA [1]. Cultural characteristics of TN262 strain were compared on the basis of observations made after 7, 14, and 21 days incubation on nutrient agar, Sabouraud agar, and yeast malt agar media [1]. To investigate the influence of the medium on antimicrobial production, spores at 107/ml were used to inoculate 500-ml Erlenmeyer flasks with four indents, containing 100 ml of TSB medium (30 g/l). After incubation at 30 °C for 24 h, this preculture was used to inoculate at 1/10 (v/v) 1,000-ml Erlenmeyer flasks with four indents, containing 200 ml of modified Bennett medium (peptone 2 g/l, yeast extract 1 g/l, beef extract 1 g/l) supplemented at 1% (w/v) with one of the five tested carbon sources (starch, fructose, glycerol, glucose, and saccharose). After incubation at 30 °C for 72 h in an orbital incubator with shaking at 200 rpm, biological activities were assayed for each culture supernatant. Influence of magnesium, potassium, and trace mineral oligoelements on active molecules production was also investigated by addition of these chemical additives to the culture medium, additive by additive, the combination of two additives, and the addition of all three additives. The final magnesium and potassium concentration was 2 and 1 mmol/l, respectively. For trace mineral oligoelements (40 mg ZnCl2, 200 mg FeSO4·7H2O, 6.5 mg H3BO3, and 13.5 mg MoNa2O4·2H2O per 100 ml distilled water), 1.5 ml were added to 200 ml of growth medium. Eight different concentrations (% w/v) of glycerol were tested: 0.2; 0.5; 0.75; 1; 1.25; 1.5; 1.75; and 2. The effect of culture conditions like different incubation temperatures (25, 30, 35, and 40 °C), initial pH (6, 6.5, 7, 7.2, 7.5, and 8), incubation times (24, 36, 48, 60, 72, 84, 96, 110, 120, and 144 h), and rotary shaker (100, 150, 200, 250, and 300 rpm) on growth and antimicrobial activities production was studied. Biomass of the Streptomyces sp. TN262 strain was determined by measuring the dry weight after drying at 105 °C.
DNA Isolation and Manipulation
Total DNA preparation from TN262 strain was carried out according to Hopwood et al. [7]. Small-scale plasmid preparations from E. coli were performed as described by Sambrook et al. [8]. Digestion with restriction endonucleases, separation of DNA fragments by agarose gel electrophoresis, dephosphorylation with alkaline calf intestinal phosphatase, ligation of DNA fragments, and transformation of E. coli were done according to Sambrook et al. [8].
PCR Amplification of the 16S rRNA Gene of TN262 Strain
PCR amplification of the 16S rRNA gene of TN262 strain was performed using two primers 5′-AGAGTTTGATCCTGGCTCAG-3′ and 5′-AAGGAGGTGATCCAGCCGCA-3′ as described by Edwards et al. [9]. Approximately 200 ng genomic template DNA was used with 150 pmol of each primer per 50 μl reaction volume. To improve the denaturation of the DNA, 5% (v/v) DMSO was added to the reaction mixture. Amplification was performed in an automated thermocycler (Perkin Elmer) using 1 U Pfu DNA polymerase (Stratagene) and the recommended buffer system according to the following amplification profile: 94 °C (3 min) followed by 45 cycles of denaturation at 94 °C (30 s), annealing at 60 °C (1 min), and extension at 72 °C (3 min). The PCR reaction mix was analyzed by agarose gel electrophoresis, and the DNA of the expected size was purified and then cloned into pCR-Blunt vector yielding pLE1.
DNA Sequencing and Analysis
Nucleotide sequence of the 16S rRNA gene of TN262 strain was determined on both strands by an automated 3100 Genetic Analyzer (Applied Biosystems) using specific primers. Homology search was performed using BLAST Search algorithm. The nucleotide sequence of the whole 16S rRNA gene (1,540 bp) of TN262 strain has been assigned GenBank (EMBL) under accession number FM867598. Multiple sequence alignment was carried out using clustal W [10] at the European Bioinformatics Institute website (http://www.ebi.ac.uk/clustalw/). Phylogenetic analyses were performed using programs from the PHYLIP package [11], and phylogenetic tree was constructed by the neighbor joining (NJ) algorithm [12] using Kimura 2-parameter distance. The robustness of the inferred tree was evaluated by bootstrap (100 replication).
Fermentation and Working Up
Spores at 107/ml of Streptomyces strain TN262 were used to inoculate 1,000-ml Erlenmeyer flasks, each containing 200 ml of TSB medium at 30 g/l. After incubation at 30 °C for 24 h in an orbital incubator with shaking at 200 rpm; the preculture was used to inoculate 20 l of modified Bennett medium, supplemented at 1% (w/v) with glycerol. For this realization, we have used two fermenters having each one a total capacity of 15 l (working capacity of 10 l). After 3 days fermentation (30 °C), the fermenter culture broth was subjected to filtration over celite using a filter press. The filtrate was exhaustively extracted by ethyl acetate (2×) and the obtained organic extract concentrated in vacuo to dryness, affording 2.4 g of a reddish brown crude extract.
Isolation of the Bioactive Constituents
The obtained crude extract (2.4 g) was fractionated by Sephadex LH-20 column chromatography (100 × 3 cm, DCM/40% MeOH). After TLC monitoring and visualization by UV light and anisaldehyde/sulfuric-acid-spraying reagent, four fractions were collected. The middle polar fraction III (600 mg), containing numerous components, was applied to a silica gel column and afforded three subfractions; IIIa (120 mg), IIIb (270 mg), and IIIc (150 mg). The subfraction IIIa was further purified using a silica gel column (DCM/40% MeOH) followed by Sephadex (DCM/40% MeOH) to afford 1-acetyl-β-carboline (1) as a faint yellow solid (2.3 mg) and colorless tryptophol (2, 1.9 mg). The major subfraction IIIb delivered three further components on a silica gel column: The first of them was further purified on RP-18 (20 × 1.5 cm, MeOH/H2O) followed by Sephadex LH-20 (MeOH) to give a colorless solid (21.1 mg) of cineromycin B (3) containing traces of 2,3-dihydrocineromycin B (4). The remaining two components delivered in a similar way on Sephadex LH-20 (DCM/40% MeOH) colorless tryptophol (2) and cyclo-(tyrosylprolyl) (5, 28.7 mg). Similarly, the subfraction IIIc was further purified to afford 3-(hydroxyacetyl)-indole (6, 5.1 mg) as colorless solid. Finally, the polar fraction IV (400 mg) gave brevianamide F (7, 2.3 mg) and cis-cyclo-(l-prolyl-l-leucyl) (8, 15.3 mg) as two colorless solids.
The unpolar two fractions I (250 mg) and II (80 mg) displayed similar TLC profiles and were combined. Spraying with anisaldehyde/sulfuric acid displayed numerous components, some of which turned blue. Three components were tentatively assigned by GC–MS analysis as benzophenone (9), N-butyl-benzenesulfonamide (10), and hexanedioic acid-bis(2-ethylhexyl)ester (11). Details of the GC–MS conditions were described under the section of “Spectroscopic Measurements.”
Cineromycin B (3) C17H26O4 (294); colorless solid, turned dark violet by anisaldehyde/sulfuric acid; Rf = 0.26 (CH2Cl2/10% MeOH); [α]25D −111 (c = 0.1, MeOH); 1H NMR (300 MHz, CDCl3) and 13C NMR (125 MHz, CDCl3): see Table 1; CI MS (NH3) m/z (%) = 608 ([2 M+NH4]+, 2), 312 ([M+NH4]+, 56), 294 ([M-H2O+NH4]+, 14); (+)-ESI MS m/z (%) = 613 ([2 M+Na]+, 6), 317 ([M+Na]+, 49); (+)-HRESI MS m/z 317.17236 ([M+Na]+, calcd: 317.17234 for C17H26O4Na).
Table 1.
13C and 1H NMR data (CDCl3, J in Hz) of Cineromycin B (3), compared with literature values.
| No. | Cineromycin B (3), exp. | Cineromycin B (3), lit. [23] | ||
|---|---|---|---|---|
| δC | δH | δC | δH | |
| 1 | 166.1 | – | 166.3 | – |
| 2 | 115.1 | 5.87 (d, 15.5) | 115.0 | 5.86 (d, 16) |
| 3 | 155.2 | 6.93 (d, 15.5) | 155.5 | 6.91 (d, 16) |
| 4 | 73.4 | – | 73.4 | – |
| 5 | 135.2 | 5.87 (d, 15.5) | 135.3 | 5.85 (d, 16) |
| 6 | 133.6 | 5.73 (dd, 16.3, 4.8) | 133.6 | 5.71 (dd, 16, 5) |
| 7 | 74.6 | 4.55 (m) | 74.5 | 4.53 (d, 5) |
| 8 | 137.9 | – | 137.9 | – |
| 9 | 129.5 | 5.23 (m) | 129.4 | 5.21 (brm) |
| 10 | 25.1 | 2.10 (m), 1.80 (m) | 25.0 | 2.10 (brm), 1.84 (d, 16.5) |
| 11 | 34.8 | 1.21 (m), 1.16(m) | 34.3 | 1.22 (m), 1.17 (m) |
| 12 | 39.5 | 1.25 (m) | 39.5 | 1.40 (brq, 6) |
| 13 | 75.5 | 4.55 (m) | 75.6 | 4.55 (d, 7) |
| CH3-4 | 27.1 | 1.55 (s) | 27.0 | 1.53 (s) |
| CH3-8 | 15.0 | 1.73 (s) | 15.0 | 1.71 (s) |
| CH3-12 | 16.1 | 0.89 (d, 6.6) | 16.1 | 0.88 (d, 6) |
| CH3-13 | 18.2 | 1.22 (d, 6.3) | 18.1 | 1.21 (d, 7) |
2,3-Dihydrocineromycin B (4) C17H28O4 (296); colorless solid with same color reaction as 3; Rf = 0.42 (CH2Cl2/10% MeOH); CI MS (NH3) m/z (%) = 610 ([2 M+NH4]+, 6), 314 ([M+NH4]+, 100), 296 ([M-H2O+NH4]+, 66); (+)-ESI MS m/z (%) = 615 ([2 M+Na]+, 12), 319 ([M+Na]+, 100); (+)-HRESI MS m/z 319.18798 ([M+Na]+, calcd: 319.18799 for C17H28O4Na). The NMR data were identical with recently published values [13].
Antimicrobial Activities
Agar diffusion tests against M. luteus LB 14110, S. aureus ATCC 6538 (Gram-positive bacteria), E. coli ATCC 8739, S. enterica ATCC43972 (Gram-negative bacteria), and a Fusarium sp. were used. For the determination of antibacterial activities in solid media, after incubation of the selected actinomycete strains for 7 days at the appropriate growth temperature, an agar disk was recuperated and picket in LB plates covered by 3 ml of top agar containing 50 μl of a 5-h culture of one of the four indicator bacteria. After 2 h at 4 °C, plates containing M. luteus, LB 14110, and S. enterica ATCC43972 were incubated overnight at 30 °C, and those inoculated with E. coli ATCC 8739 and S. aureus ATCC 6538 were incubated overnight at 37 °C. For antifungal activities determination, plates were covered by 3 ml of top agar containing 50 µl of spore suspension prepared from the Fusarium sp. After 2 h at 4 °C, plates were incubated at 30 °C for 48 h. For the determination of the antimicrobial activities from liquid media, a paper disk was impregnated with 50 μl of the corresponding sample and then laid on the surface of an agar plate containing 3 ml of top agar inseeded with the indicator microorganism using the same conditions as for the solid medium analysis. The antimicrobial activities of the pure compounds were determined against the same indicator cells under the same conditions as those from liquid media. Plates were examined for antimicrobial activities represented by an inhibition zone of the corresponding indicator microorganisms.
Results and Discussion
Isolation, Identification, and Phylogenetic Analysis of TN262 Strain
A new actinomycete strain isolate TN262 was obtained from south Tunisian soil, exhibiting antimicrobial activities against Gram-positive and Gram-negative bacteria and fungi (Fig. 1). Strain TN262 grew well in nutrient agar medium (Table 2), and the colors of the vegetative and aerial mycelia were yellowish and grayish, respectively. The spore chains were grayish-yellow. Comparison of these characteristics with those of actinomycete species described in Bergey’s manual of systematic bacteriology [14] suggested that TN262 strain belongs to the genus Streptomyces. Total nucleotide sequence of 1,540 bp (accession no. FM867598), of the whole 16S rRNA gene of the TN262 strain was determined in both strands. The alignment of this sequence through matching with reported 16S rRNA gene sequences in gene bank shows high similarity (97–98%) to the Streptomyces 16S rRNA genes. The organism most similar to the new isolate TN262 strain was Streptomyces flavogriseus AJ494864 (Fig. 2), which had been described as a cellulolytic activity producer [15]. Based on the cultural characteristics of TN262 and the nucleotide sequence of the corresponding 16S rRNA gene and the phylogenetic analysis, we propose the assignment of our new isolate bacterium as Streptomyces sp. TN262 strain.
Fig. 1.
Antimicrobial activities of TN262 strain in solid media against M. luteus LB 14110 (A1), E. coli ATCC 8739 (A2), and Fusarium sp. (A3), and in liquid media against: M. luteus LB 14110 (B1), S. aureus ATCC 6538 (B2), E. coli ATCC 8739 (B3), S. enterica ATCC43972 (B4), and Fusarium sp. (B5)
Table 2.
Cultural characteristics of TN262 strain.
| Medium | Growth | Vegetative mycelia | Aerial mycelia | Spores |
|---|---|---|---|---|
| Nutrient agar | Well, spreading | Abundant, yellowish | Abundant, grayish | Moderate, grayish-yellow |
| Sabouraud agar | Weak | Weak, yellowish | Weak, yellowish | Weak, elevated, yellowish |
| Yeast malt agar | Moderate | Detachable, yellowish | Moderate, grayish-brown | Moderate, brown |
Fig. 2.
Phylogenetic tree of the Streptomyces sp. isolate TN262 strain
Optimization of Nutritional and Cultural Conditions
Production of secondary metabolites by microorganisms is often connected with and influenced by primary metabolism. The composition and concentrations of the constituents of the media are closely linked with the metabolic capacities of the producing organism and greatly influence the biosynthesis of the bioactive molecules. Equally, the effect of other factors including medium volume, oxygen transfer rate, temperature, initial pH of the medium, and incubation time on secondary metabolites production were frequently reported [4].
Nutritional Conditions
Five carbohydrate sources (starch, fructose, glycerol, glucose, and saccharose) were tested as sole carbon source at 1% (w/v) in the modified Bennett liquid medium. The studied strain exhibited the ability to grow on all tested carbon sources. The maximum biological activity production was afforded when glycerol served as carbon source. To further optimize the culture conditions, three compounds (potassium, magnesium, and trace mineral oligoelements) were tested using glycerol at 1% (w/v) as carbon source. These chemicals were added to the culture medium, additive by additive, by the combination of two additives, and by addition of all three additives. A culture without chemical additives was used as control. The obtained results showed that maximal antimicrobial activities against the five test microorganisms were observed with the control culture. It was noticed that the addition of these chemical compounds did not alter the bioactive molecules production.
Effect of Physical Parameters; Temperature, pH, Incubation, and Time Agitation Rate
The effect of culture conditions, namely incubation temperature (25–40 °C), initial pH (6–8), incubation time (24–144 h), and rotary shaker speed (100–300 rpm) on growth and antimicrobial activity production was studied. Streptomyces sp. TN262 strain showed a narrow range of incubation temperature for good growth and active molecules production. The optimum temperature was 30 °C. The studied strain appeared to be mesophilic. The highest antimicrobial activities against the five tested microorganisms were obtained at an initial pH 7 of the culture medium. Concerning incubation time, antimicrobial activities appeared to be pronounced after 36 h of growth with a maximum at 72 h of incubation. This activity remained stable between 72 and 96 h and then decreased and disappeared after 140 h of incubation. For Streptomyces sp. TN262, agitation rates of 100, 150, and 300 rpm gave a low production of active molecules, while the best result was noticed at 200 and 250 rpm.
To further enhance the antibiotic production by Streptomyces sp. TN262, we have investigated the effect of the glycerol concentration. Eight concentrations (% w/v) were tested (0.2, 0.5, 0.75, 1, 1.25, 1.5, 1.75, and 2.0). Eight flasks, each containing 200 ml broth, were inoculated at 5% (v/v) with a 24-h preculture of Streptomyces TN262 prepared in TSB medium (107/ml of spores). After 3 days of incubation, the obtained broths were harvested. Biomass of mycelium of each culture was determined after drying at 105 °C, and the supernatants were exhaustively extracted with ethyl acetate, and the extracts concentrated in vacuo to dryness and then dissolved in 250 μl of ethyl acetate. Thereafter, the extracts were examined for their antimicrobial activity against bacteria and fungi (Fig. 3). In summary, the biological activities were correlated with biomass production. This biomass increased with increasing glycerol concentrations up to 0.75% (w/v) and remained nearly constant up to 1.25% (w/v) with a maximum at 1% (w/v). Any further increase of this substrate level resulted in a decrease in biomass and consequently in active molecules production. The same observation has been reported by Fourati et al. [16] for the antifungal compounds produced by Streptomyces sp. US80. In that case, the biomass increased with increasing glucose concentration (carbon source) and magnesium (chemical additive which influence positively the antifungal compounds production) concentration up to 5 g/l and 3.5 mM, respectively.
Fig. 3.
Effect of glycerol concentration on the growth, productivity, and antimicrobial activity of Streptomyces sp TN262 extract against: M.luteus LB 14110 (a); E.coli ATCC 8739 (b); and Fusarium sp. (c)
Isolation and Structural Elucidation of the Active Constituents
Structures of the isolated compounds were elucidated as:
*1-acetyl-β-carboline [17, 18] (1). This compound has been reported as plant metabolite [17] and from Streptomyces bacteria [18]. β-carboline derivatives were reported as herbicidal and fungicidal agents.
*tryptophol [19–21] (2), a well-known constituent of terrestrial plants and microorganisms, has so far been isolated from tryptophan fermentations, plant seedlings, Aspergillus niger, fungi Balansia epichloe, Dreschslera nodulosum, Agrobacterium tumefaciens, Ceratocystis spp., Rhizobium spp., marine sponge Ircinia spinulosa, etc. This molecule was reported to have antibacterial activity against Gram-positive bacteria and Candida albicans [19].
*cineromycin B [13, 22, 23] (3) and 2,3-dihydrocineromycin B [13, 24] (4). These two active molecules have been already described from Streptomyces species, as two 14-membered ring macrolide antibiotics. Most macrolides are produced by Streptomyces species, and these compounds are mainly bioactive against Gram-positive bacteria by the inhibition of their protein synthesis by binding to the 50s subunits of 70s ribosomes.
*3-(hydroxyacetyl)-indole [20, 25] (5) has been reported from the marine red alga Prionitis lanceolata, fungus Lactarius deliciosus, and a terrestrial Streptomyces sp. bacterium [20]. The compound was found to inhibit the growth of a minipanel of human cancer cell lines and P388 lymphocytic leukemia cells.
*cyclo-(tyrosylprolyl) [20, 26] (6) is a diketopiperazine (DKP) derivative. DKP molecules constitute a family of secondary metabolites with diverse and interesting biological activities such as antibacterial, fungicidal, herbicidal, immunosuppressor, antitumors, and antiviral [20]. This compound was previously described from Pseudomonas aeruginosa and some Streptomyces strains.
*brevianamide F [20, 27] (7) and cis-cyclo-l-prolyl-l-leucyl [20, 28, 29] (8) using 1D and 2D NMR spectra, mass spectrometry, and by comparison with the literature. Brevianamide F is a tryptophan–proline derivative. Prolinated indole derivatives: tryprostatins, cyclotryprostatins, spirotryprostatins, fumitremorgins, verruculogen, austamides, paraherquamides, and brevianamides are a large class of alkaloids containing a tryptophan moiety substituted with isoprenic moieties and are mostly found in fungi of the genera Aspergillus and Penicillium [20]. Compound (8), the cis-cyclo-l-prolyl-l-leucyl, is a DKP derivative.
The complex mixtures present in the unpolar fractions I and II were analyzed by GC–MS, and benzophenone [30] (9), N-butyl-benzenesulfonamide [31] (10), and the natural product hexanedioic acid-bis-(2-ethylhexyl)ester (11) were identified by comparison with the NIST MS database (Table 3). Benzophenone (9) is an important intermediate in the production of medicines and cosmetics but is also known as a xenoestrogen in its effect on mammals. This molecule has been produced by chemical synthesis and previously isolated from the plant Tovomita longifolia and the sea Streptomyces bacterium GWS-BW-H5 [30]. N-butyl-benzenesulfonamide (10) exhibits antifungal activities and has been described from the bacterium Pseudomonas sp. AB2 [31]. The hexanedioic acid-bis-(2-ethylhexyl)ester compound (11) is known as plasticizer derivatives [32–34], but it has never been described from natural sources.
Table 3.
GC–MS analysis of the combined unpolar fractions I and II.
| Name | Rt (min) | Relative abundance (%) | M. F. | M.Wt. |
|---|---|---|---|---|
| Benzophenone (9) | 16.67 | 17.2 | C13H10O | 182 |
| N-Butyl-benzenesulfonamide (10) | 18.32 | 60.2 | C10H15 NO2S | 213 |
| Hexanedioic acid-bis(2-ethylhexyl)ester (11) | 23.84 | 100 | C22H42O4 | 370 |
Cineromycin B
Compound 3, a colorless solid, was obtained from fraction 3 after a series of chromatographic steps including separation on RP-18. It was a weakly UV absorbing substance, exhibiting a dark violet coloration on spraying with anisaldehyde/sulfuric acid. The molecular weight of 3 was established as 294 Da according to CI MS and ESI MS, and the corresponding molecular formula was deduced as C17H26O4 by HRESI MS. This delivered five double-bond equivalents in 3.
The 1H NMR spectrum of 3 revealed the existence of five olefinic protons, among them four signals of trans-double bonds (d, J~15.5) located in the region δ 6.93–5.23. This confirmed the existence of three double bonds in the molecule. Moreover, four methyls were visible, two of them gave singlets (δ 1.73, 1.55), while the others appeared as doublets (δ 1.22, 0.89, J~6.Hz). Two oxygenated methines were observed as multiplets at δ 4.55, along with other five multiplet signals in the region of δ 1.80~1.21, corresponding to two methylenes and one methine. Based on the 13C NMR/HMQC spectra (Table 1), compound 3 entailed 17 carbon signals (as requested by the molecular formula), one carbonyl of an ester or acid (C-1, δ 166.1), and six olefinic carbons, among them five methines (δ 155.2~115.1) and one quaternary signal (C-8, δ 137.9). In the aliphatic region, there were three oxygenated carbons, two methines (δ 75.5 and 74.6), and a quaternary one (C-4) at δ 73.4. The remaining seven carbons were attributed to four methyls (δ 27.1~15.0), two non-oxygenated methylenes (δ 34.2, 25.1), and one methine (C-12, δ 39.5).
Based on the revealed NMR and MS data and the molecular formula, a search in AntiBase [35], the Dictionary of Natural Products [36], and the Chemical Abstract, referred to cineromycin B (3) as the only fitting structure.
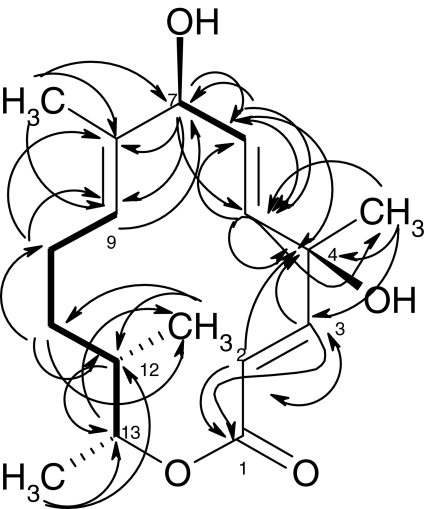
The compound was further confirmed by HMBC and H–H COSY experiments (Fig. 4). In accordance, the two olefinic protons H-2 (5.87, δC = 115.1) and H-3 (6.93, δC = 155.2) showed 2J and 3J couplings, respectively, with the lactone carbonyl C-1 (δ 166.1), confirming their direct attachment as ―CH═CH―COO. The two olefinic protons displayed further two cross signals with the quaternary oxy-carbon C-4 (δ 73.4), which in turn was connected to a methyl singlet (4-CH3) located at δ 1.55 (δC = 27.1) and an olefinic methine carbon (C-5; δ 135.2, δH 5.87) on the other side. The latter connection (C4–C5) was further established by a 3J correlation directing from 4-CH3 towards C-5. Additionally, the C-5 methine gave an H–H COSY cross signal with H-6 (5.73, δC = 133.6). To the latter (C-6), a 2J coupling was found via the oxygenated H-7 (4.55, δC = 74.6) and vice versa, confirming their direct attachment. A methyl group (1.73, δC = 15.0) was connected with a further olefinic double bond, as the 2J coupling with C-8 (137.9) showed; the methine partner (C-9) was found at δ 5.23 (129.5). At the other side, C-8 is attached to C-7 (δH = 4.55, δC = 74.6) as indicated by a cross signal from H-7 to C-8 and C-9. Finally, the remaining six carbons, two methylenes and two CH-Me fragments, were confirmed to connect the last olefinic carbon C-9 (129.5) and the lactonic carbonyl (C-1, 166.1), resulting in the structure of cineromycin B (3) (Fig. 5). This was further confirmed by comparison with recently published data [23] (Table 1).
Fig. 4.
HMBC (right arrow), and H,H COSY (thick line, left–right arrow) connectivities of Cineromycin B (3)
Fig. 5.
Structural formulas of compounds 1–11
2,3-Dihydrocineromycin B
Compound 4 was obtained in a similar way as for 3 as colorless solid, revealing the same chromatographic color reaction. The molecular weight of 4 was determined as 296 Da by CI and ESI MS, i.e., 2 amu higher than 3, corresponding to a reduction of one of the three olefinic double bonds present in 3. The corresponding molecular formula (C17H28O4) of 4 was established by HRESI MS, indicating four equivalent bonds. The 1H NMR spectra of 4 showed a similar pattern as for 3, with exception of the olefinic H-2 and H-3 signals in 4 (δ 5.87, 6.93). This pointed to the reduction of the double bond in conjugation with the lactone carbonyl, delivering two methylene groups appearing between δ 2.43~1.80. Correspondingly, the lactone carbonyl was shifted downfield shifted to δ 174.0 [13]. So, structure of 4 was established as 2,3-dihydrocineromycin B, an isomer of the alternative 5,6-dihydrocinermycin-B (Fig. 5) [13, 24].
Biological Activity of the Isolated Constituents
As shown in Table 4, the unpolar mixture of fractions I and II showed no antimicrobial activity at concentrations of 20 and 30 μg/disk, while at concentration of 40 μg/disk, potent activities against Fusarium sp., M. luteus LB 14110, and S. aureus ATCC 6538 were shown; no inhibitory activities against E. coli ATCC 8739 and S. enterica ATCC43972 (Gram-negative bacteria) were displayed. Biological activity of Tryptophol (2) is more important against M. luteus LB 14110 and S. aureus ATCC 6538 than against Fusarium sp. and S. enterica ATCC43972. Cineromycin B (3) and its dihydro-analog (4) showed weak activity against Gram-positive bacteria [24].
Table 4.
The antimicrobial activity of the unpolar fractions (UnF) and the isolated compounds 2, 3 and 4.
| Compound | Test microorganism (inhibition zones (mm)) | ||||
|---|---|---|---|---|---|
| Fusarium sp. | M. luteus LB 14110 | S. aureus ATCC 6538 | E. coli ATCC 8739 | S. enterica ATCC 43972 | |
| UnF (20 µg/disk) | – | – | – | – | – |
| UnF (30 µg/disk) | – | – | – | – | – |
| UnF (40 µg/disk) | 15 | 16 | 15 | – | – |
| UnF (50 µg/disk) | 18 | 20 | 17 | – | – |
| 2 (30 µg/disk) | 12 | 19 | 17 | – | 12 |
| 3, 4 (30 µg/disk) | – | 13 | 12 | – | – |
For each concentration and indicator microorganism, the experiment was carried out simultaneously three times under same conditions. The obtained diameters of inhibition zones were quite similar and the reported inhibition zones (mm) are the average of the three experiments
ND activity not detected
Conclusions
A new aerobic bacterium TN262 was isolated from Tunisian soil and has been selected for its antimicrobial activity against Gram-positive and Gram-negative bacteria and fungi. Based on the results of the cultural characteristic studies, analysis of the nucleotide sequence (1,540 pb) of the whole 16S rRNA gene (accession no. FM867598) of the TN262 strain and the phylogenetic analysis, this isolate has been assigned as Streptomyces sp. TN262 strain. Addition of glycerol (at 1%, w/v) as carbon source in modified Bennett liquid medium (72 h, 30 °C, 250 rpm) yielded the highest antimicrobial activity. A large-scale fermentation lead to isolation of eight components, among them two macrolides, cineromycin B (3), and 2,3-dihydrocineromycin B (4). Their structures were established by our spectroscopic techniques and comparison with reference data. It should be noted that in the active supernatant of the TN262 culture, three further metabolites were detected in the unpolar fractions using GC–MS and tentatively assigned as benzophenone (9), N-butyl-benzenesulfonamide (10), and hexanedioic acid-bis-(2-ethylhexyl) ester (11). This last compound is known as plasticizer derivatives [32–34], but it has never been described from natural sources.
Acknowledgments
This work was supported by the Tunisian–Egyptian project “MELLOULI/SHAABAN.” The authors are thankful to R. Machinek for the NMR spectra, Dr. H. Frauendorf for the mass spectra, and A. Kohl for technical assistance. Thanks are also due to Pr. A. Rebai and Dr. I. Ayadi for the phylogenetic determination and analysis and to M. B. Neili for his useful contribution to the fermentation experiments.
References
- 1.Williams ST, Goodfellow M, Alderson G, Wllington EM, Sneath PH, Sacki MJ. Journal of General Microbiology. 1983;129:1747–1813. doi: 10.1099/00221287-129-6-1743. [DOI] [PubMed] [Google Scholar]
- 2.Miyadoh S. Actinomycetologica. 1993;9:100–106. doi: 10.3209/saj.7_100. [DOI] [Google Scholar]
- 3.Tanaka YT, Mura SO. Annual Review of Microbiology. 1993;47:57–87. doi: 10.1146/annurev.mi.47.100193.000421. [DOI] [PubMed] [Google Scholar]
- 4.Fourati Ben Fguira L, Fotso S, Ben Ameur Mehdi R, Mellouli L, Laatsch H. Research in Microbiology. 2005;156:341–347. doi: 10.1016/j.resmic.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D. Journal of Molecular Biology. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 6.Mellouli L, Ghorbel R, Kammoun A, Mezghani M, Bejar S. Biotechnological Letters. 1996;18:809–814. doi: 10.1007/BF00127894. [DOI] [Google Scholar]
- 7.Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, et al. Genetic manipulation of Streptomyces. A laboratory manual. Norwich: The John Innes Foundation; 1985. [Google Scholar]
- 8.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 9.Edwards U, Rogall T, Bocker H, Emde M, Bottger E. Nucleic Acids Research. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felsenstein J. PHYLIP-phylogeny inference package, version 3.2. Cladistics. 1989;5:164–166. [Google Scholar]
- 12.Saitou N, Nei M. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Schiewe HJ, Zeeck A. Journal of Antibiotics. 1999;52:635–642. doi: 10.7164/antibiotics.52.635. [DOI] [PubMed] [Google Scholar]
- 14.Lechevalier HA, Williams ST, Sharpe ME, Holt JG. Bergey’s manual of systematic bacteriology. Baltimore: Williams & Wilkins; 1989. [Google Scholar]
- 15.Wirth S, Ulrich A. Applied Microbiology. 2002;25:584–591. doi: 10.1078/07232020260517724. [DOI] [PubMed] [Google Scholar]
- 16.Fourati-Ben Fguira L, Karray-Rebai I, Smaoui S, Bejar S, Mellouli L. Journal of Biotechnology. 2008;3:1058–1066. doi: 10.1002/biot.200700155. [DOI] [PubMed] [Google Scholar]
- 17.Zhou TC, Ye WC, Wang ZT, Che CT, Zhou RH, Xu GJ. Phytochemistry. 1998;49:1807–1809. doi: 10.1016/S0031-9422(98)00232-5. [DOI] [PubMed] [Google Scholar]
- 18.Shaaban M, Schröder D, Shaaban KA, Helmke E, Grün-Wollny I, Wagner-Döbler I, et al. Revista Latinoamericana de Química. 2007;35:58–67. [Google Scholar]
- 19.Erdogan I, Sener B, Higa T. Biochemical Systematics and Ecology. 2000;28:793–794. doi: 10.1016/S0305-1978(99)00111-8. [DOI] [PubMed] [Google Scholar]
- 20.Shaaban, M. (2004). PhD thesis, University of Göttingen, Germany.
- 21.Shaaban M, Maskey RP, Wagner-Doebler I, Laatsch H. Journal of Natural Products. 2002;65:1660–1663. doi: 10.1021/np020019a. [DOI] [PubMed] [Google Scholar]
- 22.Schneider A, Spaeth J, Breiding-Mack S, Zeeck A, Grabley S, Thiericke R. Journal of Antibiotics. 1996;49:438–446. doi: 10.7164/antibiotics.49.438. [DOI] [PubMed] [Google Scholar]
- 23.Terekhova LP, Galatenko OA, Kulyaeva VV, Malkina ND, Boikova YV, Katrukha GS, et al. Russian Chemical Bulletin. 2007;56:815–818. doi: 10.1007/s11172-007-0121-1. [DOI] [Google Scholar]
- 24.Burkhardt K, Fiedler H, Grabley S, Thiericke R, Zeeck A. Journal of Antibiotics. 1996;49:432–437. doi: 10.7164/antibiotics.49.432. [DOI] [PubMed] [Google Scholar]
- 25.Ayer WA, Trifonov LS. Journal of Natural Products. 1994;57:839–841. doi: 10.1021/np50108a026. [DOI] [PubMed] [Google Scholar]
- 26.Barrow CJ, Sun HH. Journal of Natural Products. 1994;57:471–476. doi: 10.1021/np50106a005. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi M, Aoki S, Gato K, Matsunami K, Kurosu M, Kitagawa I. Chemical and Pharmaceutical Bulletin. 1994;42:2449–2451. doi: 10.1248/cpb.42.2449. [DOI] [PubMed] [Google Scholar]
- 28.Ayer WA, Browne LM, Feng M, Orszanska H, Saeedi-Ghomi H. Canadian Journal of Chemistry. 1986;64:904–909. doi: 10.1139/v86-149. [DOI] [Google Scholar]
- 29.Schiebel, M. (2002). PhD thesis, University of Göttingen, Germany.
- 30.Dickschat JS, Martens T, Brinkhoff T, Simon M, Schulz S. Chemistry and Biodiversity. 2005;2:837–865. doi: 10.1002/cbdv.200590062. [DOI] [PubMed] [Google Scholar]
- 31.Kim KK, Kang JG, Moon SS, Kang KY. Journal of Antibiotics. 2000;53:131–136. doi: 10.7164/antibiotics.53.131. [DOI] [PubMed] [Google Scholar]
- 32.Goulas AE, Anifantaki KI, Kolioulis DG, Kontominas MG. Journal of Dairy Science. 2000;83:1712–1718. doi: 10.3168/jds.S0022-0302(00)75040-5. [DOI] [PubMed] [Google Scholar]
- 33.Dalgaard M, Hass U, Vinggaard AM, Jarfelt K, Lam HR, Sørensen IK, et al. Reproductive Toxicology. 2003;17:163–170. doi: 10.1016/S0890-6238(02)00149-1. [DOI] [PubMed] [Google Scholar]
- 34.Ghisari M, Bonefeld-Jorgensen EC. Toxicology Letters. 2009;189:67–77. doi: 10.1016/j.toxlet.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 35.Laatsch, H. (2007). AntiBase. Available from: www.user.gwdg.de/∼ucoc/laatsch/AntiBase.htm.
- 36.Dictionary of Natural Products on CD-ROM. Boca Raton: Chapman & Hall; 2004. [Google Scholar]