Abstract
In this study, we have investigated the transcriptome of Ralstonia eutropha H16 during cultivation with gluconate in presence of 3,3′-thiodipropionic acid (TDP) or 3,3′-dithiodipropionic acid (DTDP) during biosynthesis of poly(3-hydroxybutyrate-co-3-mercaptopropionate). Genome-wide transcriptome analyses revealed several genes which were upregulated during cultivation in presence of the above-mentioned compounds. Obtained data strongly suggest that two ABC-type transport system and three probable extracytoplasmic solute receptors mediate the uptake of TDP and DTDP, respectively. In addition, genes encoding the hydrolase S-adenosylhomocysteinase AhcY and the thiol-disulfide interchange proteins DsbA, DsbD, and FrnE were upregulated during cultivation on DTDP and, in case of AhcY and FrnE, on TDP as well. It is assumed that the corresponding enzymes are involved in the cleavage of TDP and DTDP. Several genes of the fatty acid metabolism exhibited increased expression levels: genes encoding two acetyltransferases, a predicted acyltransferase, the acetoacetyl-CoA reductase phaB3, an enoyl-CoA hydratase as well as an acyl dehydratase, an acetyl-CoA synthetase, two acyl-CoA dehydrogenases, the methylmalonyl-CoA mutase encoded by sbm1 and sbm2 and phaY1 were detected. Furthermore, ORF H16_A0217 encoding a hypothetical protein and exhibiting 54% amino acids identical to an acyl-CoA thioesterase from Saccharomonospora viridis was found to be highly upregulated. As the 2-methylcitrate synthase PrpC exhibited a three- to fourfold increased activity in cells grown in presence of TDP or DTDP as compared to gluconate, metabolization of the cleavage products 3MP and 3-hydroxypropionate to propionyl-CoA is proposed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00253-010-2915-6) contains supplementary material, which is available to authorized users.
Keywords: 3,3′-dithiodipropionic acid; 2-methylcitric acid cycle; Poly(3-hydroxybutyrate); Polythioester; Ralstonia eutropha H16; 3,3′-thiodipropionic acid
Introduction
Polyhydroxyalkanoates (PHAs) are linear polyesters which occur in many bacteria as storage compounds for carbon and energy. They are accumulated as water-insoluble inclusions in the cytoplasm in response to physiological stress such as oxygen or nitrogen deficiency and if a carbon source is present in excess at the same time. Poly(3-hydroxybutyrate), poly(3HB), was the first identified PHA (Lemoigne 1926) and is the best-studied storage compound in prokaryotes. In addition to 3HB, approximately 150 other hydroxyalkanoic acids have been identified as PHA constituents (Steinbüchel and Valentin 1995; Pötter and Steinbüchel 2005). Since 2001, polythioesters (PTEs) consisting of mercaptoalkanoic acids, are known as a new class of structurally related biopolymers with thioester instead of oxoester linkages in the backbone (Lütke-Eversloh et al. 2001a). The physical characteristics of PTEs such as thermal properties, solubility, and crystallinity deviate considerably from those of the corresponding oxoesters (Lütke-Eversloh et al. 2002; Kawada et al. 2003). One remarkable difference to PHAs is the non-biodegradability of PTE homopolymers. Whereas PHAs are degraded by many microorganisms by PHA depolymerases into simple organic compounds (Jendrossek and Handrick 2002), PTE homopolymers are persistent (Kim et al. 2005). Only investigations on the degradation of a copolymer of 3-hydroxybutyrate and 3-mercaptopropionate (3MP), poly(3HB-co-3MP), revealed new bacterial strains able to grow with this heteropolymer as sole carbon source by cleaving the oxoester bonds but not the thioester bonds (Elbanna et al. 2004).
The Gram-negative and facultative chemolithoautotrophic bacterium Ralstonia eutropha is probably the best-studied microorganism regarding PHA metabolism. Poly(3HB) biosynthesis is mediated by the PHA operon, comprising the genes for a β-ketothiolase (phaA), an acetoacetyl-CoA reductase (phaB) and the PHA synthase (phaC). The latter is the key enzyme of PHA biosynthesis and exhibits broad substrate specificity (Rehm 2003). Whereas phaC is essential for PHA biosynthesis, phaA and phaB can be replaced by isoenzymes (Slater et al. 1998; Lindenkamp et al. 2010). When cultivating cells of R. eutropha H16 under storage conditions with 3-mercaptoalkanoic acids in addition to fructose or gluconate as carbon source, polyesters containing sulfur in the backbone of the polymer are synthesized. With 3MP as precursor substrate, the accumulation of copolymers consisting of 3-hydroxybutyrate and 3-mercaptopropionate, poly(3HB-co-3MP), was shown, consisting of up to 40 mol% of 3MP (Lütke-Eversloh et al. 2001a, b; Lütke-Eversloh and Steinbüchel 2003b). Variations of the copolymer composition could be obtained by supplying 3-mercaptobutyrate (3MB) or 3-mercaptovalerate (3MV) as precursor substrates. In addition to 3MP, R. eutropha H16 is also able to use 3,3′-thiodipropionic acid (TDP) and 3,3′-dithiodipropionic acid (DTDP) as precursor substrates for biosynthesis of poly(3HB-co-3MP). Whereas 3MP was shown to inhibit growth of R. eutropha at a concentration of 0.1% (v/v) in the medium, TDP and DTDP exerted no inhibitory effects at concentrations of up to 1% (w/v) (Lütke-Eversloh and Steinbüchel 2003b).
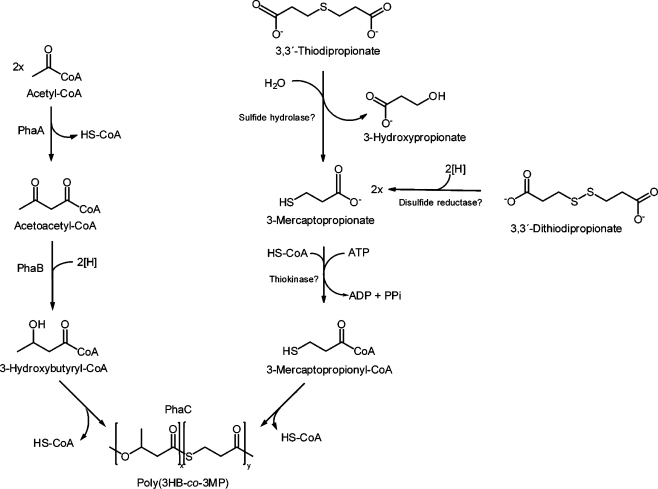
Until now, relatively little is known about the metabolic pathway for degradation of the organic sulfur compounds mentioned above and biosynthesis of PTEs, respectively. Due to the incorporation of 3MP in the copolymer, cleavage of TDP into 3MP and 3HP by a yet unknown sulfide hydrolase is proposed, whereas DTDP is probably cleaved in two molecules 3MP by an unknown disulfide reductase (Fig. 1) (Lütke-Eversloh et al. 2001a; Lütke-Eversloh and Steinbüchel 2003a).
Fig. 1.
Overview of the biosynthesis pathways of heteropolythioester in R. eutropha H16 as proposed by Lütke-Eversloh and Steinbüchel (2004)
Very recently, the conversion of 3MP into 3-sulfinopropionate (3SP) was shown for the cysteine dioxygenase CdoA from R. eutropha H16 in recombinant Escherichia coli cells (Bruland et al. 2009). In addition to R. eutropha, the catabolism of TDP and DTDP was also intensely investigated in the recently described bacteria Variovorax paradoxus strain TBEA6 and Advenella mimigardefordensis strain DPN7T (Bruland et al. 2009; Wübbeler et al. 2006, 2008). These studies led to a proposed degradation pathway from DTDP and TDP via 3MP, 3SP, and 3SP-CoA towards propionyl-CoA. The catabolism of propionyl-CoA via the 2-methylcitric acid cycle was strongly proposed for A. mimigardefordensis (Wübbeler et al. 2008).
Non-biodegradable, persistent polymers are valuable for certain applications such as permanent implants in medical applications or special technical applications in construction and automobiles (Steinbüchel 2005). For this reason, an in-depth elucidation of PTE biosynthesis and of the catabolism of various organic sulfur compounds is required. To obtain more insights into the catabolism of the sulfur-containing precursor substrates and in the uptake of these compounds into the cells as well as in the enzymes involved in the biosynthesis of PTE homo- and copolymers, genome-wide transcriptome analyses were performed.
Material and methods
Bacterial strains and growth conditions
Cells of R. eutropha H16 were cultivated at 30°C in 2 l Erlenmeyer flasks with baffles containing 400 ml mineral salts medium (Schlegel et al. 1961), with 1% (w/v) sodium gluconate as sole carbon source. The ammonia concentration was 0.05% (w/v) to promote PHA accumulation. In addition, TDP or DTDP (1% w/v) were used as second, sulfur-containing carbon source for poly(3HB-co-3MP) biosynthesis. Growth of cells was measured in a Klett–Summerson photometer using filter no. 54 (520–580 nm). Cells were harvested in the stationary growth phase after 24, 48, 72, and 96 h by centrifugation (15 min, 4,000 rpm, 4°C).
RNA isolation and cDNA synthesis
Harvested cells were washed in 0.9% (w/v) sodium chloride, directly shock-frozen in liquid nitrogen, and stored at −70°C. RNA was isolated by using the RNeasy mini kit (Qiagen, Hilden, Germany) and zirconia–silica beads (Roth, Karlsruhe, Germany) for mechanical disruption (Peplinski et al. 2010). Total RNA (25 μg) were used for random hexamer-primed synthesis of fluorescence-labeled cDNA by reverse transcription using the CyScribe First-strand cDNA labeling kit and the fluorescent nucleotide analogous FluoroLink Cy3-dUTP and Cy5-dUTP (GE Healthcare).
Microarray hybridization and scanning
Full genomic R. eutropha H16 oligonucleotide microarrays were used for transcription analyses (Peplinski et al. 2010). For each sample, Cy5-dUTP-labeled cDNAs were mixed with a common reference, consisting of a pool of all Cy3-dUTP-labeled cDNAs which were obtained in the first biological experiment during cultivations in presence of gluconate, gluconate plus TDP, and gluconate plus DTDP, and subsequently hybridized to a microarray (Conway and Schoolnik 2003). Before hybridization, samples were denatured by incubation at 98°C for 5 min. Hybridization was done in Tom Freeman hybridization buffer (Fitzpatrick et al. 2005) for 15 h at 58°C with cDNA containing approximately 80 pmols of Cy-3 and Cy-5 in an automatic Lucidea slide processor (GE-Healthcare, Munich, Germany). Two consecutive washes each with 1× SSC buffer containing 0.2% SDS and then with 0.1× SSC were applied to the slides. At the end, the hybridization chambers were flushed with isopropanol, and the slides were dried by evaporating the isopropanol with air. A GenePix 4000B microarray scanner (Molecular Devices, Canada) and GenePix Pro 6.0 software were used for scanning.
Data normalization and filtering
Data were normalized by multiplication of a constant factor so that the mean of the ratio of medians of all features became equal to 1. Arrays that required a normalization factor larger than 2 were excluded from further analysis to eliminate experiments with major deviations in the applied dye incorporated in the labeled cDNA. After determining the fluorescence intensities of both channels, a background correction was made by subtracting the local background value from the foreground intensity. For each sample, the ratio signal intensity (Cy5)/reference (Cy3) was calculated and was taken to represent the relative RNA abundance. The extent of the gene expression levels between samples provided a measure of the overall gene expression pattern between samples. Two independent biological experiments were performed. Data were generated from the first biological experiment, resulting in averaged RNA ratios of two replicates. Only genes, which exhibited at least twofold higher signal intensities in at least the first three time points at which samples were withdrawn, were considered here. Furthermore, only those genes, whose direction of expression could be reproduced in the second biological experiment, were considered. The original raw data files have been deposited in the ArrayExpress database.
Preparation of crude extracts
Cell-free extracts were obtained by mechanical disruption. For this, 50 ml of cell cultures were washed in 0.9% (w/v) sodium chloride, resuspended in 100 mM Tris/HCl (pH 7.5) containing 10 μg DNase I and passed three times through a chilled french press cell (Amicon, Silver Spring, USA) at 1,000 Mpa. Soluble protein fractions of the crude extracts were obtained in the supernatants after 1 h centrifugation at 100,000×g and 4°C.
Synthesis of propionyl-CoA
Propionyl-CoA was synthesized according to Simon et al. (1983). Trilithium salt (10 mg) of coenzyme A were dissolved in 0.5 M K2CO3 (pH 8.0), and aliquots of 2 μl of propionic anhydride were added while stirring on ice until no free coenzyme A was detectable with 5,5′-dithiobis-(2-nitrobenzoate) (DTNB) (Ellman 1958). The pH of the propionyl-CoA solution was adjusted to 4.5 by addition of 1 N HCl and was then stored at −20°C. The concentration of propionyl-CoA was calculated in consideration of complete substrate conversion.
Enzyme assay
Activity of 2-methylcitrate synthase was measured as previously described (Brämer and Steinbüchel 2001) by the method of Srere (1966). The cuvette (d = 1 cm) was contained in a total volume of 1 ml 2 mM oxaloacetate, 250 μM propionyl-CoA, and 2 mM DTNB in 10 mM Tris/HCl (pH 7.5). After addition of the crude extract, the increase of the absorbance at 412 nm (ε = 13.6 cm2/μmol) was measured with a spectrophotometer (Ultrospec 2000, Amersham-Pharmacia Biotech, Freiburg, Germany).
One unit of enzyme activity was defined as the conversion of 1 μmol substrate/min. The amount of soluble protein was determined by the method of Bradford (1976), using crystalline bovine serum albumin as standard.
Analysis of PHA content
Cells were harvested by centrifugation (15 min, 6,000×g, 4°C), washed in 0.9% (w/v) sodium chloride, and lyophilized. Samples were subjected to methanolysis in the presence of 85% (v/v) methanol and 15% (v/v) sulfuric acid. The resulting methyl esters of 3HB and 3MP were analyzed by gas chromatography (Brandl et al. 1988; Timm and Steinbüchel 1990).
Results
Polymer accumulation in R. eutropha H16
To analyze the accumulation of poly(3HB) or of the sulfur-containing copolymer poly(3HB-co-3MP), cells of R. eutropha H16 were cultivated in liquid MSM containing 1% (w/v) sodium gluconate as carbon source for growth alone or in addition to either TDP or DTDP (1% w/v) as second sulfur-containing carbon source to enable biosynthesis of poly(3HB-co-3MP). Concentration of ammonium chloride was reduced to 0.05% (w/v) to promote PHA accumulation. Samples were taken in the stationary growth phase after 24, 48, 72, and 96 h from each culture to analyze the polymer contents and compositions (Fig. 2). In addition, samples were subjected to transcriptome analyses.
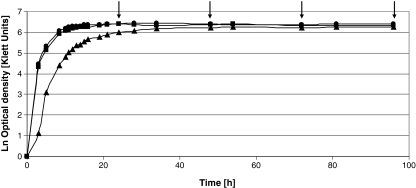
Fig. 2.
Growth curves of R. eutropha H16 cultivated in presence of 1% (w/v) sodium gluconate (filled diamond), 1% (w/v) sodium gluconate plus 1% (v/v) TDP (filled square) or DTDP (filled triangle). Samples were withdrawn after 24, 48, 72, and 96 h of cultivation, as indicated by the arrows
During cultivation with gluconate as sole carbon source as well as in presence of gluconate plus TDP, similar growth behavior of cells of R. eutropha H16 was observed. In contrast, cultivation in presence of gluconate plus DTDP resulted in a slightly diminished cell growth (Fig. 2). However, after about 34 h of cultivation, all cultures had reached the same density.
When cultivated with gluconate, gas chromatographic analyses revealed after 24 h of cultivation, a poly(3HB) content of about 73% (w/w) of the cell dry weight (CDW), which decreased to 68% (w/w, of CDW) after 96 h (Fig. 3). The decrease of the amount of stored poly(3HB) indicated degradation of small amounts of the polymer by PHB depolymerases.
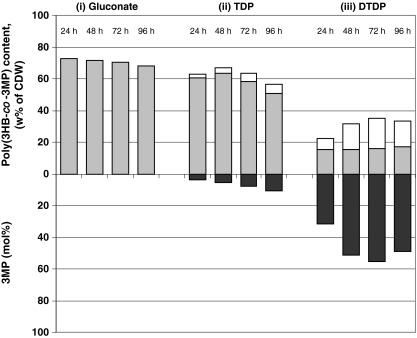
Fig. 3.
Poly(3HB) and poly(3HB-co-3MP) accumulation in R. eutropha H16. Cells were cultivated in MSM under storage conditions containing (i) 1% (w/v) sodium gluconate, (ii) 1% (w/v) sodium gluconate plus 1% (v/v) TDP, or (iii) DTDP. After 24, 48, 72, and 96 h of cultivation, the poly(3HB) or poly(3HB-co-3MP) contents of the cells were analyzed by gas chromatography. Upper y-axis: Gray bars indicate the 3HB content (weight percent of CDW) in the polymer, white bars indicate the 3MP content (weight percent of CDW) in the copolymer. Lower y-axis reflects the molar 3MP content of the corresponding bars above
When TDP or DTDP was supplemented to the medium, the copolymer poly(3HB-co-3MP) was synthesized. With TDP as second carbon source, copolymer contents of approximately 63% or 67% (w/w of CDW) were obtained after 24 and 48 h, consisting of 3.8 and 5.3 mol% 3MP (w/w), respectively. Thereafter, the copolymer content of the cells decreased to 63% and 57% (w/w of CDW) after 72 and 96 h, respectively, whereas the molar content of 3MP increased at the same time to 8.1% and 10.6 mol% (w/w), respectively.
Cultivation in presence of gluconate plus DTDP resulted in much lower copolymer contents, but the molar fractions of 3MP in the copolymers were three- to fivefold higher as compared to cells cultivated in the presence of gluconate plus TDP: after 24 h of cultivation, the cells had stored 22% (w/w of CDW) of the copolymer with 31.7 mol% (w/w) incorporated 3MP. The copolymer content increased to 35% (w/w of CDW), exhibiting 55 mol% 3MP (w/w) after 72 h of cultivation. After 96 h cultivation, the copolymer content was 34% (w/w of CDW), consisting of 49 mol% 3MP (Fig. 3).
Studies regarding the biodegradability of PTE revealed new bacterial strains that were able to grow with poly(3HB-co-3MP) as sole source for carbon and energy. Until now, intracellular degradation of this heteropolymer has not yet been investigated, but the decreasing contents of poly(3HB-co-3MP) in cells of R. eutropha H16 indicate a degradation of this copolymer.
Gene expression analyses
Genome-wide transcriptome analyses were employed to detect genes that are involved in the catabolism of TDP and DTDP. For this purpose, cells of R. eutropha H16 were cultivated in liquid MSM containing 0.05% (w/v) ammonium chloride to promote PHA accumulation, 1% (w/v) sodium gluconate as carbon source for growth, and either TDP or DTDP (1% w/v) as second sulfur-containing carbon source to enable biosynthesis of poly(3HB-co-3MP). Samples were withdrawn in the stationary growth phase after 24, 48, 72, and 96 h from each culture and were subjected to transcriptome analyses. Here, the ratios of Cy5/Cy3 intensities were compared to each other, to measure the overall gene expression changes between samples. Only genes, which exhibited at least twofold higher signal intensities in at least the first three time points (24, 48, and 72 h), were considered here, and only those genes whose direction of expression could be reproduced in the second biological experiment.
To identify genes which may be involved in the catabolism of the sulfur compounds TDP and/or DTDP, the following comparisons were carried out: cells grown on gluconate were compared to cells which were cultivated with (a) gluconate plus TDP or (b) gluconate plus DTDP. (c) In addition, differences in gene expression during growth on TDP and DTDP as second carbon source were determined, consulting the data obtained in transcriptome comparisons (a) and (b).
-
Gluconate versus TDP
During cultivation in presence of TDP, 40 genes exhibited an at least twofold higher gene expression level in comparison to cells grown in presence of gluconate (see Online Resource 1 for details). This group includes four genes which are involved in or related to fatty acid metabolism: H16_B0782 encoding an acyltransferase exhibited increased expression level ranging from eightfold (after 24 h of cultivation) to 80-fold (48 h). Two further genes encoding acyltransferases (H16_B1292; three- to sixfold, H16_A1954; two- to fourfold) which catalyze the transfer of acyl groups to a substrate, and phaB3 encoding an acetoacetyl-CoA reductase (H16_A2171; two- to fivefold) were found to be induced during cultivation in presence of TDP (Table 1A). phaB3 represents a homolog to phaB1 which is part of the phaCAB operon of R. eutropha H16. phaB1 is known to mediate the second step in the PHB biosynthesis, the reduction of acetoacetyl-CoA to R-3-hydroxybutyryl-CoA, which is then further polymerized to poly(3HB) by the PHB synthase PhaC1. Whereas phaC1 is essential for PHB biosynthesis in R. eutropha H16, phaB1 can be replaced by isoenzymes.
ORF H16_A0217 was 3- to 40-fold upregulated during cultivation in presence of TDP in comparison to gluconate. The hypothetical protein showed 54% amino acids identical to an acyl-CoA thioesterase from Saccharomonospora viridis (YP_003134187) and comprises a HotDog domain. This domain was first observed in the structure of an E. coli thiol ester dehydratase (FabA) by Leesong et al. (1996) and is found in a diverse range of enzymes, including acyl-ACP thioesterases, acyl-CoA thioesterases, and the 4HBT (4-hydroxybenzoate thioesterase) superfamily (Dillon and Bateman 2004).
Moreover, frnE encoding a dithiol-disulfide isomerase (H16_B2322; three- to sixfold) was detected (Table 1A). FrnE is a DsbA-like protein and is presumed to be a thiol oxidoreductase. DsbA is a monomeric thiol-disulfide oxidoreductase and is known as the strongest thiol oxidant being involved in the oxidative protein-folding pathway in prokaryotes.
A strong induction at all four time points investigated was found for genes encoding an ABC-type transporter including the permease components and the periplasmic solute-binding domain (H16_A3658-A3660; three- to ninefold).
Furthermore, two genes encoding a glutathione S-transferases were detected (H16_A1090, H16_B0571; three- to sixfold). The corresponding enzymes are generally known as detoxifying enzymes and are involved in the metabolism of a wide range of xenobiotic compounds. They catalyze the conjugation of these compounds with glutathione, which can then be excreted (Hayes and Pulford 1995). One further gene, encoding the peroxiredoxin bcp (H16_A0306; threefold) exhibited higher gene expression levels at all four time points. Peroxiredoxins are a ubiquitous family of thiol-specific antioxidant enzymes, sharing a redox-active cysteine in the active site.
-
Gluconate versus DTDP
111 genes exhibited increased expression levels during cultivation in presence of DTDP in comparison to cells cultivated in presence of gluconate (details are given in Online Resource 2). As also observed above (in (a)), genes known to be involved in fatty acid metabolism exhibited higher expression levels at all four time points. They code for an enoyl-CoA hydratase (H16_A0179, three- to fivefold), an acyl-dehydratase (H16_A1070, three- to fivefold) and an acetyltransferase (H16_B0782, 25- to 428-fold). Furthermore, H16_A0217 encoding the putative acyl-CoA thioesterase, which was already mentioned above, was also induced during cultivation in presence of DTDP (6- to 46-fold).
ahcY, encoding the S-adenosylhomocysteinase (H16_A0244, 6- to 13-fold) which catalyzes the reversible hydrolysis of S-adenosylhomocysteine to adenosine and l-homocysteine, exhibited increased expression levels. It has to be mentioned that this gene was detected in comparison (a) as well, but exhibited an at least twofold increased transcription level only in the last three time points (here, seven- to tenfold), hence, it is not listed. One further upregulated gene encodes for a rhodanese-related sulfurtransferase (H16_A0333, four- to eightfold) which is putatively catalyzing the sulfur transfer from thiosulfate to thiophilic acceptors (Ray et al. 2000).
dsbA and dsbD (H16_A0161, three- to fivefold; H16_A3455, six- to ninefold) encode for thiol-disulfide interchange proteins and are known to be involved in the oxidative protein-folding pathway in prokaryotes. DsbA is a monomeric thiol-disulfide oxidoreductase and is the strongest thiol oxidant known. The reduced and thereby active form is maintained by the membrane protein DsbD. A further gene encoding a DsbA-like protein exhibited increased expression level during cultivation on DTDP: frnE (H16_B2322, 7- to15-fold) is already mentioned above (Table 1B).
One group of genes, which were found to be upregulated during cultivation in presence of DTDP, encode for enzymes which can be classified as detoxifying or defense enzymes. First of all three genes encoding the two glutathione S-transferases (H16_A1090, four- to eightfold; H16_B0451, two- to fivefold) and the peroxiredoxin encoding bcp (H16_A0306, three- to fivefold) exhibited increased expression levels. These enzymes were found in transcriptome comparison (a) as well. Moreover, genes encoding molecular chaperones and co-chaperones were detected (groES, H16_A0705, 3- to 24-fold; groEL, H16_A0706, two- to sixfold; dnaK, H16_A3089, four- to ninefold and grpE, H16_A1137, five- to ninefold). In general, chaperones are known as proteins that are involved in folding nascent proteins; furthermore, many chaperones protect polypeptides from denaturation and aggregation in the face of physical or chemical stress (Macario 1995).
The biotin operon bioFADB (H16_A0180-A0183, 2- to 48-fold) was upregulated at all four time points (Table 1B). Biotin serves as a covalently bound coenzyme for several carboxylase enzymes including acetyl-CoA carboxylase, propionyl-CoA carboxylase, and pyruvate carboxylase (Hassan and Zempleni 2006). These carboxylases play an important role in fatty acid metabolism, gluconeogenesis, and other metabolic pathways.
Finally, two genes encoding probable extracytoplasmic solute receptors (H16_A2779, H16_A0337; two- to fourfold each) exhibited increased expression levels at the first three time points during cultivation in presence of DTDP. Both genes comprise a bug domain, known from representatives of Bordetella uptake genes. The genome sequence of R. eutropha H16 revealed 154 representatives of this family (Pohlmann et al. 2006; Reinecke and Steinbüchel 2009).
-
TDP versus DTDP
The comparison of cells cultivated in presence of TDP to cells cultivated in presence of DTDP revealed 72 genes with a higher expression level in TDP cells (for more details, see Online Resource 3) and 42 genes which were upregulated in DTDP cells (Online Resource 4).
The first group included four genes involved in fatty acid metabolism: H16_A0234 (threefold), H16_B1192 (3- to 77-fold) encoding acyl-CoA dehydrogenases and the 3-hydroxybutyrate oligomer hydrolase phaY1 (H16_A2251, three- to tenfold) (Table 1C). The latter enzyme is known to be involved in the degradation of poly(3HB) in R. eutropha H16. Furthermore, H16_B1102 encoding an acetyl-CoA synthetase was detected (2- to 20-fold), which has an important role in glycolysis and also in the pyruvate as well as the propionate metabolism. Two further genes involved in propionate metabolism were detected, encoding a methylmalonyl-CoA mutase (sbm1, H16_A0280, four to tenfold; sbm2, H16_A1949, 3- to 248-fold). This enzyme converts succinyl-CoA to methylmalonyl-CoA. Finally, genes for an ABC-type transporter of the NitT family (H16_A0357-A0359, 6- to 130-fold) exhibited increased expression levels at the first three time points (Table 1C).
The comparison of TDP cells to DTDP cells revealed 42 genes which were upregulated in presence of DTDP, as mentioned above. This group comprises genes encoding the rhodanese-related sulfurtransferase (H16_A0333, three- to fivefold), the thiol-disulfide interchange protein encoded by dsbA (H16_A0161, three- to fourfold), the biotin synthase BioB (H16_A0183, three-to sevenfold), and four chaperone (groES, H16_A0705, 3- to 55-fold; groEL, H16_A0706, 2- to 21-fold; dnaK, H16_A3089, four- to sevenfold, and grpE, H16_A1137, three- to ninefold) (Table 1D). These seven genes have been detected in transcriptome comparison (b) as well and are mentioned above. Moreover, genes encoding two probable extracytoplasmic solute receptors (H16_A2779, three- to fivefold; H16_A3718, two- to fivefold) exhibited increased expression levels. The former one has already been detected in transcriptome comparison (in (b)).
Table 1.
Fold change of genes which were detected as upregulated during cultivation with (A) gluconate plus TDP in comparison to gluconate, (B) gluconate plus DTDP in comparison to gluconate, (C) gluconate plus TDP in comparison to gluconate plus DTDP, and (D) gluconate plus DTDP in comparison to gluconate plus TDP
| ID | Gene name | Annotation | Fold change | |||
|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 96 h | |||
| A | ||||||
| Genes putatively involved in the uptake of TDP | ||||||
| H16_A3658 | ABC-type transporter, permease component | 6.28 | 3.66 | 3.64 | 3.60 | |
| H16_A3659 | ABC-type transporter, permease component | 6.90 | 3.34 | 6.58 | 3.44 | |
| H16_A3660 | ABC-type transporter, permease component | 6.90 | 4.48 | 7.86 | 9.37 | |
| Genes putatively involved in the catabolism of TDP | ||||||
| H16_B0782 | Acetyltransferase (GNAT) family | 8.16 | 80.25 | 30.38 | 34.75 | |
| H16_A0217 | Hypothetical membrane-associated protein | 3.16 | 3.11 | 40.29 | 30.94 | |
| H16_A1954 | Predicted acyltransferase | 4.09 | 2.04 | 2.61 | x | |
| H16_A2171 | phaB3 | Acetoacetyl-CoA reductase | 4.68 | 2.20 | 3.97 | x |
| H16_B1292 | phnT | Acetyltransferase (GNAT) family | 6.36 | 2.74 | 3.46 | x |
| H16_B2322 | frnE | Predicted dithiol-disulfide isomerase | 2.99 | 6.25 | 5.80 | 5.26 |
| Genes putatively involved in detoxification | ||||||
| H16_A0306 | bcp | Peroxiredoxin | 3.53 | 2.99 | 3.11 | 2.70 |
| H16_A1090 | Glutathione S-transferase | 2.89 | 6.15 | 3.82 | 3.77 | |
| H16_B0571 | Glutathione S-transferase | 2.78 | 5.00 | 3.56 | 2.94 | |
| B | ||||||
| Genes putatively involved in the uptake of DTDP | ||||||
| H16_A2779 | Probable extracytoplasmic solute receptor | 3.35 | 3.96 | 2.12 | x | |
| H16_A0337 | Probable extracytoplasmic solute receptor | 2.62 | 3.06 | 4.44 | x | |
| Genes putatively involved in the catabolism of DTDP | ||||||
| H16_A0180 | bioA | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase | 17.16 | 47.67 | 8.12 | 5.55 |
| H16_A0181 | bioF | 8-amino-7-oxononanoate synthase | 5.98 | 8.22 | 2.60 | 3.57 |
| H16_A0182 | bioD | Dethiobiotin synthetase | 5.16 | 6.49 | 4.68 | 2.15 |
| H16_A0183 | bioB | Biotin synthase | 8.02 | 5.86 | 6.73 | 3.39 |
| H16_A0333 | Rhodanese-related sulfurtransferase | 3.59 | 6.58 | 7.55 | 7.00 | |
| H16_A0161 | dsbA | Thiol:disulfide interchange protein | 3.78 | 3.34 | 4.53 | 3.25 |
| H16_A3455 | dsbD | Thiol:disulfide interchange protein | 8.19 | 5.70 | 8.79 | x |
| H16_B2322 | frnE | Predicted dithiol-disulfide isomerase | 6.58 | 14.50 | 10.00 | 6.60 |
| H16_A0179 | Enoyl-CoA hydratase/carnithine racemase | 3.25 | 4.35 | 5.07 | 2.98 | |
| H16_A0217 | Hypothetical membrane-associated protein | 5.77 | 13.37 | 46.08 | 21.53 | |
| H16_A0244 | ahcY | Adenosylhomocysteinase | 5.55 | 10.25 | 10.90 | 13.39 |
| H16_A1070 | Acyl dehydratase | 4.44 | 4.70 | 2.79 | 2.59 | |
| H16_B0782 | Acetyltransferase, GNAT-family | 38.41 | 428.83 | 27.49 | 25.35 | |
| H16_B1784 | Signal transduction histidine kinase | 4.17 | 8.26 | 3.62 | 2.67 | |
| H16_B1785 | Response regulator, OmpR-family | 4.00 | 4.15 | 3.73 | 3.88 | |
| Genes putatively involved in detoxification | ||||||
| H16_A0705 | groES | Co-chaperonin (HSP10) | 23.51 | 9.41 | 3.03 | x |
| H16_A0706 | groEL | Chaperoning (HSP60 family) | 15.68 | 2.94 | 2.49 | x |
| H16_A1137 | grpE | Molecular chaperone | 6.84 | 9.17 | 6.51 | 5.46 |
| H16_A3089 | dnaK | Molecular chaperone (HSP70) | 4.26 | 4.34 | 8.64 | 5.66 |
| H16_B0451 | Glutathione S-transferase | 4.77 | 3.68 | 2.44 | x | |
| H16_A1090 | Glutathione S-transferase | 7.50 | 6.88 | 3.97 | 3.87 | |
| H16_A0306 | bcp | Peroxiredoxin | 3.45 | 5.02 | 3.60 | 4.47 |
| C | ||||||
| Genes putatively involved in the uptake of TDP | ||||||
| H16_A0357 | ABC-type transporter, ATPase component: NitT family | 79.34 | 13.73 | 17.89 | x | |
| H16_A0358 | ABC-type transporter, permease component: NitT family | 129.50 | 15.34 | 11.60 | x | |
| H16_A0359 | ABC-type transporter, periplasmic component: NitT family | 107.15 | 45.76 | 6.24 | x | |
| Genes putatively involved in the catabolism of TDP | ||||||
| H16_B1102 | Acetyl-CoA synthetase | 17.01 | 19.93 | 3.29 | 2.31 | |
| H16_A0234 | aidB | Acyl-CoA dehydrogenase, short-chain specific | 2.78 | 2.65 | 3.35 | x |
| H16_A0280 | sbml | Methylmalonyl-Coa mutase | 9.72 | 6.11 | 4.05 | x |
| H16_A1949 | sbm2 | Methylmalonyl-Coa mutase | 248.35 | 2.87 | 7.00 | x |
| H16_A2251 | phaY1 | d-(−)-3-hydroxybutyrate oligomer hydrolase | 3.07 | 3.79 | 9.93 | x |
| H16_B1192 | Acyl-CoA dehydrogenase | 25.89 | 77.24 | 2.92 | x | |
| D | ||||||
| Genes putatively involved in the uptake of DTDP | ||||||
| H16_A2779 | Probable extracytoplasmic solute receptor | 5.46 | 4.13 | 2.63 | x | |
| H16_A3718 | Probable extracytoplasmic solute receptor | 5.37 | 2.01 | 2.29 | x | |
| Genes putatively involved in the catabolism of DTDP | ||||||
| H16_A0333 | Rhodanese-related sulfurtransferase | 2.74 | 3.92 | 5.22 | 3.72 | |
| H16_A0161 | dsbA | Thiol:disulfide interchange protein | 3.51 | 3.88 | 2.70 | x |
| Genes putatively involved in detoxification | ||||||
| H16_A0705 | groES | Co-chaperonin (HSP10) | 54.77 | 8.00 | 2.85 | x |
| H16_A0706 | groEL | Chaperonin (HSP60 family) | 21.04 | 2.99 | 2.11 | x |
| H16_A1137 | grpE | Molecular chaperone | 9.46 | 9.24 | 7.60 | 2.79 |
| H16_A3089 | dnaK | Molecular chaperone (HSP70) | 7.40 | 4.03 | 6.14 | 6.34 |
| H16_A0183 | bioB | Biotin synthase | 7.45 | 3.32 | 4.70 | 2.59 |
x data were excluded from further analyses due to the adjusted software filter as outlined in the “Material and methods” section
PrpC enzyme activity assay
The 2-methylcitric acid cycle (MCC) mediates the conversion of propionyl-CoA to pyruvate and is the predominant pathway for the catabolism of propionic acid in bacteria and fungi (Brämer and Steinbüchel 2001; Brock et al. 2001). The key enzyme of this pathway is the methylcitrate synthase PrpC, which condensed propionyl-CoA and oxalacetate to methylcitric acid in a claisen condensation (Brock et al. 2000). The catabolism of TDP- and DTDP-degradation products via propionyl-CoA should lead to an increased activity of PrpC. Transcriptome analyses revealed no significant changes in transcription for prpC during cultivation in the presence of TDP or DTDP in comparison to gluconate as fold changes of approximately 1 were detected in all four time points (data not shown). Hence, a PrpC enzyme activity assay was performed. For this purpose, cells of R. eutropha H16 were cultivated in MSM under storage conditions with either 1% (w/v) TDP or DTDP as second, sulfur-containing carbon source to produce poly(3HB-co-3MP). After 24, 48, 72, and 96 h of cultivation, samples were withdrawn, and the activity of 2-methylcitrate synthase was determined.
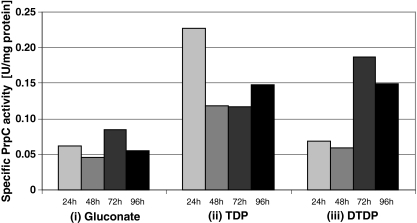
Whereas crude extracts of cells grown on gluconate revealed an average specific PrpC activity of only 0.06 U/mg of protein, an approximately fourfold higher specific PrpC activity (0.23 U/mg of protein) was measured in the soluble fractions of cells cultivated in presence of TDP after 24 h of cultivation (Fig. 4). After 48 and 72 h, the specific PrpC activity decreased but increased slightly after 96 h of cultivation. In contrast, crude extracts of DTDP-grown cells of R. eutropha H16 revealed a specific PrpC activity after 24 and 48 h of cultivation as low as gluconate-grown cells (0.06 U/mg of protein). However, an approximately threefold higher specific PrpC activity was measured after 72 and 96 h of cultivation (0.19 and 0.15 U/mg of protein), respectively. Specific PrpC activities of cells of R. eutropha H16 cultivated in presence of gluconate, TDP, or DTDP are summarized in Fig. 4.
Fig. 4.
Specific enzyme activity of PrpC measured in crude extracts of R. eutropha H16. Cells were cultivated in MSM under storage conditions containing (i) 1% (w/v) sodium gluconate, (ii) 1% (w/v) sodium gluconate plus 1% (v/v) TDP, or (iii) DTDP. After 24, 48, 72, and 96 h of cultivation, cells were harvested, cell-free crude extracts were prepared, and the activity of 2-methylcitrate synthase was measured. Data were generated from two independent biological experiments resulting in mean values of six replicates
A specific PrpC activity of only 0.04 U/mg of protein in cells of R. eutropha H16 cultivated in the presence of gluconate was reported previously (Ewering et al. 2006). The low activity is most probably due to low substrate specificity of other acyl-CoA thioester-forming enzymes. Brämer and Steinbüchel (2001) revealed a specific PrpC activity of 0.354 U/mg of protein in cells of R. eutropha HF39, a streptomycin-resistant mutant of the wild-type H16, when cultivated in presence of propionate.
Discussion
PHB and poly(3HB-co-3MP) accumulation by R. eutropha H16
PTEs are only known since 2001 as sulfur-containing polymers with thioester linkages in the backbone. R. eutropha H16 uses 3MP, TDP, and DTDP as precursor substrates for the biosynthesis of the heteropolythioester poly(3HB-co-3MP), when a second carbon source is available for growth (Lütke-Eversloh et al. 2001b). To provide a basis to further investigate the catabolism of the above-mentioned precursor substrates and to identify genes that are putatively involved in this metabolism, genome-wide transcriptome analyses were employed. For this purpose, cells of R. eutropha H16 were cultivated in the presence of only gluconate as carbon source for growth or in the presence of gluconate plus TDP or DTDP as precursor substrates for the incorporation of 3MP.
Diminished cell growth was obtained only during cultivation with gluconate plus DTDP (Fig. 2). As mentioned above, cleavage of DTDP into two molecules 3MP is proposed, whereas cleavage of TDP resulted in the release of only one molecule 3MP (Lütke-Eversloh et al. 2001a; Lütke-Eversloh and Steinbüchel 2003a). Inhibitory effects of higher concentrations of 3MP-CoA on enzymes of β-oxidation have been described previously (Sabbagh et al. 1985), possibly explaining the growth inhibition of R. eutropha during cultivation in presence of DTDP due to 3MP concentrations twice as high as in presence of TDP.
Cultivation with gluconate yielded poly(3HB) contents up to 73% (w/w) of CDW after 24 h of cultivation; henceforward, the polymer content decreased slowly, indicating weak poly(3HB) degradation (Fig. 3). The intracellular degradation of PHAs has been studied in detail. Different PHA depolymerases (phaZ) and oligomer hydrolases (phaY), which hydrolyze PHAs and the cleavage products, have been identified in R. eutropha (Jendrossek et al. 1996; Jendrossek 2002; Saegusa et al. 2001, 2002). Several studies have indicated that PHA synthesis and degradation can occur simultaneously in R. eutropha (Doi et al. 1990, Taidi et al. 1995), and a constitutive expression of both kinds of enzymes has been reported (Lawrence et al. 2005). In fact, synthesis and simultaneous hydrolysis of PHB make physiologically no sense as this would constitute a futile cycle, but the degradation of small amounts of PHB to maintain the basal metabolism was proposed (Peplinski et al. 2010).
Using TDP as second carbon source, a copolymer content of up to 67% was measured with up to 10.6 mol% of incorporated 3MP (Fig. 3). Cultivation in the presence of DTDP resulted in lower copolymer contents of up to 35% (w/w) of CDW only, but with a fivefold higher molar 3MP content of the copolymer as compared to cells cultivated in presence of TDP (Fig. 3). In this case, an increase of both the molar 3MP fraction and the copolymer content was obtained. Only after 96 h of cultivation, the copolymer as well as the molar 3MP content decreased. Studies regarding the extracellular biodegradability of PTE revealed new bacterial strains that were able to grow with poly(3HB-co-3MP) as sole source for carbon and energy, whereas no microorganisms capable of degrading PTE homopolymers could be isolated. The studies indicated that 3MP containing heteropolymers are cleaved only at the oxoester bond, resulting in the release of 3HB moieties which are then utilized as carbon source. The remaining 3MP oligomers were not utilized (Elbanna et al. 2003, 2004). However, R. eutropha H16 is not able to grow with 3MP, TDP, or DTDP as sole source for carbon and energy, and the mobilization of stored poly(3HB-co-3MP) in this bacterium has not yet been investigated. The results of this work, however, indicate an intracellular degradation of this copolymer in R. eutropha H16 and confirm the assumption of Elbanna et al. (2004), that only 3HB units of the copolymer were mobilized, whereas 3MP oligomers are left.
Uptake of TDP and DTDP
As a prerequisite for biosynthesis of poly(3HB-co-3MP), the sulfur-containing precursors TDP and DTDP must be transported into the cells. During cultivation with TDP, two genes encoding ABC-type transport systems (H16_A3658–3660; H16_A0357–0359) exhibited increased transcription levels in comparison to cells cultivated in presence of gluconate or gluconate plus DTDP, respectively (Fig. 5). The latter exhibits the conserved domain cd03293 which is related to NrtD and SsuB being the ATP-binding subunits of ABC-type nitrate and sulfonate transport systems. In E. coli, the proteins encoded by the gene cluster ssuEADCB mediate the utilization of sulfur from aliphatic sulfonates. ssuABC constitutes an ABC-type transport system, mediating the uptake of aliphatic sulfonates, whereas ssuD and ssuE encode the key enzymes for the desulfonation, an FMNH2-dependent monooxygenase and an NAD(P)H-dependent FMN reductase, respectively (Eichhorn et al. 1999). Although the ABC-type transporter encoded by H16_A0357–0359 exhibited increased expression level only as compared to DTDP, transcriptome data strongly suggest that both transport systems are involved in the uptake of TDP.
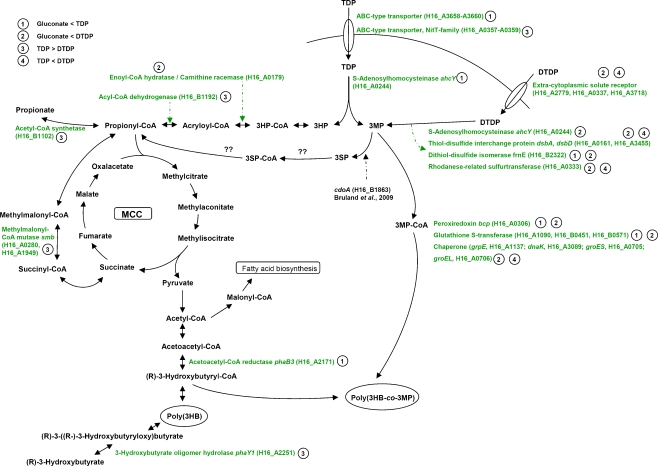
Fig. 5.
Overview of the genes putatively involved in the catabolism of TDP and DTDP. The ratio Cy5 (sample)/Cy3 (common reference) was calculated and was taken to represent the relative RNA abundance. The extent of the gene expression levels between samples provided a measure of the overall gene expression pattern between samples. (1) Genes were detected as upregulated during cultivation with TDP in comparison to gluconate. (2) Genes were detected as upregulated during cultivation with DTDP in comparison to gluconate. (3) Genes were detected as upregulated during cultivation with TDP in comparison to DTDP. (4) Genes were detected as upregulated during cultivation with DTDP in comparison to TDP. The complete lists of all studied genes are given in the Online Resources 1–4. MCC 2-methylcitric acid cycle
During cultivation in presence of gluconate plus DTDP, three genes encoding extracytoplasmic solute receptors exhibited increased expression levels in comparison to cells cultivated in presence of gluconate or gluconate plus TDP (Fig. 5). The amino acid sequences of these genes comprise sequence similarities to representatives of the gene family of Bordetella uptake genes known as “bug receptors”, which are strongly redundant in several β-Proteobacteria (Antoine et al. 2003). Some representatives of this gene family are part of operons encoding tripartite uptake transporters of the TTT family, while others are “orphan” genes (Thomas 2010). In Bordetella, Bug proteins were identified as receptor proteins for dicarboxylic compounds, and the Bug homologs BctC of Bordetella pertussis was shown to be involved in a signaling cascade as well (Antoine et al. 2005). In this context, an interesting finding was the detection of a histidine kinase (H16_B1784, three- to eightfold) and a response regulator (H16_B1785, fourfold), which exhibited increased expression levels during cultivations in the presence of gluconate plus TDP or DTDP in comparison to cells cultivated solely in presence of gluconate (Online Resource 1 and 2). These results indicate a putative signal transduction cascade with the signaling function for at least one of the cytoplasmic solute receptors and the signal transduction function for the histidine kinase and the response regulator mentioned above.
Cleavage of TDP and DTDP
As previously described, 3MP containing copolymers are synthesized in R. eutropha when TDP is supplemented to the media as second carbon source. Thus, an enzymatic cleavage of TDP into 3MP and 3HP by a hitherto unknown sulfide hydrolase was proposed (Lütke-Eversloh et al. 2001a). Several enzymes are described which catalyze the cleavage of structural analogs of TDP. The hydrolase S-adenosylhomocysteinase AhcY mediates the conversion of S-adenosylhomocysteine to adenosine and l-homocysteine (Palmer and Abeles 1979). In this study, ahcY was found to be upregulated during cultivation in the presence of gluconate plus DTDP but also during cultivation in the presence of gluconate plus TDP in the last three time points investigated (48, 72, and 96 h). Although the enzyme was characterized as highly specific for the substrates, the upregulation of the corresponding gene is an interesting finding and requires further investigations.
Cultivation of R. eutropha H16 with DTDP as sulfur-containing carbon source, resulted in the accumulation of less poly(3HB-co-3MP), while the molar 3MP fractions of the copolymers were much higher than with TDP. Very recently, data were obtained indicating the cleavage of DTDP into two molecules of 3MP by the dihydrolipoamide dehydrogenase (pdhL) in R. eutropha (Wübbeler et al. 2010). Although the specific enzyme activity was quite low for DTDP as substrate, the reduction was unequivocally detected. These results were not supported by transcriptome data presented in this study, but further evidence for the proposal of a cleavage of the disulfide bond of DTDP into two molecules 3MP by a reductive mechanism (Lütke-Eversloh and Steinbüchel 2003a) was provided. The transcription analyses revealed genes coding for thiol-disulfide interchange proteins (DsbA, DsbD) and a DsbA-like putative dithiol-disulfide isomerase (FrnE). DsbA and DsbD are known to be involved in correct folding of nascent proteins in the periplasm. Whereas DsbA is involved in the oxidative folding pathway, which is responsible for the formation of disulfide bonds, DsbD is involved in catalyzing intramolecular disulfide isomerization in proteins containing more than one disulfide bond. DsbA is the most oxidative oxidoreductase known, and dsbA mutants of E. coli are hypersensitive to the reductant dithiothreitol and some metal ions (Missiakas et al. 1993; Grauschopf et al. 1995; Stafford et al. 1999). Zheng et al. (1997) suggested a chaperone function of DsbA in addition to its function as an oxidoreductase. However, the data obtained by transcriptome analyses of this study clearly indicate that DsbA, DsbD, and the DsbA-like dithiol-disulfide isomerase FrnE are somehow involved in the cleavage of DTDP and/or TDP. However, their direct involvement has to be further investigated.
In addition, the obtained data led to the assumption, that the rhodanese-related sulfurtransferase encoded by H16_A0333, is putatively involved in the catabolism of DTDP. This kind of enzyme catalyzes the sulfur transfer from thiosulfate to thiophilic acceptors (Ray et al. 2000), and in Pseudomonas aeruginosa, the rhodanese RhdA is involved in cyanide detoxification (Cipollone et al. 2008).
Catabolism of TDP and DTDP
The fate of the cleavage products 3MP and 3HP is still unknown in R. eutropha. During poly(3HB) biosynthesis in R. eutropha, the provided gluconate is catabolized to acetyl-CoA, which serves as a precursor substrate for synthesis of this polymer. An adequate activation of 3MP and 3HP to the corresponding CoA thioester, which can subsequently serve as a substrate for the PHA synthase, was proposed (Lütke-Eversloh et al. 2001a). The PHA synthase of R. eutropha H16 exhibits a very broad substrate specificity (Haywood et al. 1989) and catalyzes the biosynthesis of a wide range of different PHAs and PTEs. 3HP can further be catabolized to acetyl-CoA, and furthermore, it has been detected as a constituent of PHAs in R. eutropha H16 as well (Nakamura et al. 1991).
Only very recently, new information regarding the fate of 3MP were obtained by Bruland et al. (2009). The cystein dioxygenase CdoA from R. eutropha H16 was shown to convert 3MP into 3SP in recombinant E. coli cells. In addition, investigations of the recently described bacteria V. paradoxus strain TBEA6 and A. mimigardefordensis strain DPN7T led to a proposed degradation pathway from DTDP and TDP via 3MP, 3SP, and 3SP-CoA towards propionyl-CoA, respectively (Bruland et al. 2009; Wübbeler et al. 2008). Catabolism of propionyl-CoA via the 2-methylcitric acid cycle is very likely to occur in A. mimigardefordensis. A similar pathway in R. eutropha H16 is feasible. In addition to the incorporation into the heteropolymer, 3MP is most probably metabolized via 3SP and 3SP-CoA towards propionyl-CoA, which is further catabolized via the 2-methylcitric acid cycle (Fig. 5). Enzyme activity assay of 2-methylcitrate synthase PrpC, the key enzyme of 2-methylcitric acid cycle, revealed a three- to fourfold increased specific PrpC activity in cells grown in the presence of TDP or DTDP, whereas transcriptome analyses revealed no significant upregulation of prpC. However, an overall low specific activity for PrpC was detected, and difficulties to correlate transcriptome and proteome data has been discussed previously: differing half-life times of RNA and protein molecules and differing RNA and protein turnover can have a large impact on the obtained data (Hedge et al. 2003). As the 2-methylcitric acid cycle is the predominant pathway for the catabolism of propionic acid in bacteria and fungi, and since it occurs also in R. eutropha (Brämer and Steinbüchel 2001; Brock et al. 2001), conversion of 3MP and 3HP to propionyl-CoA is supported by this study.
Genes of fatty acid metabolism are most probably involved in the catabolism of the cleavage products 3MP and 3HP in R. eutropha. Transcriptome analyses revealed several genes known to be involved in fatty acid metabolism exhibiting increased expression levels during cultivation in the presence of TDP or DTDP. In particular, the acyltransferase encoded by H16_B0782 and the acyl-CoA thioesterase homolog encoded by H16_A0217 attracted attention due to the huge upregulation during both cultivation with TDP and DTDP.
Stress response and detoxification
Chemical stress occurs widely in nature. Microorganisms react, e.g., to anoxia, changes in pH or towards different xenobiotics by increasing the expression of stress proteins. Heat-shock proteins are a common example. Many chaperones are also stress proteins and protect polypeptides from denaturation and aggregation when facing physical or chemical stress (Macario 1995). 3MP, TDP, and DTDP exert toxic effects on cells of R. eutropha at concentrations higher than 0.1% or 1.5%, respectively, thereby diminishing growth (Lütke-Eversloh et al. 2001a). Transcriptome analyses revealed several genes coding for chaperones induced during cultivation with DTDP as compared to gluconate and TDP, respectively. Further hints for the involvement of chaperones in the degradation of DTDP were obtained. Wübbeler et al. (2008) characterized mutants of A. mimigardefordensis exhibiting insertions in genes encoding chaperones as fully or partially impaired in utilization of DTDP. In addition, chaperone activity is obviously important for degradation of dimethylsulfoniopropionate (DMSP) (Bürgmann et al. 2007). The increased expression of genes coding for glutathione S-transferases (GST) and peroxiredoxin is a further stress response to the cultivation with TDP and DTDP. GSTs are involved in the metabolism of a wide range of xenobiotic compounds by catalyzing the conjugation of these compounds with glutathione (Armstrong 1991). Peroxiredoxin is a representative of a ubiquitous family of thiol-specific antioxidant enzymes (Wood et al. 2003). The results of the transcriptome analyses are summarized and graphically presented in Fig. 5.
Genome-wide transcriptome analyses revealed several interesting genes with increased expression level during cultivation in presence of TDP and/or DTDP. The data strongly suggest that they are somehow involved in the catabolism of these compounds in R. eutropha H16. Therefore, these transcriptome analyses provided valuable hints for proteins that are required for the uptake and catabolism of these organic sulfur compounds. Extensive further investigations are now required to characterize these genes in detail and to reveal their functions in R. eutropha H16 and other bacteria. This will of course also include the generation of knockout mutants defective in the genes found in these transcriptome analyses and the characterization of the phenotypes of these mutants.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(DOC 536 kb)
Acknowledgments
The project was carried out within the framework of the Competence Network Göttingen “Genome research on bacteria” (GenoMik and Genomik Plus) financed by the German Federal Ministry of Education and Research (BMBF, FKZ-0313751) which is gratefully acknowledged. Support of this study by Jan Hendrik Wübbeler and Nadine Stöveken is gratefully acknowledged.
Footnotes
Accession numbers
Microarray data are available in the ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MEXP-2744.
References
- Antoine R, Jacob-Dubuisson F, Drobecq H, Willery E, Lesjean S, Locht C. Overrepresentation of a gene family encoding extracytoplasmic solute receptors in Bordetella. J Bacteriol. 2003;185:1470–1474. doi: 10.1128/JB.185.4.1470-1474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoine R, Huvent I, Chemlal K, Deray I, Raze D, Locht C, Jacob-Dubuisson F. The periplasmic binding protein of a tripartite tricarboxylate transporter is involved in signal transduction. J Mol Biol. 2005;351:799–809. doi: 10.1016/j.jmb.2005.05.071. [DOI] [PubMed] [Google Scholar]
- Armstrong RN. Glutathione S-transferases: reaction mechanism, structure and function. Chem Res Toxicol. 1991;4:131–140. doi: 10.1021/tx00020a001. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brämer CO, Steinbüchel A. The methylcitric acid pathway in Ralstonia eutropha: new genes identified involved in propionate metabolism. Microbiology. 2001;147:2203–2214. doi: 10.1099/00221287-147-8-2203. [DOI] [PubMed] [Google Scholar]
- Brandl H, Gross RA, Lenz RW, Fuller RC. Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl Environ Microbiol. 1988;54:1977–1982. doi: 10.1128/aem.54.8.1977-1982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock M, Fischer R, Linder D, Buckel W. Methylcitrate synthase from Aspergillus nidulans: implications for propionate as an antifungal agent. Mol Microbiol. 2000;35:961–973. doi: 10.1046/j.1365-2958.2000.01737.x. [DOI] [PubMed] [Google Scholar]
- Brock M, Darley D, Textor S, Buckel W. 2-Methylisocitrate lyase from the bacterium Escherichia coli and the filamentous fungus Aspergillus nidulans: characterization and comparison of both enzymes. Eur J Biochem. 2001;268:3577–3586. doi: 10.1046/j.1432-1327.2001.02262.x. [DOI] [PubMed] [Google Scholar]
- Bruland N, Wübbeler JH, Steinbüchel A. 3-Mercaptopropionate dioxygenase, a cysteine dioxygenase homologue, catalyzes the initial step of 3-mercaptopropionate catabolism in the 3, 3′-thiodipropionic acid degrading bacterium Variovorax paradoxus. J Biol Chem. 2009;284:660–672. doi: 10.1074/jbc.M806762200. [DOI] [PubMed] [Google Scholar]
- Bürgmann H, Howard EC, Ye W, Sun F, Sun S, Napierala S, Moran MA. Transcriptional response of Silicibacter pomeroyi DSS-3 to dimethylsulfoniopropionate (DMSP) Environ Microbiol. 2007;9:2742–2755. doi: 10.1111/j.1462-2920.2007.01386.x. [DOI] [PubMed] [Google Scholar]
- Cipollone R, Ascenzi P, Tomao P, Imperi F, Visca P. Enzyme detoxification of cyanide: clues from Pseudomonas aeruginosa rhodanese. J Mol Microbiol Biotechnol. 2008;15:199–211. doi: 10.1159/000121331. [DOI] [PubMed] [Google Scholar]
- Conway T, Schoolnik GK. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol Microbiol. 2003;47:879–889. doi: 10.1046/j.1365-2958.2003.03338.x. [DOI] [PubMed] [Google Scholar]
- Dillon SC, Bateman A. The HotDog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinform. 2004;5:109. doi: 10.1186/1471-2105-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi Y, Segawa A, Kawaguchi Y, Kunioka M. Cyclic nature of poly(3-hydroxyalkanoate) metabolism in Alcaligenes eutrophus. FEMS Microbiol Lett. 1990;55:165–169. doi: 10.1111/j.1574-6968.1990.tb13856.x. [DOI] [PubMed] [Google Scholar]
- Eichhorn E, Ploeg JR, Leisinger T. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J Biol Chem. 1999;274:26639–26646. doi: 10.1074/jbc.274.38.26639. [DOI] [PubMed] [Google Scholar]
- Elbanna K, Lütke-Eversloh T, Trappen S, Mergaert J, Swings J, Steinbüchel A. Schlegelella thermodepolymerans gen. nov., sp. nov., a novel thermophilic bacterium that degrades poly(3-hydroxybutyrate-co-3-mercaptopropionate) Int J Syst Evol Microbiol. 2003;53:1165–1168. doi: 10.1099/ijs.0.02562-0. [DOI] [PubMed] [Google Scholar]
- Elbanna K, Lütke-Eversloh T, Jendrossek D, Luftmann H, Steinbüchel A. Studies on the biodegradability of polythioester copolymers and homopolymers by polyhydroxyalkanoate (PHA)-degrading bacteria and PHA depolymerases. Arch Microbiol. 2004;182:212–225. doi: 10.1007/s00203-004-0715-z. [DOI] [PubMed] [Google Scholar]
- Ellman GL. A colorimetric method for determining low concentrations of mercaptans. Arch Biochem Biophys. 1958;74:443–450. doi: 10.1016/0003-9861(58)90014-6. [DOI] [PubMed] [Google Scholar]
- Ewering C, Heuser F, Benölken JK, Brämer CO, Steinbüchel A. Metabolic engineering of strains of Ralstonia eutropha and Pseudomonas putida for biotechnological production of 2-methylcitric acid. Metab Eng. 2006;8:587–602. doi: 10.1016/j.ymben.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JM, Johnston DA, Williams GW, Williams DJ, Freeman TC, Dunnea DW, Hoffmann KF. An oligonucleotide microarray for transcriptome analysis of Schistosoma mansoni and its application/use to investigate gender-associated gene expression. Mol Biochem Parasitol. 2005;141:1–13. doi: 10.1016/j.molbiopara.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Grauschopf U, Winther JR, Korber P, Zander T, Dallinger P, Bardwell JCA. Why is DsbA such an oxidizing disulfide catalyst? Cell. 1995;83:947–955. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- Hassan YI, Zempleni J. Epigenetic regulation of chromatin structure and gene function by biotin. J Nutr. 2006;136:1763–1765. doi: 10.1093/jn/136.7.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JD, Pulford DJ. The glutathione S-transferase supergene family: regulation of GST and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- Haywood G, Anderson A, Dawes E. The importance of PHB-synthase substrate-specificity in polyhydroxyalkanoate synthesis by Alcaligenes eutrophus. FEMS Microbiol Lett. 1989;57:1–6. doi: 10.1111/j.1574-6968.1989.tb03210.x. [DOI] [Google Scholar]
- Hedge PS, White IR, Debouck C. Interplay of trancriptomics and proteomics. Curr Opin Biotechnol. 2003;14:647–651. doi: 10.1016/j.copbio.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Jendrossek D. Extracellular polyhydroxyalkanoate depolymerases: the key enzymes of PHA degradation. In: Doi Y, Steinbüchel A, editors. Biopolymers, volume 3b: polyesters II—properties and chemical synthesis. Weinheim: Wiley-VCH; 2002. pp. 41–84. [Google Scholar]
- Jendrossek D, Handrick R. Microbial degradation of polyhydroxyalkanoates. Annu Rev Microbiol. 2002;56:403–432. doi: 10.1146/annurev.micro.56.012302.160838. [DOI] [PubMed] [Google Scholar]
- Jendrossek D, Schirmer A, Schlegel HG. Biodegradation of polyhydroxyalkanoic acids. Appl Microbiol Biotechnol. 1996;46:451–463. doi: 10.1007/s002530050844. [DOI] [PubMed] [Google Scholar]
- Kawada J, Lütke-Eversloh T, Steinbüchel A, Marchessault RH. Physical properties of microbial polythioesters: characterization of poly(3-mercaptoalkanoates) synthesized by engineered Escherichia coli. Biomacromolecules. 2003;4:1698–1702. doi: 10.1021/bm0341327. [DOI] [PubMed] [Google Scholar]
- Kim DY, Lütke-Eversloh T, Elbanna K, Thakor N, Steinbüchel A. Poly(3-mercaptopropionate): a nonbiodegradable biopolymer? Biomacromolecules. 2005;6:897–901. doi: 10.1021/bm049334x. [DOI] [PubMed] [Google Scholar]
- Lawrence AG, Schoenheit J, He A, Tian J, Liu P, Stubbe J, Sinskey AJ. Transcriptional analysis of Ralstonia eutropha genes related to poly-(R)-3-hydroxybutyrate homeostasis during batch fermentation. Appl Microbiol Biotechnol. 2005;68:663–672. doi: 10.1007/s00253-005-1969-3. [DOI] [PubMed] [Google Scholar]
- Leesong M, Henderson BS, Gillig JR, Schwab JM, Smith JL. Structure of a dehydratase-isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site. Structure. 1996;4:253–264. doi: 10.1016/S0969-2126(96)00030-5. [DOI] [PubMed] [Google Scholar]
- Lemoigne M. Produits de deshydration et de polymerization de lácide β-oxybutyrique. Bull Soc Chim Biol. 1926;8:770–782. [Google Scholar]
- Lindenkamp N, Peplinski K, Volodina E, Ehrenreich A, Steinbüchel A. Multiple β-ketothiolase deletion mutants of Ralstonia eutropha: impact on the composition of 3-mercaptopropionic acod-containing copolymer. Appl Environ Microbiol. 2010 doi: 10.1128/AEM.01058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütke-Eversloh T, Steinbüchel A. Microbial polythioester. Macromol Biosci. 2004;4:165–174. doi: 10.1002/mabi.200300084. [DOI] [PubMed] [Google Scholar]
- Lütke-Eversloh T, Steinbüchel A. Novel precursor substrates for polythioesters (PTE) and limits of PTE biosynthesis in Ralstonia eutropha. FEMS Microbiol Lett. 2003a;221:191–196. doi: 10.1016/S0378-1097(03)00185-X. [DOI] [PubMed] [Google Scholar]
- Lütke-Eversloh T, Steinbüchel A. Polythioesters. In: Matsumura S, Steinbüchel A, editors. Biopolymers, vol. 9. Weinheim: Wiley-VCH; 2003b. pp. 63–80. [Google Scholar]
- Lütke-Eversloh T, Bergander K, Luftmann H, Steinbüchel A. Identification of a new class of biopolymer: bacterial synthesis of a sulfur-containing polymer with thioester linkages. Microbiology. 2001a;147:11–19. doi: 10.1099/00221287-147-1-11. [DOI] [PubMed] [Google Scholar]
- Lütke-Eversloh T, Bergander K, Luftmann H, Steinbüchel A. Biosynthesis of poly(3-hydroxybutyrate-co-3-mercaptobutyrate) as a sulfur analogue to poly(3-hydroxybutyrate) (PHB) Biomacromolecules. 2001b;2:1061–1065. doi: 10.1021/bm015564p. [DOI] [PubMed] [Google Scholar]
- Lütke-Eversloh T, Kawada J, Marchessault RH, Steinbüchel A. Characterization of biological polythioesters: physical properties of novel copolymers synthesized by Ralstonia eutropha. Biomacromolecules. 2002;3:159–166. doi: 10.1021/bm015603x. [DOI] [PubMed] [Google Scholar]
- Macario AJL. Heat-shock proteins and molecular chaperones: implications for pathogenesis, diagnostics, and therapeutics. Int J Clin Lab Res. 1995;25:59–70. doi: 10.1007/BF02592359. [DOI] [PubMed] [Google Scholar]
- Missiakas D, Georgopoulos C, Raina S. Identification and characterization of the Escherichia coli gene dsbB, whose product is involved in the formation of disulfide bonds in vivo. Proc Natl Acad Sci USA. 1993;90:7084–7088. doi: 10.1073/pnas.90.15.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Kunioka M, Doi Y. Biosynthesis and characterization of bacterial poly(3-hydroxybutyrate-co-3-hydroxypropionate) Macromol Rep. 1991;A28:15–24. [Google Scholar]
- Palmer JL, Abeles RH. The mechanism of action of S-adenosylhomocysteinase. J Biol Chem. 1979;254:1217–1226. [PubMed] [Google Scholar]
- Peplinski K, Ehrenreich A, Döring C, Bömeke M, Reinecke F, Hutmacher C, Steinbüchel A. Genome-wide transcriptome analyses of the ‘Knallgas’ bacterium Ralstonia eutropha H16 with regard to polyhydroxyalkanoate metabolism. Microbiology (SGM) 2010;156:2136–2152. doi: 10.1099/mic.0.038380-0. [DOI] [PubMed] [Google Scholar]
- Pohlmann A, Fricke WF, Reinecke F, Kusian B, Liesegang H, Cramm R, Eitinger T, Ewering C, Pötter M, Schwartz E, Strittmatter A, Voß I, Gottschalk G, Steinbüchel A, Friedrich B, Bowien B. Genome sequence of the bioplastic-producing “Knallgas” bacterium Ralstonia eutropha H16. Nat Biotechnol. 2006;24:1257–1262. doi: 10.1038/nbt1244. [DOI] [PubMed] [Google Scholar]
- Pötter M, Steinbüchel A. Poly(3-hydroxybutyrate) granule-associated proteins: impacts on poly(3-hydroxybutyrate) synthesis and degradation. Biomacromolecules. 2005;6:552–560. doi: 10.1021/bm049401n. [DOI] [PubMed] [Google Scholar]
- Ray WK, Zeng G, Potters MB, Mansuri AM, Larson TJ. Characterization of a 12-kilodalton rhodanese encoded by glpE of Escherichia coli and its interaction with thioredoxin. J Bacteriol. 2000;182:2277–2284. doi: 10.1128/JB.182.8.2277-2284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm BHA. Polyester synthase: natural catalysts for plastics. Biochem J. 2003;376:15–33. doi: 10.1042/BJ20031254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinecke F, Steinbüchel A. Ralstonia eutropha strain H16 as model organism for PHA metabolism and for biotechnological production of technically interesting biopolymers. J Mol Microbiol Biotechnol. 2009;16:91–108. doi: 10.1159/000142897. [DOI] [PubMed] [Google Scholar]
- Sabbagh E, Cuebas D, Schulz H. 3-Mercaptopropionic acid, a potent inhibitor of fatty acid oxidation in rat heart mitochondria. J Biol Chem. 1985;260:7337–7342. [PubMed] [Google Scholar]
- Saegusa H, Shiraki M, Kanai C, Saito T. Cloning of an intracellular poly [d(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J Bacteriol. 2001;183:94–100. doi: 10.1128/JB.183.1.94-100.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saegusa H, Shiraki M, Saito T. Cloning of an intracellular d(−)-3-hydroxybutyrate oligomer hydrolase gene from Ralstonia eutropha H16 and identification of the active site serine residue by site-directed mutagenesis. J Biosci Bioeng. 2002;94:106–112. doi: 10.1263/jbb.94.106. [DOI] [PubMed] [Google Scholar]
- Schlegel HG, Gottschalk G, Bartha R. Formation and utilization of poly-β-hydroxybutyric acid by Knallgas bacteria (Hydrogenomonas) Nature. 1961;191:463–465. doi: 10.1038/191463a0. [DOI] [PubMed] [Google Scholar]
- Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- Slater T, Houmiel KL, Tran M, Mitsky TA, Taylor NB, Padgette SR, Gruys K. Multiple β-ketothiolases mediate poly(β-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J Bacteriol. 1998;180:1979–1987. doi: 10.1128/jb.180.8.1979-1987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srere PA. Citrate-condensing enzyme-oxaloacetate binary complex. J Biol Chem. 1966;241:2157–2165. [PubMed] [Google Scholar]
- Stafford SJ, Humphreys DP, Lund PA. Mutations in dsbA and dsbB, but not in dsbC, lead to an enhanced sensitivity of Escherichia coli to Hg2+ and Cd2+ FEMS Microbiol Lett. 1999;174:179–184. doi: 10.1111/j.1574-6968.1999.tb13566.x. [DOI] [PubMed] [Google Scholar]
- Steinbüchel A. Non-biodegradable biopolymers from renewable resources: perspectives and impacts. Curr Opin Biotechnol. 2005;16:607–613. doi: 10.1016/j.copbio.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Steinbüchel A, Valentin HF. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol Lett. 1995;128:219–228. doi: 10.1016/0378-1097(95)00125-O. [DOI] [Google Scholar]
- Taidi B, Mansfield D, Anderson AJ. Turnover of poly(3-hydroxybutyrate) (PHB) and its influence on the molecular mass of the polymer accumulated by Alcaligenes eutrophus during batch culture. FEMS Microbiol Lett. 1995;129:201–206. [Google Scholar]
- Thomas GH. Homes for the orphans: utilization of multiple substrate-binding proteins by ABC transporter. Mol Microbiol. 2010;75:6–9. doi: 10.1111/j.1365-2958.2009.06961.x. [DOI] [PubMed] [Google Scholar]
- Timm A, Steinbüchel A. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl Environ Microbiol. 1990;56:3360–3367. doi: 10.1128/aem.56.11.3360-3367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood ZA, Schröder E, Harris JR, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/S0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- Wübbeler JH, Lütke-Eversloh T, Trappen S, Vandamme P, Steinbüchel A. Tetrathiobacter mimigardefordensis sp. nov., isolated from compost, a betaproteobacterium capable of utilizing the organic disulfide 3, 3′-dithiodipropionic acid. Int J Syst Evol Microbiol. 2006;56:1305–1310. doi: 10.1099/ijs.0.64126-0. [DOI] [PubMed] [Google Scholar]
- Wübbeler JH, Bruland N, Kretschmer K, Steinbüchel A. A novel pathway for the catabolism of the organic sulfur compound 3, 3′-dithiodipropionic acid via 3-mercaptopropionic acid and 3-sulfinopropionic acid to propionyl-CoA by the aerobic bacterium Tetrathiobacter mimigardefordensis strain DPN7. Appl Environ Microbiol. 2008;74:4028–4035. doi: 10.1128/AEM.00422-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wübbeler JH, Raberg M, Brandt U, Steinbüchel A (2010) Dihydrolipoamide dehydrogenases of Advenella mimigardefordensis and Ralstonia eutropha catalyze cleavage of 3,3′-dithiodipropionic acid into 3-mercaptopropionic acid. Appl Environ Microbiol. doi:10.1128/AEM.01706-10 [DOI] [PMC free article] [PubMed]
- Zheng WD, Quan H, Song JL, Wang CC. Does DsbA have chaperone-like activity? Arch Biochem Biophys. 1997;337:326–331. doi: 10.1006/abbi.1996.9783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
(DOC 536 kb)