Abstract
The vacuolar H+ ATPase (v-ATPase) is crucial for endosome acidification, endocytosis, and trafficking in essentially all eukaryotic cells. Recent studies have shown that inhibition of the v-ATPase also leads to downregulation of important signaling pathways, including Notch and Wnt, which are key regulators of cell differentiation and tissue homeostasis across the animal kingdom. However, the requirement of endosome acidification and endocytosis in the transduction of Notch signaling is still highly debated. Moreover, no study has yet investigated the role of the v-ATPase during mammalian development. Here we show that expression of a dominant-negative subunit of the v-ATPase in neural precursors of the developing mouse cortex depleted neural stem cells by promoting their differentiation and the generation of neurons. Moreover, inhibition of the v-ATPase reduced endogenous Notch signaling and prevented the proliferative effect of a transmembrane, γ-secretase-dependent, active Notch without blocking the effects of its cytoplasmic intracellular domain (NICD). Our data are consistent with recent reports in Drosophila in which the v-ATPase has been suggested to be important for the transduction of Notch signaling. By extending these reports to mammalian embryos, our data may contribute to a better understanding of the role of the v-ATPase, endosome acidification, and endocytosis in signal transduction during neural stem cell differentiation and brain development.
Introduction
The multimeric protein complex known as the vacuolar ATPase (v-ATPase) is an ancient and extremely well-conserved proton pump responsible for the acidification of intracellular compartments and the extracellular space, which are important for endocytosis and vesicle transport in essentially all eukaryotic cells, from yeast to human [1–3].
More recently, 3 laboratories have independently shown that inhibition of the v-ATPase also affects the transduction of important signaling molecules, such as Notch [4,5] and Wnt [6], that are key determinants controlling proliferation, tissue patterning, embryonic development, and organ homeostasis throughout the animal kingdom [7–10]. While it is still unclear to which extent the effects of the v-ATPase on signaling are due to its direct function on endosome acidification as opposed to indirect effects on endocytosis and trafficking [11], these pioneering studies [4–6] indicate that this particular proton pump is a key player in signal transduction of cell fate determinants.
Endosome acidification and endocytosis were suggested to control Notch signaling even before manipulations of the v-ATPase were reported. This is because the γ-secretase, whose activity is essential for Notch processing, has been shown to have a higher proteolytic activity in acidic compartments [12] and because proteins involved in endocytosis, including dynamin and Rab5, revealed to be essential for Notch activation [5,13,14]. On the other hand, these data may also suggest that ligand presentation by the signal-presenting cell, rather than signal-transduction in the receiving cell, may be influenced by endocytosis [7,15]. The intrinsic difficulties in distinguishing between cell-autonomous versus extrinsic effects and reports showing that Notch signaling may not require endocytosis [16–18] have led to a long debate in the field.
Although the interplay between the v-ATPase and Notch signaling has been shown in Drosophila, evidence for such a link in other organisms, remarkably mammals, is still completely lacking. Considering that both the v-ATPase and Notch are well conserved across phyla, we decided to inhibit the v-ATPase in neural precursors of developing mouse embryos by tissue-specific expression of the YCHE78 peptide, a well-characterized dominant negative, truncated version of the V1e1 subunit of the v-ATPase complex [19].
YCHE78 has been successfully used to analyze the role of the proton pump in left–right asymmetry [20], appendage regeneration [21], planar cell polarity, and Wnt signaling [6] of nonmammalian vertebrates. In this context, YCHE78 expression is preferred to competing technologies such as RNAi because (1) it directly interferes with the existing proteins and is independent from their turnover kinetics that may delay a loss of function, (2) it is effective despite compensation by multiple isoforms of v-ATPase subunits while multiple shRNAs would have to be combined raising serious concerns about off-targets effect, and (3) it does not require the dicer-mediated dsRNA degradation machinery that may lead to confounding effects during development. Finally, YCHE78 expression is superior to pharmacological approaches that are difficult to perform in vivo, in particular while studying mammalian embryonic development.
Importantly, the moderate levels of YCHE78 afforded by transfection in vivo do not induce cell death. As shown in previous studies in Xenopus tadpoles [6,20,21], a reduction in v-ATPase activity is quite compatible with life and normal development. This is not surprising given the many studies in which subtle patterning and regulatory functions of ion flows can be functionally manipulated and dissected separately from their housekeeping roles [22,23], which is particularly true in stem cells [24].
Thus, we decided to investigate the role of the v-ATPase, and its possible effects on Notch signaling, during mammalian development by expressing YCHE78 in neural stem cells using in utero electroporation, a technique widely used to acutely and tissue-specifically manipulate genes in neural stem cells of the mouse brain [25]. Cell autonomous effects on proliferation versus differentiation were investigated in targeted cells of the developing neocortex, one of the best characterized Notch-dependent model systems in mammals [26–28].
Specifically, during early development of the mouse cerebral cortex, neuroepithelial and radial glial cells undergo proliferative divisions at the apical boundary of the ventricular zone (VZ), hence the name apical progenitors (APs). With the onset of differentiation, AP increasingly switch to self-renewing divisions that generate neurons or, most frequently, committed neurogenic progenitors that migrate toward the basal boundary of the VZ, hence the name basal progenitors (BP), to form the subventricular zone (SVZ) [28,29]. Since Notch maintains the identity of AP while limiting the generation of BP and neurons [27,28,30], we decided to investigate the effects of YCHE78 expression on the generation of AP, BP, and neurons during brain development.
Materials and Methods
Plasmids
pCAGGS-mRFPnls, pCS2-YCHE78, and pBS-Hes5-GFP have been described elsewhere [21,31,32]. pEF1-BOS-caNotch1 and pEF1-BOS-NICD [33] were kindly provided by Dr. Masato Nakafuku and Dr. Hideyuki Okano, respectively. pcDNA3.1 (Invitrogen) was used as empty plasmid.
In utero electroporation
In utero electroporation was performed on isofluorane-anaesthetized C57BL/6J pregnant mice at embryonic day (E) 12.5 by injecting 1–3 μL of phosphate-buffered saline containing 1–3 μg/μL of plasmids into the lumen of the embryonic telencephalon followed by 6 pulses of 30 V, 50 ms each at 1 s interval delivered through platinum electrodes (1 mm diameter) using a square-shaped electroporator (BTX-830; Genetronics), as previously described [34,35]. After surgery, gestation was allowed to continue for additional 16 or 24 h. When coelectroporation was performed without YCHE78, pCS2-YCHE78 was replaced with pcDNA3.1 at the same molarity.
Immunohistochemistry
Brains were fixed overnight at 4°C in 4% paraformaldehyde, equilibrated in 30% sucrose, and embedded in Tissue-Tek. Immunolabeling with Tbr2 (AB 9618, 1:500; Millipore), Tbr1 (AB9616, 1:250; Millipore), or RFP (125A13-5, 1:250, MPI-CBG antibody facility) antibodies on 10-μm-thick cryosections was performed as previously described [34] to identify AP (Tbr2− cells within the VZ), BP (Tbr2+ cells), and neurons [Tbr2− and/or Tbr1+ cells within the SVZ or intermediate zone (IZ)]. Sections were analyzed with a conventional Axioscope or confocal LSM510 Axiovert 200M microscopes (Carl Zeiss) as indicated in figure legends. Images were acquired using Axiovision or ZEISS LSM 4.2 software (Carl Zeiss) and processed with ImageJ 1.33 (www.imagej.nih.gov) or Photoshop CS3 (Adobe).
Quantification of endogenous Notch signaling
Quantification of endogenous Notch signaling was performed after electroporation of a Hes5GFP reporter plasmid [32]. Particular attention was given to inject identical amounts of a cytoplasmic RFP and Hes5GFP expression vectors; thus, in controls the fluorescent reporter plasmids were supplemented with an equimolar amount of empty vectors to compensate for the lack of YCHE78. Brains were collected 16 h later to reduce the chance of saturating the levels of GFP fluorescence at longer survival times and pictures of the individual channels taken on cryosections by one investigator. To reduce bias, RFP+ cells within the VZ were chosen on electronic files by a second investigator and their somata outlined digitally. A total of 60 cells were randomly selected from 3 different brains per condition, which was sufficient to provide a representative sample of cell distributions along the apical-to-basal boundaries of the VZ and RFP fluorescence intensities. Finally, the GFP channel was digitally added and green fluorescence quantified in the area previously outlined using ImageJ.
Statistical analyses
Statistical analyses were performed by pulling together the counts from 1 to 4 cryosections chosen among brains targeted in field 40 of the lateral cortex [34] until a total count of at least 200 cells was obtained. Brains from 3 litters were used for calculation of mean and standard deviation. Significance was evaluated by Student's t-test. For testing the positive correlation of GFP and RFP fluorescence, the Spearman's rank correlation coefficient (rS) was calculated for control and YCHE78 on a significance level of α = 5% using the SPSS statistical package 15.0 (SPSS Inc.). Due to non-normal distribution of the data points, the nonparametric Mann–Whitney U-test was used to assess significance of the respective medians.
Results
YCHE78 expression promotes neurogenesis
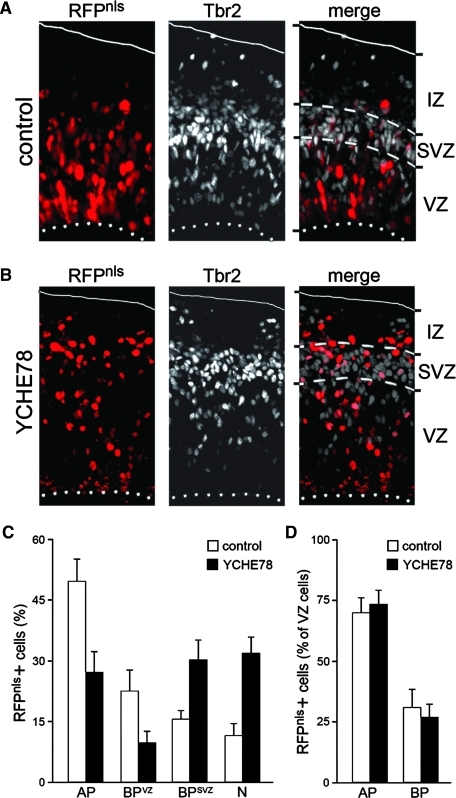
We transfected mice at embryonic day (E) 12.5 by in utero coelectroporation with either control or YCHE78 expression vectors together with a plasmid encoding nuclear-localized RFP (RFPnls) to label and reliably count targeted cells and their progeny. One day later, brains were collected and sectioned through the electroporated portion of the lateral cortex. Immunohistochemistry for the BP marker Tbr2 and/or the neuronal marker Tbr1 [36] was used to identify the VZ, SVZ, and IZ and to quantify the proportion of AP, BP, and neurons within the population of RFPnls+ cells. Specifically, within the VZ, AP were identified by the absence of Tbr2 immunoreactivity, which was due to difficulties in detecting Tbr2 and RFP in combination with the AP marker Pax6. This approach was legitimate despite the fact that neurons downregulate Tbr2 [36] because at this stage of development most neurons are generated by BP in the SVZ [37,38], thus representing a negligible proportion of all cells within the VZ (<3%) [39,40].
In controls we found that the majority of RFPnls+ cells were Tbr2− AP (50.0 ± 5.2) and Tbr2 + newborn BP still residing in the VZ (22.6% ± 5.3%), whereas a minority were Tbr2+ BP in the SVZ (15.6% ± 2.2%) and neurons in the IZ (11.7% ± 2.9%) (Fig. 1A, C). In contrast, expression of YCHE78 induced a 50% decrease in the proportion of AP (27.4% ± 5.1%) and newborn BP in the VZ (10.0% ± 2.8%) with a 2-fold increase in the proportion of Tbr2+ BP in the SVZ (30.5% ± 4.7%) and neurons in the IZ (32.2% ± 3.8%) (Fig. 1B, C). Importantly, upon expression of YCHE78 the vast majority of RFPnls+ cells in the IZ were Tbr1+ (data not shown), indicating an increase in neurogenesis rather than just delamination of neural progenitors. No significant difference was observed with regard to the efficiency of electroporation (∼15%–30% of RFPnls+ cells over DAPI) in the 2 conditions (data not shown).
FIG. 1.
YCHE78 induces differentiation of apical progenitor (AP). (A, B) Epifluorescence pictures of the mouse cortex 24 h after electroporation with control (A) or YCHE78 (B) plasmids at E12.5 and immunohistochemistry for RFP (left) and Tbr2 (middle). Apical and pial boundaries of cortex and subventricular zone (SVZ) were identified by DAPI counterstaining (not shown) and Tbr2 labeling and indicated by dotted, continuous, or dashed lines, respectively. Note the increase in the proportion of RFPnls+ cells in the intermediate zone (IZ) after YCHE78 expression (B), indicating an increase in neurogenesis. (C, D) Quantification of AP, basal progenitor (BP) in the VZ (BPVZ), BP in the SVZ (BPSVZ), and neurons (N) as a percentage of all RFPnls+ cells (C) or only those within the VZ (D). Bars = standard deviation; P < 0.01 for all cell populations in (C); differences in (D) are not significant.
Moreover, the proportion of RFPnls+ cells in the VZ was reduced upon expression of YCHE78, whereas the proportion of AP and newborn BP within the VZ was similar. In fact, when Tbr2+ BP in the VZ were expressed as a proportion of all RFPnls+ cells in the VZ, a similar value was found in mice electroporated with control (31.1% ± 7.1%) or YCHE78 (26.7% ± 5.7%) plasmids (Fig. 1D). Since BP in the VZ are a transient population of migrating cells, this suggests that the phenotype induced by YCHE78 is transitory, which may be explained by the reduction in plasmid levels upon cell division and the dose-dependent effect of YCHE78 on the transduction of signaling pathways, as already reported for Wnt [6].
Consistent with previous results during Xenopus development [6,20,21], YCHE78 expression did not seem to have any effect neither on cell survival nor on cell migration as judged by immunostaining for cleaved caspase-3, absence of pycnotic nuclei counterstained with DAPI, and distribution of Tbr2+ or Tbr1+ nuclei in the different cortical layers, respectively (data not shown).
In essence, these data are reminiscent of the effect induced by inhibition of Notch signaling leading to an increased differentiation of AP and generation of BP and neurons [30,41] and are consistent with a role of the v-ATPase in Notch signaling recently reported in flies [4,5].
YCHE78 expression inhibits endogenous Notch signaling
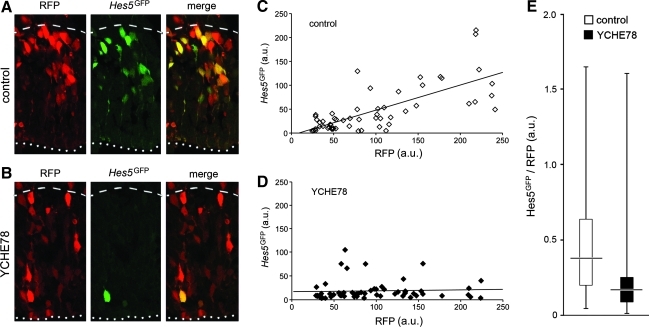
To directly investigate the effect of YCHE78 on endogenous Notch signaling, we used a Notch reporter plasmid in which GFP is under the transcriptional control of the Hes5 regulatory elements (Hes5GFP) [32]. Hes5GFP was coelectroporated at E12.5, with or without YCHE78, together with a cytoplasmic RFP, which allowed us to identify somata of transfected cells.
In control transfected embryos, ratio of GFP intensity relative to RFP fluorescence was variable (Fig. 2A, C) ranging from almost 0 (ie, no detectable GFP) to up to 1.6 (Fig. 2E; white bar), likely reflecting the different levels of Notch signaling in individual VZ cells as previously described [27,30,32]. Moreover, GFP and RFP fluorescence intensities showed a strong and significant positive correlation [Spearman's rank correlation coefficient (rS) = 0.70; P < 0.001] (Fig. 2C) with a median GFP/RFP ratio of 0.38 (Fig. 2E; white) and 33% of cells displaying values above 0.5. In contrast, when RFP and Hes5GFP were electroporated together with YCHE78 we found a substantial overall decrease in GFP fluorescence, loss of GFP/RFP positive correlation (rS = 0.24, not significantly different from rS = 0 but significantly different from rS = 0.70 of control, P < 0.01) (Fig. 2B, D) and a reduction by 50% in the median GFP/RFP ratio to 0.16 (Fig. 2E; black; P < 0.0001). Moreover, <10% of cells displayed values above 0.5.
FIG. 2.
YCHE78 autonomously inhibits Notch signaling. (A, B) Confocal fluorescence pictures of the mouse cortex 16 h after coelectroporation of Hes5GFP with control (A) or YCHE78 (B) plasmids at E12.5 showing endogenous RFP (left) or GFP (middle) fluorescence from a confocal 0.8-μm-thick optical section. Apical and basal boundaries of the VZ were identified by cell morphology and nuclear orientations after DAPI counterstaining (not shown) and indicated by dotted or dashed lines, respectively. Note the colocalization of RFP and GFP (merge; right) in control (A) but the almost complete absence of GFP in YCHE78 targeted cells (B), indicating a strong inhibition of endogenous Notch activity upon YCHE78 expression. (C, D) Plots showing the correlation between GFP and RFP fluorescence intensity (from 0 to a max of 250 arbitrary units [a.u]) after quantification of 60, randomly chosen, RFP+ somata. Lines indicate the best-fit of the recorded values outlining a strong positive correlation of controls (rS = 0.70) (C) but not YCHE78 (rS = 0.24) (D) electroporated brains. (E) Box plot showing the distribution of GFP/RFP fluorescence ratio of cells shown in (C) and (D) in 4 quartiles. Note the decreased median (horizontal lines) in YCHE78 (black) as compared with control (white) transfected cells; U-test P < 0.0001.
Thus, expression of YCHE78 autonomously inhibits endogenous Notch signaling, which may, at least in part, explain the depletion of AP and increased generation of BP and neurons previously described (Fig. 1).
YCHE78 expression rescues the phenotype induced by activated Notch but not of NICD
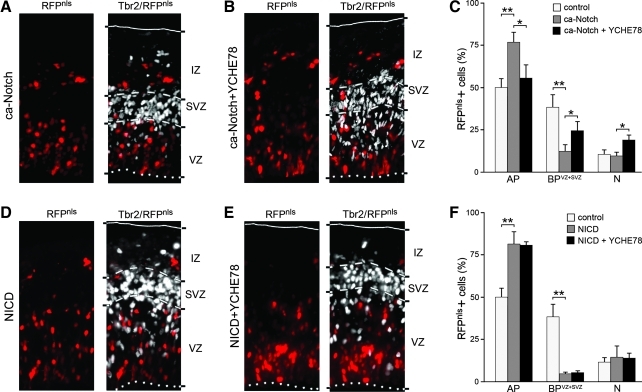
To further characterize the effect of YCHE78 on regulation of Notch activity and neural stem cell differentiation, we expressed, with or without YCHE78, 2 different activators of the Notch pathway, that is, the plasma membrane, γ-secretase-dependent, constitutively active Notch (ca-Notch), or its cytoplasmic, γ-secretase-independent, active domain NICD. We reasoned that if the v-ATPase should be important for processing of Notch to NICD, coexpression of YCHE78 together with ca-Notch might partly rescue the phenotype induced by expression of ca-Notch alone. Conversely, coexpression of YCHE78 together with NICD may have no effect on the phenotype induced by expression of NICD alone.
One day after electroporation at E12.5, both ca-Notch and NICD expression (Fig. 3A, D) resulted in a similar increase in the proportion of RFPnls+ AP in the VZ (76.6% ± 6.0% and 81.1 ± 7.5 for ca-Notch and NICD, respectively) as compared with control brains electroporated only with RFPnls plasmids (50.0% ± 5.2%), which was concomitant to a decrease in Tbr2+ BP in the VZ and SVZ (sum in the 2 areas = 12.0% ± 4.2% and 4.3% ± 0.8% for ca-Notch and NICD, respectively, as compared with 38.3% ± 7.4% for control) (Fig. 3C, F; gray bars). Therefore, in agreement with previous reports, Notch activation inhibits the generation of the main neurogenic progenitor type, which over the long term has been shown to cause a reduction in neuronal output [30,42].
FIG. 3.
YCHE78 rescues the constitutively active Notch-induced (ca-Notch), but not NICD-induced, effects. (A, B, D, E) Epifluorescence pictures of the mouse cortex 24 h after coelectroporation at E12.5 with ca-Notch (A, B) or NICD (D, E) with (A, D) or without (B, E) YCHE78 and immunohistochemistry for RFP (left) and Tbr2 (right; shown as merge). Apical, pial, and SVZ boundaries were identified by DAPI counterstaining (not shown) and Tbr2 immunoreactivity and identified by dotted, continuous, or dashed lines, respectively. (C, F) Quantification (%) of RFPnls+ AP, BP in the VZ and SVZ (BPVZ+SVZ), and neurons (N). Note the rescue of the ca-Notch (C), but not NICD (F), phenotype upon expression of YCHE78 (gray vs. black bars), suggesting that the vacuolar ATPase (v-ATPase) is essential for Notch signaling downstream of ca-Notch but upstream of NICD. Values for control (white bars) are taken from Fig. 1C and added for better comparison. Bars = standard deviation; *P < 0.05; **P < 0.005.
Compared with ca-Notch alone, coelectroporation of YCHE78 plus ca-Notch reduced the proportion of RFPnls+ AP (55.6% ± 7.7%) and increased that of BP in the VZ and SVZ (24.2% ± 5.7%) to levels that were comparable to control (Fig. 3B, C). In contrast, when YCHE78 was coelectroporated together with NICD the proportion of RFPnls+ AP (80.6% ± 2.2%) and BP in the VZ and SVZ (5.0% ± 1.2%) were undistinguishable from that observed after electroporation of NICD, or ca-Notch, alone (Fig. 3E, F).
Thus, the phenotype caused by activation of Notch signaling by ca-Notch was largely rescued by coexpression of YCHE78, whereas the NICD effect was not. This in turn suggests that the effect of the v-ATPase on neural stem cell differentiation is downstream of ligand activation of Notch but upstream of its γ-secretase-dependent cleavage and production of NICD.
Discussion
Here we found that expression of YCHE78 during mouse brain development (1) triggers the differentiation of neural precursors, (2) cell-autonomously inhibits endogenous Notch signaling, and (3) rescues the proliferative effect of ca-Notch, but not that of NICD, expression.
YCHE78 inhibited both endogenous and ectopic Notch signaling, and its effect on neurogenesis was dominant over ca-Notch suggesting that, at least in our conditions, v-ATPase-independent pathways of Notch processing play a minor role in physiological Notch signaling in the embryonic mammalian cortex. Moreover, the effect of YCHE78 on other signaling molecules still need to be investigated in mammalian neural stem cells, as recently done in Xenopus for Wnt [6].
Beyond endosome acidification and endocytosis the v-ATPase regulates H+ flows at the level of the plasma membrane, thereby contributing to the tissue bioelectrical gradients [21]. While the bioelectrical gradients per se can influence cell differentiation and regeneration [43,44], in our conditions only a minor fraction of all cells in the targeted area express YCHE78, making it unlikely that this would induce a major change in the electric potential throughout the whole neuroepithelium. Moreover, our data seem inconsistent with a γ-secretase cleavage at the inner surface of the plasma membrane because the v-ATPase pumps protons out of the cytoplasm, which would be incompatible with the optimum γ-secretase activity in an acidic milieu [12].
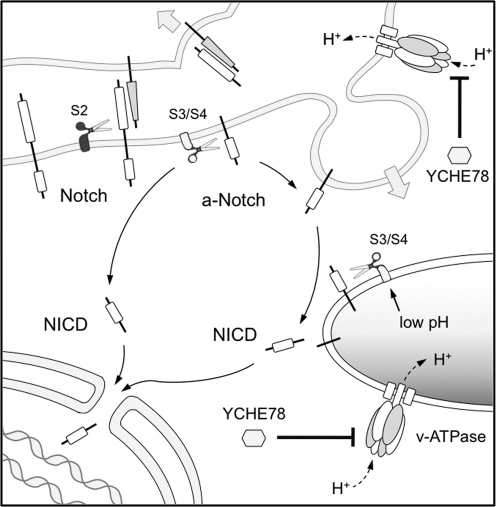
While other studies have concluded that endocytosis and endosomes are not involved in Notch signaling [16–18], our data showing a reduction in both the endogenous and ectopic Notch signaling upon YCHE78 expression seem more consistent with the opposite view, which would extend recent works in flies [4,5,13] to the developing mammalian brain (Fig. 4). While validation of this hypothesis would still require further investigation, we hope that our observation may contribute to a better understanding of this highly debated field. Moreover, our finding that inhibition of the v-ATPase can influence the proliferation of mammalian neural stem cells and Notch signaling opens up the possibility of using v-ATPase inhibitors [45] for therapeutic intervention in Notch-dependent brain tumors, which are thought to originate from cancer stem cells [46,47].
FIG. 4.
Notch pathway in a signal-receiving cell. Notch activation occurs at the plasma membrane upon ligand-mediated S2 cleavage (black scissor). Further processing of active Notch (a-Notch) occurs upon S3/S4 cleavage by γ-secretase (white scissors), resulting in the generation of NICD. While γ-secretase cleavage at the level of the plasma membrane (left route) has been originally proposed, recent studies [4,5] suggest that an alternative route through endosomes (right route) is also possible. Our data suggest that in the mouse cortex the latter is the prominent route. Note the inhibition of the v-ATPase by YCHE78 (hexagon, bottom, right) in endosomes (bottom; right) and at the plasma membrane (top; right), which is known to affect endosome acidification and endocytosis, respectively.
Finally, it is interesting to note that membrane potential and cellular pH can influence cell cycle progression and differentiation [22,43] and that inhibition of Notch has been shown to lengthen the cell cycle, in particular G1, of neural precursors [48], which is probably due to its effects on cyclin D1 expression [49]. The fact that lengthening of G1 may alone cause the differentiation of neural [50], and likely also other [51], stem cells reveals an elegant interplay between the cell biophysical state, signaling molecules, cell cycle progression, and differentiation during animal development, which should be better explored in future studies.
Acknowledgments
We are very grateful to Dr. Masato Nakafuku and Dr. Hideyuki Okano for providing essential material to perform this study and to Änne Glass for helping us with statistical analyses. Animal experiments were approved by local authorities (249168.11-9-2007-1/2). Financial support: This work was supported by the DFG-funded Center for Regenerative Therapies, the Medical Faculty of the Technical University Dresden, and the Collaborative Research Center SFB655 of the DFG (subproject A20). VT was supported by the DFG (TA-310-1 and -2) and ML by the NIH (R01 GM078484) and the Telemedicine and Advanced Technology Research Center, U.S. Army Medical Research and Materiel Command (W81XWH-10-2-0058).
Author Disclosure Statement
The authors declare no competing interest.
References
- 1.Harvey WR. Wieczorek H. Animal plasma membrane energization by chemiosmotic H+ V-ATPases. J Exp Biol. 1997;200:203–216. doi: 10.1242/jeb.200.2.203. [DOI] [PubMed] [Google Scholar]
- 2.Forgac M. Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 3.Marshansky V. Futai M. The V-type H+ -ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol. 2008;20:415–426. doi: 10.1016/j.ceb.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan Y. Denef N. Schupbach T. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. Dev Cell. 2009;17:387–402. doi: 10.1016/j.devcel.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaccari T. Duchi S. Cortese K. Tacchetti C. Bilder D. The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Development. 2010;137:1825–1832. doi: 10.1242/dev.045484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruciat CM. Ohkawara B. Acebron SP. Karaulanov E. Reinhard C. Ingelfinger D. Boutros M. Niehrs C. Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science. 2010;327:459–463. doi: 10.1126/science.1179802. [DOI] [PubMed] [Google Scholar]
- 7.Kopan R. Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fortini ME. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Tien AC. Rajan A. Bellen HJ. A Notch updated. J Cell Biol. 2009;184:621–629. doi: 10.1083/jcb.200811141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Amerongen R. Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 11.Niehrs C. Boutros M. Trafficking, acidification, and growth factor signaling. Sci Signal. 2010;3:pe26. doi: 10.1126/scisignal.3134pe26. [DOI] [PubMed] [Google Scholar]
- 12.Pasternak SH. Bagshaw RD. Guiral M. Zhang S. Ackerley CA. Pak BJ. Callahan JW. Mahuran DJ. Presenilin-1, nicastrin, amyloid precursor protein, and gamma-secretase activity are co-localized in the lysosomal membrane. J Biol Chem. 2003;278:26687–26694. doi: 10.1074/jbc.m304009200. [DOI] [PubMed] [Google Scholar]
- 13.Seugnet L. Simpson P. Haenlin M. Requirement for dynamin during Notch signaling in Drosophila neurogenesis. Dev Biol. 1997;192:585–598. doi: 10.1006/dbio.1997.8723. [DOI] [PubMed] [Google Scholar]
- 14.Gupta-Rossi N. Six E. LeBail O. Logeat F. Chastagner P. Olry A. Israel A. Brou C. Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor. J Cell Biol. 2004;166:73–83. doi: 10.1083/jcb.200310098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fortini ME. Bilder D. Endocytic regulation of Notch signaling. Curr Opin Genet Dev. 2009;19:323–328. doi: 10.1016/j.gde.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Struhl G. Adachi A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol Cell. 2000;6:625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- 17.Shaye DD. Greenwald I. Endocytosis-mediated downregulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature. 2002;420:686–690. doi: 10.1038/nature01234. [DOI] [PubMed] [Google Scholar]
- 18.Sorensen EB. Conner SD. Gamma-secretase-dependent cleavage initiates notch signaling from the plasma membrane. Traffic. 2010;11:1234–1245. doi: 10.1111/j.1600-0854.2010.01090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu M. Vergara S. Zhang L. Holliday LS. Aris J. Gluck SL. The amino-terminal domain of the E subunit of vacuolar H(+)-ATPase (V-ATPase) interacts with the H subunit and is required for V-ATPase function. J Biol Chem. 2002;277:38409–38415. doi: 10.1074/jbc.M203521200. [DOI] [PubMed] [Google Scholar]
- 20.Adams DS. Robinson KR. Fukumoto T. Yuan S. Albertson RC. Yelick P. Kuo L. McSweeney M. Levin M. Early, H+-V-ATPase-dependent proton flux is necessary for consistent left-right patterning of non-mammalian vertebrates. Development. 2006;133:1657–1671. doi: 10.1242/dev.02341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams DS. Masi A. Levin M. H+ pump-dependent changes in membrane voltage are an early mechanism necessary and sufficient to induce Xenopus tail regeneration. Development. 2007;134:1323–1335. doi: 10.1242/dev.02812. [DOI] [PubMed] [Google Scholar]
- 22.Blackiston DJ. McLaughlin KA. Levin M. Bioelectric controls of cell proliferation: ion channels, membrane voltage and the cell cycle. Cell Cycle. 2009;8:3519–3528. doi: 10.4161/cc.8.21.9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin M. Bioelectric mechanisms in regeneration: unique aspects and future perspectives. Semin Cell Dev Biol. 2009;20:543–556. doi: 10.1016/j.semcdb.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sundelacruz S. Levin M. Kaplan DL. Membrane potential controls adipogenic and osteogenic differentiation of mesenchymal stem cells. PLoS ONE. 2008;3:e3737. doi: 10.1371/journal.pone.0003737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LoTurco J. Manent JB. Sidiqi F. New and improved tools for in utero electroporation studies of developing cerebral cortex. Cereb Cortex. 2009;19(Suppl. 1):i120–i125. doi: 10.1093/cercor/bhp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoon K. Gaiano N. Notch signaling in the mammalian central nervous system: insights from mouse mutants. Nat Neurosci. 2005;8:709–715. doi: 10.1038/nn1475. [DOI] [PubMed] [Google Scholar]
- 27.Kageyama R. Ohtsuka T. Shimojo H. Imayoshi I. Dynamic regulation of Notch signaling in neural progenitor cells. Curr Opin Cell Biol. 2009;21:733–740. doi: 10.1016/j.ceb.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 28.Miyata T. Kawaguchi D. Kawaguchi A. Gotoh Y. Mechanisms that regulate the number of neurons during mouse neocortical development. Curr Opin Neurobiol. 2010;20:22–28. doi: 10.1016/j.conb.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Götz M. Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 30.Mizutani K. Yoon K. Dang L. Tokunaga A. Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- 31.Fish JL. Kosodo Y. Enard W. Paabo S. Huttner WB. Aspm specifically maintains symmetric proliferative divisions of neuroepithelial cells. Proc Natl Acad Sci U S A. 2006;103:10438–10443. doi: 10.1073/pnas.0604066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basak O. Taylor V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high Notch activity and Hes5 expression. Eur J Neurosci. 2007;25:1006–1022. doi: 10.1111/j.1460-9568.2007.05370.x. [DOI] [PubMed] [Google Scholar]
- 33.Tokunaga A. Kohyama J. Yoshida T. Nakao K. Sawamoto K. Okano H. Mapping spatio-temporal activation of Notch signaling during neurogenesis and gliogenesis in the developing mouse brain. J Neurochem. 2004;90:142–154. doi: 10.1111/j.1471-4159.2004.02470.x. [DOI] [PubMed] [Google Scholar]
- 34.Lange C. Huttner WB. Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5:320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 35.De Pietri Tonelli D. Calegari F. Fei JF. Nomura T. Osumi N. Heisenberg CP. Huttner WB. Single-cell detection of microRNAs in developing vertebrate embryos after acute administration of a dual-fluorescence reporter/sensor plasmid. Biotechniques. 2006;41:727–732. doi: 10.2144/000112296. [DOI] [PubMed] [Google Scholar]
- 36.Englund C. Fink A. Lau C. Pham D. Daza RA. Bulfone A. Kowalczyk T. Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haubensak W. Attardo A. Denk W. Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kowalczyk T. Pontious A. Englund C. Daza RA. Bedogni F. Hodge R. Attardo A. Bell C. Huttner WB. Hevner RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calegari F. Haubensak W. Haffner C. Huttner WB. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci. 2005;25:6533–6538. doi: 10.1523/JNEUROSCI.0778-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attardo A. Calegari F. Haubensak W. Wilsch-Brauninger M. Huttner WB. Live imaging at the onset of cortical neurogenesis reveals differential appearance of the neuronal phenotype in apical versus basal progenitor progeny. PLoS ONE. 2008;3:e2388. doi: 10.1371/journal.pone.0002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoon KJ. Koo BK. Im SK. Jeong HW. Ghim J. Kwon MC. Moon JS. Miyata T. Kong YY. Mind bomb 1-expressing intermediate progenitors generate notch signaling to maintain radial glial cells. Neuron. 2008;58:519–531. doi: 10.1016/j.neuron.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 42.Kato TM. Kawaguchi A. Kosodo Y. Niwa H. Matsuzaki F. Lunatic fringe potentiates Notch signaling in the developing brain. Mol Cell Neurosci. 2010;45:12–25. doi: 10.1016/j.mcn.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 43.Levin M. Large-scale biophysics: ion flows and regeneration. Trends Cell Biol. 2007;17:261–270. doi: 10.1016/j.tcb.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 44.Funk RH. Monsees TK. Effects of electromagnetic fields on cells: physiological and therapeutical approaches and molecular mechanisms of interaction. A review. Cells Tissues Organs. 2006;182:59–78. doi: 10.1159/000093061. [DOI] [PubMed] [Google Scholar]
- 45.Perez-Sayans M. Somoza-Martin JM. Barros-Angueira F. Rey JM. Garcia-Garcia A. V-ATPase inhibitors and implication in cancer treatment. Cancer Treat Rev. 2009;35:707–713. doi: 10.1016/j.ctrv.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 46.Yang ZJ. Wechsler-Reya RJ. Hit 'em where they live: targeting the cancer stem cell niche. Cancer Cell. 2007;11:3–5. doi: 10.1016/j.ccr.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 47.Ischenko I. Seeliger H. Schaffer M. Jauch KW. Bruns CJ. Cancer stem cells: how can we target them? Curr Med Chem. 2008;15:3171–3184. doi: 10.2174/092986708786848541. [DOI] [PubMed] [Google Scholar]
- 48.Borghese L. Dolezalova D. Opitz T. Haupt S. Leinhaas A. Steinfarz B. Koch P. Edenhofer F. Hampl A. Brustle O. Inhibition of notch signaling in human embryonic stem cell-derived neural stem cells delays G1/S phase transition and accelerates neuronal differentiation in vitro and in vivo. Stem Cells. 2010;28:955–964. doi: 10.1002/stem.408. [DOI] [PubMed] [Google Scholar]
- 49.Das D. Lanner F. Main H. Andersson ER. Bergmann O. Sahlgren C. Heldring N. Hermanson O. Hansson EM. Lendahl U. Notch induces cyclin-D1-dependent proliferation during a specific temporal window of neural differentiation in ES cells. Dev Biol. 2010;348:153–166. doi: 10.1016/j.ydbio.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 50.Salomoni P. Calegari F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 2010;20:233–243. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Lange C. Calegari F. Cdks and cyclins link G(1) length and differentiation of embryonic, neural and hematopoietic stem cells. Cell Cycle. 2010;9:1893–1900. doi: 10.4161/cc.9.10.11598. [DOI] [PubMed] [Google Scholar]