Abstract
The nuclear factor κB (NF-κB) transcription factor regulates the expression of genes involved in cell survival and immune responses. We have identified a novel interferon (IFN)-activated signaling pathway that leads to NF-κB activation and demonstrate that a subset of IFN-stimulated genes and microRNAs that play key roles in cellular response to IFN is regulated by NF-κB. This review focuses on the IFN-induced NF-κB activation pathway and the role of NF-κB in the expression of IFN-induced coding and noncoding genes, antiviral activity and apoptosis, and the therapeutic application of IFN in cancer and infectious disease.
Introduction
Type I interferons (IFNs) regulate diverse cellular functions by modulating the expression of IFN-stimulated genes (ISGs) through the activation of the well-established signal transduction pathway of the Janus tyrosine kinases and signal transducers and activators of transcription (STAT) proteins. Recent studies have shown that IFNs also regulate the expression of small noncoding RNAs, microRNAs (miRNAs), which mediate the cellular response to IFNs. Although the JAK/STAT signal transduction pathway is critical in mediating IFN's antiviral and antiproliferative activities, other signaling pathways are activated by IFNs and regulate cellular response to IFN. This review will summarize the recent finding that the nuclear factor κB (NF-κB) transcription factor also plays an important role in regulating the cellular response to IFNs. NF-κB clearly plays a critical role in the signal transduction pathway that senses viral nucleic acids during pathogenic infection. These pathways lead to the induction of the IFNα/β genes, which have been the subject of many reviews. However, less is known about the role of this transcription factor family in the induction of IFN-induced coding genes, ISGs, and noncoding genes, miRNAs.
Nuclear Factor κB
The NF-κB transcription factor was first discovered as a factor in the nucleus of B cells that binds to the kappa light chain of immunoglobulin (Ig) gene enhancer (Baeurele and Baltimore 1988). The family of NF-κB transcription factors bind to the cis-acting κB consensus sequence [GGRNNN(N)YCC] and regulate the expression of genes that play important roles in immunity, inflammation, cell growth, and cell survival, which are also IFN-regulated processes. The most interesting connection between NF-κB and IFN is that NF-κB is a master regulator of cell survival, and we, and most investigators in the field, had found that IFN is a rather poor apoptosis inducer, despite its induction of a number of apoptotic genes (Chawla-Sarkar and others 2003). In mammals the NF-κB family of related proteins includes NFKB1 (p105 processed to p50), NFKB2 (p100 processed to p52), RelA (p65), RelB, and cRel. p50 and p52 lack a transcription activation domain and function as repressors. In contrast, p65, cRel, and RelB have a transcription activation domain, and, thus, when complexed with p50 or p52, are capable of activating transcription. The dimers formed by members of this family differ with regard to their DNA-binding specificity and transactivation potentials. The p50:p65 and p52:RelB heterodimers are the most commonly observed NF-κB complexes in cells. The proteolytic processing of NFKB1 (p105) into p50 is constitutive and largely cotranslational. In contrast, the processing of NFKB2 (p100) into p52 is tightly controlled through phosphorylation induced by various agents and subsequent ubiquitination. Under most circumstances, NF-κB homodimers or heterodimers are bound to IκB inhibitory proteins in the cytoplasm of unstimulated cells. Many cytokines promote the classical NF-κB pathway, in which the serine phosphorylation and degradation of IκB proteins leads to the dissociation of the cytosolic inactive NF-κB/IκB complexes, and subsequent NF-κB translocation to the nucleus for DNA binding. The degradation of IκB proteins requires the activation of IκB kinase (IKK), a multiprotein complex consisting of IKKα and IKKβ catalytic subunits and the IKKγ/NEMO regulatory subunit (Baeuerle 1998). Targeted gene disruption of individual IKK proteins has determined that IKKβ and IKKγ (but not IKKα) are the major mediators of the classical NF-κB signal transduction (Li and others 2005). In addition, a growing number of NF-κB inducers activate an alternative NF-κB pathway that does not involve IκB degradation, but rather involves the linkage of TNF receptor-associated factors (TRAFs) to the activation of the MAP3K-related kinase, the NF-κB-inducing kinase (NIK). The alternative pathway results in the ubiquitination and proteolytic processing of p100/NFKB2 protein and nuclear translocation of p52:RelB dimers to regulate specific NF-κB target genes (Pomerantz and Baltimore 2002; Bonizzi and others 2004). The alternative pathway for NF-κB activation is strictly dependent on IKKα (Senftleben and others 2001), but independent of IKKβ and IKKγ (Dejardin and others 2002).
The activation of NF-κB is a double-edged sword: while normal functions of NF-κB are needed for proper innate and adaptive immune responses, dysregulation of NF-κB can lead to inflammatory diseases and tumorigenesis. For example, constitutive NF-κB activity results in overexpression of proinflammatory genes, which is associated with acute and chronic inflammatory diseases, including ulcerative colitis, rheumatoid arthritis, and Crohn's disease. Moreover, high constitutive NF-κB activity has been found in many tumor cell lines and in certain types of cancers, including acute myelogenous leukemia, acute lymphocytic leukemia, chronic myelogenous leukemia, multiple myeloma, prostate cancer, ovarian cancer, and breast cancer. Inhibitors targeting components of the NF-κB signaling pathway effectively suppress NF-κB activity, protect and relieve inflammatory symptoms, and induce the apoptosis of tumor cells. Thus, NF-κB represents an attractive drug target for therapy of inflammatory and autoimmune diseases, as well as for cancer (Karin 2006; Du and others 2007).
MicroRNAs
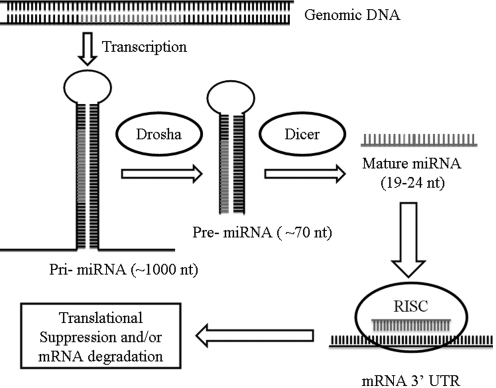
Mature miRNAs are small single-stranded noncoding RNAs that are highly conserved between different eukaryotic species (Bartel 2004). Following the synthesis of the hundred or thousand nucleotide (nt) long primary miRNA by RNA polymerase II or III, nuclear processing by the enzyme Drosha produces a pre-miRNA transcript of ∼70 nucleotides forming a hairpin structure that is shuttled into the cytoplasm (Fig. 1). Final production of the mature miRNA species in the cytoplasm requires processing by an RNase called Dicer, producing a 19- to 24-nucleotide product, capable of being incorporated into the RNA-induced silencing complex. The RNA-induced silencing complex, in turn, is able to use the ∼7-nucleotide “seed sequence” of the miRNA to recognize complementary mRNA transcripts in its 3′ untranslated region for translational suppression and/or degradation of the mRNA target. Over 800 human miRNA genes have been identified in the human genome that may regulate an estimated 30% of all human genes (Lim and others 2005). Each miRNA appears to regulate the expression of tens to hundreds of genes, thereby functioning as “master-switches” that fine-tune gene expression post-transcriptionally and coordinate multiple cellular pathways in important processes such as embryonic development, immune response, inflammation, and oncogenesis, as well as cellular growth and proliferation.
FIG. 1.
Schematic of microRNA (miRNA) biogenesis.
The IFN system is critical in the innate immune response to pathogens, and thus it is not surprising that IFN-regulated miRNAs are involved in this defense pathway. For example, the liver-specific miRNA, miR-122, targets the 5′ noncoding region of the hepatitis C virus genome, acting to enhance virus RNA replication as opposed to enhancing viral mRNA translation or stability (Jopling and others 2005). IFNα/β treatment of the Huh7 human hepatocyte line reduces miR-122 expression by 20%–40%, which may play a role in inhibitory action of IFN on hepatitis C virus (HCV) replication (Pedersen and others 2007; Sarasin-Filipowicz and others 2009). Moreover, IFNβ induces a number of miRNAs, which display seed sequences present in the HCV genome and specific mimics of these IFN-induced miRNAs were found to inhibit HCV replication in vitro (Pedersen and others 2007). The expression of several of these IFN-induced miRNAs (miR-1, miR-30, miR-128, miR-196, and miR-296) are basally present as well as induced by IFN to varying degrees in peripheral blood mononuclear cells from healthy individuals and from chronic HCV-infected patients (Scagnolari and others 2010). These results clearly demonstrate that IFN-induced miRNAs are critical to the host defense in certain viral infections. However, future studies will elucidate whether individual IFN-induced miRNAs are specific for infection by certain pathogens.
The Activation of NF-κB Signaling Pathway by IFNα/β, and the Roles of STAT3, PI3K, and Akt
By analyzing the promoter sequences within a number of ISGs, we identified potential NF-κB binding sites. This led us to assay nuclear extracts from IFN-stimulated cells for NF-κB activity by DNA-binding assays (electrophonetic mobility shift assay [EMSA]) with a consensus oligonucleotide probe based on the κB binding site in the Ig light chain enhancer (Yang and others 2000). Within 30 min of IFN stimulation, NF-κB DNA-binding activity was induced in diverse cell types (lymphoblastoid, fibrosarcoma, renal carcinoma, and normal fibroblasts). Moreover, the kinetics and dose dependence of IFN-promoted NF-κB activation was similar to that of the IFN-induced JAK-STAT signaling pathway. The induction of NF-κB DNA binding activity paralleled the IFN-induced increase in transcriptional activity dependent on NF-κB as determined with a κB-driven luciferase construct.
In the classical signaling pathway, NF-κB activity is tightly controlled by inhibitory IκB proteins, which bind to NF-κB complexes and sequester them in the cytoplasm. To determine whether IFN induces classical NF-κB activation, IκBα levels were determined by immunoblotting with anti-IκBα at various times after IFN addition to the highly IFN-sensitive Daudi lymphoblastoid cell line. IFN induced a progressive decrease in cellular IκBα levels (Yang and others 2000). The kinetics of induction of NF-κB activation in Daudi cells paralleled that of IκBα degradation, indicating that IFN promotes the dissociation of IκBα/NF-κB complexes through induction of IκBα degradation. Since serine phosphorylation of IκBα leads to its degradation, IκBα proteins with mutations (IκBαM) or deletions of serine phosphorylation sites (IκBαΔN) function as super-repressors of NF-κB activation as determined by EMSA or by κB-dependent reporter assays. We found that expression of super-repressor IκBα (SR-IκBα) mutants significantly blocked IFN-induced DNA binding and transcriptional activity dependent on NF-κB.
Previous studies demonstrated that blockage of the classical NF-κB pathway by expression of suppressor constructs sensitized cells to TNF-induced cell death (Beg and Baltimore 1996; Van Antwerp and others 1996; Wang and others 1996), suggesting that NF-κB plays a key role in protecting cells against apoptosis. We examined whether IFN could protect cells against apoptosis through an NF-κB dependent pathway, and found that IFN prevented apoptosis induced by virus infection with vescular stomatitis virus (VSV) or by crosslinking of cell surface receptors with anti-Ig (Yang and others 2000). Previous studies showed that IFNα/β promotes the survival of activated T cells in vitro (Marrack and others 1999). Expression of SR-IκBα constructs markedly sensitized Daudi cells to IFN-induced death as determined by TUNEL assays or by highly sensitive DNA fragmentation assays (Yang and others 2000). Sensitization to IFN-induced apoptosis by expression of SR-IκBα was also observed in other tumor lines, including renal cancer and squamous carcinoma cells. These results suggested that the IFN-induced NF-κB pathway protects cells against apoptosis that is induced by multiple apoptotic agents, including IFN itself.
Moreover, we found that there was crosstalk between the IFN-activated STAT3 pathway and the NF-κB pathway. While IFN induced prominent NF-κB activation in Daudi cells, IFN did not induce NF-κB activation in a subclone of Daudi cells with a STAT3 defect (Yang and others 1998). Expression of STAT3 in this subclone restored STAT3 activation and IFN-induced NF-κB activation. In addition, we previously established that STAT3 acts as an adapter to couple PI3K to the IFNAR1 subunit of the IFN receptor (Pfeffer and others 1997). A phosphopeptide corresponding to the STAT3-docking site of IFNAR1 was found to block IFN-induced NF-κB activation, indicating that NF-κB activation is directed through the tyrosine phosphorylation-dependent formation of the STAT3/PI3K signaling complex with the IFNAR1 chain (Yang and others 2001). While expression of dominant-negative (DN) PI3K blocked the IFN-induced IκB degradation and NF-κB activation, constitutively active (CA) PI3K promoted NF-κB activity, demonstrating that PI3K is involved in NF-κB activation by IFN. In addition, DN-Akt blocked IFN-promoted NF-κB activation and CA-Akt promoted NF-κB activity in Daudi cells, demonstrating that Akt is also involved in NF-κB activation by IFN. Further, while IFN stimulated κB-dependent transcription in empty vector-transfected cells, expression of DN-PI3K suppressed IFN-activated κB-dependent transcription but not ISRE-dependent transcription. These results indicate that NF-κB activation by IFN via the PI3K pathway is distinct from the ISRE-driven mechanism in regulating gene expression. Activation of PI3K/AKT by IFN has also been described through the insulin receptor substrate 1 (Uddin and others 1997) and through the direct interaction of PI3K with IFNAR1, which also leads to induction of NF-κB activity (Rani and others 2002). IFN-induced NF-κB activation was found to require TYK2 but not JAK1 (Yang and others 2005b). IFNs induce NF-κB activation in JAK1-deficient cells, an event blocked by the expression of a kinase dead TYK2 construct. Protein kinase R, an ISG, appears to directly activate the IKK complexes leading to the phosphorylation and degradation of IκB and resulting in NF-κB activation (Deb and others 2001). Taken together these results suggest that, since the NF-κB pathway is essential to IFN-induced cell survival, there are multiple IFN-stimulated pathways leading to NF-κB activation.
IFN Activates an Alternative NF-κB Pathway That Involves NIK and TRAF Proteins
The alternative NF-κB pathway is dependent on NIK and TRAF proteins. To define the role of NIK in IFN-promoted NF-κB activation, a DN kinase-inactive NIK mutant (DN-NIK) was expressed in Daudi cells and was found to block IFN-promoted NF-κB DNA binding activity and κB-dependent transcription (Yang and others 2005a). However, while expression of DN-PI3K blocked IκBα degradation (Yang and others 2001), expression of DN-NIK had no effect on IFN-promoted IκBα degradation (Yang and others 2005a), demonstrating that NIK and PI3K may play distinct roles in IFN-induced NF-κB activation. TRAF2 functions as an adaptor molecule for various members of the TNF superfamily to activate NIK and subsequently mediate NF-κB activation (Au and Yeh 2007). To assess the role of TRAF2 in IFN-induced NF-κB signal transduction pathway, cells were transfected with an expression plasmid for DN-TRAF2, which lacks the NH2-terminal 86 amino acids of the RING finger domain of TRAF2 and functions as a DN for TRAF2-dependent signaling events (Rothe and others 1995). Expression of DN-TRAF2 in Daudi cells blocked IFN-promoted NF-κB DNA binding activity, as well as the stimulation by IFN of κB-dependent transcription. These results indicate that both TRAF2 and its downstream effector NIK are involved in the IFN-induced alternative NF-κB signal transduction pathway. The alternative NF-κB pathway mediates p100 processing into p52 and the formation of p52 heterodimers, which translocate into the nucleus to regulate gene transcription (Pomerantz and Baltimore 2002). IFN induced the appearance of p52 in both nuclear and cytoplasmic extracts prepared from Daudi cells, as determined by pull-down assays with glutathione S-transferase fused p65 (RelA), although untreated Daudi cells contained no detectable p52 (Yang and others 2005a). We also found that both DN-NIK and TRAF2 constructs blocked IFN-induced p52 appearance in nuclear extracts, demonstrating the roles of NIK and TRAF2 in IFN-induced p100 processing. In contrast, DN-PI3K expression or treatment with the pharmacological PI3K inhibitor LY294002 had no effect on basal or IFN-promoted appearance of p52, although they inhibited IFN-induced NF-κB activation and NF-κB dependent gene transcription. Thus, these results demonstrate that IFN-induced p100 processing to p52 is dependent on NIK/TRAF signaling but is independent of PI3K/Akt signaling.
To assess the role of NIK and TRAF2 in IFN-mediated cell survival, Daudi cells were transfected with DN expression plasmids, and assessed for apoptosis. Expression of DN-NIK or TRAF2, as well as PI3K, sensitized Daudi cells to IFN-induced cell death as determined by TUNEL or DNA fragmentation assays, while expression of the empty vector alone had no effect. These results indicate that the NIK/TRAF-dependent pathway leading to NF-κB activation protects cells against IFN's proapoptotic action. Thus, IFN generates cell survival signals through both the alternative NF-κB pathway dependent on NIK/TRAF, and the classical NF-κB pathway dependent on Akt/PI3K (Yang and others 2001).
These results indicate that there is a dynamic equilibrium between the ability of IFN to promote apoptosis and the ability of IFN to generate cell survival signals through an NF-κB pathway. Inhibiting the NF-κB pathway drives IFN-promoted apoptosis. The understanding of the pathways that limit IFN-induced apoptosis may provide new avenues to develop therapeutics to enhance the apoptotic action of IFN. For example, the effectiveness of IFN in cancer therapy is often limited by its inability to induce significant cell death. Better characterization of such pathways could lead to new strategies that improve IFN's efficacy as an antitumor agent.
The Role of NF-κB in Antiviral Activity and ISG Expression
Since the hallmark of IFNs is their antiviral activity, we examined the role of NF-κB in the induction of IFN's antiviral action by comparing mouse embryo fibroblasts (MEFs) that had normal NF-κB function (wild type [WT]-MEFs) with MEFs that had a germline disruption of both p50 and p65 NF-κB proteins (p50/p65-DKO MEFs). While IFN induced NF-κB activation in WT-MEFs as determined by EMSA with the consensus NF-κB oligonucleotide probe, IFN treatment did not induce NF-κB activation in p50/p65-DKO MEFs. However, p50/p65-DKO MEFs were more sensitive than WT-MEFs to the antiviral action of IFN against influenza and VSV virus (Pfeffer and others 2004; Wei and others 2006). Thus, the NF-κB pathway suppressed IFN-induced antiviral activity. The IκB kinase family member, IKKɛ, also appears to play an important role in IFN-mediated antiviral activity by directly regulating STAT1 phosphorylation (Tenoever and others 2007).
IFNs produce many of their biological effects through the induction of the ISG family of early response genes. To identify NF-κB-regulated ISGs, gene expression profiling was performed on RNA from IFN-treated WT-MEFs and p50/p65-DKO MEFs (Pfeffer and others 2004). The subset of NF-κB-regulated ISGs included genes encoding GTP-binding and antigen presentation proteins (Wei and others 2006). The NF-κB-regulated GTP-binding ISGs have antiviral, antimicrobial, and antiproliferative properties, and include the 65–67 kDa guanylate-binding proteins (Gbp 1 and Gbp2), the Mx proteins (Mx1 and Mx2), and the 47-kDa GTPase Ifi47/Irg47 (Vestal and Jeyaratnam 2011). The NF-κB-regulated antigen presentation ISGs include Tap1, Tap2, Psmb9/Lmp2, and Psmb8/Lmp7, and are involved in degrading intracellular proteins into antigenic peptides, and contribute to the transport of these peptides to endoplasmic reticulum where they bind to the assembled major histocompatability complex (MHC) class I molecules (Ploegh 2000). While NF-κB suppresses the IFN-induced expression of Gbp1, Ifi47, Mx1, Mx2, Tap1, Psmb9/Lmp2, and Psmb8/Lmp7, NF-κB enhances the IFN-induced expression of Gbp2 and Tap2 (Pfeffer and others 2004). Thus, the IFN-activated NF-κB pathway not only counterbalances the ability of IFN to induce apoptosis and antiviral activity, but also differentially regulates the expression of specific ISGs.
The Binding of NF-κB Proteins to the Promoters of a Subset of ISGs
To define specific transcription factors that may directly regulate the expression of Ifi47, Tap1, and Mx1, we performed chromatin immunoprecipitation (ChIP) assays on the promoters of NF-κB-regulated ISGs (Wei and others 2006). IFN treatment rapidly (between 15 and 60 min) induced STAT1 and STAT2 binding to Ifi47, Tap1, and Mx1 promoters in both WT and p50/p65-DKO MEFs. Importantly, p50 was basally bound to Mx1, Ifi47, and Tap1 promoters in WT cells and remained bound for up to 2 h after IFN addition. Remarkably, the time course of expression of these ISGs correlated precisely to when p50 detached from these promoters. Interestingly, in p50/p65-DKO MEFs IFN induced the recruitment of IRF1 to the promoters of all 3 ISGs, and IRF1 recruitment closely correlated with the more rapid and enhanced gene induction upon IFN treatment. These results taken together demonstrate that p50 homodimers are basally bound to the promoters of these ISGs and may suppress their induction by IFN, perhaps by impairing the recruitment of IRF1 to the promoter. Most interesting, the finding that p50 homodimers inhibit the expression of a subset ISGs has recently been confirmed (Cheng and others 2011).
To further characterize the role that specific NF-κB proteins play in IFN-induced gene expression, we examined the binding to the promoter of IFN-induced chemokine, CXCL11. IFNα/β induces CXCL11 expression in IFN-sensitive Daudi cells but not in an IFN-resistant Daudi subclone (DRST3) that has a defective STAT3-dependent signaling pathway (Yang and others 2007). The p50 NF-κB subunit was found basally bound to the CXCL11 promoter in IFN-sensitive Daudi cells but was displaced from the promoter after IFN treatment (1 h). In contrast, while p65 was not basally promoter-bound, IFN induced its binding. In DRST3 cells, while p50 was not basally bound to the CXCL11 promoter, IFN induced p50 binding within 1 h after addition, and p65 was neither basally promoter-bound nor was its binding IFN-induced. Moreover, IFN also induced the binding of the transcriptional activator IRF1 to the CXCL11 promoter in Daudi cells. WT-STAT3 expression in DRST3 cells rescued the recruitment of the p65 NF-κB subunit and IRF1 to the CXCL11 promoter by IFN. Similar to the findings in MEFs, p50 dimers are bound to ISG promoter to suppress their expression, and are displaced upon IFN-induced ISG expression. The pattern of transcription factor binding in WT-STAT3-expressing DRST3 cells was indistinguishable from that in IFN-sensitive Daudi cells. These results indicate that the defective STAT3 signaling pathway in DRST3 cells affects the binding of NF-κB and IRF proteins, which cooperate in the transcription regulation of CXCL11.
The Role of NF-κB in the Induction miR-21 by IFN
We examined the miRNA database to identify potential IFN-induced miRNAs by focusing on miRNAs with putative STAT3 and NF-κB sites within their promoters. One such potential IFN-induced miRNA was miR-21, which is found overexpressed in many human cancers (glioblastoma, breast cancer, and prostate cancer) and accumulating evidence indicates it functions as an oncogene (Loffler and others 2007; Folini and others 2010). IFN induced miR-21 expression in a variety of human and mouse cells with a time course and dose dependence similar to the IFN induction of an ISG, ISG15 (Yang and others 2010). A role for STAT3 was implicated in the induction by IFN of miR-21 based on the lack of an IFN effect on miR-21 expression in the PC-3 prostate cancer cell line, which has a genomic deletion of the STAT3 gene (Clark and others 2003). As further evidence of the role of STAT3 in this pathway, STAT3 knockdown or expression of a DN-STAT3 mutant (the canonical Y705 phosphorylation site mutated to phenylalanine) was found to block IFN-induced miR-21 expression (Yang and others 2010). ChIP analysis showed that IFN induced the binding of STAT3 to the most distal putative STAT3 binding site in the miR-21 promoter. Because of our interest in the crosstalk between STAT3 and the NF-κB pathway in IFN action, experiments were performed in MEFs with a genetic deletion of the p65 NF-κB subunit, which showed that IFN-induced miR-21 expression was also dependent on NF-κB. STAT3 knockdown blocked both IFN-induced p65 binding to the miR-21 promoter and p65 nuclear translocation, suggesting that STAT3 and p65 are linked together in this IFN signaling pathway. Thus, IFN-induced miR-21 expression is regulated by both STAT3 and NF-κB at the level of the miR-21 promoter. Because miR-21 may promote cell survival (Chan and others 2005; Si and others 2007), we examined whether miR-21 plays a critical role in suppressing IFN-induced apoptosis (Yang and others 2010). We found that miR-21 was inversely correlated with the ability of IFN to promote tumor cell apoptosis in prostate cancer cell lines. Moreover, miR-21 knockdown also sensitized cells to other inducers of apoptosis (camptothecin and staurosporine).
Concluding Remarks
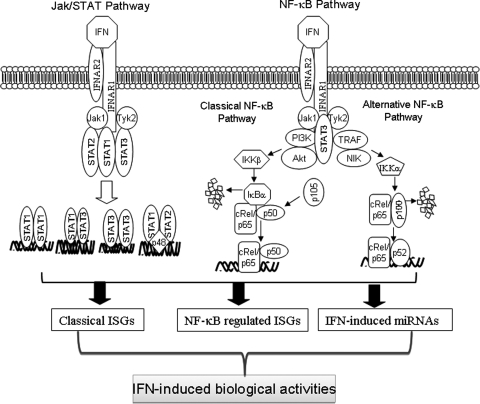
As illustrated in Fig. 2, IFNs activate not only the JAK/STAT signaling pathway but also the NF-κB signaling pathway. NF-κB regulates the expression of coding (mRNA) and noncoding (miRNA) genes as well as the cellular response to IFNs. Although STAT3, TYK2, PI3K, AKT, IKK, TRAF, and NIK have been identified in the IFN-activated NF-κB signaling pathway, the components of this signaling pathway have not been fully elucidated and could be cell context dependent. ChIP assays on the promoters regulated by NF-κB indicate that the outcome is highly dependent on the interplay between different STAT, NF-κB, and IRF proteins. The therapeutic effectiveness of IFN in cancer is often limited by IFN's inability to induce significant cell death because of the high constitutive NF-κB activity in tumor cells. Since there is a close relationship of NF-κB between inflammation, tumorigenesis, and the cellular response to IFN, we hypothesize that selectively targeting the NF-κB pathways may represent a novel strategy for enhancing IFN's therapeutic effectiveness and/or diminishing IFN's undesirable side effects. Inhibiting NF-κB activity in vitro sensitizes cells to IFN's apoptotic and antiviral activities. However, it will be important to determine whether inhibiting NF-κB activity in vivo will enhance IFN's antiviral and antitumor clinical activity.
FIG. 2.
Schematic of interferon (IFN)-activated signaling pathways.
Acknowledgments
Research in the author's laboratory was supported by National Institutes of Health Grants CA133322, CA73753, and AI66316, and by funds from the Muirhead Chair Endowment at the University of Tennessee Health Science Center. The author thanks the present and former members of the Pfeffer laboratory for their contributions to this research.
Author Disclosure Statement
No competing financial interests exist.
References
- Au PY. Yeh WC. Physiological roles and mechanisms of signaling by TRAF2 and TRAF5. Adv Exp Med Biol. 2007;597:32–47. doi: 10.1007/978-0-387-70630-6_3. [DOI] [PubMed] [Google Scholar]
- Baeuerle PA. IkB-NFkB structures: at the interface of inflammation control. Cell. 1998;95:729–731. doi: 10.1016/s0092-8674(00)81694-3. [DOI] [PubMed] [Google Scholar]
- Baeurele PA. Baltimore D. IkB: a specific inhibitor of NFkB transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Beg AA. Baltimore D. An essential role for NF-κB in preventing TNF-α-induced cell death. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- Bonizzi G. Bebien M. Otero DC. Johnson-Vroom KE. Cao Y. Vu D. Jegga AG. Aronow BJ. Ghosh G. Rickert RC. Karin M. Activation of IKKalpha target genes depends on recognition of specific kappaB binding sites by RelB:p52 dimers. EMBO J. 2004;23(21):4202–4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA. Krichevsky AM. Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chawla-Sarkar M. Lindner DJ. Liu YF. Williams BR. Sen GC. Silverman RH. Borden EC. Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis. 2003;8(3):237–249. doi: 10.1023/a:1023668705040. [DOI] [PubMed] [Google Scholar]
- Cheng CS. Feldman KE. Lee J. Verma S. Huang DB. Huynh K. Chang M. Ponomarenko JV. Sun SC. Benedict CA. Ghosh G. Hoffmann A. The specificity of innate immune responses is enforced by repression of interferon response elements by NF-{kappa}B p50. Sci Signal. 2011;4(161):ra11. doi: 10.1126/scisignal.2001501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. Edwards S. Feber A. Flohr P. John M. Giddings I. Crossland S. Stratton MR. Wooster R. Campbell C. Cooper CS. Genome-wide screening for complete genetic loss in prostate cancer by comparative hybridization onto cDNA microarrays. Oncogene. 2003;22(8):1247–1252. doi: 10.1038/sj.onc.1206247. [DOI] [PubMed] [Google Scholar]
- Deb A. Haque SJ. Mogensen T. Silverman RH. Williams BR. RNA-dependent protein kinase PKR is required for activation of NF-kappa B by IFN-gamma in a STAT1-independent pathway. J Immunol. 2001;166(10):6170–6180. doi: 10.4049/jimmunol.166.10.6170. [DOI] [PubMed] [Google Scholar]
- Dejardin E. Droin NM. Delhase M. Haas E. Cao Y. Makris C. Li ZW. Karin M. Ware CF. Green DR. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity. 2002;17(4):525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- Du Z. Wei L. Murti A. Pfeffer SR. Fan M. Yang CH. Pfeffer LM. Non-conventional signal transduction by type 1 interferons: the NF-kappaB pathway. J Cell Biochem. 2007;102(5):1087–1094. doi: 10.1002/jcb.21535. [DOI] [PubMed] [Google Scholar]
- Folini M. Gandellini P. Longoni N. Profumo V. Callari M. Pennati M. Colecchia M. Supino R. Veneroni S. Salvioni R. Valdagni R. Daidone MG. Zaffaroni N. miR-21: an oncomir on strike in prostate cancer. Mol Cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling CL. Yi M. Lancaster AM. Lemon SM. Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309(5740):1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441(7092):431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Li Q. Withoff S. Verma IM. Inflammation-associated cancer: NF-kappaB is the lynchpin. Trends Immunol. 2005;26(6):318–325. doi: 10.1016/j.it.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Lim LP. Lau NC. Garrett-Engele P. Grimson A. Schelter JM. Castle J. Bartel DP. Linsley PS. Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Loffler D. Brocke-Heidrich K. Pfeifer G. Stocsits C. Hackermuller J. Kretzschmar AK. Burger R. Gramatzki M. Blumert C. Bauer K. Cvijic H. Ullmann AK. Stadler PF. Horn F. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110(4):1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- Marrack P. Kappler J. Mitchell T. Type I interferons keep activated T cells alive. J Exp Med. 1999;189:521–530. doi: 10.1084/jem.189.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen IM. Cheng G. Wieland S. Volinia S. Croce CM. Chisari FV. David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449(7164):919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer LM. Kim JG. Pfeffer SR. Carrigan DJ. Baker DP. Wei L. Homayouni R. The role of NF-kappaB in the antiviral action of interferon and interferon-regulated gene expression. J Biol Chem. 2004;279(30):31304–31311. doi: 10.1074/jbc.M308975200. [DOI] [PubMed] [Google Scholar]
- Pfeffer LM. Mullersman JE. Pfeffer SR. Murti A. Shi W. Yang CH. STAT3 as an adapter to couple phosphatidylinositol-3 kinase to the IFNAR-1 chain of the type I IFN receptor. Science. 1997;276:1418–1420. doi: 10.1126/science.276.5317.1418. [DOI] [PubMed] [Google Scholar]
- Ploegh HL. Immune and other responses to viral infections. Nutr Rev. 2000;58(2 Pt 2):S25–S30. doi: 10.1111/j.1753-4887.2000.tb07800.x. discussion S63–S73. [DOI] [PubMed] [Google Scholar]
- Pomerantz JL. Baltimore D. Two pathways to NF-kappaB. Mol Cell. 2002;10(4):693–695. doi: 10.1016/s1097-2765(02)00697-4. [DOI] [PubMed] [Google Scholar]
- Rani MR. Hibbert L. Sizemore N. Stark GR. Ransohoff RM. Requirement of phosphoinositide 3-kinase and Akt for interferon-beta-mediated induction of the beta-R1 (SCYB11) gene. J Biol Chem. 2002;277(41):38456–38461. doi: 10.1074/jbc.M203204200. [DOI] [PubMed] [Google Scholar]
- Rothe M. Sarma V. Dixit VM. Goeddel DV. TRAF2-mediated activation of NF-kappa B by TNF receptor 2 and CD40. Science. 1995;269(5229):1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M. Krol J. Markiewicz I. Heim MH. Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15(1):31–33. doi: 10.1038/nm.1902. [DOI] [PubMed] [Google Scholar]
- Scagnolari C. Zingariello P. Vecchiet J. Selvaggi C. Racciatti D. Taliani G. Riva E. Pizzigallo E. Antonelli G. Differential expression of interferon-induced microRNAs in patients with chronic hepatitis C virus infection treated with pegylated interferon alpha. Virol J. 2010;7:311. doi: 10.1186/1743-422X-7-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senftleben U. Cao Y. Xiao G. Greten FR. Krahn G. Bonizzi G. Chen Y. Hu Y. Fong A. Sun SC. Karin M. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science. 2001;293(5534):1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- Si ML. Zhu S. Wu H. Lu Z. Wu F. Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Tenoever BR. Ng SL. Chua MA. McWhirter SM. Garcia-Sastre A. Maniatis T. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315(5816):1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- Uddin S. Fish EN. Sher DA. Gardziola C. White MF. Platanias LC. Activation of the phosphatidylinositol 3-kinase serine kinase by IFN-alpha. J Immunol. 1997;158:2390–2397. [PubMed] [Google Scholar]
- Van Antwerp DJ. Martin SJ. Kafri T. Green DR. Verma IM. Suppression of TNF-α-induced apotosis by NF-κB. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- Vestal DJ. Jeyaratnam JA. The guanylate-binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J Interferon Cytokine Res. 2011;31(1):89–97. doi: 10.1089/jir.2010.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-Y. Mayo MW. Baldwin AS., Jr TNF- and cancer therapy-induced apoptosis: potentiation by inhibition of NF-κB. Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- Wei L. Sandbulte MR. Thomas PG. Webby RJ. Homayouni R. Pfeffer LM. NFkappaB negatively regulates interferon-induced gene expression and anti-influenza activity. J Biol Chem. 2006;281(17):11678–11684. doi: 10.1074/jbc.M513286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH. Murti A. Basu L. Kim JG. Pfeffer LM. IFNalpha/beta promotes cell survival by activating NF-kappaB. Proc Natl Acad Sci U S A. 2000;97:13631–13636. doi: 10.1073/pnas.250477397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH. Murti A. Pfeffer LM. STAT3 complements defects in an interferon-resistant cell line: evidence for an essential role for STAT3 in interferon signaling and biological activities. Proc Natl Acad Sci U S A. 1998;95:5568–5572. doi: 10.1073/pnas.95.10.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH. Murti A. Pfeffer LM. IFN induces NIK/TRAF dependent NF-kappaB activation to promote cell survival. J Biol Chem. 2005a;280(36):31530–31536. doi: 10.1074/jbc.M503120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH. Murti A. Pfeffer SR. Kim JG. Donner DB. Pfeffer LM. Interferonalpha/beta promotes cell survival by activating NF-kappaB through phosphatidylinositol-3 kinase and Akt. J Biol Chem. 2001;276:13756–13761. doi: 10.1074/jbc.M011006200. [DOI] [PubMed] [Google Scholar]
- Yang CH. Murti A. Valentine WJ. Du Z. Pfeffer LM. Interferon alpha activates NF-kappaB in JAK1-deficient cells through a TYK2-dependent pathway. J Biol Chem. 2005b;280(27):25849–25853. doi: 10.1074/jbc.M413721200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CH. Wei L. Pfeffer SR. Du Z. Murti A. Valentine WJ. Zheng Y. Pfeffer LM. Identification of CXCL11 as a STAT3-dependent gene induced by IFN. J Immunol. 2007;178(2):986–992. doi: 10.4049/jimmunol.178.2.986. [DOI] [PubMed] [Google Scholar]
- Yang CH. Yue J. Fan M. Pfeffer LM. IFN induces miR-21 through a signal transducer and activator of transcription 3-dependent pathway as a suppressive negative feedback on IFN-induced apoptosis. Cancer Res. 2010;70(20):8108–8116. doi: 10.1158/0008-5472.CAN-10-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]