Abstract
Using a group of structurally related cytofectins, the effects of different vehicle constituents and mixing techniques on the physical properties and biological activity of lipoplexes were systematically examined. Physical properties were examined using a combination of dye accessibility assays, centrifugation, gel electrophoresis and dynamic light scattering. Biological activity was examined using in vitro transfection. Lipoplexes were formulated using two injection vehicles commonly used for in vivo delivery (PBS pH 7.2 and 0.9% saline), and a sodium phosphate vehicle previously shown to enhance the biological activity of naked pDNA and lipoplex formulations. Phosphate was found to be unique in its effect on lipoplexes. Specifically, the accessible pDNA in lipoplexes formulated with cytofectins containing a γ-amine substitution in the headgroup was dependent on alkyl side chain length and sodium phosphate concentration, but the same effects were not observed when using cytofectins containing a β-OH headgroup substitution. The physicochemical features of the phosphate anion, which give rise to this effect in γ-amine cytofectins, were deduced using a series of phosphate analogs. The effects of the formulation vehicle on transfection were found to be cell type-dependent; however, of the formulation variables examined, the liposome/pDNA mixing method had the greatest effect on transgene expression in vitro. Thus, though predictive physical structure relationships involving the vehicle and cytofectin components of the lipoplex were uncovered, they did not extrapolate to trends in biological activity.
INTRODUCTION
Lipoplexes form when liposomes containing positively-charged cationic lipids (cytofectins) are mixed with negatively-charged polynucleic acids. Lipoplexes have been widely used as a safe non-viral delivery method of nucleic acids in vitro and in vivo. Functional genes have been delivered in vivo to a variety of different tissues, including lung (1–3), catheterized blood vessels (4–5), brain (6) and tumor (7–11). There have also been several human clinical trials using lipoplexes for treatment of cancer (12–16) and cystic fibrosis (17). Lipoplexes have also been used to enhance immune responses against several pDNA-encoded antigens (18–24).
The large number of cytofectins currently in use for lipoplex-mediated gene delivery reflects that the optimal cytofectin varies with the application. For example, DMRIE [()-N-(2-hydroxyethyl)-N,N-dimethyl-2,3-bis(tetradecycloxy)-1-propanaminium bromide] has been shown to be very effective for in vivo transfection of a variety of tumor types, while GAP-DLRIE [()-N-(3-aminopropyl)-N,N-dimethyl-2,3-bis(dodecyloxy)-1-propanaminium bromide] is better for functional gene delivery to the lung following intranasal administration (2). Cytofectin formulations have also been optimized for vaccination applications. Vaxfectin, a new cytofectin formulation, has recently shown significant enhancement of humoral immune responses against several antigens compared to naked pDNA (19).
The basis for the observed tissue and application specificity of lipoplex-mediated gene delivery is complex and not well understood. Multiple factors influence the delivery process: the physical structure of the lipoplex and its interaction with the specific tissue environment are both important (25,26). The physical nature of lipoplexes is highly dependent on the formulation components. Each cytofectin and colipid combination results in unique liposome properties and lipoplex structure. The method used for liposome and nucleic acid mixing (27,28) and the formulation vehicle (27,29,30) also affect lipoplex structure and stability. Once the lipoplex is administered, interactions with the target tissue and extracellular environment occur, further altering the lipoplex structure. Thus, it is unclear to what extent one should anticipate that the physical structure of the lipoplexes prior to administration will relate to biological activity.
Using a group of structurally related cytofectins, the effects of vehicle constituent and preparation technique on the pDNA binding properties and in vitro transfection of lipoplexes were systematically examined.
MATERIALS AND METHODS
Reagents
All chemicals were USP grade. All solutions were prepared using sterile water for injection (Baxter Health Care, Deerfield, IL) and sterile filtered through 0.2 µm nylon chamber filters (Nalgene, Inc., Rochester, NY).
Plasmid DNA
The plasmid VR1412 used for these studies contains the cytoplasmic β-galactosidase gene and was constructed, produced and purified as described previously (31,32).
Cytofectin components
Synthesis of the (±)-N-(2-hydroxyethyl)- and (±)-N-(3-aminopropyl)-N,N-dimethyl-2,3-bis(alkyloxy)-1-propanaminium bromides. Racemic 1-dimethylamino-2,3-propanediol (0.96 g; Janssen Chimica, Geel, Belgium) was converted to the disodium salt in situ by treatment with sodium hydride (60% in oil, 0.8 g) in tetrahydrofuran (70 ml). Condensation with the appropriate saturated alkyl methane sulfonate (2.2 equiv.; NuChek Prep, Elysian, MN) afforded the crude (±)-N,N-dimethyl-2,3-bis(alkyloxy)propylamine. This material was purified to homogeneity by filtration through celite followed by silica gel chromatography employing a step gradient of ether in hexane (from 10 to 50%), and finally neat ether, as the eluents. The structure and purity of this common intermediate were confirmed by 1H-NMR and IR.
The N,N-dimethyl-2,3-bis(alkyloxy)-1-propanamine thus prepared (2.4 g) was then treated with either 2-bromoethanol (3 equiv.) or with N-(3-bromopropyl)phthalimide (2 equiv.) in dimethylformamide (15 ml) at 105°C to effect quaternization of the amine. Removal of the dimethylformamide in vacuo followed by silica gel chromatography using chloroform/methanol/aqueous ammonia (90/10/0.5) as the eluent yielded the (±)-N-(2-hydroxyethyl) and (±)-N-(3-phthalimido) compounds, respectively. The N-(2-hydroxyethyl) compounds were recrystallized from hexanes to afford white solids. The purified N-(3-phthalimido) intermediates (2.1 g) were treated with anhydrous hydrazine (20 equiv.) in absolute ethanol (40 ml) to effect deprotection of the primary amine. Filtration, evaporative removal of the ethanol and basic extraction afforded the (±)-N-(3-aminopropyl) derivatives, which were also recrystallized from hexanes to afford white solids. The structure and purity of both series of cationic lipids were confirmed by 1H-NMR, IR and TLC.
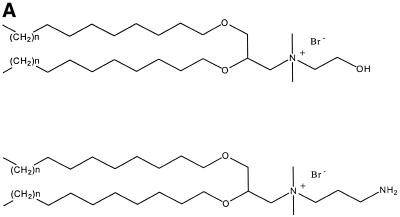
The molecular structures of the cytofectins used in this study are shown in Figure 1A. All have two saturated hydrophobic chains attached to a quaternary ammonium moiety via a polar dioxypropyl group.
Figure 1.
(A) Cytofectin structural components. The chemical structures of the cytofectins used in this paper where n = 1, 3, 5 or 7. The cytofectins contained either a primary alcohol or primary amine group. (B) Structures of the divalent anions used in this study.
1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE)
Material of >99% purity was purchased from Avanti Polar Lipids, Inc. (Alabaster, AL) as a chloroform solution and used without further purification.
Cytofectin:DOPE mixtures were prepared by mixing chloroform solutions of cationic lipid and DOPE. Dried films were prepared in 2 ml sterile glass vials by evaporating the chloroform under a stream of argon, and placing the vials under vacuum overnight to remove solvent traces. Each lipid film contained 1.5 µmol each of cytofectin and DOPE.
Lipoplex formulation
Liposomes were prepared by adding 1 ml sterile water for injection (SWFI; VWR) to a vial containing 1.5 µmol cytofectin:DOPE (1:1), followed by continuous vortexing for 5 min on the highest setting of a Genie Vortex Mixer (Fisher Scientific). The resulting liposome solution contained 1.5 mM cytofectin.
Liposomes and pDNA were prepared at twice the final formulation concentration. Lipoplexes were formed by adding an equal volume of liposomes to pDNA using a syringe and a 28 gauge needle. Liposomes were added in a steady stream, followed by brief, gentle vortex to mix (a few seconds on setting number 4 of a Fisher Genie vortex mixer).
For experiments examining the molar ratio dependence of the accessible pDNA, formulations were prepared at final cytofectin/pDNA (phosphate) molar ratios of 1:8, 1:6, 1:4 and 1:2, and a final pDNA concentration of 0.5 mg/ml. The molar concentration of pDNA phosphate was calculated by dividing the pDNA concentration (in mg/ml) by 330, the average nucleotide molecular mass.
For experiments comparing mixing technique, formulations were prepared using either the ‘2X’ or ‘jump’ mixing method at 1:2 cytofectin/pDNA molar ratio. For the 2X mixing method, liposomes (in SWFI) were added to pDNA in 2X vehicle. For the jump method, liposomes and pDNA were prepared and mixed in SWFI. An equal volume of 2X vehicle (jump method) or 1X vehicle (2X method) was added to yield a pDNA concentration of 50 µg/ml. Formulations were incubated for 30 min at room temperature, then diluted 1:1 with serum-free OptiMEM and used for in vitro transfection (see below).
DNA accessibility assays
The PicoGreen assay for measuring accessible pDNA in lipoplex formulations was adapted for use in the 96-well format (33). All dilutions were made in polypropylene tubes to avoid PicoGreen absorption to container surfaces. The PicoGreen reagent (Molecular Probes, Eugene, OR) was diluted (1:200) into the appropriate vehicle. Diluted PicoGreen reagent (500 µl) was added to 500 µl of formulation or DNA control containing 0.5 µg pDNA. Blanks contained PicoGreen reagent with or without liposomes, as appropriate. Samples were prepared in duplicate and incubated at room temperature for 10 min in the dark. Three hundred microliters of sample was transferred into each of three wells of a 96-well plate. The fluorescence was measured with a Cytofluor fluorescence plate reader (PerSeptive BioSystems, Framingham, MA) using a 485 nm excitation and 530 nm emission filter (gain = 55).
Ethidium bromide (EthBr) assays were performed by diluting each formulation or pDNA control into 1 ml of the appropriate vehicle to 8 µg/ml, followed by addition of EthBr to a final concentration of 5 µM. Blanks contained EthBr with or without the appropriate concentration of liposomes. Samples were incubated and analyzed as described above using a 430 nm excitation and 620 nm emission filters (gain = 55). The percentage of accessible pDNA was calculated by normalizing the fluorescence signal of the formulation to the corresponding pDNA control: Fformulation × 100/Fcontrol.
In vitro transfection
C2C12 (mouse muscle myoblasts, #CRL 1772) and B16F10 cell lines (mouse melanoma #CRL6322) were purchased from American Type Tissue Culture Collection. On day 0, 1 × 104 cells (100 µl) were plated into each well of a 96-well plate (Nunc) and incubated overnight at 37°C. On day 1, cell culture medium was removed by aspiration and cells were transfected with 100 µl of serially-diluted lipoplexes containing 0.5 vol serum-free OptiMEM culture medium. Cells were incubated for 1.5 h at 37°C, followed by addition of 50 µl/well of OptiMEM supplemented with 30% fetal calf serum. On day 2 (24 h post-transfection), cells received 100 µl/well of OptiMEM supplemented with 10% fetal calf serum. On day 3 (48 h post-transfection), cell culture medium was removed and 50 µl of lysis buffer (0.1% Triton X-100 in 250 mM Tris pH 8.0) was added to each well. Plates were frozen at –80°C for at least 2 h, and assayed for β-galactosidase (34).
Analysis of free pDNA by agarose gel electrophoresis
Samples were loaded on 1% agarose gels prepared in TAE buffer containing 1 µg/ml EthBr and run at 90 V for 2 h. Gels were visualized and photographed under UV light. Photographs were scanned using a Hewlett Packard ScanJet 6200C (Hewlett Packard, Palo Alto, CA).
Lipoplex particle size analysis
Particle sizing was performed by photon correlation spectroscopy on a Malvern Zetasizer 4 equipped with a series 7032 Multi-8 correlator (Malvern, PA), a ZET 5104 cell and a 400 µm photomultiplier aperture. Measurements were taken in the sizing mode at a 90° scattering angle. The particle refraction index was set to 1.45 (35), the solution refractive index to 1.33 and the temperature to 25°C. Samples were prepared by adding 1 ml of 0.15 mM cytofectin in SWFI to 1 ml of 2X vehicle (free liposomes) or 0.2 mg/ml DNA (lipoplexes) followed by gentle vortexing. Software provided by the manufacturer was used to analyze the data in CONTIN mode. The instrument was standardized using 204 nm latex NanosphereTM size standards (Duke Scientific Corp., Palo Alto, CA). One drop of standard (∼25 µl) was mixed with 50 ml of 25 mM NaCl + 10 mM sodium phosphate pH 7.2, resulting in 200–500 kCps during measurement.
RESULTS
Effects of vehicle on pDNA accessibility in lipoplexes
The effect of PBS pH 7.2 and 0.9% NaCl, two commonly used injection vehicles, on accessible pDNA in lipoplexes was examined. Lipoplexes were prepared at four different cytofectin/pDNA molar ratios varying from 1:8 to 1:2 with GAP-DMRIE and DMRIE, which both contain C = 14 alkyl side chains but different headgroups: GAP-DMRIE contains a γ-amine-substituted headgroup, and DMRIE a β-OH substitution.
A linear dependence of accessible pDNA on cytofectin/pDNA molar ratio was observed for both cytofectins in PBS [10 mM sodium phosphate pH 7.2, 150 mM (0.9%) NaCl] and 150 mM (0.9%) NaCl and no difference was observed between the two vehicles (Fig. 2A and B). The results show that the addition of 10 mM sodium phosphate as a buffering agent does not significantly alter the accessible pDNA. The accessible pDNA shows a greater dependence on cytofectin/pDNA molar ratio for complexes prepared with GAP-DMRIE:DOPE (1:1) relative to DMRIE:DOPE (1:1) (Fig. 2A and B, respectively). No significant difference was observed between the accessible pDNA determined using either PicoGreen or EthBr (data not shown), suggesting that the results are not dye-specific and reflect the physical state of the lipoplexes.
Figure 2.
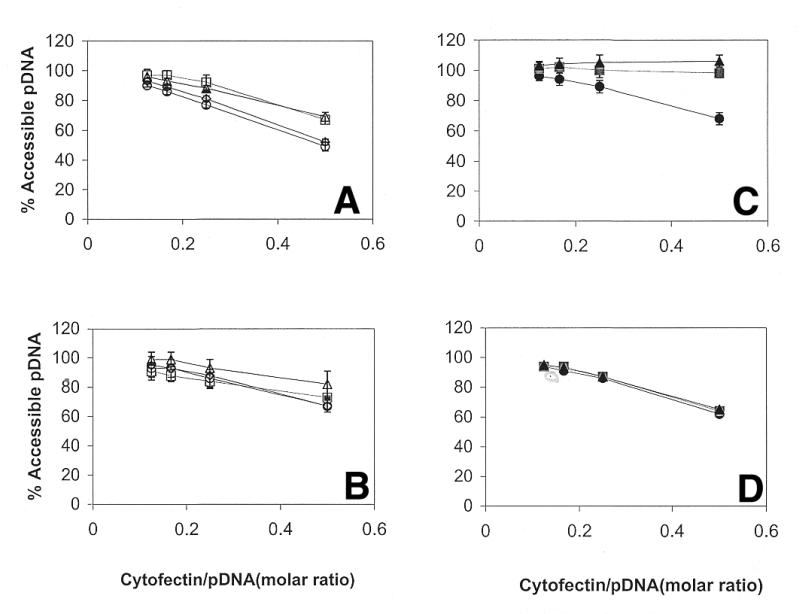
The effect of sodium phosphate pH 7.2 and sodium chloride on the accessible pDNA at different cytofectin/pDNA molar ratios. Lipoplexes were prepared with GAP-DMRIE:DOPE (1:1) (A and C) or DMRIE:DOPE (1:1) (B and D) at different vehicle concentrations. Sodium chloride (A and B) at 150 mM (open circles), 300 mM (open squares) and 450 mM (open triangles); PBS (150 mM NaCl + 10 mM sodium phosphate pH 7.2) (open diamonds). Sodium phosphate pH 7.2 (C and D) at 50 mM (closed circles), 100 mM (closed squares) and 150 mM (closed triangles). pDNA accessibility was determined using PicoGreen assays as described in Materials and Methods.
Sodium phosphate has been shown to enhance biological activity of naked pDNA and lipoplexes (18,19,29,30). Based on these findings, it was of interest to compare the effects of sodium phosphate with 150 mM NaCl and PBS pH 7.2. Accessible pDNA was measured in 50, 100 and 150 mM sodium phosphate pH 7.2. Figure 2 summarizes the comparison of the accessible pDNA in formulations prepared with GAP-DMRIE:DOPE (1:1) and DMRIE:DOPE (1:1) (Fig. 2C and D, respectively). A linear dependence of accessible pDNA on cytofectin/pDNA molar ratio was observed for both cytofectins at 50 mM sodium phosphate pH 7.2. However, at higher sodium phosphate concentrations (100 and 150 mM), nearly all of the pDNA in GAP-DMRIE:DOPE (1:1) lipoplexes was accessible at all cytofectin/pDNA molar ratios examined (Fig. 2C). In contrast, accessible pDNA was independent of sodium phosphate concentration for lipoplexes containing DMRIE:DOPE (1:1) and decreased linearly, as anticipated, with increasing cytofectin/pDNA molar ratio (Fig. 2D). To determine if the vehicle ionic strength contributed to the observed effect of sodium phosphate on accessible pDNA, measurements were performed at NaCl concentrations 300 and 450 mM, to bracket the ionic strength range of sodium phosphate. Accessible pDNA in lipoplexes prepared with DMRIE:DOPE (1:1) depends linearly on cytofectin/pDNA molar ratio, independent of NaCl concentration (Fig. 2B). In lipoplexes containing GAP-DMRIE:DOPE (1:1), a linear dependence of accessible pDNA on cytofectin/pDNA molar ratio is observed at all NaCl concentrations. The accessible pDNA is comparable at 300 and 450 mM NaCl though higher than observed in 150 mM NaCl (Fig. 2A). At a comparable ionic strength, the NaCl concentration affects the accessible pDNA in lipoplexes prepared with GAP-DMRIE:DOPE (1:1), but to a lesser degree than sodium phosphate. Thus, the effect of sodium phosphate on accessible pDNA is not due simply to an increase in ionic strength.
Effect of cytofectin alkyl chain length on accessible pDNA
The length of the alkyl side chain is known to affect liposome structure, and longer saturated alkyl chains generally result in stiffer liposome structures (34). The extent of headgroup exposure to solvent, charge and hydration on the liposome surface, will all be affected by liposome structure, and consequently influence interactions with pDNA.
The accessible pDNA was measured as a function of cytofectin alkyl chain lengths (C = 10, 12, 16 or 18) for lipoplexes prepared with cytofectins containing either β-OH or γ-amine headgroup substitutions in 50, 100 or 150 mM sodium phosphate pH 7.2. No effect of chain length was observed in formulations prepared with cytofectins containing a β-OH-substituted headgroup (data not shown). However, for the γ-amine materials, the accessible pDNA in 50 and 100 mM sodium phosphate increased with increasing chain length, while lipoplexes prepared in 150 mM sodium phosphate showed 100% accessible pDNA for cytofectins of alkyl chain length >10 (C > 10) (Fig. 3). Similar results were obtained using EthBr (data not shown).
Figure 3.

The effect of alkyl chain length on accessible pDNA. Lipoplexes were prepared with cytofectins containing a γ-amine substitution in the headgroup at different cytofectin/pDNA molar ratios. (A) C = 10, GAP-DDRIE:DOPE (1:1); (B) C = 12, GAP-DLRIE:DOPE (1:1); (C) C = 16, GAP-DPRIE:DOPE (1:1); (D) C = 18, GAP-DSRIE:DOPE (1:1). Sodium phosphate pH 7.2 concentrations of 50 mM (closed circles), 100 mM (closed squares) and 150 mM (closed triangles) were examined. pDNA accessibility was determined using PicoGreen assays as described in Materials and Methods.
Structural elements of phosphate: effect on accessible pDNA
Several features of the phosphate anion may contribute to the observed effects of sodium phosphate on accessible pDNA. These include the anion protonation state (formal charge), the number of oxygens and the geometry of the phosphate anion. The phosphate anion has four oxygens arranged in a tetrahedral configuration; three of the four oxygens can bear titratable protons (pKa1 = 2.2; pKa2 = 7.2; pKa3 = 12.3) with a formal charge of about –1.5 at pH 7.2. Several different structural analogs were examined to determine the effect of individual structural elements on the pDNA–cytofectin interaction (Fig. 1B). Phosphonate and methyl phosphonate were examined to determine whether the number of oxygens and the overall anion charge affects accessible pDNA. Both analogs are tetrahedral phosphorous anions with three oxygens and two titratable protons (pKa1 ≈ 1.3; pKa2 ≈ 6.7), and have a formal charge of about –0.75 at pH 7.2. In marked contrast to sodium phosphate, a linear decrease in accessible pDNA was observed with increasing GAP-DMRIE:DOPE (1:1)/pDNA molar ratio for both analogs at all three phosphonate concentrations (Fig. 4A and B). Sodium sulfate was used to examine the effect of titratable protons, since the sulfate anion also has four oxygens in a tetrahedral geometry, but no titratable protons, and has a net charge of –2 at biologically relevant pH. The effect of sodium sulfate concentration on the accessible pDNA in GAP-DMRIE:DOPE (1:1)/pDNA complexes is shown in Figure 4C. Increasing the sodium sulfate concentration from 50 to 150 mM increased the accessibility of pDNA to PicoGreen. The dependence of accessible pDNA on cytofectin/pDNA molar ratio in sodium sulfate was more similar to sodium phosphate than to the phosphonates, suggesting that the number of oxygens as well as the overall anion charge may be the most important structural elements contributing to the observed effect of sodium phosphate. The effects of anion geometry were further examined using succinate dianion (pKa1 = 4.19; pKa2 = 5.57; formal charge of about –2 at pH 7.2), which also contains four oxygens but as two flexibly linked, planar, delocalized anions, rather than in a tetrahedral geometry around a central atom. Compared to the other vehicles, the accessible pDNA showed a steeper linear decrease with increasing GAP-DMRIE:DOPE (1:1)/pDNA molar ratio (Fig. 4D), suggesting that anion geometry contributes to the effect of sodium phosphate on the accessible pDNA, and that charge alone is insufficient to rationalize the effect.
Figure 4.

The effect of different divalent anions on accessible pDNA. Lipoplexes were prepared with GAP-DMRIE:DOPE (1:1) at different cytofectin/pDNA molar ratios. The sodium salts of methyl phosphonate (A), phosphonate (B), sulfate (C) and succinate (D) were examined at 50 mM (closed circles), 100 mM (closed squares) and 150 mM (closed triangles) at pH 7.2.
Effect of mixing method on accessible pDNA
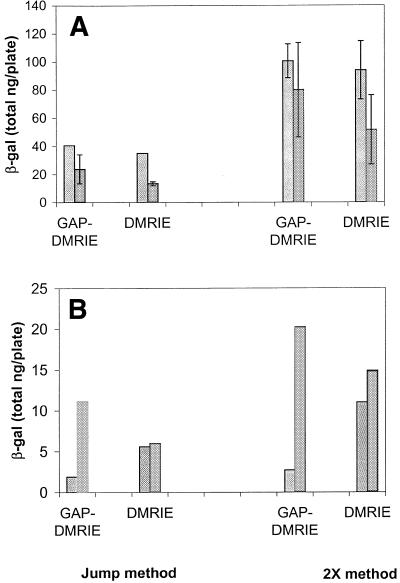
Experiments were performed to determine whether the presence of vehicle anions at the time of mixing affects the accessible pDNA in lipoplex formulations. Vehicle anions were present either at the initial binding step when cytofectin was added to pDNA (2X method), or after complex formation (jump method). Formulations were prepared at a 1:2 cytofectin/pDNA molar ratio using either GAP-DMRIE:DOPE (1:1) or DMRIE:DOPE (1:1) and then assayed for accessible pDNA. The results in Table 1 show that the mixing method had little (<10%) effect on the accessible pDNA for formulations prepared in 150 mM NaCl regardless of the cytofectin. However, when 450 mM NaCl was used in lipoplex preparation, the accessible pDNA was ∼20% lower using the jump method compared with the 2X method for both DMRIE:DOPE (1:1) and GAP-DMRIE:DOPE (1:1). These results suggest that, at high concentration, NaCl may influence the initial lipoplex formation rather than affecting the preexisting complex. No effect of mixing method was observed with 150 mM sodium phosphate. However, a decrease of ∼25% accessible pDNA was observed when 150 mM sodium sulfate was used to prepare GAP-DMRIE:DOPE (1:1) lipoplexes using the jump method compared to the 2X method. Sodium sulfate appears to affect initial complex formation of GAP-DMRIE:DOPE (1:1) lipoplexes, but not DMRIE:DOPE (1:1) complexes.
Table 1. Effect of formulation mixing technique and vehicle on the percentage of accessible pDNA in GAP-DMRIE:DOPE (1:1) and DMRIE:DOPE (1:1) lipoplexes.
| Vehicle | GAP-DMRIE:DOPE (1:1) | DMRIE:DOPE (1:1) | ||
| Mixing technique | Mixing technique | |||
| |
2X |
Jump |
2X |
Jump |
| 150 mM sodium phosphate pH 7.2 | 98 (±9) | 89 (±5) | 62 (±2) | 55 (±2) |
| 150 mM sodium sulfate | 75 (±4) | 47 (±1) | 62 (±1) | 51 (±2) |
| 150 mM sodium chloride | 40 (±2) | 30 (±0) | 58 (±2) | 49 (±0) |
| 450 mM sodium chloride | 67 (±4) | 43 (±2) | 82 (±1) | 64 (±1) |
| PBS pH 7.2 | 48 (±3) | 40 (±1) | 62 (±1) | 54 (±1) |
The numbers in parentheses indicate the standard deviation of triplicate measurements.
In vitro transfection
Lipoplexes were prepared with cytofectins containing a γ-amine or β-OH headgroup and alkyl side chains of C = 14, 16 or 18. Cytofectins and pDNA were mixed using the jump or 2X methods at a 0.5 cytofectin/pDNA molar ratio in 150 mM sodium phosphate pH 7.2, PBS pH 7.2 or 150 mM NaCl. Lipoplexes were diluted with an equal volume of OptiMEM prior to transfection, reducing the formulation vehicle concentrations by one-half (e.g. lipoplexes formulated in 150 mM sodium phosphate and diluted to 75 mM sodium phosphate pH 7.2 and 50% OptiMEM for transfection). When transfections were performed using undiluted formulation vehicle concentrations (e.g. lipoplexes formulated in 300 mM sodium phosphate and diluted to final concentrations of 150 mM sodium phosphate pH 7.2 and 50% OptiMEM for transfection), β-gal expression levels were significantly decreased (data not shown).
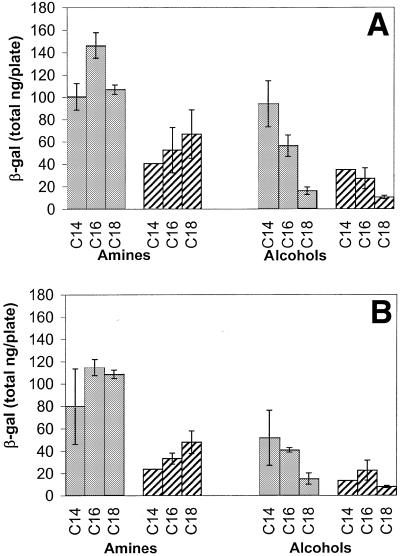
Figure 5 shows that the lipoplex mixing method affected β-gal expression in B16F10 cells transfected with lipoplexes containing cytofectins bearing either a γ-amine or β-OH headgroup substitution. Higher expression levels were observed when cells were transfected with lipoplexes prepared using the 2X mixing method. Expression levels did not show a systematic cytofectin chain length-dependence when lipoplexes were prepared with γ-amine cytofectins. However, transfection with lipoplexes containing a β-OH headgroup substituent showed lower expression levels for C18 cytofectins side chains compared to C14 and C16. The effects of formulation vehicle on in vitro expression were cell type-dependent. β-Galactosidase expression levels in B16F10 cells did not differ significantly between cells transfected with formulations prepared in 150 mM sodium phosphate compared to PBS (Fig. 5A and B, respectively). Results for 0.9% NaCl were similar to those obtained with PBS (data not shown). In contrast with B16F10 cells, C2C12 cells transfected with GAP-DMRIE:DOPE (1:1)/pDNA prepared in PBS showed higher β-gal expression levels compared to complexes prepared in 150 mM sodium phosphate pH 7.2 (Fig. 6). The results for NaCl were similar to PBS (data not shown). The 2X method resulted in higher transfection levels compared to the jump method in C2C12 cells (Fig. 6B).
Figure 5.
The effect of headgroup substituent, alkyl chain length and mixing techniques on β-galactosidase expression in B16F10 cells. Lipoplexes were prepared at a 0.5 cytofectin/pDNA molar ratio in (A) 150 mM sodium phosphate or (B) PBS pH 7.2 using the 2X (solid bars) or jump (striped bars) mixing technique.
Figure 6.
Comparison of β-galactosidase expression in two cell types. (A) B16F10 or (B) C2C12 cells were transfected with lipoplexes prepared at a 1:2 cytofectin/pDNA molar ratio with either GAP-DMRIE:DOPE (1:1) or DMRIE:DOPE (1:1) in PBS (darker bars) or 150 mM sodium phosphate pH 7.2 (lighter bars). Two mixing techniques were compared (noted on the figure).
Since culture medium was added to the cells during the course of transfection, it was of interest to determine the accessible pDNA in the presence of OptiMEM. Formulations were diluted with an equal volume of serum-free OptiMEM culture medium or 1X vehicle (controls), then incubated at 37°C for 1.5 h, as in transfection experiments. A 30% difference in accessible pDNA was observed between GAP-DMRIE:DOPE (1:1)/pDNA complexes prepared in 150 mM sodium phosphate and PBS pH 7.2 (Table 2). However, in the presence of OptiMEM, the difference decreased to 10%. Similar results were obtained if complexes were diluted into 0.9% saline instead of OptiMEM (data not shown), suggesting that simply diluting sodium phosphate restores accessible pDNA to the levels observed for PBS. Dilution with OptiMEM had no effect on the accessible pDNA when lipoplexes were prepared with DMRIE:DOPE (1:1).
Table 2. Effect of OptiMEM on the percentage of accessible pDNA in GAP-DMRIE:DOPE (1:1) and DMRIE:DOPE(1:1) lipoplexes.
| GAP-DMRIE:DOPE (1:1) | DMRIE:DOPE (1:1) | |||
| |
(–) OptiMEM |
(+) OptiMEM |
(–) OptiMEM |
(+) OptiMEM |
| 150 mM sodium phosphate pH 7.2 | 93 (±2) | 67 (±2) | 54 (±1) | 55 (0) |
| PBS pH 7.2 | 60 (±1) | 52 (±2) | 67 (±1) | 63 (0) |
The numbers in parentheses indicate the standard deviation of triplicate measurements.
Macroscopic features of lipoplexes
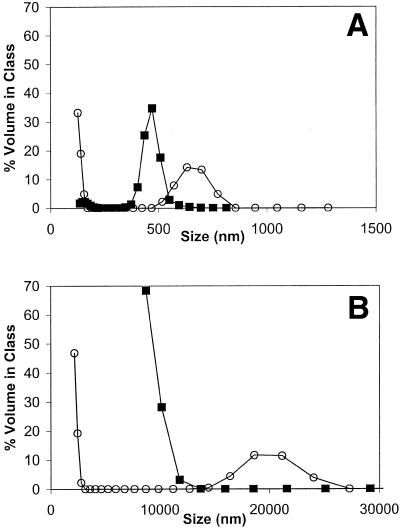
The high degree of accessible pDNA in sodium phosphate could result from either a configuration of the complex, in which more DNA is available for dye binding, or lack of complex formation. To address this issue, particle sizing, visual analysis, centrifugation and agarose gel electrophoresis were performed. GAP-DMRIE:DOPE (1:1)/pDNA complexes were prepared in PBS or 150 mM sodium phosphate pH 7.2 using the 2X mixing method at a final ratio of 1:4 cytofectin:pDNA. Clear differences were observed between the particle size distributions of lipoplexes and liposomes (Fig. 7A and B, respectively). A narrower and more homogeneous size distribution was observed for lipoplexes compared to the corresponding liposomes in PBS and 150 mM sodium phosphate pH 7.2. Visual observation of lipoplex formulations 30 and 60 min after mixing showed slight turbidity, but no aggregation, while snowflake-like particles were present in the liposome solution after 60 min. Centrifugation was used to separate lipoplexes from unbound liposomes and pDNA. Lipoplexes were prepared in 150 mM sodium phosphate (1:4 molar ratio cytofectin/pDNA) and centrifuged at 20 800 g for 30 min at 20°C: a clearly observable pellet was formed. The complexes (prior to centrifugation) and supernates collected after centrifugation were analyzed by agarose gel electrophoresis (data not shown). The band observed in the wells containing complexes is absent from the naked pDNA control and the supernatant, as would be anticipated only for lipoplex formation in the presence of sodium phosphate. The intensity of the band in the wells decreased with increasing sodium phosphate concentration, consistent with the trend in pDNA accessibility results. The transfection results also inherently argue in favor of complex formation in the presence of sodium phosphate since naked pDNA does not transfect cells in vitro.
Figure 7.
Comparison of particle size distributions for lipoplexes and liposomes. Lipoplexes (A) and liposomes (B) were prepared in final vehicles of PBS (squares) or 150 mM sodium phosphate (circles) pH 7.2 with GAP-DMRIE:DOPE (1:1) at a 0.25 cytofectin/pDNA molar ratio. Note the difference in the x-axis scale between (A) and (B).
DISCUSSION
The surface interactions of liposomes composed of cytofectins and DOPE are complex, and several cytofectin structural components influence the resulting liposome structure. An increase in aliphatic chain length and degree of saturation increases the phase transition temperature and bilayer ‘stiffness’ (34), which may result in tighter lipid packing. Conversely, when shorter chain-saturated cytofectins are combined with DOPE (C18:1) a ‘mismatch’ between the aliphatic regions may result, causing defects in lipid packing. The orientation of cytofectin headgroups on the liposome surface may also be affected by lipid packing. The nature of the cytofectin headgroup will also affect the physicochemical relationships between lipids at the bilayer surface as well as influence solvent–lipid interactions. For example, the cytofectin headgroup may interact with DOPE to form ion pairs (34), and this pairing will likely be a function of the chemical moieties present in the cytofectin headgroup. Lipoplex structure depends not only on cytofectin structure variation, but also on the formulation vehicle. Indeed, variations in cytofectin alkyl chain length and headgroup resulted in vehicle-dependent effects on the degree of accessible pDNA. Lipoplexes prepared in PBS or normal saline resulted in comparable levels of accessible pDNA, and showed the anticipated decrease in accessible pDNA with increasing cytofectin/pDNA molar ratio. The dependence of accessible pDNA on cytofectin/pDNA molar ratio was greater for lipoplexes prepared with cytofectins containing a γ-amine headgroup substituent compared to a β-OH headgroup substituent. The difference may be due to the additional charge contributed by protonation of the γ-amine, which could allow greater pDNA binding at a given molar ratio. An increase in sodium chloride concentration resulted in an increased accessible pDNA, consistent with greater screening of charge interactions between cytofectin and pDNA at higher salt concentration, and a lower equilibrium constant for the pDNA–cytofectin binding reaction. Similar results were observed by others using EthBr accessibility studies, which showed that in lipoplexes prepared with DMRIE:DOPE (1:1), pDNA accessibility increased with increasing NaCl concentration (36). A rationale was proposed in which pDNA binds tightly to the liposome surface at low NaCl concentration, and as the salt concentration increases, pDNA remains bound to the cytofectin but extends into solution. Thus, the cytofectin headgroup and the solution environment both influence the overall lipoplex structure. In the present study, sodium phosphate had the greatest effect of any vehicle examined. When lipoplexes were prepared in 150 mM sodium phosphate with cytofectins of C > 10 containing a γ-amine in the headgroup, all of the pDNA was accessible to dye binding, independent of the cytofectin/pDNA molar ratio. This is unusual, since lipoplexes generally show a degree of accessible pDNA varying proportionally with the cytofectin/pDNA molar ratio, except at molar NaCl concentrations (36). Indeed, lipoplexes containing β-OH cytofectins showed the expected degree of accessible pDNA, regardless of the sodium phosphate concentration. Increasing the cytofectin chain length may result in tighter lipid packing, less mismatch between lipid side chains and altered surface properties. Tighter lipid packing may facilitate interactions between sodium phosphate and the γ-amine functional group on the liposome surface, thereby allowing greater dye accessibility.
Sodium phosphate appears to affect both initial complex formation and preexisting lipoplexes, since the degree of accessible pDNA was the same using the 2X and jump mixing methods. Thus, it appears that a high sodium phosphate concentration significantly alters the lipoplex structure, conceivably by affecting the interactions between the bilayer surface and pDNA. Based on the results presented here, a structure–activity relationship (SAR) between elements of the phosphate anion and the cytofectin γ-amine headgroup can be proposed, which has broader implications for the effects of solution conditions on lipoplex structure. The unique combination of the geometry, structure and charge of the phosphate anion may allow the phosphate anion to bridge between γ-amine headgroups on the liposome surfaces through electrostatic and hydrogen bonding interactions. Studies with model compounds support the notion of bridging between sodium phosphate and the γ-amine headgroups (37). The formation of stable chelation complexes between multiple amine groups and the phosphate anion (H2PO4–) was demonstrated using model polyamine compounds. The liposome surface may be viewed as a functional polyamine, in which amine spacing depends on the packing of lipids in the bilayer. Tighter packing imposed by longer chain cytofectins may provide a more preferred orientation of amine groups on the surface to bind with the phosphate. This is consistent with the data, which show that sodium phosphate has a greater effect on the accessible pDNA in lipoplexes prepared with longer chain γ-amine cytofectins. There is evidence that sodium phosphate may form ‘bridges’ between bilayer surfaces from liposome mixing studies. Fluorescent probes have been employed to specifically examine the effect of di- and multivalent anions, including pDNA, on liposome mixing. Multivalent anions, as well as pDNA, were shown to promote liposome mixing, and further support the notion that sodium phosphate significantly alters the structure of lipoplex and may act by facilitating interactions between lipid bilayers (38,39).
Phosphate may also affect solvent structure around the lipoplex and interactions between lipoplexes. Phosphate is the most effective anion in the Hofmeister series and is thought to act by increasing the surface tension of solvent around macromolecules, resulting in dehydration and excluded volume effects that force molecules closer together (40). By analogy, phosphate may increase interactions between lipid bilayers. The end result may be to limit the number of binding sites available for pDNA at the bilayer surface. For example, if pDNA interacts with x γ-amine headgroups on the liposome surface, then in the presence of phosphate, there will be less than x binding sites available. The binding of pDNA to the liposome would be weaker and may result in loosely associated pDNA, possibly extending from the surface of the bilayer, thus increasing the accessibility of the pDNA to the dye.
In summary, the effects of formulation vehicle on accessible pDNA are consistent with an external model in which pDNA is associated with the bilayer surface. The specific combination of structural elements in the phosphate anion may promote interactions between lipid bilayers in the lipoplex at moderately elevated concentrations. Accessible pDNA would increase as the enhanced association between lipid bilayers causes displacement of the pDNA the close association with the bilayer surface to a more extended configuration.
It is important to note that for the transfection experiments reported here, lipoplexes were diluted with an equal volume of OptiMEM prior to transfection, effectively reducing the vehicle concentration by one-half. Dilution of lipoplexes prepared in 150 mM sodium phosphate or PBS with an equal volume of OptiMEM showed more comparable levels of accessible pDNA. A key finding of this work is that changes in formulation variables that affect the physical properties of lipoplex formulations are not predictive of in vitro biological activity. Formulations are best viewed as an independent ‘whole’, and not simply as the sum of the individual components and preparation procedures. Thus, changing a formulation requires reexamination ‘in toto’ for each application to determine the biological consequences of the change, since simple extrapolation of the effect of changing a ‘single’ variable may lead to an unanticipated outcome. For example, the SAR of formulation vehicle and cytofectin on in vitro reporter gene expression was found to be cell type-dependent. Differences in transfection between cell types may be due to numerous factors. Thus, the limiting step for transfection is likely to be different for different cell types. For example, B16F10 cells are more rapidly growing and more easily transfected than C2C12 cells. During cell division, the nuclear membrane ‘dissolves’, which is thought to facilitate entry of pDNA into the nucleus, and may therefore be relatively less sensitive to formulation changes.
The effects of chain length and mixing method illustrate the interrelated nature of formulation variables and the effect on biological activity, and the difficulty in making simple predictions of the changes in biological response based on formulation changes. For example, β-gal expression in B16F10 cells was dependent on alkyl side chain length when cells were transfected with lipoplexes containing cytofectins with a β-OH substituted headgroup, which is consistent with previous results (34). A SAR was also observed in the present study in both PBS and 150 mM sodium phosphate when formulations were prepared using the 2X mixing method. By contrast, when the jump method was used, the alkyl chain length-dependence in the β-OH series was less pronounced. The length of the cytofectin side chain has previously been shown to affect luciferase expression for lipoplexes delivered in vivo (41). It is possible that tighter packing afforded by the longer alkyl chains results in less release of pDNA and lower transfection levels (34). A different relationship between mixing method and alkyl chain length was observed for lipoplexes prepared using β-OH- versus γ-amine-substituted cytofectins. No systematic effect of alkyl chain length was observed when B16F10 cells were transfected with lipoplexes prepared using γ-amine-substituted cytofectins and the 2X mixing method. However, when the jump method was used, a trend towards increasing transfection levels with increasing chain length was observed. When using the jump method, the γ-amine substitution may allow the headgroup to extend further from the liposome surface, resulting in a less stable lipoplex structure, hence faster, more efficient dissociation of pDNA within the cell. Similarly, the amine moiety could function as a buffer to enhance endosomal escape (42), thus delivering more active plasmid into the cell interior.
The method of mixing pDNA and liposomes had the greatest effect of any formulation variable on expression in both C2C12 and B16F10 cells. Lipoplexes prepared using the 2X mixing method gave higher transfection levels than those prepared using the jump method, even though the final formulation vehicle was the same in both cases. The fact that using the 2X mixing method resulted in higher levels of transfection is interesting in light of the studies where preincubation of DOTMA:DOPE (29) or DOGS:DOPE (1:1) (30) liposomes in phosphate was required to observe enhancement of biological activity. Liposomes precipitated when mixed with phosphate vehicles at the concentrations used in the present studies, thus precluding pre-incubation in sodium phosphate. The differences in the results may be a function of cytofectin structure, or the concentrations of liposomes used.
The studies in this report present a systematic characterization of the effects of formulation vehicle, preparation methodology, and cytofectin structure on the physical and biological properties of lipoplexes. The results illustrate that the interplay of the different formulation variables on lipoplex structure and biological activity, and the dynamic nature of lipoplexes, tend to preclude easy ‘predictive’ extrapolation between assay systems. Lipoplexes prepared in PBS pH 7.2 afford an example of physically robust, generally applicable formulation conditions. The addition of 10 mM sodium phosphate to 0.9% saline buffers the solution, and aids in protecting the lipid and pDNA from long-term degradation. In addition, the biological activity in different cell types was not compromised. However, more active lipoplex formulations, such as those prepared in 150 mM sodium phosphate, can be developed for specific applications by systematically varying components of the lipoplex and the formulation methods. Studies are currently underway to examine the effect on the physical and biological properties of lipoplexes when headgroup, chain length and degree saturation of both the cytofectin and colipid are varied.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank L.Sukhu and J.Stupack for tissue culture support, J.Meek and the Vical Manufacturing staff for preparation of plasmid DNA, and Dr A.Vilalta and P.Ramsey for useful discussions and editing of this manuscript.
References
- 1.Brigham K.L., Meyrick,B., Christman,B., Magnuson,M., King,G. and Berry,L.C.,Jr (1989) In vivo transfection of murine lungs with a functioning prokaryotic gene using a liposome vehicle. Am. J. Med. Sci., 298, 278–281. [DOI] [PubMed] [Google Scholar]
- 2.Wheeler C.J., Felgner,P.L., Tsai,Y.J., Marshall,J., Sukhu,L., Doh,S.G., Hartikka,J., Nietupski,J., Manthorpe,M., Nichols,M. et al. (1996) A novel cationic lipid greatly enhances plasmid DNA delivery and expression in mouse lung. Proc. Natl Acad. Sci. USA, 93, 11454–11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Logan J.J., Bebok,Z., Walker,L.C., Peng,S., Felgner,P.L., Siegal,G.P., Frizzell,R.A., Dong,J., Howard,M. and Matalon,S. et al. (1995) Cationic lipids for reporter gene and CFTR transfer to rat pulmonary epithelium. Gene Ther., 2, 38–49. [PubMed] [Google Scholar]
- 4.Nabel E.G., Plautz,G. and Nabel,G.J. (1990) Site-specific gene expression in vivo by direct gene transfer into the arterial wall. Science, 249, 1285–1288. [DOI] [PubMed] [Google Scholar]
- 5.Stephan D.J., Yang,Z.Y., San,H., Simari,R.D., Wheeler,C.J., Felgner,P.L., Gordon,D., Nabel,G.J. and Nabel,E.G. (1996) A new cationic liposome DNA complex enhances the efficiency of arterial gene transfer in vivo. Hum. Gene Ther., 7, 1803–1812. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz B., Benoist,C., Abdallah,B., Scherman,D., Behr,J.P. and Demeneix,B.A. (1995) Lipospermine-based gene transfer into the newborn mouse brain is optimized by a low lipospermine/DNA charge ratio. Hum. Gene Ther., 6, 1515–1524. [DOI] [PubMed] [Google Scholar]
- 7.Nabel G.J., Chang,A.E., Nabel,E.G., Plautz,G.E., Ensminger,W., Fox,B.A., Felgner,P., Shu,S. and Cho,K. (1994) Immunotherapy for cancer by direct gene transfer into tumors. Hum. Gene Ther., 5, 57–77. [DOI] [PubMed] [Google Scholar]
- 8.Nabel G.J., Nabel,E.G., Yang,Z.Y., Fox,B.A., Plautz,G.E., Gao,X., Huang,L., Shu,S., Gordon,D. and Chang,A.E. (1993) Direct gene transfer with DNA–liposome complexes in melanoma: expression, biologic activity and lack of toxicity in humans. Proc. Natl Acad. Sci. USA, 90, 11307–11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parker S.E., Khatibi,S., Margalith,M., Anderson,D., Yankauckas,M., Gromkowski,S.H., Latimer,T., Lew,D., Marquet,M., Manthorpe,M. et al. (1996) Plasmid DNA gene therapy: studies with the human interleukin-2 gene in tumor cells in vitro and in the murine B16 melanoma model in vivo. Cancer Gene Ther., 3, 175–185. [PubMed] [Google Scholar]
- 10.Saffran D.C., Horton,H.M., Yankauckas,M.A., Anderson,D., Barnhart,K.M., Abai,A.M., Hobart,P., Manthorpe,M., Norman,J.A. and Parker,S.E. (1998) Immunotherapy of established tumors in mice by intratumoral injection of interleukin-2 plasmid DNA: induction of CD8(+) T-cell immunity. Cancer Gene Ther., 5, 321–330. [PubMed] [Google Scholar]
- 11.Horton H.M., Dorigo,O., Hernandez,P., Anderson,D., Berek,J.S. and Parker,S.E. (1999) IL-2 plasmid therapy of murine ovarian carcinoma inhibits the growth of tumor ascites and alters its cytokine profile. J. Immunol., 163, 6378–6385. [PubMed] [Google Scholar]
- 12.Felgner P.L., Zaugg,R.H. and Norman,J.A. (1995) Synthetic recombinant DNA delivery for cancer therapeutics. Cancer Gene Ther., 2, 61–65. [PubMed] [Google Scholar]
- 13.Gao X. and Huang,L. (1995) Cationic liposome-mediated gene transfer. Gene Ther., 2, 710–722. [PubMed] [Google Scholar]
- 14.Nabel G.J., Gordon,D., Bishop,D.K., Nickoloff,B.J., Yang,Z.Y., Aruga,A., Cameron,M.J., Nabel,E.G. and Chang,A.E. (1996) Immune response in human melanoma after transfer of an allogeneic class I major histocompatibility complex gene with DNA–liposome complexes. Proc. Natl Acad. Sci. USA, 93, 15388–15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rubin J., Galanis,E., Pitot,H.C., Richardson,R.L., Burch,P.A., Charboneau,J.W., Reading,C.C., Lewis,B.D., Stahl,S., Akporiaye,E.T. and Harris,D.T. (1997) Phase I study of immunotherapy of hepatic metastases of colorectal carcinoma by direct gene transfer of an allogenic histocompatibility antigen, HLA-B7. Gene Ther., 4, 419–425. [DOI] [PubMed] [Google Scholar]
- 16.Stopeck A.T., Hersh,E.M., Akporiaye,E.T., Harris,D.T., Grogan,T., Unger,E., Warneke,J., Schluter,S.F. and Stahl,S. (1997) Phase I study of direct gene transfer of an allogeneic histocompatibility antigen, HLA-B7, in patients with metastatic melanoma. J. Clin. Oncol., 15, 341–349. [DOI] [PubMed] [Google Scholar]
- 17.Caplen N.J., Alton,E.W., Middleton,P.G., Dorin,J.R., Stevenson,B.J., Gao,X., Durham,S.R., Jeffery,P.K., Hodson,M.E., Coutelle,C. et al. (1995) Liposome-mediated CFTR gene transfer to the nasal epithelium of patients with cystic fibrosis. Nat. Med., 1, 39–46. [DOI] [PubMed] [Google Scholar]
- 18.Hartikka J., Bozoukova,V., Jones,D., Mahajan,R., Wloch,M.K., Sawdey,M., Buchner,C., Sukhu,L., Barnhart,K.M., Abai,A.M. et al. (2000) Sodium phosphate enhances plasmid DNA expression in vivo. Gene Ther., 7, 1171–1182. [DOI] [PubMed] [Google Scholar]
- 19.Hartikka J., Bozoukova,V., Ferrari.,M., Sukhu,L., Enas,J., Sawdey,M., Wloch,M.K., Tonsky,K., Norman,J., Manthorpe,M. and Wheeler,C.J. (2001) Vaxfectin enhances the humoral response to pDNA encoded antigens. Vaccine, 19, 1911–1923. [DOI] [PubMed]
- 20.Gregoriadis G., Saffie,R. and de Souza,J.B. (1997) Liposome-mediated DNA vaccination. FEBS Lett., 402, 107–110. [DOI] [PubMed] [Google Scholar]
- 21.Ishii N., Fukushima,J., Kaneko,T., Okada,E., Tani,K., Tanaka,S.I., Hamajima,K., Xin,K.Q., Kawamoto,S., Koff,W. et al. (1997) Cationic liposomes are a strong adjuvant for a DNA vaccine of human immuno-deficiency virus type 1. AIDS Res. Hum. Retroviruses, 13, 1421–1428. [DOI] [PubMed] [Google Scholar]
- 22.Toda S., Ishii,N., Okada,E., Kusakabe,K.I., Arai,H., Hamajima,K., Gorai,I., Nishioka,K. and Okuda,K. (1997) HIV-1-specific cell-mediated immune responses induced by DNA vaccination were enhanced by mannan-coated liposomes and inhibited by anti-interferon-γ antibody. Immunology, 92, 111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okada E., Sasaki,S., Ishii,N., Aoki,I., Yasuda,T., Nishioka,K., Fukushima,J., Miyazaki,J., Wahren,B. and Okuda,K. (1997) Intranasal immunization of a DNA vaccine with IL-12- and granulocyte-macrophage colony-stimulating factor (GM-CSF)-expressing plasmids in liposomes induces strong mucosal and cell-mediated immune responses against HIV-1 antigens. J. Immunol ., 159, 3638–3647. [PubMed] [Google Scholar]
- 24.Yokoyama M., Zhang,J. and Whitton,J.L. (1996) DNA immunization: effects of vehicle and route of administration on the induction of protective antiviral immunity. FEMS Immunol. Med. Microbiol., 14, 221–230. [DOI] [PubMed] [Google Scholar]
- 25.Li S., Tseng,W.-C., Stolz,D.B., Wu,S.-P., Watkins,S.C. and Huang,L. (1999) Dynamic changes in the characteristics of cationic lipidic vectors after exposure to mouse serum: implications for intravenous lipofection. Gene Ther., 6, 585–594. [DOI] [PubMed] [Google Scholar]
- 26.Monkkonen J. and Urtti,A. (1998) Lipid fusion in oligonucleotide and gene delivery with cationic lipids. Adv. Drug Del. Rev., 34, 37–49. [DOI] [PubMed] [Google Scholar]
- 27.Zelphati O., Nguyen,C., Ferrari,M., Felgner,J., Tsai,Y. and Felgner,P.L. (1998) Stable and monodisperse lipoplex formulations for gene delivery. Gene Ther., 5, 1272–1282. [DOI] [PubMed] [Google Scholar]
- 28.Zuidam N.J. and Barenholz,Y. (1999) Characterization of DNA–lipid complexes commonly used for gene delivery. Int. J. Pharmacol., 183, 43–46. [DOI] [PubMed] [Google Scholar]
- 29.Kariko K., Kuo,A., Barnathan,E.S. and Langer,D.J. (1998) Phosphate-enhanced transfection of cationic lipid-complexed mRNA and plasmid DNA. Biochim. Biophys. Acta, 1369, 320–334. [DOI] [PubMed] [Google Scholar]
- 30.Kichler A., Zauner,W., Ogris,M. and Wagner,E. (1998) Influence of the DNA complexation medium on the transfection efficiency of lipospermine/DNA particles. Gene Ther., 5, 855–860. [DOI] [PubMed] [Google Scholar]
- 31.Wheeler C.J., Sukhu,L., Yang,G., Tsai,Y., Bustamente,C., Felgner,P., Norman,J. and Manthorpe,M. (1996) Converting an alcohol to an amine in a cationic lipid dramatically alters the co-lipid requirement, cellular transfection activity and the ultrastructure of DNA–cytofectin complexes. Biochim. Biophys. Acta, 1280, 1–11. [DOI] [PubMed] [Google Scholar]
- 32.Horn N.A., Meek,J.A., Budahazi,G. and Marquet,M. (1995) Cancer gene therapy using plasmid DNA: purification of DNA for human clinical trials. Hum. Gene Ther., 6, 565–573. [DOI] [PubMed] [Google Scholar]
- 33.Ferrari M.E., Nguyen,C.M., Zelphati,O., Tsai,Y. and Felgner,P.L. (1998) Analytical methods for the characterization of cationic lipid–nucleic acid complexes. Hum. Gene Ther., 9, 341–351. [DOI] [PubMed] [Google Scholar]
- 34.Felgner, J., Kumar,R., Sridhar,C.N., Wheeler,C.J., Tsai,Y.T., Border,R. Ramsey,P., Martin,M. and Felgner,P.L. (1994) Enhanced gene delivery and mechanism studies with a novel series of cationic lipid formulations. J.Biol. Chem., 269, 2550–2561. [PubMed]
- 35.Blessing T., Remy,J.S. and Behr,J.P. (1998) Monomolecular collapse of plasmid DNA into stable virus-like particles. Proc. Natl Acad. Sci. USA, 95, 1427–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eastman S.J., Siegel,C., Tousignant,J., Smith,A.E., Cheng,S.H. and Scheule,R.K. (1997) Biophysical characterization of cationic lipid:DNA complexes. Biochim. Biophys. Acta, 1325, 41–62. [DOI] [PubMed] [Google Scholar]
- 37.Dietrich B., Fyles,D.L., Fyles,T.M. and Lehn,J.-M. (1979) Anion coordination chemistry: polyguanidinium salts as anion complexones. Helv. Chim. Acta, 62, 2763–2787. [Google Scholar]
- 38.Mok K.W. and Cullis,P.R. (1997) Structural and fusogenic properties of cationic liposomes in the presence of plasmid DNA. Biophys. J., 73, 2534–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Düzgünes N., Goldstein,N., Friend,D.S. and Felgner,P.L. (1989) Fusion of liposomes containing a novel cationic lipid, N-(2,3-(dioleyloxy)propyl)-N,N,N-trimethylammonium: induction by multivalent anions and asymmetric fusion with acidic phospholipid vesicles. Biochemistry, 28, 9179–9184. [DOI] [PubMed] [Google Scholar]
- 40.Scopes R.K. (1994) in Protein Purification:Principles and Practice, 3rd edn. Springer-Verlag, New York, NY. Ch. 4, pp. 71–100..
- 41.Ren T., Song,Y.K., Zhang,G. and Liu,D. (2000) Structural basis of DOTMA for its high intraveneous transfection activity in mouse. Gene Ther., 7, 764–768. [DOI] [PubMed] [Google Scholar]
- 42.Lee R.J. and Huang,L. (1997) Lipidic vector systems for gene transfer. Crit. Rev. Ther. Drug Carrier Syst., 14, 173–206. [PubMed] [Google Scholar]