Abstract
MTF-1 is a zinc finger transcription factor that mediates the cellular response to heavy metal stress; its targeted disruption in the mouse leads to liver decay and embryonic lethality at day E14. Recently, we have sequenced the entire MTF-1 gene in the compact genome of the pufferfish Fugu rubripes. Here we have defined the promoter sequences of human and mouse MTF-1 and the genomic structure of the mouse MTF-1 locus. The transcription unit of MTF-1 spans 42 kb (compared to 8.5 kb in Fugu) and is located downstream of the gene for a phosphatase (INPP5P) in mouse, human, and fish. In all of these species, the MTF promoter region has the features of a CpG island. In both mouse and human, the 5′ untranslated region harbors conserved short reading frames of unknown function. RNA mapping experiments revealed that in these two species, MTF-1 mRNA is transcribed from a cluster of multiple initiation sites from a TATA-less promoter without metal-responsive elements. Transcription from endogenous and transfected MTF-1 promoters was not affected by heavy metal load or other stressors, in support of the notion that MTF-1 activity is regulated at the posttranscriptional level. Tissue Northern blots normalized for poly A+ RNA indicate that MTF-1 is expressed at similar levels in all tissues, except in the testes, that contain more than 10-fold higher mRNA levels.
INTRODUCTION
Most organisms from fungi to humans respond to heavy metal stress by increased transcription of genes that encode detoxifying proteins, notably metallothioneins (Kägi and Kojima 1987; Heuchel et al 1995). We have previously identified a zinc finger protein, termed MTF-1, as a key transcription factor in mediating this stress response (Westin and Schaffner 1988; Radtke et al 1993). MTF-1, which is highly conserved among vertebrates, was eliminated by targeted gene disruption in the mouse and was thereby identified as an essential protein as null mutant embryos died in utero at E14 due to liver decay (Heuchel et al 1994; Günes et al 1998). Interestingly, elimination of either of 2 other stress-responsive transcription factors, c-Jun and NF-κB/relA, yields a similar phenotype of embryonic liver decay (Hilberg et al 1993; Beg et al 1995). Because MTF-1 is expressed ubiquitously, it seems likely that other organs besides the liver are also affected. However, no other target tissues where MTF-1 is essential could be identified thus far. Neural grafting experiments revealed that, at least for development of the basic central nervous system cell types, MTF-1 is dispensable (Lichtlen et al 1999). Fibroblasts obtained from day 12.5 embryos (ie, before liver crisis) could be grown in culture and did display increased sensitivity toward both heavy metal toxicity and oxidative stress due to hydrogen peroxide treatment (Günes et al 1998). The expression of the cellular detoxifying proteins, metallothionein I and II, was barely detectable in MTF-1 knockout cells and embryos. Furthermore, a number of MTF-1 target genes are affected, including gamma glutamyl cysteine synthetase (γ-GCS) (Günes et al 1998; P.L., Y.W., and W.S., unpublished).
Recently, we isolated and sequenced the MTF-1 gene from a cold-blooded vertebrate, the Japanese pufferfish Fugu rubripes (Auf der Maur et al 1999). Fugu is a useful model for the comparative analysis of vertebrate genomes, as it contains a full set of vertebrate genes, yet has a haploid DNA content of a mere 400 Mb, only 13% as much as that of the mammalian genome. Genome compaction in Fugu is the result of shorter introns and intergenic spacers and a paucity of repetitive DNA, a feature common among the bonyfish family of Tetraodontidae (Elgar et al 1996). The selective pressure for this compaction is presently unknown. We demonstrated that the Fugu MTF-1 is closely related to counterparts in mouse and human, and that the transcription unit spans about 8.5 kb of genomic DNA. A tentative comparison of the mouse and Fugu promoters for MTF-1 revealed that both loci are preceded by a phosphatase gene (inositol polyphosphate 5-phosphatase II), have the properties of a so-called CpG island (Auf der Maur et al 1999), but otherwise do not exhibit any sequence similarity in this region. Here we have sequenced the entire gene of mouse MTF-1. We also sequenced the human MTF-1 promoter and determined the transcription start sites in mouse and human. In accordance with the larger genome size of the mouse relative to Fugu, the mouse MTF-1 transcription unit is 5 times longer than that of Fugu. Remarkably, the intron/exon boundaries are conserved to the nucleotide between Fugu and mouse, even in regions of low protein similarity. MTF-1 is expressed to a similar level in all mouse tissues analyzed, except in testes that curiously contain a much higher level. In accordance with the general notion that housekeeping promoters are often associated with CpG islands and are transcribed with scattered starts from TATA-less promoters, we find scattered starts for both mouse and human MTF-1 mRNA.
MATERIALS AND METHODS
Plasmids and constructs
All constructs were engineered according to standard DNA cloning procedures (Sambrook et al 1989).
The human MTF-1 promoter was polymerase chain reaction (PCR)–amplified in 3 rounds of nested PCR using Ready To Go™ PCR beads from Pharmacia. Two hundred nanograms of human genomic DNA was used for the first round of PCR, and 0.1% of the PCR product served as template for the subsequent round of PCR, performed as follows. One denaturation cycle at 94°C for 4 minutes, followed by 35 cycles of 1 minute at 94°C, 1 minute at 65°C, and 3 minutes at 72°C, and a final incubation time of 30 minutes at 72°C. The 3, nested upstream primers (hINPP5P-trailer 1: 5′-CCAGTTCCTCTGCAACCCACTC-3′, hINPP5P-trailer 2: 5′-CCAGCCCCACCTGTTTCAGCTC-3′, hINPP5P-trailer 3: 5′-GCCCACCATTGGTTAGCACACTC-3′) anneal in the 3′ untranslated region (UTR) of the human inositol polyphosphate 5-phosphatase gene (accession number M74161), and the 2 downstream primers (hMTF-1 first exon 1: 5′-GGCTCCCGGCAACGGCGGCAGC-3′, hMTF-1 first exon 2: 5′-TTCTCCCCTCCCCAGCGGCACC-3′) anneal in the first exon of the human MTF-1 gene. The 2.6-kb PCR product obtained after the third round of PCR was blunt-end ligated into Bluescript pBS SK(-) and sequenced from both ends.
Cell culture
Wildtype mouse fibroblasts were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco) supplemented with 10% fetal calf serum (Gibco), 100 U/mL penicillin and 100 U/mL streptomycin and 2 mM l-glutamine. Hela cells were maintained in DMEM medium with 2.5% fetal calf serum and 2.5% newborn calf serum (Gibco) as described for previous studies on heavy metal induction (Westin and Schaffner 1988).
Mapping of the mouse MTF-1 transcription start site
Plasmid transfections and S1 nuclease mapping of resultant transcripts were performed as previously described (Kemler et al 1991; Radtke et al 1993). Wildtype mouse fibroblasts were transfected with 10 μg of the mMTF-1aOVEC promoter construct along with 10 μg of an expression plasmid carrying mMTF-1cDNA by calcium phosphate methodology. For heavy metal induction, ZnSO4 was added to a final concentration of 100 μM for 2 hours before harvest. For S1 mapping, cytoplasmic total RNA was hybridized overnight with 32P-labeled, single-stranded DNA oligonucleotide (mMTF-1-S1 oligo-2: 5′-GGCTCCCGGCAACGGCGGCAGCAGCAGCTTCTCCCCTCCCAAGCGGCACCATGATTGCCCCCACCTGTCCCTAGCGTCACTTCCGCTTCCGCTTTCCGGAGGAAGGAGTCAGATG-3′) and digested with 300 U S1 nuclease at RT for 1 hour. The samples were electrophoresed through a 10% polyacrylamide sequencing gel, and S1 nuclease data were collected and quantified using a PhosphorImager.
For reliable mapping of MTF-1 transcripts, we found that the digestion conditions usually applied to a β-globin reporter gene transcripts were too mild. S1 nuclease digestion had to be done at elevated temperature and/or higher concentration of S1 nuclease, typically room temperature or 37°C with 150–450 units of S1 nuclease in order to allow for complete degradation of probe oligo not hybridized to MTF-1 mRNA.
o-Nitrophenyl-β-d-galactopyranoside and luciferase assay
Hela cells were transiently transfected with 5 μg of the uORF lacZ fusions and 1 μg of the control luciferase construct pGL3 (Promega) using the calcium phosphate method. For zinc induction, cells were incubated for 4 hours with tissue culture medium at 100 μM ZnSO4.
Forty-eight hours after transfection, cells were scraped, and the resultant pellet from each 10-cm dish was resuspended in 120 μL 100 mM, pH 7.5, Tris-Cl. Cells were lysed by 3 freeze-thaw cycles, frozen in liquid nitrogen for 5 minutes, then thawed at 37°C for 10 minutes. Samples were clarified for 5 minutes by centrifugation, and 30-μL supernatant aliquots were incubated with 500-μL β-Gal buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgCl2) and 100 μL o-nitrophenyl-β-d-galactopyranoside (ONPG) buffer (60 mM Na2HPO4, 40 mM NaH2PO4, and 0.1 M ONPG) at 30°C for 30 minutes, after which the reaction was stopped via addition of 500 μL of 1 M Na2CO3. Absorbance was measured spectrometrically at 420 nm. As an internal reference for transfection efficiency, 30 μL of cell extract was also assayed for luciferase activity (Promega) with a Lumat LB 9507 luminometer (EG&G Berthold).
Synthesis of uORF1 and uORF2 peptides
Upstream open reading frame (uORF) peptides were synthesized with a Perkin Elmer/Applied Biosystems model 433A peptide synthesizer, and purified over Sephadex G-10 column (Dr S. Klauser, Institute of Biochemistry, University of Zürich).
RESULTS
Isolation of the complete mouse MTF-1 gene and comparison of MTF-1 promoters from human and mouse
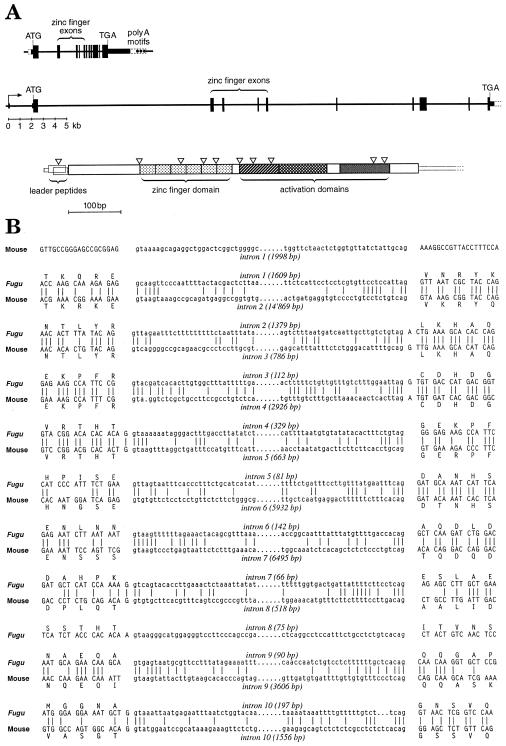
Recently, we isolated and sequenced the entire MTF-1 gene from the pufferfish Fugu rubripes and found the 3′ end of a phosphatase gene (INPP5P) immediately upstream of the MTF-1 gene (Auf der Maur et al 1999). Working under the assumption that in mammals the same gene might precede MTF-1, we designed primer pairs annealing forward from the INPP5P 3′ UTR and backward from the 5′ UTR of human MTF-1. Sequence analysis of the major DNA segment amplified by PCR revealed that it indeed represented the 3′ end of INPP5P, the intergenic region, and the 5′ end of MTF-1. In similar fashion, we also isolated the corresponding intergenic spacer region from the mouse. Additionally, we isolated and sequenced the entire mouse MTF-1 transcription unit (accession number AJ251880). It spans 42 kb, which is 5 times the size of the corresponding gene in the pufferfish (8.5 kb) (Fig 1A). While the overall amino acid identity of mouse and Fugu proteins is 57%, the 6 putative zinc fingers are extremely conserved (94%). In both species, 11 exons were identified, although the 5′ UTR in the mouse is split by an extra intron, and the protein-coding region bears a small deletion that removes a few amino acids plus an intron relative to Fugu in this position. These differences notwithstanding, the splice sites are conserved to the nucleotide between mouse and Fugu (Fig 1B).
Fig 1.
Genomic structure of Fugu and mouse MTF-1 genes. (A) In the pufferfish (on top) and in the mouse, the transcription unit of MTF-1 spans 8.5 and 42 kb, respectively. The exons (black boxes), introns, and spacers (black lines) are drawn to approximate scale and clearly illustrate the compaction of the Fugu MTF-1 gene relative to MTF-1 of the mouse and probably most other vertebrates. Shown underneath the genomic structure is a schematic view of the mouse cDNA with protein reading frames indicated, including the 2 short overlapping uORFs (uORF1 and 2), followed by the MTF-1 ORF with its 6 zinc fingers for DNA binding and 3 activation domains. The position of introns is indicated by inverted open triangles. (B) Comparison of the Fugu and mouse MTF-1 splice junctions. Both genes contain 11 exons; 1 intron and flanking exon sequences are deleted in the mouse relative to the Fugu, while the mouse leader sequence contains an additional intron. Splice junction sequences are conserved to the nucleotide, even if located in a region of low similarity between the 2 genes (notably at intron 7 of Fugu vs intron 8 of the mouse). Accession numbers pufferfish: AJ131394; accession number mouse: AJ251880
MTF-1 promoters of mouse, human, and fish are TATA-less CpG islands
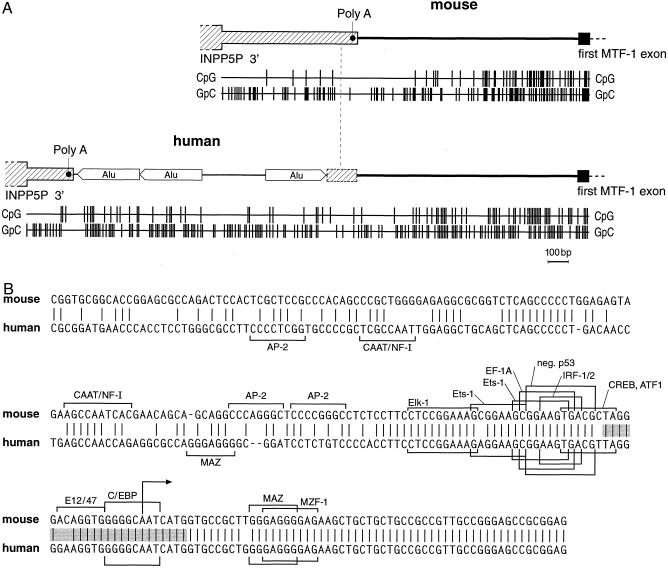
When comparing the intergenic region between INPP5P and MTF-1 of human and mouse, that of human is considerably longer (2.5 kb vs 1.0 kb, respectively). The spacer is poorly conserved between the 2 species and harbors, in human, 3 Alu repeats that account for most of the size difference with the mouse (Fig 2A). For a relatively short stretch of 60 bp upstream of the transcription starts, the MTF-1 promoter/initiator region is very similar between mouse and human (86% identity) but diverges further upstream (Fig 2B). In addition we have determined [G+C] content and CpG dinucleotide distribution in mouse and human. In both species, as in the pufferfish analyzed before (Auf der Maur et al 1999), the MTF-1 promoter displays the typical features of a CpG island, namely a CpG dinucleotide frequency that is approaching or even exceeding the statistically expected value, rather than the depression to about one fifth of expectation typically observed in coding sequences (Fig 2A). In none of the 3 species did the MTF-1 promoter region contain a TATA box, a feature not unusual to promoters of housekeeping genes (Smale and Baltimore 1989).
Fig 2.
Structure of the MTF-1 promoter region. (A) Genomic structure of the last exon of INPP5P, subsequent intergenic segment, and the promoter region of the MTF-1 gene in mouse (accession number AJ251880), and human (accession number AJ251881). In human, the polyadenylation site is shifted upstream relative to the mouse, probably as a consequence of the insertion of 3 Alu repeats. CpG-density and [G + C] content, as represented by the GpC-density, is also indicated by the bar graph below. In mammals (shown here) and pufferfish (Auf der Maur et al 1999), the MTF-1 promoter is embedded in a CpG island. In mouse and human, not only the CpG density but also the overall [G + C] content is increased, typical for warmblooded vertebrates, but not fish. Note that the CpG island features extend quite far upstream of the first MTF-1 exon in mouse and human, yet only about 60 bp of exon-proximal promoter sequences are conserved (Fig 2B). (B) Comparison of mouse and human MTF-1 promoters. While the typical CpG island properties extend further upstream (Fig 2A), the region of strong sequence similarity is confined to the immediate upstream region and the mRNA leader (5′ UTR). Putative transcription factor binding sites, as found using the TRANSFAC database (Heinemeyer et al 1998), are indicated by brackets if present in either 1 or both species. Putative binding sites correspond to the ones of previously characterized transcription factors but have not been verified experimentally for the mouse or human MTF-1 promoters. The hatched region denotes the region of multiple transcription starts, as shown in Figure 4, with the major start indicated by an arrow within the sequence CAATC
Conserved ORFs in the 5′ UTR of mammals but not in fish
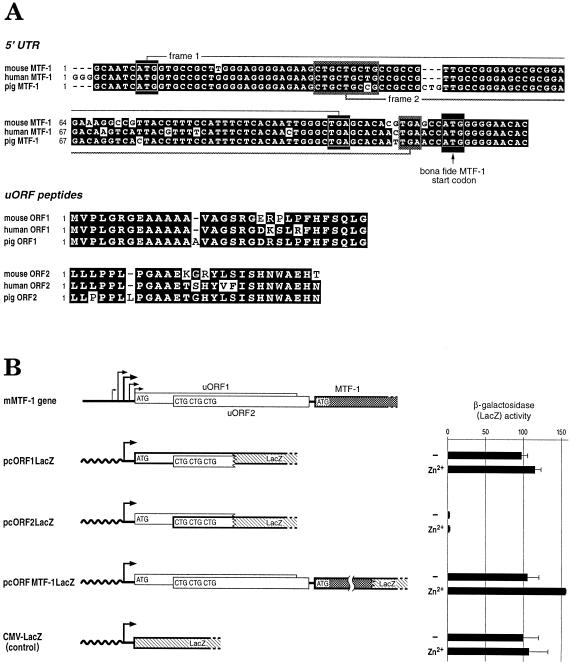
Unlike the less conserved 5′ UTR sequences of other mammalian genes, the 140-nt leader of mouse MTF-1 is highly similar to the 5′ UTR of its human counterpart but not to that of the pufferfish Fugu. We also noted that the leader sequence contains two short overlapping ORFs of 31 and 26 amino acids, termed uORF1 and uORF2, respectively (Fig 3A). The first one, starting very close to the major transcription start sites, begins with a near perfect Kozak sequence AtcAUGG (Kozak 1986), while the second one begins with 3 consecutive CUG triplets. The uORFs and/or additional initiation codons in the 5′ UTR are not uncommon among mammalian mRNAs, notably in those coding for regulatory proteins such as transcription factors and proto-oncogenes (see Discussion).
Fig 3.
Sequence of the 5′ UTR region of the MTF-1 gene. (A) Alignment of mouse, human, and pig MTF-1 5′ UTR sequences and peptide ORFs. Two ORFs (uORF1 and uORF2) are located upstream of the actual MTF-1 ORF. For uORF1, AUG start and TGA stop are demonstrated by white rectangles, and uORF2 triple CUG and TGA by shadowed rectangles. The start codon of the bona fide MTF-1 ORF is marked with an arrow. Shown below is the alignment of uORF1 and 2 peptide sequences. Conserved nucleotides and amino acids are highlighted as white letters on a black background. The 5′ UTR of pig MTF-1 represents an EST (accession number Z84162). (B) Translation initiation at uORF start codons. To assess whether the uORFs are translated, recombinant plasmids were constructed wherein the start codon from either uORF1, uORF2, or hMTF-1 was fused in frame to the β-galactosidase gene ORF. Plasmids pcORF1lacZ, with β-Gal fused with the uORF1 AUG, pcORF2lacZ, with 3 CUGs from uORF2, and pcMTF1ORFlacZ, with AUG of the MTF-1 gene ORF, were separately transfected into HeLa cells. Zinc induction was performed with 100 μM ZnCl2 in the medium for 4 hours. The expression level of β-galactosidase from the start codon of each construct was quantified by the ONPG assay (see Materials and Methods). The expression level of the control CMV-lacZ plasmid is set as 100%. Note that the ORF fused to lacZ is indicated in bold. The standard deviation for the zinc-treated pcORFMTF-1lacZ is so small that the error bar is hardly visible. Shown on top for comparison is the genomic structure of the mouse MTF-1 gene with its multiple transcription starts, and the 2 uORFs (uORF1 and uORF2), followed by the bona fide start of the MTF-1 ORF
We have made the following attempts to determine the function of the uORFs: (1) To test whether the MTF-1 uORFs could be translated at all, each of the 3 ORFs (uORFs 1 and 2 and MTF-1) were fused to the lacZ ORF, and transfected into HeLa cells along with a plasmid encoding the luciferase gene as a transfection reference (Fig 3B). Quantification of β-galactosidase expression by the ONPG assay revealed that the AUG codon of uORF1 was indeed very efficiently utilized, while the triple CUG start yielded very low β-galactosidase activity. The construct where the bona fide ORF of MTF-1 was fused to lacZ was also efficiently used, in spite of the presence of uORF1 and uORF2. (2) The nuclear concentration of MTF-1 protein had previously been shown to increase within a few hours of zinc treatment (Heuchel et al 1994). This increase might have been due to a more efficient translation of MTF-1 mRNA under zinc conditions. Therefore we tested the above constructs, containing or not containing uORFs, in transient transfection assays, and compared resultant β-galactosidase levels of untreated and zinc-treated cells. As shown in Figure 3B we did not observe any strong differences.
(3) Because CUG triplets have been reported to be utilized with reasonable efficiency as translational starts in mammalian cells, in particular under stress conditions (Vagner et al 1996), we investigated whether utilization of the potential CUG start of uORF2 could be boosted by stressors, such as heavy metals, oxidative stress, heat shock, serum starvation, or UV irradiation. However, after transfection of HeLa cells with the uORF2-lacZ fusion constructs, there was only marginal β-galactosidase activity under all conditions tested, indicating that the CUG triplets were poorly used to initiate translation (not shown).
(4) To test whether the mRNA leader could affect cellular concentration/activity of MTF-1, MTF expression constructs with or without peptide-encoding leader sequences were transfected into the 2 mouse MTF-1 knockout cell lines KO1.1 and KO1.9. Irrespective of the presence or absence of zinc, MTF-1 protein concentration was found to be at about the same level, as determined by electrophoretic mobility shift assays (bandshift) with transfected cell lysates (not shown). (5) We also wished to investigate whether 1 of the uORF peptides might regulate transcriptional activation by MTF-1. Human MTF-1 expression clones containing or lacking the uORFs were transfected into the MTF-1 knockout cell line DKO7 (Heuchel et al 1994) along with a β-globin reporter gene under the control of tandem metal-regulated enhancer (MRE) sequences (4× MREd). Reporter gene activity, as quantified by S1 mapping, was the same among all MTF-1 expression clones irrespective of the presence of uORFs (not shown). (6) To see whether the uORF1 or uORF2 peptides affected translation efficiency of MTF-1 mRNA, we had both of the peptides synthesized and added them to a reticulocyte lysate system for cell-free coupled transcription/translation. However, the synthetic peptides, alone or in combination, and tested at various concentrations, had no effect on translation efficiency, as determined by bandshift experiments (not shown). (7) To see whether the peptides affected DNA-binding of MTF-1, a bandshift assay was done in presence of synthetic uORF1 and/or uORF2 peptides in the binding buffer; no effect was observed. (8) Finally, we tested not only transfections but also cell-free transcription reactions of a metal-responsive promoter under the control of MTF-1 in the presence of synthetic uORF1 or 2 peptides. Again no effect on reporter gene expression was observed in S1 mapping experiments. Also, the peptides did not affect cell-free transcription from MTF-1's own promoter (not shown). In spite of all these negative findings, we still consider it likely that the conserved uORFs contribute to some, as yet unknown, functional aspects of MTF-1.
Mapping of the transcript starts of MTF-1
We also performed a number of transcription start mapping experiments of MTF-1 in mouse and human cells. These studies revealed a multitude of scattered transcription initiation sites, upstream and around the putative initiator AUG of uORF1. Such scattered starts are typical for TATA-less promoters and occur in many housekeeping genes (Grosschedl et al 1981; Smale and Baltimore 1989 and references therein). The DNA probe for transcript mapping extended to nucleotide −100 from the main starts (see Materials and Methods for details). The same pattern of multiple transcription starts was observed with transfected MTF-1 promoters and with endogenous MTF-1 transcripts from mouse and human cells, suggesting that the complete information for transcription initiation was embodied within the subcloned promoter segments (Fig 4 and data not shown).
Fig 4.
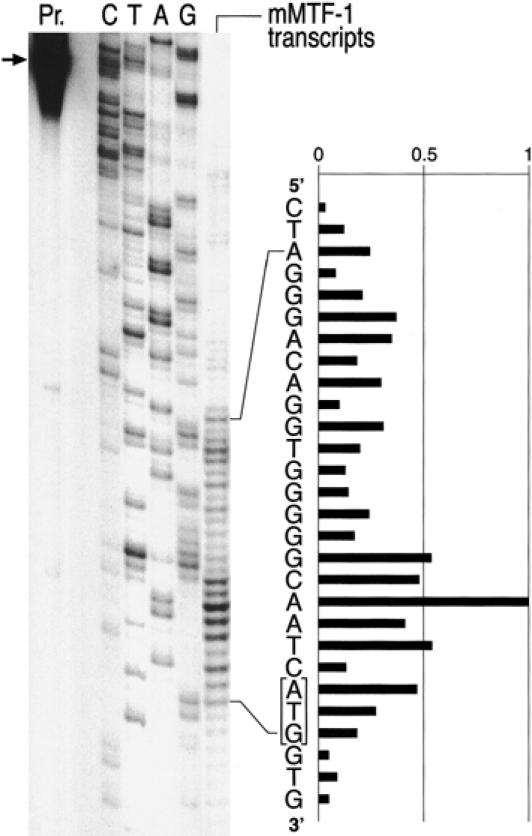
Precise mapping of the multiple transcription start sites in the MTF-1 gene. RNA was isolated from cells and subjected to S1 nuclease mapping with a labeled DNA oligonucleotide corresponding to the entire promoter region of MTF-1. While our β-globin reporter gene under these assay conditions yields a strong, narrowly defined site of transcription initiation (Westin et al 1987, and not shown), the MTF-1 promoter shows the scattered multiple starts characteristic of TATA-less, CpG-island type promoters. At least the longer transcripts are expected to allow for translation initiation from the AUG of uORF1 (framed) (see also Fig 3). Pr., position of input oligonucleotide probe (for details see Materials and Methods); C/T/A/G, sequencing reaction of the MTF-1 promoter region used as reference; “mMTF-1 transcripts” indicates the pattern of protection from S1 nuclease digest after hybridization of cellular mRNA to the probe oligonucleotide
MTF-1 expression is not inducible by heavy metal and other selected stressors
The budding yeast Saccharomyces cerevisiae and the opportunistic pathogenic fungus Candida glabrata harbor very similar heavy metal inducible transcription factors, designated Ace1 (Thiele 1988) and AMT1 (Zhou and Thiele 1991), respectively. Ace1/AMT1 are completely different from MTF-1 and likely evolved separately. While in S. cerevisiae the Ace1 factor gene itself is not stimulated by metal stress, in C. glabrata the AMT1 transcription factor gene is activated by heavy metal load, resulting in a positive feedback. In order to test whether the promoter of MTF-1 is responsive to cellular stress, we tested several stress conditions in transfected mammalian cells and also tested for a possible induction of the endogenous MTF-1 transcripts. Neither zinc, nor cadmium, nor H2O2 significantly elevated the level of MTF-1 transcripts (not shown). These data indicate that MTF-1 is constitutively expressed and that the induction of its activity under heavy metal stress is likely largely post-transcriptional. Indeed, evidence is accumulating that MTF-1 activity is induced at a post-translational level; N. Saydam and W.S., unpublished).
MTF-1 is ubiquitously expressed
In order to assess tissue expression patterns of MTF-1 mRNA and functional protein, we carried out Northern blot and bandshift analyses with a number of mouse tissues. A commercially available, tissue Northern blot was hybridized with a full-length MTF-1 cDNA probe. We observed quite uniform levels of expression of MTF-1 transcripts in all tissues tested, with the exception of testis. Under conditions normalized for poly A+ RNA, analysis of testes revealed at least a 10-fold higher level of MTF-1 transcripts in this tissue (Fig 5). When MTF-1 protein itself was analyzed by bandshift, in all tissue extracts of mice and in all cell types tested so far, we found a strong band characteristic for MTF-1, and again detected extremely high expression levels of MTF-1 in testes (not shown).
Fig 5.
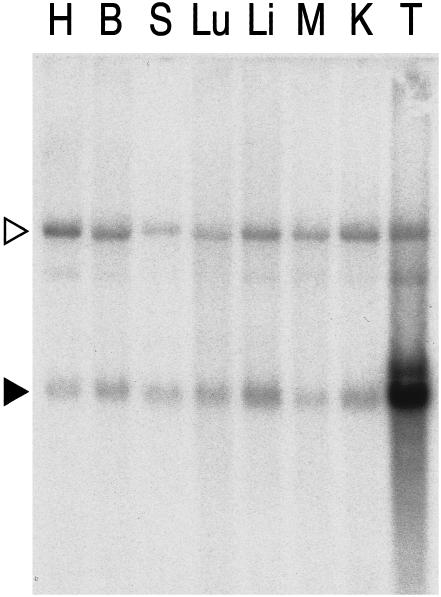
Similar levels of MTF-1 mRNA in all tissues except testes. A tissue Northern blot containing a normalized amount of poly A+ RNA from various mouse organs was hybridized with a cDNA probe of mouse MTF-1. The autoradiograph reveals the presence of MTF-1 mRNA (black triangle) in all tissues: heart (H), brain (B), spleen (S), lung (Lu), liver (Li), skeletal muscle (M), kidney (K), and testes (T). An upper band (open triangle) that was not seen in a previous Northern blot from liver (not shown) was not further investigated. The extremely high representation of MTF transcripts in testicular tissue was confirmed by quantification of protein by electrophoretic mobility shift (not shown). Similar high testicular transcript levels have been previously reported for other components of the RNA Pol II transcriptional apparatus (Schmidt and Schibler 1995)
DISCUSSION
The zinc finger transcription factor MTF-1 is a key player in the response to cellular stress, notably to heavy metal load and to hydrogen peroxide, and works by activating metallothionein and other genes involved in control of cellular redox balance and/or stress response (Westin and Schaffner 1988; Radtke et al 1993; Labbé et al 1993; Heuchel et al 1994; Otsuka et al 1994; Bittel et al 1998; Günes et al 1998; Murphy et al 1999). Here we have determined the promoter sequences for the human and mouse MTF-1 gene and elucidated the complete genomic structure of the mouse MTF-1 locus for purposes of comparison with its counterpart locus in the Japanese pufferfish Fugu rubripes genome, described previously (Auf der Maur et al 1999). The transcription unit of MTF-1 in the mouse spans some 42 kb, compared to 8.5 kb in Fugu. This is yet another illustration of the extreme compaction of the Fugu genome that contains a full set of vertebrate genes but, due to shortening of introns and spacers, is collectively only 13% the size of a typical mammalian genome (Elgar et al 1996). Even though most of the mouse introns are much longer than the corresponding ones of Fugu, we find that the actual splice junctions are conserved to the nucleotide between the 2 species, even in segments where the protein sequence is poorly conserved. Conservation of splice junctions could be most readily explained by selection pressure for a complex pattern of alternative splicing. However, MTF-1 mRNA and protein are expressed in all tissues analyzed so far, without any evidence for developmental or cell type–specific alternative splicing (Fig 5 and not shown). Furthermore, we have argued before that introns, whose sequences can diverge rapidly, are important to prevent homologous recombination between related members of a gene family (Matsuo et al 1993). Again, however, MTF-1 does not fulfill a necessary criterium: the gene encoding it seems to be unique, ie, without further members of a transcription factor family. For these reasons, the high conservation of splice junctions remains enigmatic.
For comparison with mouse and Fugu, the intergenic region preceding the MTF-1 transcription start was isolated from human as well. Not unexpectedly, like in the other species, the same phosphatase gene (INPP5P) was also found to precede the human MTF-1 locus. Curiously, a large portion of the 3′ UTR of the mouse INPP5P gene appears to have been made obsolete during human evolution by the insertion of 3 Alu repeats, with a concomitant upstream shift of the polyadenylation site (Fig 1). An analysis of CpG-density vs overall [G + C] content shows that the intergenic region and leader sequence in both human and mouse possess the typical features of a CpG island. Interestingly, the CpG island extends far upstream of the sequence conserved between human and mouse, suggesting that a high CpG density and [G + C] content are more important than the actual sequence. It might be that the mere features of a CpG island is to ensure an open promoter structure; Cross and Bird (1995) have shown that CpG islands are associated with open, DNAse-sensitive, and hence transcriptionally accessible chromatin. Accordingly, in the pufferfish Fugu, the promoter region is also organized as a CpG island (Auf der Maur et al 1999), but no primary sequence is conserved between mammals and fish whatsoever. Both mouse and human, but not fish, 5′ UTRs contain 2 conserved short reading frames (uORFs). Our studies indicate that in spite of a very short leader sequence, at least uORF1 with a near perfect Kozak rule initiation context is likely to be expressed. Upstream ORFs or additional start codons in frame with the bona fide protein are particularly frequent in genes controlling cell growth, including transcription factors and kinases. A common denominator of such upstream initiation sites seems to be a downregulation of the translation efficiency of the bona fide ORF, perhaps by feedback mechnisms (Geballe 1996; Brown et al 1999). A well-characterized example is the transcription factor GCN4 in yeast that contains no less than 4 uORFs in the leader that can be bypassed under conditions of amino acid starvation to allow for more efficient translation of GCN4 (Grant et al 1995). Another case is represented by the mammalian AdoMetDC gene transcript, where translation of a 7-amino acid leader peptide has been shown to downregulate translation from the downstream ORF for the AdoMetDC enzyme (Hill and Morris 1992). In our case, however, several experiments have not yielded any evidence for an effect of the upstream ORFs on MTF-1 concentration or function, neither in a positive nor negative way. As mentioned above, fungal metallothionein genes are activated by a copper-containing transcription factor unrelated to MTF-1. In the pathogenic yeast C. glabrata, AMT1 is subject to positive autoregulation by an MRE in its promoter, while in the budding yeast S. cerevisiae, the closely related ACE1 factor is constitutively expressed. The latter situation seems be the case for MTF-1. First, we found no signature MRE motifs in its promoter. Second, neither transfection of the transcription factor MTF-1, or Sp1, nor various conditions of stress such as heavy metals or heat resulted in any appreciable up- or downregulation of a reporter gene driven by the MTF-1 promoter. Thus, MTF-1 seems to be ubiquitously and constitutively expressed (see also below). Curiously, unlike many CpG island promoters, the MTF-1 promoter also does not contain binding sites for the Sp1 transcription factor, and accordingly transfection of an Sp1 expression vector had no effect on transcription from an MTF-1 promoter (not shown).
MTF-1 is a key player in the cellular stress response, notably to heavy metals, but also to oxidative stress. Its target genes include the stress-induced metallothioneins MT-I and MT-II and γ-GCS, an enzyme essential for glutathione synthesis (Günes et al 1998). It is therefore not unexpected that MTF-1 was found to be expressed in all cell lines tested by us (not shown) and in all tissues assayed by Northern blot analysis (Fig 5). The exorbitant level of MTF-1 gene transcripts in testes also holds true for at least some other components of the transcription apparatus: Schmidt and Schibler (1995) have found that the TATA box–associated protein TBP and also TFIIB and the pol II machinery are all very highly expressed in spermatocyte precursor cells. It was argued that this overexpression ensures heavy duty transcription throughout spermatogenesis, even into the stages of sperm maturation. Nevertheless, the very strong MTF-1 expression in testes may well serve a specific function, because testes from sexually mature mice contain 10-fold higher mRNA levels for MT-I and -II than adult liver (De et al 1991).
Knockout of the murine MTF-1 results in embryonic lethality at day E13, with liver decay as the primary defect (Günes et al 1998). So far, it has not been possible to rescue MTF-1 knockout mice with a liver-specific promoter activity, while rescue was possible with a ubiquitous expression pattern (P.L., T. Rülicke, and W.S., unpublished). Even though MTF-1 seems not essential for the growth of fibroblasts and for the differentiation into a few basal neural cell types, it may well fulfill an essential role in cellular stress response in many tissues other than the liver.
Acknowledgments
We are indebted to Dr Lee Martin for critical reading of the manuscript, to Fritz Ochsenbein for the preparation of the diagrams and figures, to Oliver K. Clay (Marine Biology Station “Anton Dohrn,” Naples) for his support in Biocomputing, and to Drs Charles Weissmann, Richard Eckner, and Werner Lutz for providing the mouse genomic DNA library. This work was supported by the Schweizerischer Nationalfonds and by the Kanton Zürich.
REFERENCES
- Auf der Maur A, Belser T, Elgar G, Georgiev O, Schaffner W. Characterization of the transcription factor MTF-1 from the Japanese pufferfish (Fugu rubripes) reveals evolutionary conservation of heavy metal stress response. Biol Chem. 1999;380:175–185. doi: 10.1515/BC.1999.026. [DOI] [PubMed] [Google Scholar]
- Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NK-kappa B. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- Bittel D, Dalton T, Samson SL, Gedamu L, Andrews GK. The DNA binding activity of metal response element-binding transcription factor-1 is activated in vivo and in vitro by zinc, but not by other transition metals. J Biol Chem. 1998;273:7127–7133. doi: 10.1074/jbc.273.12.7127. [DOI] [PubMed] [Google Scholar]
- Brown CY, Mize GJ, Pineda M, George DL, Morris DR. Role of two upstream open reading frames in the translational control of oncogene mdm2. Oncogene. 1999;18:5631–5637. doi: 10.1038/sj.onc.1202949. [DOI] [PubMed] [Google Scholar]
- Cross SH, Bird AP. CpG islands and genes. Curr Opin Genet Dev. 1995;5:309–314. doi: 10.1016/0959-437x(95)80044-1. [DOI] [PubMed] [Google Scholar]
- De SK, Enders GC, Andrews GK. High levels of metallothionein messenger RNAs in male germ cells of adult mouse. Mol Endocrinol. 1991;5:628–636. doi: 10.1210/mend-5-5-628. [DOI] [PubMed] [Google Scholar]
- Elgar G, Sandford R, Aparicio S, Macrae A, Venkatesh B, Brenner S. Small is beautiful: comparative genomics with the pufferfish (Fugu rubripes) Trends Genet. 1996;12:145–150. doi: 10.1016/0168-9525(96)10018-4. [DOI] [PubMed] [Google Scholar]
- Geballe AP. 1996 Translational control mediated by upstream AUG codons. In: Translational Control, ed Hershey JWB, Mathews MB, Sonenberg N. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 173–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CM, Miller PF, Hinnebusch AG. Sequences 5′ of the first upstream open reading frame of GCN4 mRNA are required for efficient translational reinitiation. Nucleic Acids Res. 1995;23:3980–3988. doi: 10.1093/nar/23.19.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosschedl R, Wasylyk B, Chambon P, Birnstiel ML. Point mutation in the TATA box curtails expression of sea urchin H2A histone gene in vivo. Nature. 1981;294:178–180. doi: 10.1038/294178a0. [DOI] [PubMed] [Google Scholar]
- Günes C, Heuchel R, Georgiev O, et al. Embryonic lethality and liver degeneration in mice lacking the metal-responsive transcriptional activator MTF-1. EMBO J. 1998;15:2846–2854. doi: 10.1093/emboj/17.10.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer T, Wingender E, Reuter I, et al. Databases on transcriptional regulation: TRANSFAC, TRRD, and COMPEL. Nucleic Acids Res. 1998;26:264–370. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuchel R, Radtke F, Georgiev O, Stark G, Aguet M, Schaffner W. The transcription factor MTF-1 is essential for basal and heavy metal-induced metallothionein gene expression. EMBO J. 1994;13:2870–2875. doi: 10.1002/j.1460-2075.1994.tb06581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuchel R, Radtke F, and Schaffner W. 1995 Transcriptional regulation by heavy metals, exemplified at the metallothionein genes. In: Inducible Gene Expression, vol 1, ed Baeuerle PA. Birkhäuser, Boston, 206–240. [Google Scholar]
- Hilberg F, Aguzzi A, Howells N, Wagner EF. C-jun is essential for normal mouse development and hepatogenesis. Nature. 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- Hill JR, Morris DR. Cell-specific translation of S-adenosylmethionine decarboxylase mRNA regulation by the 5′ transcript leader. J Biol Chem. 1992;267:21886–21893. [PubMed] [Google Scholar]
- Kägi JHR, Kojima Y. Chemistry and biochemistry of metallothionein. Experientia Suppl. 1987:25–61. doi: 10.1007/978-3-0348-6784-9_3. [DOI] [PubMed] [Google Scholar]
- Kemler I, Bucher E, Seipel K, Müller-Immerglück MM, Schaffner W. Promoters with the octamer DNA motif (ATGCAAAT) can be ubiquitous or cell type-specific depending on binding affinity of the octamer site and Oct-factor concentration. Nucleic Acids Res. 1991;19:237–242. doi: 10.1093/nar/19.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eucaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- Labbé S, Larouche L, Mailhot D, Séguin C. Purification of mouse MEP-1, a nuclear protein which binds to the metal regulatory elements of genes encoding metallothionein. Nucleic Acids Res. 1993;21:1549–1554. doi: 10.1093/nar/21.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtlen P, Georgiev O, Schaffner W, Aguzzi A, Brandner S. The heavy metal-responsive transcription factor-1 (MTF-1) is not required for neural differentiation. Biol Chem. 1999;380:711–715. doi: 10.1515/BC.1999.089. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Clay O, Takahashi T, Silke J, Schaffner W. Evidence for erosion of mouse CpG islands during mammalian evolution. Somat Cell Mol Genet. 1993;19:543–555. doi: 10.1007/BF01233381. [DOI] [PubMed] [Google Scholar]
- Murphy BJ, Andrews GK, Bittel D, et al. Activation of metallothionein gene expression by hypoxia involves metal responsive elements and metal transcription factor-1. Cancer Res. 1999;59:1315–1322. [PubMed] [Google Scholar]
- Otsuka F, Iwamatsu A, Suzuki K, Ohsawa M, Hamer DH, Koizumi S. Purification and characterization of a protein that binds to metal responsive elements of the human metallothionein IIA gene. J Biol Chem. 1994;269:23700–23707. [PubMed] [Google Scholar]
- Radtke F, Heuchel R, Georgiev O, Hergersberg M, Gariglio M, Dembic Z, Schaffner W. Cloned transcription factor MTF-1 activates the mouse metallothionein-I promoter. EMBO J. 1993;12:1355–1362. doi: 10.1002/j.1460-2075.1993.tb05780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, and Maniatis T. 1989 Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Schmidt EE, Schibler U. High accumulation of components of the RNA polymerase II transcription machinery in rodent spermatids. Development. 1995;121:2373–2383. doi: 10.1242/dev.121.8.2373. [DOI] [PubMed] [Google Scholar]
- Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Thiele DJ. ACE1 regulates expression of the Saccharomyces cerevisiae metallothionein genes. Mol Cell Biol. 1988;8:2745–2752. doi: 10.1128/mcb.8.7.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner S, Touriol C, Galy B, et al. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J Cell Biol. 1996;135:1391–1402. doi: 10.1083/jcb.135.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin G, Gerster T, Müller MM, Schaffner G, Schaffner W. OVEC, a verstaile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 1987;15:6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westin G, Schaffner W. A zinc-responsive factor interacts with a metal-regulated enhancer element (MRE) of the mouse metallothionein-I gene. EMBO J. 1988;7:3763–3770. doi: 10.1002/j.1460-2075.1988.tb03260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Thiele DJ. Isolation of a metal-activated transcription factor gene from Candida glabrata by complementation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:6112–6116. doi: 10.1073/pnas.88.14.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]