Abstract
Multichannel electroencephalography (EEG) offers a non-invasive tool to explore spatio-temporal dynamics of brain activity. With EEG recordings consisting of multiple trials, traditional signal processing approaches that ignore inter-trial variability in the data may fail to accurately estimate the underlying spatio-temporal brain patterns. Moreover, precise characterization of such inter-trial variability per se can be of high scientific value in establishing the relationship between brain activity and behavior. In this paper, a statistical modeling framework is introduced for learning spatiotemporal decomposition of multiple-trial EEG data recorded under two contrasting experimental conditions. By modeling the variance of source signals as random variables varying across trials, the proposed two-stage hierarchical Bayesian model is able to capture inter-trial amplitude variability in the data in a sparse way where a parsimonious representation of the data can be obtained. A variational Bayesian (VB) algorithm is developed for statistical inference of the hierarchical model. The efficacy of the proposed modeling framework is validated with the analysis of both synthetic and real EEG data. In the simulation study we show that even at low signal-to-noise ratios our approach is able to recover with high precision the underlying spatiotemporal patterns and the evolution of source amplitude across trials; on two brain-computer interface (BCI) data sets we show that our VB algorithm can extract physiologically meaningful spatio-temporal patterns and make more accurate predictions than other two widely used algorithms: the common spatial patterns (CSP) algorithm and the Infomax algorithm for independent component analysis (ICA). The results demonstrate that our statistical modeling framework can serve as a powerful tool for extracting brain patterns, characterizing trial-to-trial brain dynamics, and decoding brain states by exploiting useful structures in the data.
Keywords: Hierarchical Bayesian, Variational Bayesian, Common spatial patterns, Spatio-temporal decomposition, Inter-trial variability, Sparse learning, Brain-computer interface
1. Introduction
Thanks to advances in data recording technologies, the past decades have witnessed widespread applications of multichannel electroencephalographs (EEG) in neuroscience studies to probe into the working mechanisms of the human brain (Ray and Cole, 1985; Miltner et al., 1999; Makeig et al., 2002), as well as in clinical applications to monitor brain states (Rampil, 1998) and assist the diagnosis of neurological abnormalities (Cichocki et al., 2005). In recent years, EEG has also been widely used in emerging fields such as neural engineering (Wolpaw et al., 2002) and neuromarketing (McClure et al., 2004) for decoding brain activity. Compared to microscopic recordings that measure the activities of only single neuron or a group of nearby neurons with a spatial scale of at most millimeters (Wilson and McNaughton, 1993), multichannel EEG has the advantage of being able to map the macroscopic dynamics across the whole brain, albeit indirectly from the scalp, with a high temporal resolution of milliseconds. On the other hand, in order to gain insights into brain function for answering relevant scientific questions or for practical purposes, it is crucial to link the spatio-temporal dynamics of EEG to the underlying neurophysiological processes or behavioral changes. However, due to volume conduction, scalp EEG hardly preserves the fidelity of the original brain dynamics, which often renders its interpretation difficult (Baillet et al., 2001). In particular, functionally distinct brain activities that may well be separated in the brain are mixed up in EEG in a simultaneous and linear manner1, leading to substantially distorted signals with high correlations between spatially adjacent data channels. The situation is made even worse by the contamination from various artifacts such as electrocardiogram (ECG), electromyogram (EMG), and electrooculogram (EOG).
A major challenge, therefore, is to decompose multichannel EEG signals into a set of source signals that represent functionally independent processes. Each source signal is associated with a spatial pattern (SP), i.e., its activation map on the scalp, which is assumed to be fixed across time under the same experimental condition. The SP reflects the spatial geometry of the source signal and thus may have important functional significance. A vast range of approaches have been proposed to perform spatio-temporal decomposition of EEG data (Parra et al., 2005). Early approaches include principal component analysis (PCA) and factor analysis (Koles et al., 1995; Lagerlund et al., 1997). More recently, the field of blind source separation (BSS) has been dedicated to similar purposes (Hyvarinen et al., 2001; Cichocki and Amari, 2002; Vigário and Oja, 2008). One unsupervised BSS methodology that has proven to be highly successful in EEG signal processing is independent component analysis (ICA) (Makeig et al., 2002), in which the non-Gaussianity of source signals is maximized. Successful biomedical applications of ICA include analysis of event-related dynamics (Makeig et al., 1997, 2002), artifact identification and removal (Vigário, 1997), and brain-computer interfaces (BCIs) (Kachenoura et al., 2008).
Despite the apparent proliferation of methods for spatio-temporal decomposition of EEG, our perspective is that there are two useful structures in EEG data yet to be fully utilized in developing new signal processing approaches. The first is the multiple-trial structure. Within an experiment it is often the case that each condition may be repeated for many trials. The inter-trial amplitude variability is a common phenomenon in part because the brain as a dynamical system, its state is constantly changing over time. Well-known examples of trial-to-trial fluctuations in EEG recordings include the habituation effects (Bruin et al., 2000), the P300 effects (Klimesch, 1999) and the event-related desynchronization/synchronization (ERD/ERS) effects (Pfurtscheller and Aranibar, 1977) (see Figure ?? for an illustration). As will be shown in this paper, ignoring inter-trial amplitude variability may result in inaccuracy in identifying the underlying spatiotemporal patterns. Furthermore, accurate characterization of inter-trial amplitude variability in the brain activity may be of high importance per se, e.g., in studies that examine the relationship between human brain activity and variability in behavior (Ergenoglu et al., 2004; Fox et al., 2007). Albeit relatively well-recognized in the channel space, to the best of our knowledge inter-trial amplitude variability has been hitherto considered by few signal processing approaches for spatio-temporal modeling of multichannel EEG data. A few studies of this line are aimed at solving the EEG inverse problem (Friston et al., 2006; Limpiti et al., 2009), which is different from our current setting, where the structural information of the brain is unavailable.
The second structure that has been largely ignored is the multiple-condition structure of EEG within an experiment. This structure may turn out to be highly useful if we have the knowledge that the EEG data recorded under different conditions share certain commonalities in their spatio-temporal patterns. Nonetheless, most conventional signal processing approaches (e.g., factor analysis and ICA) by design are only able to handle one condition at a time and thus their application to multiple-condition EEG data does not seem to be straightforward. To proceed, in data analysis they are either employed to deal with each condition separately, or simply applied to the entire data consisting of both conditions, which is problematic in theory as it violates the stationarity assumption of the basic models underlying these approaches without proper model extensions. Development of appropriate statistical models to take into consideration the non-stationarity and shared information, if any, between conditions may make more efficient use of the data and hence could potentially yield findings that are unable to be obtained using conventional approaches.
Motivated by the two abovementioned useful structures in EEG signals, in this paper we cast the problem of learning spatio-temporal decomposition of multichannel EEG data into a statistical modeling setting. Without loss of generality, we focus on the EEG data that are recorded under two experimental conditions, each presented over multiple trials. A two-stage hierarchical Bayesian model is developed to take account of both the aforementioned multiple-condition structure and inter-trial amplitude variability in the EEG data. Here amplitude refers to the standard deviation of each source signal at each trial. The strength of the hierarchical modeling lies in that it endows the variance of each source signal with a second-stage distribution to model its evolution across trials. For the purpose of inferring the hierarchical model, we derive a variational Bayesian learning algorithm, which enables us not only to obtain posterior distributions of the model parameters but also to automatically infer the model size (i.e., source number) via sparse learning.
The paper is organized as follows. Section 2.1 presents and elaborates the proposed hierarchical Bayesian model. Section 2.2 introduces the variational Bayesian algorithm for model inference. Section 3 demonstrates the efficacy of the proposed modeling framework using both simulated and real data experiments. Discussions and concluding comments are given in Section 4.
The notation used in this paper is listed in Table 1. Note that a few symbols might be slightly abused depending on the context. We will redefine them where necessary.
Table 1.
List of Notation
| Symbol | Definition | |
|---|---|---|
| k | index for experimental conditions | |
| i | index for trials | |
| j | index for sample points in each trial | |
| c | index for data channels | |
| m | index for source signals | |
| Nk | number of trials for each condition | |
| Jk | number of sample points in each trial | |
| C | number of data channels | |
| M | number of source signals | |
| A | mixing matrix | |
| Â | estimated mixing matrix | |
| am | the m-th column of A | |
| ãc | the transpose of the c-th row of A | |
| α(m) | precision parameter for am | |
| multichannel EEG signals at the j-th sample for condition k | ||
| source signals at the j-th sample for condition k | ||
| additive noise component at the j-th sample for condition k | ||
| Λk | source covariance matrix for condition k, with the variance for source m being | |
| Ψk | noise covariance matrix for condition k, with the variance for channel c being | |
| ℝn | real n-dimensional vectors | |
| ℝm×n | real m × n matrices | |
| Γ(y) | gamma function (y > 0) | |
| Ϝ (y) | digamma function defined as | |
| 𝒩(µ, Σ) | multivariate Gaussian distribution with mean µ and covariance Σ | |
| 𝒢a(a, b) | gamma distribution defined as | |
| 𝒬 | subspace of probability distributions | |
| q* | variational distribution | |
| ℒ(p) | variational lower bound as a functional of probability distribution p | |
| Lmax | maximal value of the log-likelihood function for an estimated model | |
| D(p ‖ q) | Kullback-Leibler (KL) divergence between probability distribution p and q | |
| 〈·〉p | mathematical expectation with respect to probability distribution p | |
| B−1 | inverse of matrix B | |
| BT | transpose of matrix B | |
| I | identity matrix | |
| tr(B) | trace of matrix B | |
| |B| | determinant of matrix B | |
| d(B, C) | Amari index between matrix A and matrix B | |
| diag(b) | diagonal matrix with vector b as diagonal entries | |
| diag(B) | diagonal matrix with diagonal entries identical to those of matrix B | |
| ‖ b ‖2 | l2 norm of vector b | |
| ln | natural logarithm function | |
| Const | constant | |
| i.i.d. | independent and identically distributed |
2. Methods
2.1. A Hierarchical Bayesian Spatio-Temporal Model for EEG
Given an EEG data set recorded under two conditions, (i, j, k are indices for trials, sample points in each trial, and experimental conditions, respectively. k = 1, 2; i = 1, …, Nk; j = 1, …, Jk), the two-stage hierarchical Bayesian model can be constructed as follows:
| (1) |
where without loss of generality we assume that observations are adjusted to have zero mean, and that M ≤ C, i.e., that there are no more sources than EEG channels.
2.1.1. First stage: modeling multiple-condition structure
The first stage of model (1) essentially consists of two factor-analysis models, with each giving a linear spatio-temporal decomposition of the data from one condition. Each factor-analysis model specifies for condition k the probability distribution of observations conditioned on parameters , which are assumed as random variables in the Bayesian framework. Here are diagonal covariance matrices2, implying that the source signals, namely , are mutually independent, and that the additive noise components, namely , are independent among channels. Ψ1 and Ψ2 are allowed to be non-isotropic to permit noise variance to differ across channels. For simplicity, it is also assumed that are i.i.d. across time.
To utilize the multiple-condition structure in EEG data, a key modeling assumption at the first-stage model is that the mixing matrix A, which contains the SPs as the columns, is identical for both conditions. The assumption is reasonable in a broad range of situations; since by designing experiments involving two contrasting conditions, experimenters often hope to discover differences in spatio-temporal patterns of brain activities between conditions, and the differences are typically observed by first fixing an SP and then comparing the source signals associated with this SP in both conditions. Furthermore, from an estimation standpoint the assumption of identical A allows us to integrate information from both conditions to improve the estimation accuracy.
2.1.2. Second stage: modeling inter-trial amplitude variability
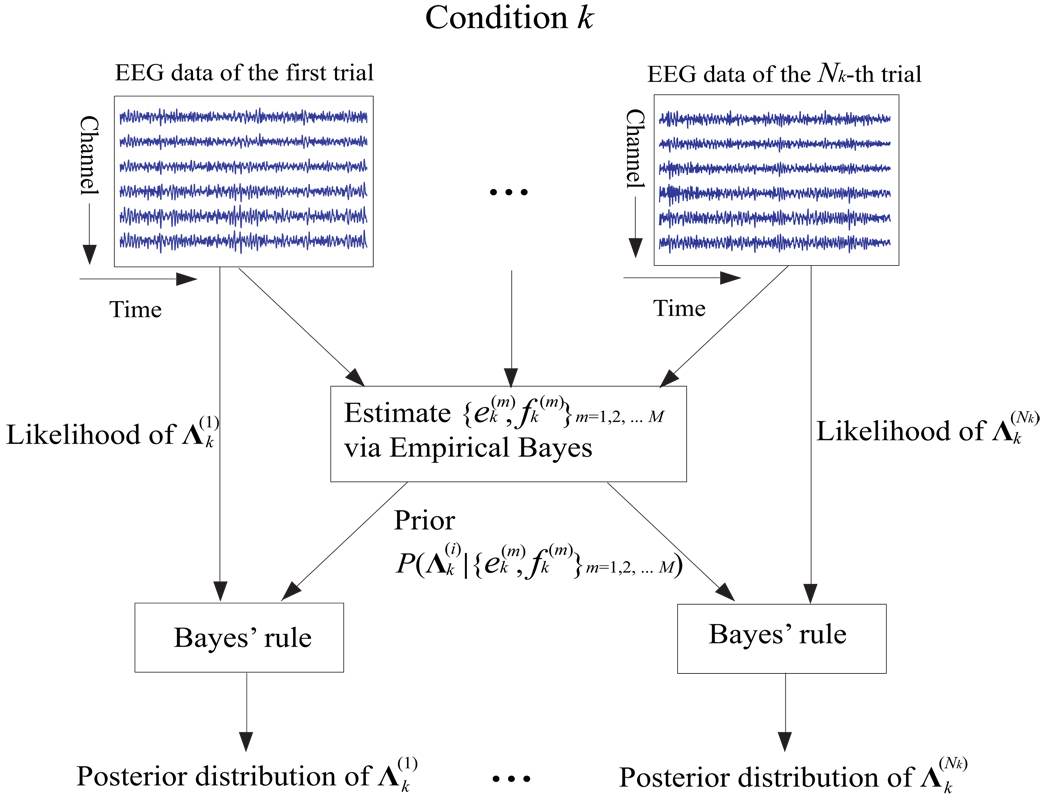
The second stage of model (1) specifies for condition k the prior probability distribution of source covariance conditioned on hyperparameters . In other words, the second-stage model assumes that within a condition each source signal’s variance across trials is multiple draws from a common unknown inverse-gamma distribution, which has two important consequences. First, each source signal’s variance is allowed to vary from trial to trial within a condition. Second, the variation is nonetheless structured rather than arbitrary since it is known a priori that for each condition the source signal’s variance across trials follows an inverse-gamma distribution parameterized by , which are to be estimated by pooling data of all trials within the condition (refer to Section 2.2 for details). As such, the second-stage model provides a useful way of sharing information among multiple trials. Figure 1 offers an intuitive illustration of how the second-stage model allows each trial to borrow strength from one another in the inference of the posterior distribution of the source signal’s covariance within the trial. We emphasize here that although the actual variability may not restrict to the variance of source signals, such simplification is especially appropriate for modeling modulations in the power of ongoing oscillatory EEG activity (Pfurtscheller and Aranibar, 1977).
Figure 1.
An illustration of how the second-stage model allows information to be shared among trials for condition k. The hyperparameters in the prior of are estimated via empirical Bayes (described in Section 2.2) by pooling information from all trials. For the i-th trial, the Bayes’ rule then combines the evidence from single-trial data (likelihood) with the prior information to yield the posterior distribution of .
To ensure the uniqueness of model inference, in model (1) we avoid the issue of scaling indeterminancy (i.e., any column of A may be scaled by a non-zero scalar as long as the corresponding columns in are multiplied by the inverse value) by constraining
| (2) |
where is the prior mean of the variance of the m-th source signal for condition k. The spatial patterns corresponding to different source signals are now placed on the same scale to be able to be compared with each other.
2.1.3. Sparse learning of the source number
To facilitate computation, in model (1) we assume conjugate gamma priors for precisions . In specifying the prior distribution for the mixing matrix A, the idea of automatic relevance determination (ARD) (MacKay, 1992) comes into play for determining the number of source signals: at the outset, without any a priori knowledge regarding the true source number we simply assume the full model (i.e., M = C). Each column of A, denoted by am, is then assigned an associated precision parameter α(m) to control its magnitude/relevance, where α(m) is again endowed with a conjugate gamma prior. The zero mean assumption for each element of A is appropriate when the elements are allowed to take both positive and negative values. After model inference, if the posterior distribution of α(m) turns out to be concentrating upon large values, am in the mixing matrix would essentially be switched off, with only the relevant columns remaining. Thus, α = [α(1), …, α(M)]T can be viewed as the index for models of varied complexities, and its posterior reflects the belief of each model generating the observed data. Model averaging can then be effectively achieved by marginalizing over α when computing the posterior of other variables of interest.
Further insights can be gained into ARD by observing that the prior distribution placed on A encourages the sparsity of the number of source signals since the marginal distribution of am by integrating out αm yields a Student-t distribution (Andrews and Mallows, 1974)
| (3) |
which has heavy tails. The Student-t prior encompasses the well-known Gaussian-Jeffreys prior as a special case when u(m) → 0 and v(m) → 0 (recall that being the parameters of a gamma distribution, u(m) and v(m) cannot attain exact zero), and in this case it is sharply peaked at zero, a hallmark of sparsity-inducing priors. The Jeffereys hyperprior for α(m) is noninformative in the sense that it is scale-invariant (Robert, 2007). By setting u(m) and v(m) to values close to zero, there is no need for tuning of hyperparameters to control the degree of sparsity in our model.
In light of the analysis in (Wipf and Rao, 2007), using an independent Student-t prior for each column of A in model (1) has the effect of enabling significant posterior mass of A to be centering on matrices with as many zero columns as possible. It is noteworthy that this idea of learning sparse representations of data by shrinking the elements within each group (in this paper each column of A) in a collective manner is similar in spirit to many popular algorithms developed for simultaneous sparse learning, e.g., group LASSO (Yuan and Lin, 2007) and M-FOCUSS (Cotter et al., 2005).
2.1.4. More insight into model (1)
The following remarks examine model (1) more closely:
The linearity of the first-stage model complies well with the physical process of volume conduction. Moreover, the assumption of an identical A resolves the issue of rotational indeterminancy that is inherent in the standard factor-analysis model (Anderson, 2003), thus rendering the spatio-temporal decomposition in model (1) unique. Indeed, a rotation of A can no longer be offset by changing all identically without altering their diagonality.
The Gaussianity assumption of source signals conditioned on each condition and trial3 is suited to modeling mildly amplitude-modulated oscillatory activities as their kurtoses are close to zero (Hyväarinen et al., 2010), a hallmark of Gaussian random variables. Spontaneous EEG signals as well as induced reponses in EEG are typically made up of such amplitude-modulated oscillatory activities across multiple frequency bands, each attributed to a different underlying physiological process (Niedermeyer and da Silva, 2004). In this case, the Gaussian distribution is a valid description of our stage of knowledge.
- In the case when there is merely a single trial of EEG recorded for each condition, by performing the maximum likelihood estimation of the following model, which is the first-stage model with all parameters viewed as fixed:
an interesting connection with the common spatial patterns (CSP) algorithm (reviewed in Appendix A, also known as Fukunaga-Koontz transform (FKT) (Fukunaga and Koontz, 1970) in the pattern recognition community) would emerge, as formalized by the following theorem (Parra and Sajda, 2003; Blankertz et al., 2008; Wu et al., 2009; Gouy-Pailler et al., 2010):(4)
Theorem 1. Let  be the ML estimate of A in model (4). The transformation matrix W in the CSP algorithm is equal to Â−T if the following three additional assumptions hold: (i) the additive noise vanishes to zero; (ii) A is a non-singular square matrix; (iii) for two distinct sources m and n (m ≠ n), their variance ratios are not equal in both conditions, i.e., .
The proof of the theorem is provided in Appendix B. Therefore, despite that CSP optimizes a discriminative criterion, Theorem 1 offers a generative perspective in terms of its formulation, which not only gives insights into the algorithm but also opens up the possibility of its further performance improvement (Wu et al., 2009). In particular, in the case when data is in fact corrupted by additive noise and the source number is significantly lower than the channel number, in estimation the noise-free model assumed by CSP may yield a substantial number of spurious sources to fit the additive noise. These spurious sources not only do not lend themselves to easy interpretation but also lower the estimation accuracy (see Section 3). In other words, CSP is prone to overfitting noisy observations due to under-constrained estimation (Hill et al., 2007; Blankertz et al., 2008). A robust noise model was introduced in (Wu et al., 2009) to extend the CSP model to address the issues of both additive noise and outliers.
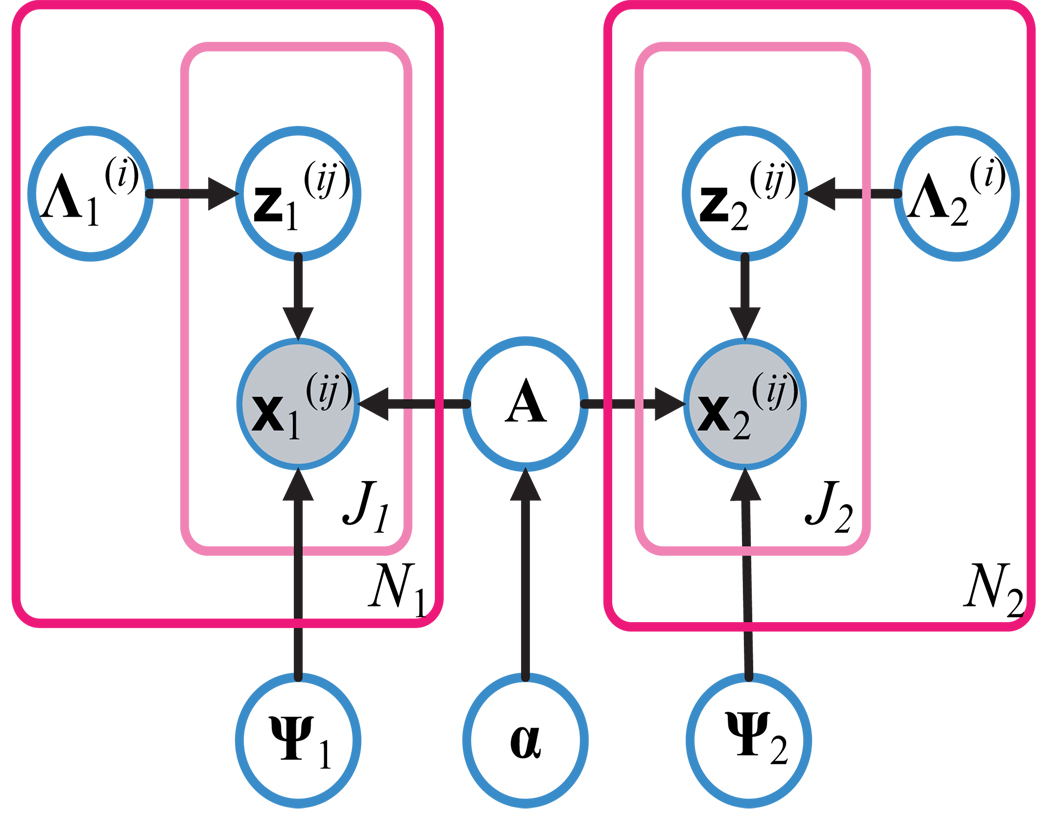
The probabilistic relationships between the variables in model (1) are shown in Figure 2 as a graphical model (Koller and Friedman, 2009).
Figure 2.
A graphical model representation of the hierarchical model (1), with arrows indicating statistical dependencies. The outer plate represents the multiple-trial data set with Nk trials for condition k (k = 1, 2), where the trial index is given by i; the inner plate represents single-trial data within the data set with Jk observations for condition k, and the observation index is given by j.
2.2. The Variational Bayesian (VB) Algorithm for Model Inference
The goal of Bayesian estimation for model (1) is to compute the posterior distribution of relevant variables using Bayes’ rule. Nonetheless, the computation of the posteriors is analytically intractable due to the marginalization operation involved in the Bayesian inference of model (1). We adopt the variational method for approximate Bayesian inference in our work as it permits fast computation and enables us to glean intuition as well as insight into the results. The VB inference is also invariant to re-parameterization of the model (MacKay, 2003).
Let Xk denote collectively all the EEG data recorded from condition k, and similarly Zk denote collectively TPs of all the sources for condition k. The key idea of VB is to seek a “simple” distribution q*, termed variational distribution, from a structured subspace of probability distributions 𝒬 to achieve the minimum Kullback-Leibler (KL) divergence between the distributions from 𝒬 and the true posterior distribution:
where . Thus, VB reformulates Bayeisian inference into an optimization problem. However, direct computation of D(q ‖ p) is again intractable because it depends on p(Θ|X1, X2), which is exactly what we seek to approximate. Fortunately, it turns out that minimizing D(q ‖ p) is equivalent to maximizing a functional ℒ thanks to the following decomposition of the log marginal probability of the data (MacKay, 2003):
where
| (5) |
Hence we can work with ℒ(q) without altering the structure of the search space of 𝒬. For the choice of 𝒬, we adopt the so-called mean-field approximation, assuming that the distributions in 𝒬 can be factorized over the elements of Θ, namely,
Given the structure of the hierarchical model (1), the lower bound ℒ in (5) can be maximized with respect to q(Θ) via a coordinate ascent optimization technique: in each iteration only the variational posterior of one variable in Θ is updated, while the variational posteriors of other variables are fixed; the update then cycles through each variable in Θ iteratively until convergence. The derivation of the update equations for the variational posteriors is provided in detail in Appendix C. The update equations for the variational posteriors are
| (6) |
where ãc denotes the transpose of the c-th row of A, and . The parameters used in (6) are given by
| (7) |
| (8) |
| (9) |
| (10) |
| (11) |
| (12) |
where 〈·〉p denotes the mathematical expectation with respect to probability distribution p. After each iteration, the lower bound ℒ can be evaluated to check the correctness of the update (with each iteration ℒ must be non-decreasing) and the convergence of the algorithm. The derivation and the form of ℒ can be found in Appendix D.
2.2.1. Learning the hyperparameters in model (1)
The hyperparameters in the model also need to be determined. In the current work, we use the nonin-formative Jeffreys prior for and α(m) by setting to a constant that is close to zero, e.g. 10−8. By contrast, for the purpose of sharing information among trials, we employ the method of empirical Bayes (Carlin and Louis, 2000) to obtain the point estimates of by maximizing the marginal likelihood of the EEG data, namely, . However, since the marginal likelihood is difficult to evaluate for the same reason why the exact Bayesian inference is intractable, we instead resort to maximizing the lower bound of ℒ with respect to , expecting that this would increase the marginal likelihood as well, or even if it does not, the bound would at least become tighter. Specifically, maximizing ℒ with respect to the hyperparameters and taking into account the constraint in (2) leads to the following update equations
| (13) |
where Ϝ denotes the digamma function, and can then be obtained by solving the above equations. Since the hyperparameters are coupled with which are estimated in VB, their computation needs to alternate with (6). In our implementation, to speed up the computation the hyperparameters are updated once after every fixed number of iterations (say, 100) for VB.
2.2.2. Initialization
The initialization of the VB algorithm is important since the lower bound ℒ is a non-convex function of the variational distribution, which means different local maxima may be reached with different initializations. In our implementation, the initial conditions are set as follows:
| (14) |
where R̂1 and R̂2 are the empirical spatial covariance of both conditions (see Equation (19) in Appendix A), and Λk, ãc, am are all estimated using the CSP algorithm. In our experience, the above choice of initial conditions works well on both simulated and real data sets. However, it is possible that certain modifications are needed when the VB algorithm is applied to a broader range of EEG data sets.
The VB algorithm is summarized in Algorithm 1. Each iteration of the VB algorithm requires 𝒪((C + N1 + N2)M3) operations. The algorithm terminates when the change of the lower bound ℒ is less than some predefined threshold (say, 10−8). Note that the VB algorithm is guaranteed to converge due to the non-decreasing property of the variational lower bound as the iteration increases.
Algorithm 1.
The V B pseudocode
| Input: Multichannel EEG data Xk (k = 1, 2) that are recorded from two experimental conditions |
| Output: The variational distributions q*(Zk), q*(A), q*(α), |
| Initialization: Use the settings in (14), and set iter = 0, ℒ(0) = −inf, (c = 1, …, C; k = 1, 2; m = 1, …, M) |
| Method: |
| repeat |
| iter = iter + 1 |
| Update the parameters in the variational distributions using (7) – (12) |
| if (iter mod 100) = 0 then |
| Compute by solving (13) |
| end if |
| Compute the variational lower bound ℒ(iter) using (19) |
| until ℒ(iter) − ℒ (iter − 1) ≤ a pre-defined threshold (e.g., 10−8) |
2.2.3. More insight into the VB algorithm
The following remarks offer insights into how the ideas of model size determination and hierarchical modeling in (1) are manifested in the VB algorithm:
- To see how VB leads to a sparse mixing matrix, applying matrix inversion lemma to the right-hand side of (8) yields
where(15)
Thus the (m, m)-th element of is calculated as
which is close to zero when 〈α(m)〉q* is sufficiently large. In light of (9) the m-th element of is close to zero as well (note that the results apply to all c’s). Hence a sufficiently large 〈α(m)〉q* results in a small . In addition, it follows from (10) that , which means that a small would in turn increase 〈α(m)〉q*. Therefore through such a re-weighting scheme, when 〈α(m)〉q* becomes sufficiently large, will keep decreasing as the iterations are continued until the convergence criterion is met. It is also clear that the hierarchical modeling has a pooling effect on the source variance across trials. Indeed, based on the variational distribution of , its variational mean can be computed as . Here play the role of pooling towards a global quantity since they are estimated using the data from all trials. On the other hand, if the priors of the source variance are instead chosen to be noninformative with , the variational mean would simply be , for which there is no pooling effect among trials as merely the data from the i-th trial are used to estimate the source variance of the i-th trial.
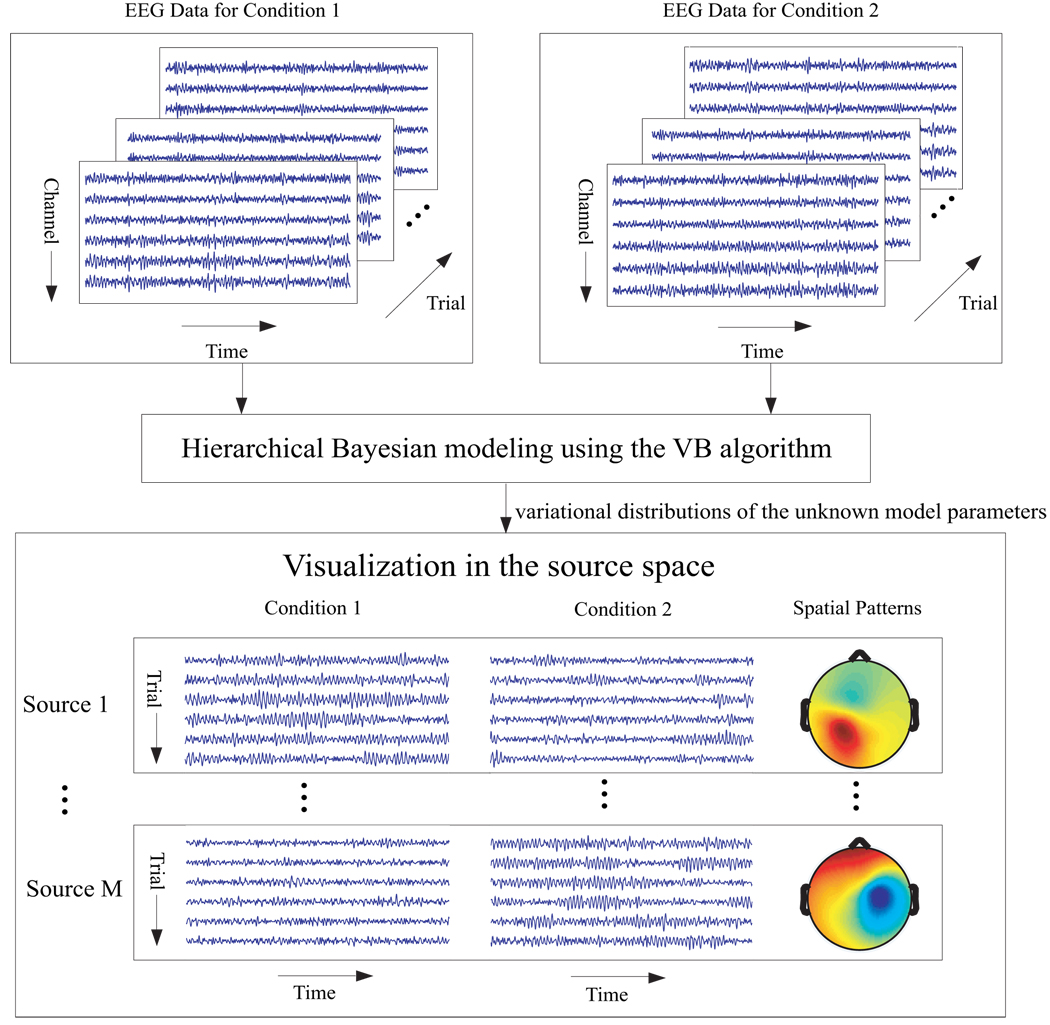
Figure 3 illustrates a scheme for exploratory data analysis by using the proposed hierachical modeling framework. Note that in the last step the variational mean of the mixing matrix and source signals is employed for visualization in the source space.
Figure 3.
A scheme for exploratory EEG data analysis by using the proposed hierarhical modeling framework. The source number M can be determined by discarding those sources whose corresponding columns in the estimated mixing matrix (variational mean of A) are with negligible l2 norms (see Section 3.1.1).
3. Experiments
A range of experiments are conducted on both simulated and real EEG data. The goal is to provide empirical evidence for verifying the aforementioned properties of the proposed statistical modeling framework, and to evaluate the performance of the VB algorithm by comparing it with the state-of-the-arts algorithms, namely CSP and Infomax (Bell and Sejnowski, 1995; Amari et al., 1996), the latter being the predominant algorithm for ICA and extensively employed for multichannel EEG data analysis.
In the simulation experiment where ground truth is available, we compare the performance of the VB, CSP, and Infomax algorithms on the reconstruction accuracy of spatio-temporal patterns and evolution of source amplitude across trials, all based on Monte Carlo simulations. Here CSP is chosen for comparison as it is shown by Theorem 1 to yield maximum likelihood estimates for generative model (4), and the fact that it has no regard for inter-trial variability makes it interesting to see how the algorithm performs in the presence of inter-trial variability in the data. Besides, given that the simulated data within each trial follow a Gaussian distribution, due to the inter-trial variability of the variance the distribution of the data, taken over conditions and trials, is a Gaussian scale mixture, which is non-Gaussian, thus justifying the use of Infomax.
For real EEG data analysis, we assess on two multiple-trial motor imagery BCI data sets whether the VB algorithm can extract physiologically meaningful task-related spatio-temporal patterns. Here motor imagery data sets are chosen for real data analysis because previous studies have shown that inter-trial amplitude variability is generally present in subjects’ EEG data during motor imagery due to fluctations in their attention, arousal, and task strategy (Pfurtscheller and Aranibar, 1977). In addition, to show that the extracted patterns are indeed useful, we employ them for predicting the unknown class labels in the test sets, and compare the prediction accuracies with those of the CSP and Infomax algorithms.
All computations are done using MATLAB (The MathWorks, Inc.). The Infomax algorithm is implemented using runica.m in the EEGLAB toolbox (Delorme and Makeig, 2004). The learning rate of Infomax is set heuristically to 0.00065/ log(C), where C is the number of channels; the algorithm converges when the weight change between consecutive iterations is smaller than 10−7.
3.1. Experimental Setup and Data Analysis
3.1.1. Simulation Study
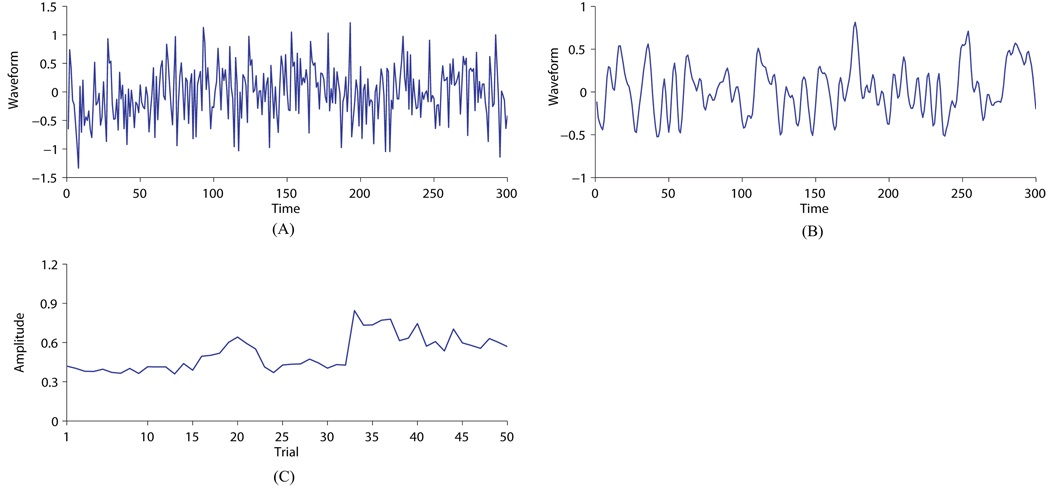
The study consists of Monte Carlo simulations of 50 runs. In each run, Nk = 50 trials of data from model (1) are generated for each of two conditions. In each trial a set of M = 10 mutually uncorrelated sources are generated. Each source signal comprises Jk = 300 data points that are generated using either of the following two settings. The first setting generates data points that are independently and identically Gaussian distributed with zero mean, which is consistent with the assumption in model (1) (Figure 4(A)). By contrast, to simulate source signals that more resemble real EEG signals, the second setting generates data points from fourth-order autoregressive (AR) models (Figure 4(B)). For each condition in this study, 8 source signals are simulated using the first setting, while the rest 2 are simulated using the second.
Figure 4.
An example of the simulated waveforms and inter-trial amplitude variability of source signals. (A) A source signal with i.i.d. data points generated from a Gaussian distribution. (B) A source signal generated from a fourth-order AR model. (C) Trial amplitude evolution of a source signal for condition 1.
To simulate inter-trial amplitude variability, for each condition the variances of each source signal across trials are random samples from a fixed gamma distribution (𝒢a(5, 0.5) for condition 1 and 𝒢a(2, 0.5) for condition 2), with a correlation coefficient between the variances in the i-th trial and the (i + d)-th trial being 1 − 0.1|d| for −9 ≤ d ≤ 9 and 0 otherwise (Figure 4(C)). Although this violates the conditional independence assumption in model (1), it is interesting to see if in this more realistic situation our VB algorithm is still able to recover the true source amplitude evolution faithfully. Finally, a 20 × 10 mixing matrix is randomly generated (thus C = 20), with each entry standard Gaussian distributed, and additive non-isotropic white Gaussian noise is simulated with varying SNRs of 20, 15, 10, 5, and 0 dB. The channel-wise SNR is defined as the ratio of the variance of the mixture signal over the noise variance at each channel.
The simulated noisy mixture signals are then presented to VB, CSP, and Infomax separately to estimate the underlying spatio-temporal source patterns. To form the inputs to the algorithms, for CSP the multiple-trial signals are concatenated across trials for each condition, while for Infomax they are concatenated across both conditions and trials.
The Amari index (Amari et al., 1996) is used as the first performance index to measure the proximity of the estimated mixing matrix  and the true one A, which is invariant to permutation and scaling of the columns of A and Â:
where bij = ((ATA)−1AT Â)ij. A smaller value of the Amari index indicates a more accurate estimate of A, with zero implying a perfect fit.
To compute the Amari index, based on the output of each algorithm it is necessary to form  that has the same dimension as the true mixing matrix (20 × 10). To achieve this, we select the 10 columns in the estimated mixing matrix (For VB, it refers to the variational mean of A computed using Equation (9)) that have the 10 largest l2 norms to form ÂVB, ÂCSP, and ÂInfomax, respectively. Note that for both CSP and Infomax, the columns of the estimated mixing matrix are normalized by the sum of the estimated source variances of the two conditions, i.e., , where are the estimated variance of the source signals associated with the m-th SP for condition 1 and 2, respectively.
The correlation coefficient between the estimated source signals and the true ones is used as the second performance index:
| (16) |
where denotes the m-th true source signal and denotes the m-th estimated source signal using either of the two algorithms. For VB, is simply the variational mean of the m-th source signal computed using Equation (7). Note that for each run of the Monte Carlo simulation the correlation coefficient is averaged over conditions, trials, and sources. To resolve beforehand the permutation indeterminancy of the estimated source signals, we pair the true source signals with the estimated ones through the following strategy: for the first true source signal, we compute the correlation coefficient of its associated SP with the SP associated with each estimated source signal in turn and pair it with the estimated source signal that achieves the largest correlation coefficient. The remaining true source signals are paired similarly except that each time the estimated source signals that have been chosen earlier are excluded from consideration.
Likewise, the third performance index is the correlation coefficient that assesses the reconstruction accuracy of the evolution of source amplitudes across trials for the two algorithms:
| (17) |
where .
3.1.2. Real EEG Data Analysis
Data Set 1
Seven healthy volunteers (s1–s7, four males and three females, all right handed, 21–24 years old) participated in our online motor imagery experiments with visual feedback. The left- or right-hand movement imagination was designated to control the vertical movement of a cursor. Figure 5 shows the paradigm of the feedback experiment. The EEG was recorded using a BioSemi ActiveTwo system. A total of 32 data channels were placed at positions according to the 10/20 international system, including C3/C4 and FCz electrodes over the primary motor area (M1) and the supplementary motor area (SMA). All the data channels were referenced to the left earlobe. Signals were sampled at 256 Hz. For each subject, a data set of 240 trials (120 trials per task) recorded in a single session is used for offline algorithmic studies, where the whole data set is split into a training set of 160 trials and a test set of 80 trials, with equal number of trials per class.
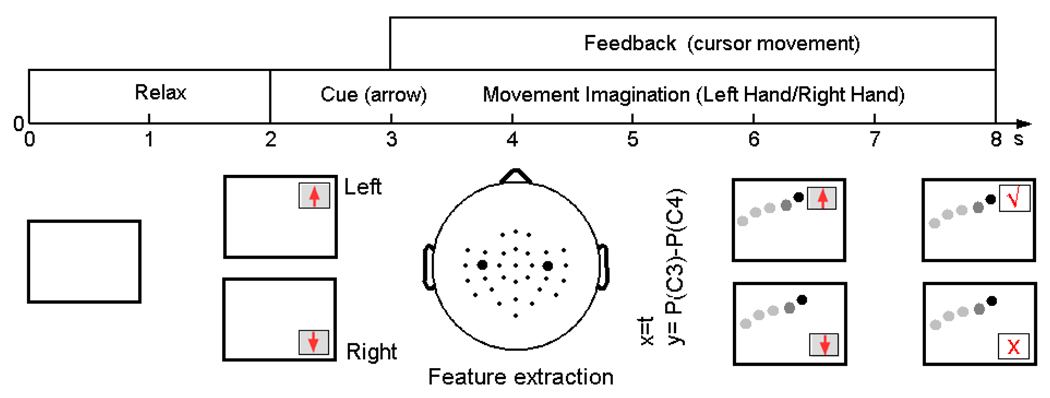
Figure 5.
Paradigm of the online motor imagery experiment for Data Set 1. In each trial, the subject was in the relaxed state for the first two seconds when the screen was blank. Starting from the 3rd second, a visual cue (an arrow) appeared on the screen, indicating the imagery task to be initiated. The arrow pointing upward and downward indicated the tasks of imagination of the left hand and the right hand movement, respectively. From the 4th second, a cursor started to move horizontally in a constant speed from the left side to the right side of the screen. The vertical position of the cursor was determined by the accumulated power difference between channel C3 and C4. At the end of the trial, a tick or cross mark appeared to indicate correctness of the classification.
Data Set 2 (Data Set IVa, BCI Competition III (Blankertz et al., 2006))
The EEG data of five healthy subjects (aa, al, av, aw, and ay) were recorded during motor imagery experiments without feedback, in which they were instructed to perform one of three motor imageries in each trial: left-hand, right-hand, or right-foot. The tasks in the data set include only the right-hand and right-foot imageries. The EEG was recorded using BrainAmp amplifiers. A total of 118 data channels were placed according to the international 10/20 system. Each trial lasted for 3.5 seconds following the visual cues. Signals were downsampled to 100 Hz for analysis. A total of 280 trials of EEG data were collected for each subject, with varying number of training and test trials per subject (see Table 2).
Table 2.
Number of training and test trials for each subject in Data Set 2
| aa | al | av | aw | ay | |
|---|---|---|---|---|---|
| training trials | 168 | 224 | 84 | 56 | 28 |
| test trials | 112 | 56 | 196 | 224 | 252 |
We compare the performance of VB, CSP, and Infomax on both real EEG data sets. To avoid any potential bias towards a specific algorithm, for each data set identical preprocessing settings (e.g., channel selection, band-pass filtering, time windowing) are applied before using any algorithm, with details as follows:
All data channels are used in the analysis for Data Sets 1 and 2, i.e., no channel selection is performed.
All EEG signals are band-pass filtered between 8 Hz and 30 Hz, which is known to encompass the ERD/ERS effects4.
The specific time windows for each trial of each data set are (0s denotes the start of each trial) (1) 2.5–6s (Data Set 1), (2) 0.5–3.5s (Data Set 2). The 0.5s delay of the window following the appearance of visual cues excludes the reaction period of subjects in the data.
The VB, CSP, and Infomax algorithms are then employed to learn spatiotemporal decompositions of each preprocessed data set from its training set. Since VB is naturally able to deal with multiple-trial and multiple-condition data, each training set is formed as a direct input to the algorithm. For CSP, because it can handle multiple conditions, within each condition the EEG data is first concatenated across trials before being fed into the algorithm. By ignoring both multiple-trial and multiple-condition structures, Infomax is applied to the EEG data concatenated across trials and conditions.
An informative source signal should carry signatures in distinguishing different conditions; to assess to what degree each estimated source signal exhibits task-related changes, we use the R-square (coefficient of determination) (Casella and Berger, 2002) as a metric of its correlation with condition labels. Specifically, for each individual source signal we compute the R-square between its variance and the condition labels across trials in the unseen test tests; the larger is the resulting R-square, the more task-related is the corresponding source signal. Note that the variance is a legitimate measure that has long been used to quantify changes in EEG power that are associated with ERD/ERS (Pfurtscheller and Aranibar, 1977).
In addition to considering each source signal separately, we can also assess how much information they collectively carry regarding the condition labels. For such a purpose, we predict the condition labels in the unseen test sets from all three data sets, using all the sources signals estimated from each algorithm. Fisher linear discriminant analysis (FLDA) (Bishop, 2006), which is simple and computationally efficient, is employed as the classifier. As the input to FLDA, the feature vector of each trial consists of features that are each defined as the log-variance of a source signal. Note that the log-transform makes the features follow a Gaussian-like distribution, which is to ensure the optimality of FLDA. Moreover, to avoid the overfitting of FLDA, it is crucial to select only part of the features that are discriminative to form the feature vector5. We opt for a simple feature selection approach based on the R-square: The estimated source signals are first ranked according to their R-squares on the training set, and then only the n features derived from the source signals with the n largest R-squares are selected; the optimal n is determined using 10-fold cross-validation on the training sets.
3.2. Results on Synthetic Data
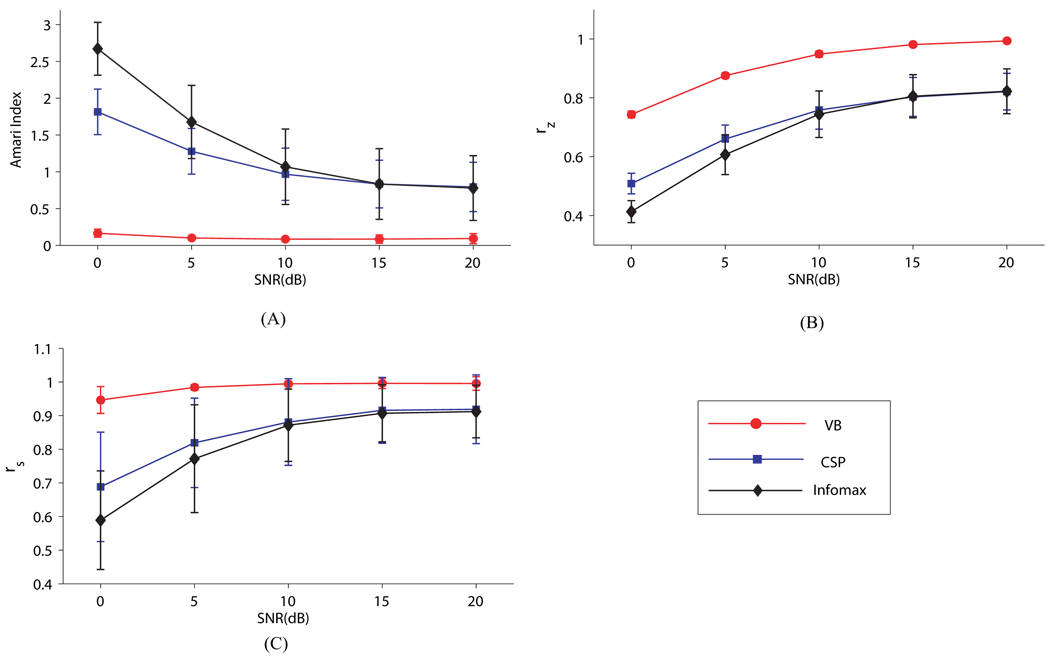
Figure 6 summarizes the results of comparison among VB, CSP, and Infomax using all three performance measures. Overall, it is clear that VB outperforms CSP and Infomax significantly under all SNR settings for all performance indices. Specifically, the fast increase of the Amari index for CSP and Infomax as the SNR decreases indicates the algorithms’ significant degrade in performance at low SNRs. By contrast, the Amari index remains quite stable around a small value for VB under all SNR settings. Figure 6(B)(C) shows similar superior performance of VB over CSP and Infomax in terms of the other two indices. Remarkably, VB recovers the trial amplitude evolution of the source signals well even under the SNR as low as 0dB, whereas CSP and Infomax performs poorly when the SNR becomes low. It is encouraging that excellent and robust performance is achieved in VB despite the presence of both strong within-trial temporal dynamics and between-trial amplitude dynamics in the data.
Figure 6.
Comparison between the results of the VB, CSP, and Infomax algorithms at different SNR settings. Each result is obtained by averaging over 50 Monte Carlo runs. (A) The Amari indices between the estimated mixing matrix and the true one. (B) The correlation coefficients between the estimated source signals and the true ones. (C) The correlation coefficients between the trial amplitude of the estimated source signals and that of the true ones.
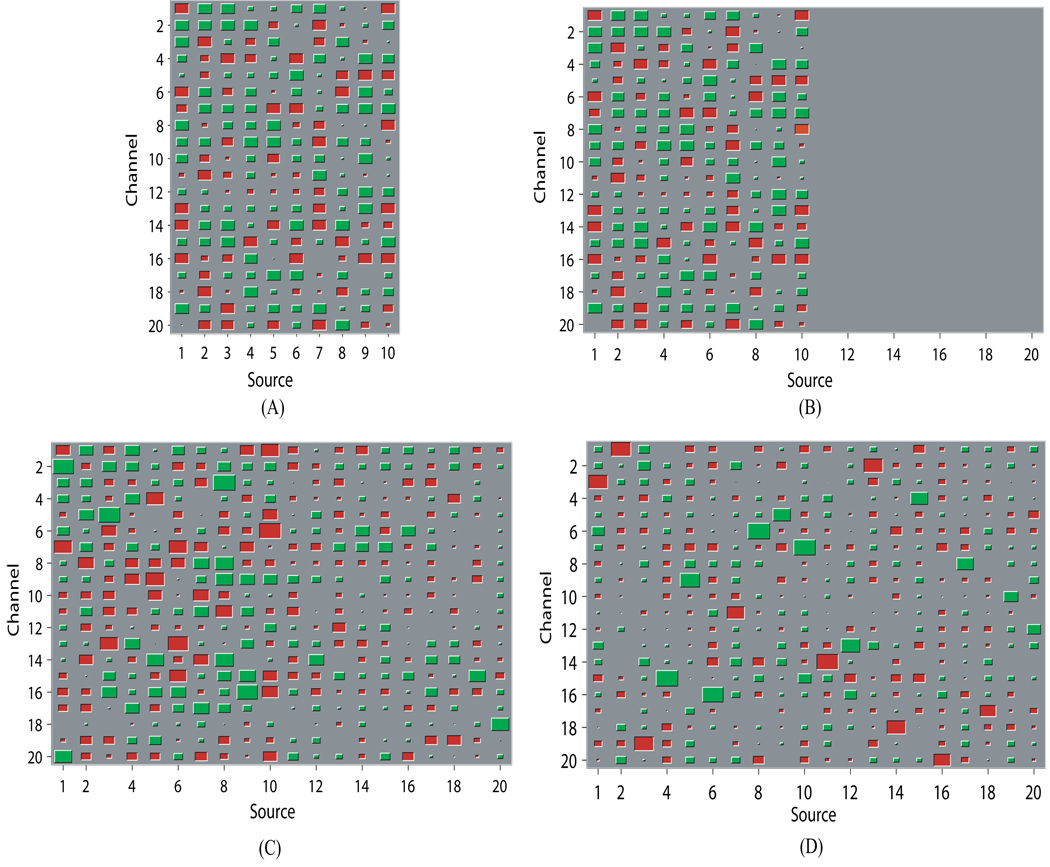
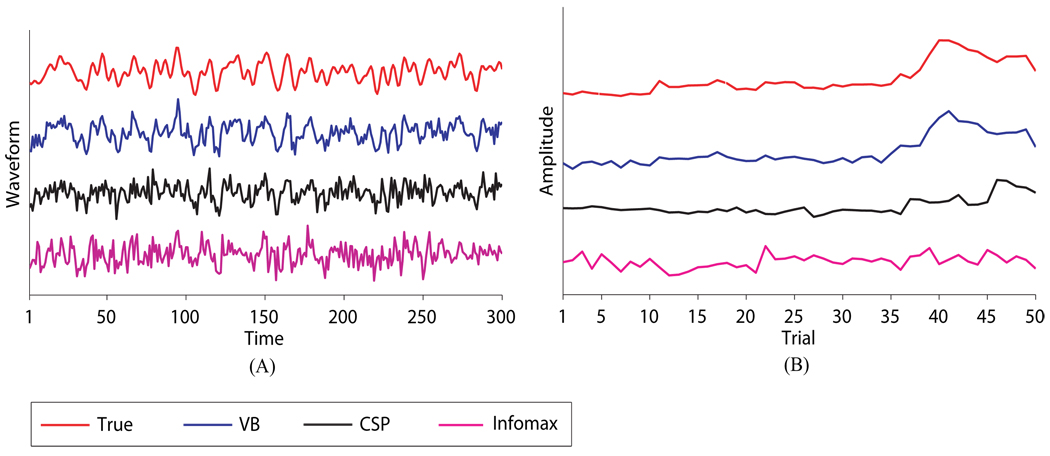
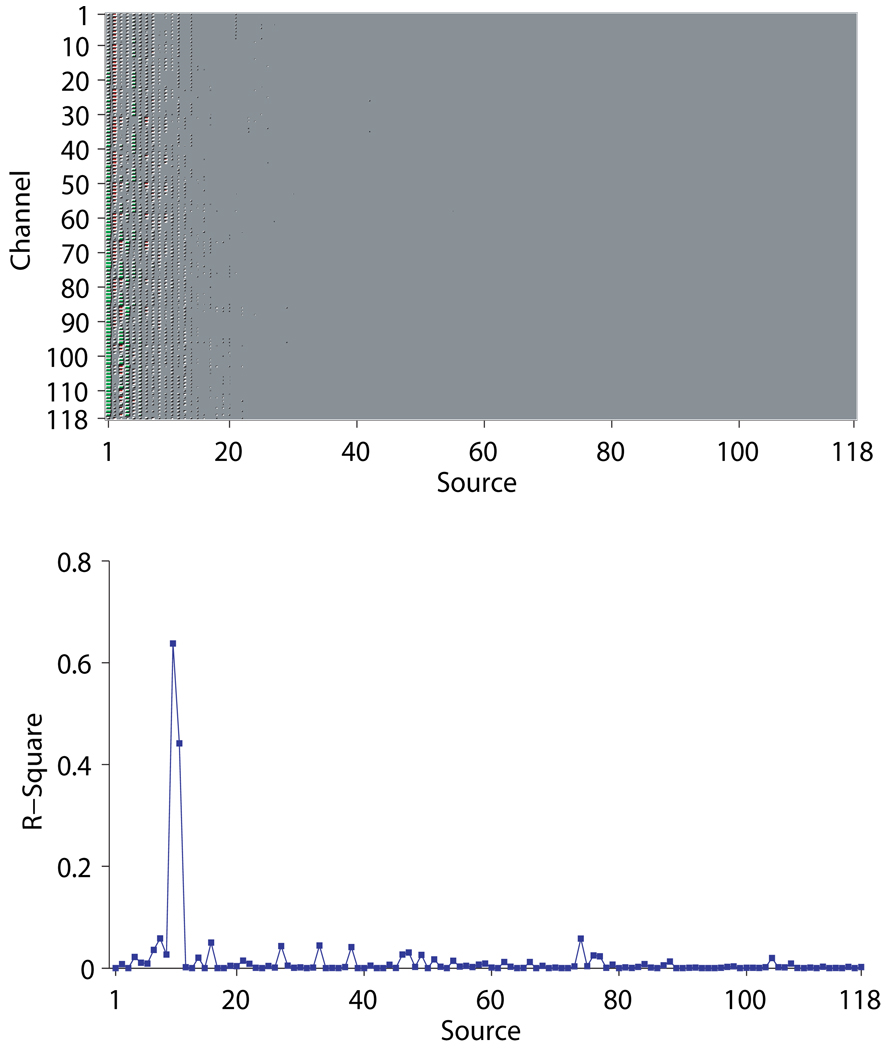
As an intuitive example, Figures 7~ 8 give results from one Monte Carlo run at 0dB. For this specific run, d(ÂVB, A) = 0.1365, rz = 0.7542, and rs = 0.9678 for VB; d(ÂCSP, A) = 2.0726, rz = 0.5107, and rs = 0.7253 for CSP; d(ÂCSP, A) = 2.4369, rz = 0.4153, and rs = 0.5654 for Infomax. Figure 7 shows the estimated mixing matrices using the Hinton diagram. It can be seen that CSP and Infomax fail to differentiate the 10 redundant columns in the mixing matrix. These redundant columns play the role of fitting the additive noise and hence cause an overfit. By contrast, VB successfully shrinks the redundant columns to negligible values that are barely discernible; the remaining columns are highly close to the true mixing matrix according to both visualization and small value of the Amari index. Figure 8 shows the estimated temporal dynamics and trial amplitude evolution of a specific source signal. It is observed that the estimates from VB result in significantly less distortion to the true ones than those from CSP and Infomax do. In particular, as shown in Figure 8(B), in estimating the trial amplitude evolution for one condition VB correctly identifies the large positive deflection of the amplitude that lasts approximately from trial 38 to trial 46; the one that CSP identifies is clearly misplaced forward in time. Worse still, the deflection is completely missed by Infomax.
Figure 7.
An example from one Monte Carlo run illustrating that ARD can effectively determine the source number. (A) Hinton diagram of the true mixing matrix A. (B) Hinton diagrams of ÂVB, whose Amari index is 0.1365. (C) Hinton diagram of ÂCSP, whose Amari index is 2.0726. (D) Hinton diagram of ÂInfomax, whose Amari index is 2.4259. Each entry in the matrices is represented by the size of the area of a red (positive value) or green (negative value) square.
Figure 8.
Temporal dynamics and trial amplitude evolution of a specific source signal at one Monte Carlo run. (A) True and estimated temporal dynamics of the source signal at a specific trial. (B) True and estimated trial amplitude evolution of the source signal across trials for condition 1.
All the above results demonstrate that ignoring inter-trial amplitude variability and misidentification of source number in the modeling phase, as in CSP and Infomax, can lead to significant estimation error for spatiotemporal patterns and trial amplitude evolution of source signals. On the other hand, by explicitly modeling the inter-trial amplitude variability in the data, VB is able to accurately recover the underlying spatio-temporal patterns and track the dynamics of source amplitude across trials.
3.3. Results on Real EEG Data
Table 3 shows the classification accuracies for Infomax, CSP, and VB on the test sets of all 12 subjects from the two BCI data sets. As can be seen, VB’s prediction performance is consistently better than Infomax and CSP’s for most of the subjects, with equal performance for others. In terms of the mean classification accuracy, VB also has a higher rate (92.27%) than Infomax (87.21%) and CSP (85.77%). One-sided paired-sample t test shows that the improvement is significant, with P-values being 0.0018 versus Infomax and 0.0353 versus CSP6. The improvement is most conspicuous for subjects ay, whose results are thus picked out for illustration below.
Table 3.
Comparison of Infomax, CSP, and VB’s classification accuracies (%) on the test sets from two BCI data sets.
| Data Set | Subject | Infomax | CSP | VB |
|---|---|---|---|---|
| s1 | 93.75 | 93.75 | 93.75 | |
| s2 | 93.75 | 100 | 100 | |
| s3 | 73.75 | 81.25 | 88.75 | |
| 1 | s4 | 86.25 | 83.75 | 90.00 |
| s5 | 96.25 | 100 | 100 | |
| s6 | 96.25 | 98.75 | 98.75 | |
| s7 | 96.25 | 98.75 | 98.75 | |
| aa | 76.79 | 74.11 | 77.68 | |
| al | 100 | 100 | 100 | |
| 2 | av | 73.47 | 57.14 | 79.08 |
| aw | 83.04 | 93.30 | 95.98 | |
| ay | 76.98 | 48.41 | 85.71 | |
| mean | 87.21** | 85.77* | 92.27 | |
P = 0.0018,
P = 0.0353
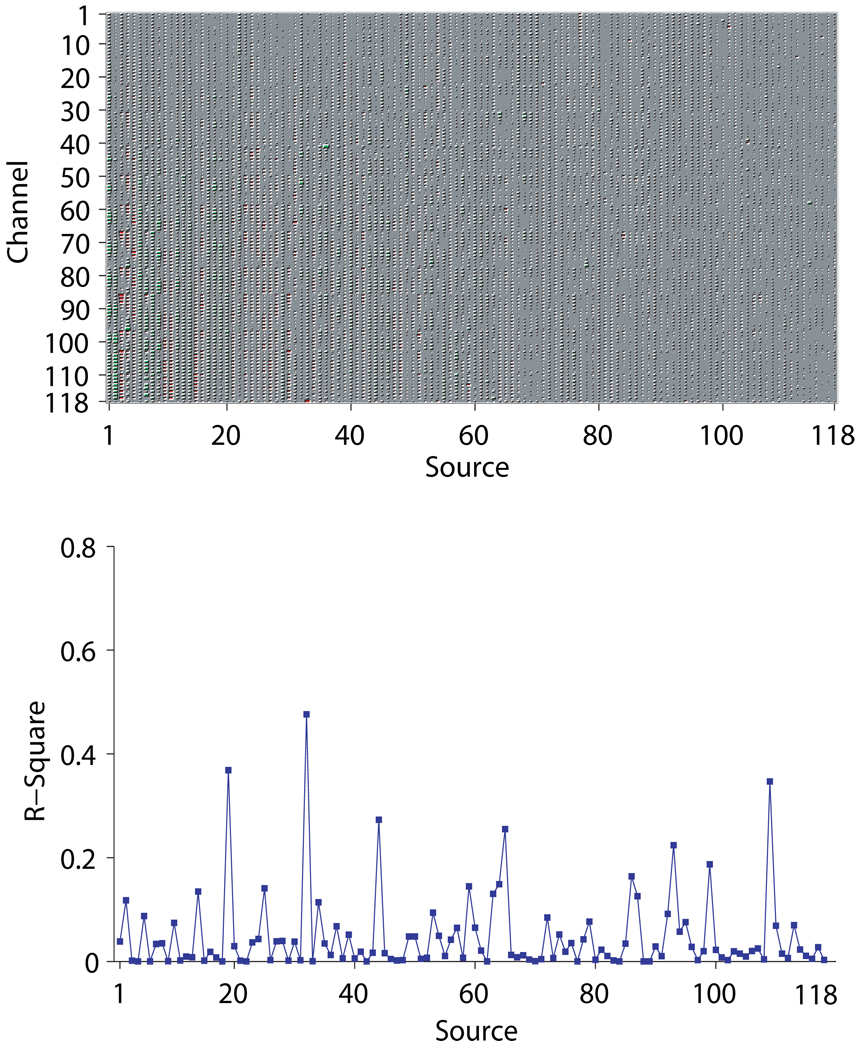
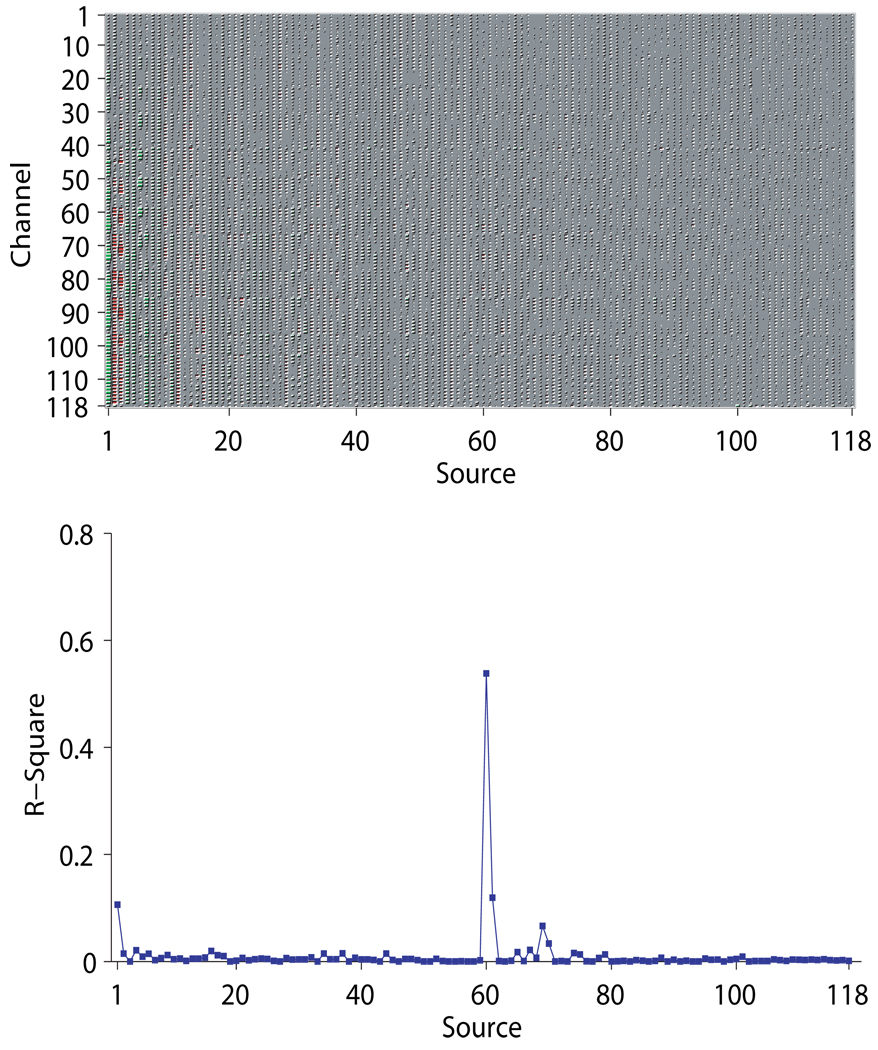
Figures 9~ 11 show for subject ay the mixing matrices estimated by the three algorithms (upper panels) and the R-squares of each estimated source signal computed on the test set (lower panels). The sparseness of the mixing matrix estimated by VB is clearly visible in Figure 9, with merely less than 30 non-zero columns remaining. CSP and Infomax by contrast have no shrinking effect on the number of sources – the mixing matrices are simply 118 × 118 full matrices, as shown in Figures 10~ 11.
Figure 9.
The mixing matrix estimated by VB and the R-squares of all the estimated source signals for subject ay. The upper pannel shows the estimated mixing matrix as a Hinton matrix; the lower pannel shows the R-squares of each source signal computed on the test set. Note that both plots are drawn such that each source is located at the same position on the horizontal axes.
Figure 11.
The mixing matrix estimated by Infomax and the R-squares of all the estimated source signals for the right-hand motor imagery for subject ay. The layout of both pannels is the same as in Figure 9.
Figure 10.
The mixing matrix estimated by CSP and the R-squares of all the estimated source signals for subject ay. The layout of both pannels is the same as in Figure 9.
In terms of the R-squares, for VB it can be seen that the number of the discriminative source signals (the 10th and 11th) are sparse as well. For Infomax, apart from the 19th and 32th ones, many of the extracted source signals have small yet non-negligible R-squares. In fact, much of the discriminative information carried by these source signals may well be redundant for classification, as evidenced by the lower classification accuracy of Infomax than that of VB on subject ay. For CSP, it is observed that there is one source signal (the 60th) with fairly high R-square, which means it correlates well with the tasks. Why in this case is CSP only able to achieve an accuracy slightly above random guessing? It should be noted that the displayed R-squares are computed ad hoc on the test set with its task labels known, whereas during classification the R-squares on which the ranking of features are based are computed on the training set, whose size is very small for subject ay. As a consequence, it is found that on the training set some ”noisy” source signals in fact yield inflated R-squares that surpass the one achieved by the 60th source signal simply due to overfitting. Thus through our feature selection procedure, the features associated with these ”noisy” source signals, rather than that associated with the 60th source signal, are selected to form the feature vector, resulting in a low classification accuracy.
Another interesting observation is that the CSP’s test accuracy for subject ay is much lower than the winning entry of the competition (which was 97.6% contributed by the authors’ group. See http://www.bbci.de/competition/iii/results/). Note that intensive manual tuning of parameters were done on the training set in achieving the winning entry. In particular, only part of the 118 channels were carefully selected to avoid the overfitting issue. The reason why overfitting does not happen to VB is that most of the redudant sources that represent noise in the data have been eliminated after the VB estimation, again stressing the importance of model size determination. That said, we note that the most discrminative source signal (the 10th) for VB still has a higher R-square value than the one for CSP. Moreover, for VB there is another source signal (the 11th) attains a R-square value larger than 0.5, which further increases the discriminability of the features.
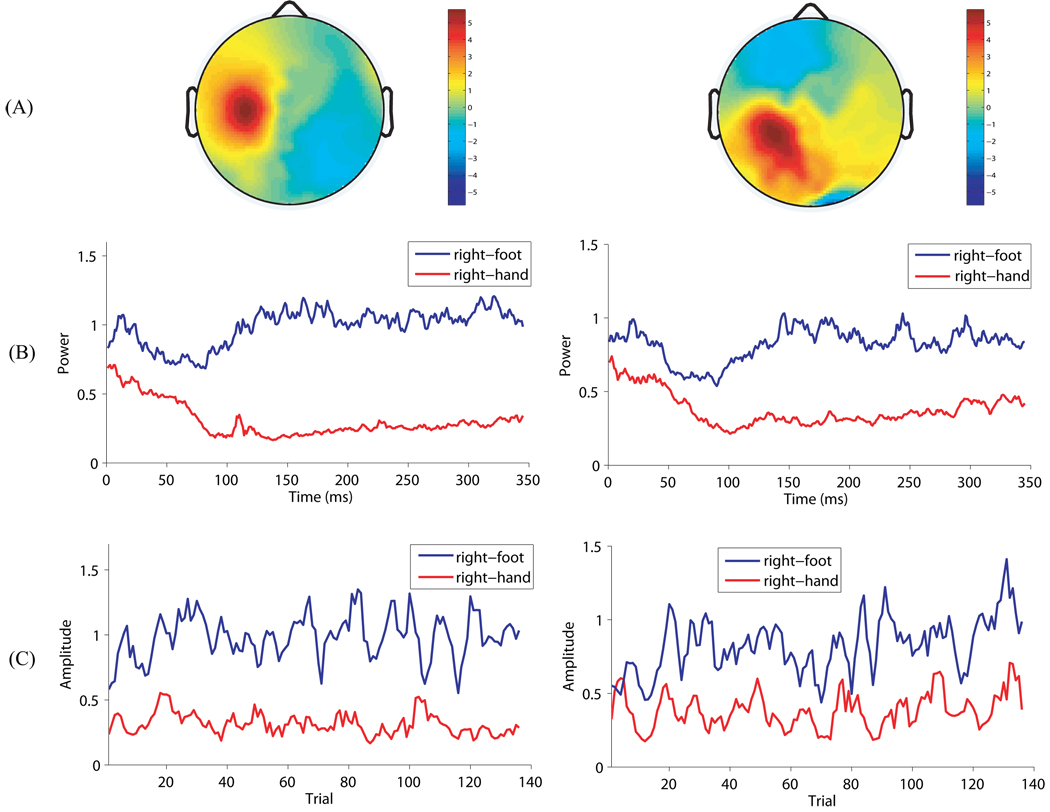
Figure 12 shows for subject ay the spatio-temporal patterns for the 10th and 11th source signals estimated by VB, with the left column for the 10th and the right column for the 11th. Figure 12(A) shows the SPs corresponding to both source signals, which are located over the left sensorimotor cortex. Figure 12(B) shows the change of instantaneous power (i.e., square of the source signal at each time point) across time for the two source signals. Each curve is obtained by averaging over the test trials associated with the corresponding condition. For both source signals, it can be observed that there is a clear ERD phenomenon, which is a short-lasting decrease of EEG power, for the right-hand movement imagery shortly after the visual cues appeared. The results are consistent with the exisiting neurophysiological knowledge that ERD typically appears in the contralateral sensorimotor cortex during movement imagination (Pfurtscheller and Aranibar, 1977). Figure 12(C) shows for each condition the evolution of trial amplitude. For both source signals the inter-trial variaiblity during the right-foot movement imagination is prominent, suggesting the need to explicitly take it into account in the modeling stage.
Figure 12.
The spatio-temporal patterns of the two most discriminative source signal estimated by VB for subject ay. The left column is for the most discriminative source signal, and the right column is for the second most discriminative source signal. (A) The spatial pattern on the scalp. (B) The change of instantaneous power across time for both conditions. Both curves are obtained by averaging over the associated test trials. (C) The evolution of trial amplitude for both conditions. The amplitude at each trial is smoothed by taking its average within the subsequent 3 trials. Note that the trial order on the horizontal axis may not be important in the interpretation of the results since the trials may have been randomized in order in the BCI competition data set.
4. Discussion and Conclusion
4.1. Discussion
By taking into account the inter-trial variability in the data, it appears that the complexity of model (1) may be too high given the large number of parameters involved. Two techniques keep us on a safe ground. First, we have shown earlier in (3) that ARD as a principled way for model size determination induces sparsity of A in model (1). Sparse learning is especially suited in situations where the source number is smaller than the channel number. Even in scenarios where sparsity is not the case for the true model, sparse learning may still be desirable since estimation of a model with a dimension as high as the true one may be unreliable due to the limited amount of data available (also known as the curse of dimensionality) – a parsimonious model would typically lead to a better generalization ability (Hastie et al., 2009). Furthermore, compared to the MAP methods for sparse learning, e.g., group-LASSO and M-FOCUSS, which aim to search for the mode in the posterior distribution, it is argued that sparse learning by means of ARD is more likely to capture significant posterior mass and has much less risk of getting stuck at local optima (Wipf and Rao, 2007). Second, the use of hierarchical modeling adds another layer of guard against model overfitting. Indeed, in model (1) the source covariance across trials is designated to follow a common unknown multivariate gamma distribution. Such a top-down constraint of the prior distribution imposes an underlying structure (via Bayes’ rule) on the posterior distribution of the source covariance and thus leads to a significantly reduced search space of covariance parameters.
The VB principle has recently attracted a fair amount of attention in the EEG signal processing community (Nagarajan et al., 2006; Hoffmann, 2007; Chatzis et al., 2008; Wipf and Nagarajan, 2009). Nonetheless, as an approximation one might wonder how close the variational distribution is to the true posterior. This remains an open question as the closeness is dependent on the structure of the specific model at hand. In general, the quality of variational approximation is good provided that the structured subspace 𝒬 is in a sufficiently small neighborhood of the true posterior distribution. (Beal, 2003) presented a simulation study on a mixture factor-analysis model, where the KL divergence between the variational and exact posterior distribution was found to be fairly small, yet it increased approximately linearly with the number of parameters in the model. This inevitably would have an unfavorable influence on model selection within the variational framework, in that simpler models might always be preferred to complex ones. However, in our Monte Carlo simulation study we do not found this to be a severe issue as in nearly all runs VB can accurately identify the correct model size. Furthermore, from a pragmatic viewpoint, the variational distribution is useful for the following intuitive reason (MacKay, 2003). The way of computing the MAP solution can be viewed as using a delta-function shape probability distribution to fit the full posterior distribution. Regardless of its geometry, the structured probability space for variational approximation is larger in size than the one containing only delta-function shape probability distributions, thus leading to a better approximation to the full posterior distribution. In fact, the modified EM algorithm for computing the MAP estimates (Dempster et al., 1977) is recovered by restricting the form of the variational distribution of parameters to a delta function or point estimate. More recently, VB has also been postulated as an inference principle implemented in the brain (Friston, 2010).
A similar Bayesian treatment of the amplitude variability in the context of EEG inverse problem can be found in (Friston et al., 2006). (Limpiti et al., 2009) has recently proposed a likelihood-based framework for modeling the inter-trial amplitude variability in event-related potentials (ERPs). In particular, two approaches, namely linear dynamical system response (LDSR) and independent response (IR), were employed for the amplitude estimation. There are three important distinctions of how inter-trial amplitude variability is modeled between our paper and (Limpiti et al., 2009). First, because ERP is a phase-locked response in EEG, that is, positive or negative deflections always occur at the same time relative to the external stimulus, it is typically modeled as a fixed effect, that is, its waveform is assumed to be constant across trials. By contrast, induced responses such as ERD/ERS reflect changes in ongoing oscillatory EEG activity that are not phase-locked to any external stimuli, and as such they are modeled as random variables in our model. Second, unlike the LDSR approach where a first-order AR model is employed to track the dynamics of the learning effect across trials, we have not modeled the learning effect because in an experiment involving multiple conditions, trials of different conditions are mingled and randomized so as to avoid the habituation confound. In this case, the dynamics of the learning effect may not be simply modeled by an AR model since the transition between inter-condition trials and the transition between intra-condition trials are likely to differ from each other, e.g., in terms of change in amplitude. Third, despite that in the IR approach the amplitude is modeled as i.i.d. random variables across trials, the negative values allowed by the underlying Gaussian distribution may lead to difficulty in interpreting results. By contrast, in our framework the variance of each source signal is drawn from an inverse-gamma distribution, which has non-zero probabilities at non-negative values only. With that said, it is true in theory that the conditional independence assumption for trial amplitude in our hierarchical model may be overly strong for modeling certain types of inter-trial amplitude variability, such as those where the amplitude changes smoothly across trials. Nonetheless, we believe our hierarchical modeling framework provides a natural basis for further development of more complex models. Besides, our simulation study has shown that our VB algorithm works satisfactorily even in the cases when there are strong correlation between the amplitude of consecutive trials.
Canonical correlation analysis (CCA) (Hotelling, 1936) is another popular tool for the joint analysis of two multivariate data sets. To avoid possible confusion, Appendix E reviews CCA and contrasts it with our current methodology.
The hierarchical Bayesian modeling framework is flexible in that it may serve as a basis for exploring potential extensions in a wide variety of settings. First, extension of the current framework to the cases where there are more than two experimental conditions is straightforward by expanding the number of sub-models in model (1). Second, similar models can be developed to learn spatio-temporal decomposition of phase-locked components in the EEG data. A crucial part of this effort will be to integrate the phase-locking information into model development. Again, it would be interesting to evaluate its prediction performance on ERP data sets, such as P300. Third, the idea of hierarchical modeling can also be applied to factor analysis and ICA to enable them to cope with the inter-trial amplitude variability in the EEG data. In fact, such a strategy has recently been applied to ICA for the fusion of multi-subject data sets in the context of fMRI data analysis (Varoquaux et al., 2010). Fourth, as opposed to the i.i.d. assumption in this paper, extending model (1) into a state-space form would allow it to capture the temporal correlations in source signals (Brockwell and Davis, 2002). However, as the model complexity increases the computational load of the associated inference algorithm will grow as well. Finally, the generative modeling framework allows us to use unlabelled data for augmenting the labelled training data to perform semi-supervised learning in classification since both types of data can be employed to jointly specify the likelihood function of the generative model (Zhu, 2005).
4.2. Conclusion
In this paper, we have introduced a hierarchical Bayesian framework for learning spatio-temporal decomposition of multichannel EEG data. The major features of the proposed framework are summarized as follows:
The hierarchical model is capable of accounting for the inter-trial amplitude variability that is prevalent in multichannel EEG data, which has rarely been brought into the attention of practitioners in EEG data analysis.
The hierarchical model provides a natural characterization for spatiotemporal decomposition of EEG data recorded under multiple conditions, which makes it appealing for the analysis of data collected in a large array of neuroscience studies.
The model can be viewed as an extension to the CSP algorithm, which has achieved great successes in BCI data analysis. Meanwhile, it precludes the overfitting issue of CSP by enforcing sparsity on the number of sources7; each estimated source now much more likely represent a physical or physiological process rather than random noise.
The VB algorithm yields approximate posterior distributions of all the variables in the model, which facilitates the potential assessment of their statistical significance in future studies.
Using both simulated and real data sets, we have demonstrated that our VB algorithm is able to produce more accurate estimates of spatio-temporal patterns as well as better prediction performance than the CSP and Infomax algorithms. The reason why our proposed method outperforms CSP and Infomax is because a proper probabilistic model is chosen to characterize the complex nature of multi-trial EEG data. In conclusion, we believe that our statistical modeling framework can serve as a powerful tool for extracting brain patterns, characterizing trial-to-trial brain dynamics, and decoding brain states by exploiting useful structures in the data.
Acknowledgements
This work was supported by NIH Grants DP1-OD003646, R01-EB006385, and the National Natural Science Foundation of China under Grant 30630022. We are grateful to YijunWang for providing Data Set 1 and to Klaus-Robert Müller, Benjamin Blankertz, and Gabriel Curio for providing the BCI competition data sets (Data Set 2). We thank Francis Bach for helpful discussions, and the anonoymous reviewers for their insightful comments and valuable suggestions.
A. A Brief Review of the CSP Algorithm
Let us introduce the CSP algorithm in the context of EEG signal processing. Consider two classes of zero-mean multichannel EEG signals Xk ∈ ℝC×Jk (k = 1, 2). The empirical spatial covariance matrices for the two classes can then be computed as
| (18) |
The aim of CSP is to find a set of spatial filters by which the ratio of variance between the two classes is maximized. Here the ratio of variance potentially serves as a measure of separability between the two classes. Mathematically, the spatial filters can be obtained in a collective manner by solving the following optimization problem
where W ∈ ℝC×C contains all C spatial filters as rows. Equivalently, the eigenvectors are given by simultaneous diagonalization of the covariance matrices R̂1 and R̂2
where Ω1 and Ω2 are both diagonal matrices.
B. CSP as ML Estimation of Model (4): Proof of Theorem 1
The proof is in line with the idea in (Pham and Cardoso, 2001).
Under the setup of the noiseless and square mixing matrix (assumptions (i) and (ii)), the log-likelihood of the observed EEG data is
where Rk = AΛkAT, and DKL(S1‖S2) denotes the KL divergence between two Gaussian distributions with covariance matrices S1 and S2, respectively. The last equality follows from the definition of KL divergence.
Because the KL divergence is invariant to invertible linear transformations and the Pythagorean decomposition holds as the involved distributions are all Gaussian, the log-likelihood is further rewritten as
where Λ1 and Λ2 are fully parameterized diagonal matrices. Therefore, regardless of A the second KL divergence in the bracket can always be made exactly to zero. The first KL divergence will be zero if and only if A−1R̂kA−T is a diagonal matrix. In other words, the log-likelihood is maximized if and only if R̂1 and R̂2 are jointly diagonalized by Â−1 and Â−T. In addition, it can be shown that under assumption (iii) the matrix that jointly diagonalizes R̂1 and R̂2 is unique (Belouchrani, 1997), hence Â−T = W.
C. Derivation of the VB updates for Model (1)
The joint probability distribution of all the random variables in model (1) is derived as
The variational posteriors can then be computed as follows by maximizing (5) with respect to q (see (Bishop, 2006) for the derivation)
where denotes all the random variables in Θ except .
where
where .
where .
where .
D. Derivation of the variational lower bound ℒ in (5)
In light of (6), the variational lower bound is expanded as
| (19) |
where each term in (19) is given by
E. Connections with Canonical Correlation Analysis (CCA)
CCA (Hotelling, 1936) is a useful data analysis tool for exploring the associations between two multivariate data sets (termed views) X1 ∈ ℝC1×J and X2 ∈ ℝC2×J (both assumed to have zero mean for simplicity). It has been successfully employed in both EEG and fMRI data analysis (Friman et al., 2003; Lin et al., 2006).
The idea of CCA is to first find the pair of linear combinations having the largest correlation, and then to find the pair of linear combinations having the largest correlation among all pairs uncorrelated with the initially selected pair, and so forth. The pairs of linear combinations are called canonical directions, and their correlations are called canonical correlations. Specifically, the canonical directions can be obtained by solving the following generalized eigenvalue problem:
where . The following generative model can be written for CCA:
| (20) |
where Ψk can be non-diagonal. It is proved in (Bach and Jordan, 2006) that the connection between the ML estimate of Ak and the canonical directions is given by
where Uk ∈ ℝCk×D contains the first D canonical directions as columns, Mk ∈ ℝD×D are arbitrary matrices such that (where P is the diagonal matrix consisting of the first D canonical correlations) and the spectral norms of Mk are smaller than one.
Similarities of models (4) and (20), and the fact that they both are able to deal with two data sets notwithstanding, important differences exist between CCA and our proposed methodology on both conceptual and technical levels. Conceptually, as clearly shown in model (20) the two data sets involved in the CCA analysis are supposed to reflect a common underlying physical process, which can be estimated by maximally correlating the two data sets. An example is simultaneously recorded EEG and fMRI signals, which are different modalities yet reflect the same brain state. By contrast, the two data sets in our analysis are regarded as being associated with two different conditions. The goal is not to correlate the data sets but typically to estimate the task-related components for each condition.
The fundamental distinction in their goals in turn helps us understand the structural differences between the generative models (4) and (20). First, the dimensions of the two data sets are allowed to differ in model (20), which again can be exemplified by the extremely high dimensionality of fMRI data (~ 104 voxels) and the comparatively moderate dimensionality of EEG data (~ 102 channels). Second, as representing the same physical process the TPs of the sources are identical for the two data sets, while the mixing matrices are different. Third, as long as they are positive semidefinite, the noise covariance matrices in model (20) are allowed to have full degrees of freedom as apposed to being restricted to be diagonal in model (4). Indeed, model (20) is formulated such that the latent space merely captures the cross-correlations between data sets, leaving the correlations within each data set to be fully accounted for by the noise covariance, which is hence modeled as being non-diagonal. By contrast, in model (4) the cross-correlations between the data sets recorded under two conditions are zero because the TPs are assumed to be uncorrelated. Thus fitting both data sets reduces to fitting each individual data set separately with the only a priori information being that they share common SPs. To avoid any trivial solutions when fitting individual data set, the noise covariance matrices are constrained to be diagonal.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The validity of simultaneity and linearity is guaranteed by the quasistatic approximation to the Maxwell equations, since the effective frequency range of EEG lies below 1 kHz.
Henceforth we assume that all covariance matrices are positive definite.
However, the distribution of source signals, taken over conditions and trials, is a Gaussian scale mixture (GSM) (Wainwright and Simoncelli, 2000), which is known to be super-Gaussian, except in the case of trivial degeneracy.
The issue of learning optimal temporal filters for classification is not addressed in this work. Refer to (Dornhege et al., 2006; Lemm et al., 2005; Wu et al., 2008) for various treatments of the topic.
This differs from the model size determination issue encountered in the generative modeling phase; a source signal may be indispensable for the generative description of the data yet the feature constructed therefrom may be useless for discrimination purpose.
Note that for some subjects (e.g., al, s5, s6, s7) since their EEG data are so strongly correlated with the tasks that there is barely any space for further performance improvement. Nonetheless we report all the results for the sake of integrity.
An elegant treatment of the same issue from a discriminative learning perspective can be found in (Tomioka and Müller, 2010).
References
- Amari S, Cichocki A, Yang HH. Advances in Neural Information Processing Systems. Vol. 8. 1996. A new learning algorithm for blind signal separation; pp. 757–763. [Google Scholar]
- Anderson TW. An Introduction to Multivariate Statistical Analysis. 3rd Edition. Wiley-Interscience; 2003. [Google Scholar]
- Andrews DF, Mallows CL. Scale mixtures of normal distributions. Journal of the Royal Statistical Society. 1974;36:99–102. [Google Scholar]
- Bach FR, Jordan MI. Technical Report 688. Berkeley: Department of Statistics, University of California; 2006. A probabilistic interpretation of canonical correlation analysis. [Google Scholar]
- Baillet S, Mosher JC, Leahy RM. Electromagnetic brain mapping. IEEE Signal Proc. Mag. 2001;18:14–30. [Google Scholar]
- Beal MJ. Ph.D. Thesis. Gatsby Computational Neuroscience Unit, University College London; 2003. Variational Algorithms for Approximate Bayesian Inference. [Google Scholar]
- Bell AJ, Sejnowski TJ. An information-maximization approach to blind separation and blind deconvolution. Neural Comp. 1995;7:1129–1159. doi: 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- Belouchrani A-M. A blind source separation technique using second order statistics. IEEE Trans. Signal Processing. 1997;45:434–444. [Google Scholar]
- Bishop CM. Pattern Recognition and Machine Learning. Springer; 2006. [Google Scholar]
- Blankertz B, Müller K-R, Krusienski D, Schalk G, Wolpaw JR, Schlögl A, Pfurtscheller G, Millán JdR, Schröder M, Birbaumer N. The BCI competition III: validating alternative approaches to actual BCI problems. IEEE Trans. Neural Sys. Rehab. Eng. 2006;14:153–159. doi: 10.1109/TNSRE.2006.875642. [DOI] [PubMed] [Google Scholar]
- Blankertz B, Tomioka R, Lemm S, Kawanabe M, Müller K-R. Optimizing spatial filters for robust EEG single-trial analysis. IEEE Signal Proc. Mag. 2008;25:41–56. [Google Scholar]
- Brockwell PJ, Davis RA. Introduction to Time Series and Forecasting. 2nd Edition. Springer; 2002. [Google Scholar]
- Bruin KJ, Kenemans JL, Verbaten MN, Van Der Heijden AH. Habituation: an event-related potential and dipole source analysis study. Int. J. Psychophysiol. 2000;36:199–209. doi: 10.1016/s0167-8760(99)00114-2. [DOI] [PubMed] [Google Scholar]
- Carlin BP, Louis TA. Bayes and Empirical Bayes Methods for Data Analysis. 2nd Edition. Chapman & Hall/CRC; 2000. [Google Scholar]
- Casella G, Berger RL. Statistical Inference. 2nd Edition. Thomson Learning; [Google Scholar]
- Chatzis SP, Kosmopoulos DI, Varvarigou TA. Signal modeling and classification using a robust latent space model based on t distributions. IEEE Trans. Signal Processing. 2008;56:949–963. [Google Scholar]
- Cichocki A, Amari S. Adaptive Blind Signal and Image Processing. John Wiley & Sons, Inc.; 2002. [Google Scholar]
- Cichocki A, Shishkin S, Musha T, Leonowicz Z, Asada T, Kurachi T. EEG filtering based on blind source separation (BSS) for early detection of Alzheimer’s disease. Clin. Neurophysiol. 2005;116:729–737. doi: 10.1016/j.clinph.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Cotter SF, Rao BD, Engan K, Kreutz-Delgado K. Sparse solutions to linear inverse problems with multiple measurement vectors. IEEE Trans. Signal Processing. 2005;53:2477–2488. [Google Scholar]
- Delorme A, Makeig S. EEGLAB: an open toolbox for analysis of single-trial EEG dyanmics including independent component analysis. Journal of Neuroscience Methods. 2004;134:9–21. doi: 10.1016/j.jneumeth.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J. Royal Stat. Soc. B. 1977;39:1–38. [Google Scholar]
- Dornhege G, Blankertz B, Krauledat M, Losch F, Curio G, Müller K-R. Combined optimization of spatial and temporal filters for improving brain-computer interfacing. IEEE Trans. Biomed. Eng. 2006;53:2274–2281. doi: 10.1109/TBME.2006.883649. [DOI] [PubMed] [Google Scholar]
- Ergenoglu T, Demiralp T, Bayraktaroglu Z, Ergen M, Beydagi H, Uresin Y. Alpha rhythm of the EEG modulates visual detection performance in humans. Brain Res. Cogn. Brain Res. 2004;20:376–383. doi: 10.1016/j.cogbrainres.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Raichle ME. Intrinsic fluctuations within cortical systems account for intertrial variability in human behavior. Neuron. 2007;56:171–184. doi: 10.1016/j.neuron.2007.08.023. [DOI] [PubMed] [Google Scholar]
- Friman O, Borga M, Lundberg P, Knutsson H. Adaptive analysis of fMRI data. Neuroimage. 2003;19:837–845. doi: 10.1016/s1053-8119(03)00077-6. [DOI] [PubMed] [Google Scholar]
- Friston K. The free-energy principle: a unified brain theory? Nature Reviews Neuroscience. 2010;11:127–138. doi: 10.1038/nrn2787. [DOI] [PubMed] [Google Scholar]
- Friston K, Henson R, Phillips C, Mattout J. Bayesian estimation of evoked and induced responses. Human Brain Mapping. 2006;27:722–735. doi: 10.1002/hbm.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga F, Koontz W. Applications of the Karhunen-Loève expansion to feature selection and ordering. IEEE Trans. Comput. 1970;19:311–318. [Google Scholar]
- Gouy-Pailler C, Congedo M, Brunner C, Jutten C, Pfurtscheller G. Non-stationary brain source separation for multi-class motor imagery. IEEE Trans. Biomed. Eng. 2010;57:469–478. doi: 10.1109/TBME.2009.2032162. [DOI] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd Edition. Springer; 2009. [Google Scholar]
- Hill NJ, Lal TN, M Schröder TH, Widman G, Elger GE, Schölkopf B, Birbaumer N. Classifying event-related desynchronization in EEG,ECoG and MEG signals. In: Dornhege G, del R Millán J, Hinterbeger T, McFarland D, Müller K-R, editors. Towards Brain-Computer Interfacing. MIT Press; 2007. in press. [Google Scholar]
- Hoffmann U. Ph.D. Thesis. Ecole Polytechnique Federale de Lausanne (EPFL); 2007. Baysian Machine Learning Applied in a Brain-Computer Interface for Disabled Users. [Google Scholar]
- Hotelling H. Relations between two sets of variables. Biometrika. 1936;28:321–377. [Google Scholar]
- Hyvarinen A, Karhunen J, Oja E. Independent Component Analysis. John Wiley & Sons, Inc.; 2001. [Google Scholar]
- Hyvärinen A, Ramkumar P, Parkkonen L, Hari R. Independent component analysis of short-time Fourier transforms for spontaneous EEG/MEG analysis. Neuroimage. 2010;49:257–271. doi: 10.1016/j.neuroimage.2009.08.028. [DOI] [PubMed] [Google Scholar]
- Kachenoura A, Albera L, Senhadji L, Comon P. ICA: a potential tool for BCI systems. IEEE Sig. Proc. Mag. 2008;25:57–68. [Google Scholar]
- Klimesch W. EEG alpha and theta oscillations reflect cognitive and memory performance: a review and analysis. Brain Res. Rev. 1999;29:169–195. doi: 10.1016/s0165-0173(98)00056-3. [DOI] [PubMed] [Google Scholar]
- Koles ZJ, Lind JC, Soong ACK. Spatio-temporal decomposition of the EEG: a general approach to the isolation and localization of sources. Clin. Neurophysiol. 1995;95:219–230. doi: 10.1016/0013-4694(95)00083-b. [DOI] [PubMed] [Google Scholar]
- Koller D, Friedman N. Probabilistic Graphical Models: Principles and Techniques. The MIT Press; 2009. [Google Scholar]
- Lagerlund TD, Sharbrough FW, Busacker NE. Spatial filtering of multichannel electroencephalographic recordings through principal component analysis by singular value decomposition. J. Clin. Neurophysiol. 1997;14:73–82. doi: 10.1097/00004691-199701000-00007. [DOI] [PubMed] [Google Scholar]
- Lemm S, Blankertz B, Curio G, Müller K-R. Spatio-spectral filters for improved classification of single trial EEG. IEEE Trans. Biomed. Eng. 2005;52:1541–1548. doi: 10.1109/TBME.2005.851521. [DOI] [PubMed] [Google Scholar]
- Limpiti T, Van Veen BD, Attias HT, Nagarajan SS. A spatiotemporal framework for estimating trial-to-trial amplitude variation in event-related MEG/EEG. IEEE Trans. Biomed. Eng. 2009;56:633–645. doi: 10.1109/TBME.2008.2008423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhang C, Wu W, Gao X. Frequency recognition based on canonical correlation analysis for SSVEP-based BCIs. IEEE Trans. Biomed. Eng. 2006;53:2610–2614. doi: 10.1109/tbme.2006.886577. [DOI] [PubMed] [Google Scholar]
- MacKay D. Bayesian interpolation. Neural Comput. 1992;4:415–447. [Google Scholar]
- MacKay D. Information Theory, Inference, and Learning Algorithms. Cambridge University Press; 2003. [Google Scholar]
- Makeig S, Jung TP, Bell A, Ghahremani D, Sejnowski T. Blind separation of auditory event-related brain responses into independent components. Proc. Nat. Acad. Sci. 1997;94:10979–10984. doi: 10.1073/pnas.94.20.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeig S, Westerfield M, Jung TP, Enghoff S, Townsend J, Courchesne E, Sejnowski T. Blind separation of auditory event-related brain responses into independent components. Proc. Nat. Acad. Sci. 2002;295:690–694. doi: 10.1073/pnas.94.20.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure SM, Li J, Tomlin D, Cypert KS, Montague LM, Montague PR. Neural correlates of behavioral preference for cultiurally familiar drinks. Neuron. 2004;44:379–387. doi: 10.1016/j.neuron.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Miltner WHR, Braun C, Arnold M, Witte H, Taub E. Coherence of gamma-band EEG activity as a basis for associative learning. Nature. 1999;397:434–436. doi: 10.1038/17126. [DOI] [PubMed] [Google Scholar]
- Nagarajan S, Attias HT, Hild KE, II, Sekihara K. A graphical model for estimating stimulus-evoked brain responses from magnetoencephalography data with large background brain activity. Neuroimage. 2006;30:400–416. doi: 10.1016/j.neuroimage.2005.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedermeyer E, da Silva FL. Electroencephalography: Basic Principles, Clinical Applications, and Related Fields. Lippincot Williams and Wilkins; 2004. [Google Scholar]
- Parra LC, Sajda P. Blind source separation via generalized eigenvalue decomposition. Journal of Machine Learning Research. 2003;4:1261–1269. [Google Scholar]
- Parra LC, Spence CD, Gerson AD, Sajda P. Recipes for the linear analysis of EEG. Neuroimage. 2005;28:326–341. doi: 10.1016/j.neuroimage.2005.05.032. [DOI] [PubMed] [Google Scholar]
- Pfurtscheller G, Aranibar A. Event-related cortical desynchronization detected by power measurements of scalp EEG. Electroencephalogr. Clin. Neurophysiol. 1977;42:817–826. doi: 10.1016/0013-4694(77)90235-8. [DOI] [PubMed] [Google Scholar]
- Pham D-T, Cardoso J-F. Blind separation of instantaneous mixtures of non-stationary sources. IEEE Trans. Signal Processing. 2001;49:1837–1848. [Google Scholar]
- Rampil IJ. A primer for EEG signal processing in anesthesia. Anesthesiology. 1998;89:980–1002. doi: 10.1097/00000542-199810000-00023. [DOI] [PubMed] [Google Scholar]
- Ray WJ, Cole HW. EEG alpha activity reflects attentional demands, and beta activity reflects emotional and cognitive processes. Science. 1985;228:750–752. doi: 10.1126/science.3992243. [DOI] [PubMed] [Google Scholar]
- Robert CP. The Bayesian Choice: From Decision-Theoretic Foundations to Computational Implementation. 2nd Edition. Springer; 2007. [Google Scholar]
- Tomioka R, Müller K-R. A regularized discriminative framework for EEG analysis with application to brain-computer interface. Neuroimage. 2010;49:415–432. doi: 10.1016/j.neuroimage.2009.07.045. [DOI] [PubMed] [Google Scholar]
- Varoquaux G, Sadaghiani S, Pinel P, Kleinschmidt A, Poline JB, Thirion B. A group model for stable multi-subject ICA on fMRI datasets. Neuroimage. 2010;51:288–299. doi: 10.1016/j.neuroimage.2010.02.010. [DOI] [PubMed] [Google Scholar]
- Vigário R. Extraction of ocular artifacts from EEG using independent component analysis. Electroenceph. Clin. Neurophysiol. 1997;103:395–404. doi: 10.1016/s0013-4694(97)00042-8. [DOI] [PubMed] [Google Scholar]
- Vigário R, Oja E. BSS and ICA in neuroinformatics: from current practices to open challenges. IEEE Reviews in Biomed. Eng. 2008;1:50–61. doi: 10.1109/RBME.2008.2008244. [DOI] [PubMed] [Google Scholar]
- Wainwright MJ, Simoncelli EP. Scale mixtures of Gaussians and the statistics of natural images. Advances in Neural Information Processing Systems. 2000;12:855–861. [Google Scholar]
- Wilson MA, McNaughton BL. Dynamics of the hippocampal ensemble code for space. Science. 1993;261:1055–1058. doi: 10.1126/science.8351520. [DOI] [PubMed] [Google Scholar]
- Wipf D, Nagarajan S. A unified Bayesian framework for MEG/EEG source imaging. Neuroimage. 2009;44:947–966. doi: 10.1016/j.neuroimage.2008.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipf D, Rao BD. An empirical Bayesian strategy for solving the simultaneous sparse approximation problem. IEEE Trans. Signal Processing. 2007;55:3704–3716. [Google Scholar]
- Wolpaw JR, Birbaumer N, McFarland DJ, Pfurtscheller G, Vaughan TM. Brain-computer interfaces for communication and control. Clin. Neurophysiol. 2002;113:767–791. doi: 10.1016/s1388-2457(02)00057-3. [DOI] [PubMed] [Google Scholar]
- Wu W, Chen Z, Gao S, Brown EN. A probabilistic framework for learning robust common spatial patterns. Proc. 31st Int. Conf. IEEE-EMBS; 2009. pp. 4658–4661. [DOI] [PubMed] [Google Scholar]
- Wu W, Gao X, Hong B, Gao S. Classifying single-trial EEG during motor imagery by iterative spatio-spectral patterns learning. IEEE Trans. Biomed. Eng. 2008;55:1733–1743. doi: 10.1109/tbme.2008.919125. [DOI] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. Journal of the Royal Statistical Society, Series B. 2007;68:49–67. [Google Scholar]
- Zhu X. Technical Report 1530. Department of Computer Sciences, University of Wisconsin-Madison; 2005. Semi-supervised learning literature survey. [Google Scholar]