Abstract
In both prokaryotes and eukaryotes, the heat shock protein ClpB functions as a molecular chaperone and plays a key role in resisting high temperature stress. ClpB is important for the development of thermotolerance in yeast and cyanobacteria but apparently not in Escherichia coli. We undertook a complementation study to investigate whether the ClpB protein from E coli (EcClpB) differs functionally from its cyanobacterial counterpart in the unicellular cyanobacterium Synechococcus sp. PCC 7942. The EcClpB protein is 56% identical to its ClpB1 homologue in Synechococcus. A plasmid construct was prepared containing the entire E coli clpB gene under the control of the Synechococcus clpB1 promoter. This construct was transformed into a Synechococcus clpB1 deletion strain (ΔclpB1) and integrated into a phenotypically neutral site of the chromosome. The full-length EcClpB protein (EcClpB-93) was induced in the transformed Synechococcus strain during heat shock as well as the smaller protein (EcClpB-79) that arises from a second translational start inside the single clpB message. Using cell survival measurements we show that the EcClpB protein can complement the Synechococcus ΔclpB1 mutant and restore its ability to develop thermotolerance. We also demonstrate that both EcClpB-93 and -79 appear to contribute to the degree of acquired thermotolerance restored to the Synechococcus complementation strains.
INTRODUCTION
All organisms synthesize heat shock proteins (Hsps) in response to increasing growth temperature (Lindquist and Craig 1988; Parsell and Lindquist 1993). Many of these Hsps are now recognized as members of distinct families of molecular chaperones, primarily based on amino acid sequence homology and similar molecular masses. Constitutive representatives of these families impact directly on protein maturation, promoting activities such as protein synthesis, translocation, folding, and assembly into multimeric structures. Chaperones induced during heat stress perform additional functions that include stabilization of protein structures, refolding of denatured polypeptides, and resolubilizing protein aggregates. They also play a role in targeting irreversibly damaged polypeptides for degradation by proteases, thereby selectively removing potentially toxic protein structures (Parsell and Lindquist 1993). It is the combined actions of these chaperones that make them essential Hsps for cell survival at high temperatures.
One relatively new class of molecular chaperones that also includes heat shock–inducible members is the Hsp100/Clp (caseinolytic protease) family. The family can be divided into 2 broad groups that are separated further into subgroups based on specific sequence signatures and other structural characteristics (Schirmer et al 1996). Proteins in the first and main group are large (79–105 kDa) and contain 2 distinct ATP-binding domains separated by a spacer region of variable length. Five subgroups have been identified to date, termed ClpA–E. ClpA is found exclusively in gram-negative bacteria like Escherichia coli, ClpB in almost all eubacteria and eukaryotes, ClpC in gram-positive bacteria, cyanobacteria and plants, ClpD exclusively in plants, and ClpE in certain gram-positive bacteria. The second group has only 2 members, ClpX and ClpY, that differ from the first group by having only 1 ATP-binding domain. ClpX is found in all eubacteria and eukaryotes, whereas ClpY (or HslU) is restricted to certain bacteria. Besides functioning as molecular chaperones, certain Hsp100 proteins (eg, E coli ClpA and ClpX) also associate with a proteolytic subunit, ClpP, to form an ATP-dependent Clp protease (Katayama-Fujimura et al 1987; Wojtkowiak et al 1993). Within this complex, the Hsp100 partner is essential for activating the proteolytic activity of ClpP, which alone is inactive due to steric restrictions and inaccessibility of the protein substrate to the proteolytic active sites (Wang et al 1997). In steps requiring ATP, the Hsp100 chaperone selectively binds and unfolds the target polypeptide and then transfers it into the degradation chamber of ClpP (Hoskins et al 1998). Once inside, the unfolded protein is quickly and indiscriminately degraded to small peptide fragments that eventually diffuse out (Thompson and Maurizi 1994).
One of the best-studied Hsp100/Clp proteins is ClpB, an Hsp found in almost all organisms studied to date. In most eubacteria, a single gene codes for 2 different-sized ClpB proteins (79 and 93 kDa) via a second translational initiation site within the clpB transcript (Squires et al 1991; Eriksson and Clarke 1996). Different-sized ClpB proteins are also synthesized in eukaryotes but are encoded by separate nuclear genes (Sanchez and Lindquist 1990; Leonhardt et al 1993). In yeast, both forms function as molecular chaperones, with cytosolic ClpB (Hsp104) resolubilizing protein aggregates that form during severe heat stress (Parsell et al 1994), and mitochondrial ClpB (78 kDa) preventing protein denaturation at high temperatures (Schmitt et al 1995). Yeast cytosolic ClpB does not influence the cells' ability to withstand direct shifts to severe high temperatures, but it is essential for the development of thermotolerance (Sanchez and Lindquist 1990), which is the resistance acquired to an otherwise lethal high temperature by being first conditioned to a more permissive high temperature. Cytosolic ClpB from higher plants can also confer thermotolerance to yeast (Lee et al 1994; Schirmer et al 1994), suggesting shared functional characteristics between eukaryotic homologues. In contrast, loss of ClpB in E coli reduces cell viability after shifts directly to severe high temperatures, but it has no effect on the acquisition of thermotolerance (Squires et al 1991). This difference between the yeast and E coli proteins suggested that the role of ClpB during heat shock might differ somewhat in bacteria and eukaryotes.
In an earlier study we reported the cloning of the clpB1 gene from the unicellular cyanobacterium Synechococcus sp. PCC 7942 (hereafter referred to as Synechococcus). We demonstrated that unlike ClpB in E coli, the Synechococcus ClpB1 protein was not involved in cell survival during direct shifts to high temperatures but was instead important for the development of thermotolerance, similar to the role of Hsp104 in yeast (Eriksson and Clarke 1996). However, like the E coli homologue, Synechococcus ClpB1 has all the characteristics typical of eubacterial ClpB proteins. Structurally it is more related to ClpB in other eubacteria as judged by primary amino acid sequence. Different-sized ClpB1 proteins are also induced during heat shock as a result of dual translational initiation sites within the single clpB1 mRNA (Eriksson and Clarke 1996). To investigate the apparent functional difference between Synechococcus and E coli ClpB proteins during high temperature stress, we examined whether E coli ClpB could restore thermotolerance to the Synechococcus clpB1 inactivation mutant (ΔclpB1). We now show that the E coli protein can indeed complement ClpB1 and confer thermotolerance to Synechococcus, and that this function seems to involve both the full-length and truncated forms of ClpB protein.
MATERIALS AND METHODS
Culture conditions
Synechococcus cultures were grown on BG-11 plates or in liquid BG-11 medium buffered with 10 mM 3-(N-morpholino) propanesulfonic acid (pH 7.5) (Rippka et al 1979). Liquid cultures were grown in 80-mL glass tubes in 37°C water tanks with continuous light of 50 μmol photons m−2 s−1. To avoid changes in antenna size the cultures were bubbled with 5% CO2 in air (1 mL s−1). The strains ΔclpB1, EcB, and EcB1 were maintained on BG-11 plates and in liquid precultures with 5 μg mL−1 of antibiotic to maintain selection; kanamycin (ΔclpB1), chloramphenicol, and kanamycin (EcB and EcB1). No antibiotics were added to the experimental cultures so as to avoid any antibiotic-induced phenotypic changes.
Construction of E coli clpB complementation plasmid
To prepare the E coli clpB complementation strains, a new plasmid was first prepared to facilitate recombination of each complementation construct into a chromosomal site of Synechococcus that tolerates genetic disruption without causing phenotypic changes. This neutral site locus (NSL) was chosen from the 2.8-kb site previously described by Bustos and Golden (1992). From this NSL, DNA fragments of 0.9 (NS 1) and 1 kb (NS 2) were polymerase chain reaction (PCR) amplified from Synechococcus chromosomal DNA and ligated in pUC-4K (Amersham Pharmacia). Incorporated into the oligonucleotides used to amplify the two NSL fragments were sites for selected restriction endonucleases, thereby creating a multicloning site between NS 1 and −2. The resulting 4.6-kb plasmid (pNSL) could therefore be used for transformation of Synechococcus via single or double recombination events.
For the first complementation construct pEcB, the entire clpB gene from E coli was isolated from the original clone (pClpB; Squires et al 1991) in 2 steps. The first 900 bp from the 5′ end was PCR amplified in order to create a KpnI site upstream of start ATG and then restricted at the KpnI and 3′ native NcoI site. The remaining 1.9-kb 3′ region of clpB was then restricted from pClpB with NcoI and HindIII. A third fragment, a 200-bp region upstream of the Synechococcus clpB1 gene that confers ∼85% heat shock inducibility of the native promoter, was also isolated by PCR and restricted with HindIII and KpnI. All 3 DNA fragments were then ligated together and inserted into the multicloning site of pNSL at the vacant HindIII site. Afterward, a 1.8-kb chloramphenicol-resistance gene (Shapira et al 1983) was ligated just upstream of the 200-bp clpB1 promoter fragment, resulting in the final pEcB complementation plasmid (Fig 1A). The second complementation plasmid pEcB1 was made as for pEcB except for PCR-created changes at the 3′ end of the 200-bp clpB1 promoter fragment so as to alter the ribosome-binding site (RBS) (Fig 4A). The E coli strain DH5α was the host for all plasmids during construction of both pEcB and pEcB1. Correct insertion of all DNA fragments in pEcB and pEcB1 was confirmed by restriction digestion, and all PCR-amplified DNA regions were sequenced to ensure no base errors.
Fig 1.
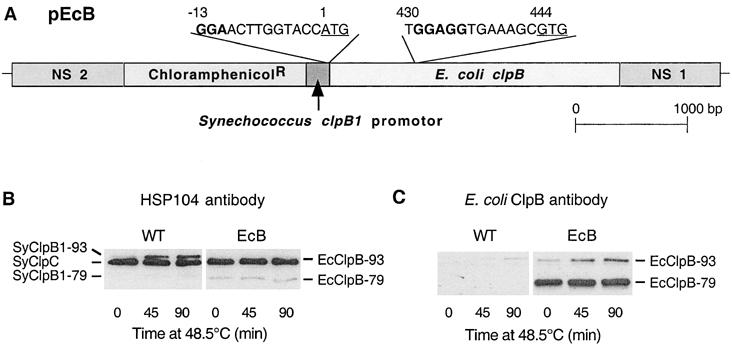
Synthesis of E coli ClpB proteins in the Synechococcus ΔclpB1 mutant. (A) Structural representation of the plasmid construct pEcB used to express the E coli clpB gene in the Synechococcus ΔclpB1 strain. The entire E coli clpB gene was cloned into pNSL under the control of the heat shock–inducible Synechococcus clpB1 promoter. The chloramphenicol-resistance gene acting as selectable marker was then inserted upstream of the clpB1 promoter. Flanking this construct were the 2 neutral-site loci (NS 1 and −2) to enable recombination of the resulting plasmid (pEcB) into a phenotypically neutral region of the Synechococcus ΔclpB1 genome (Bustos and Golden 1992). The DNA regions from the dual translational start codons (underlined) for EcClpB-79 and -93 to 13 bases upstream are shown, along with the putative RBS in bold. (B,C) Induction of ClpB proteins in wild-type Synechococcus and the complementation strain EcB. Wild-type (WT) and EcB cultures were shifted from 37 to 48.5°C for 90 minutes. Cellular proteins were isolated at selected times and separated by polyacrylamide gel electrophoresis on the basis of equal Chl content. (B) Immunoblot detection of SyClpC, SyClpB1–93, and –79 in WT, and SyClpC and EcClpB-93 and -79 in EcB using the yeast Hsp104 antibody. (C) Immunoblot detection of EcClpB proteins in EcB using a specific polyclonal antibody. Results are representative of 3 replicates
Fig 4.
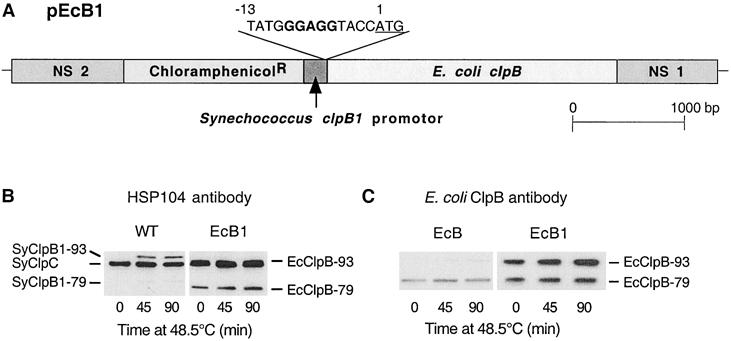
Increased synthesis of EcClpB-93 in Synechococcus ΔclpB1 mutant. (A) Structural representation of the second construct pEcB1 used to increase the amount of EcClpB-93 protein synthesized in the Synechococcus ΔclpB1 strain. The pEcB1 was made as described for pEcB in Figure 1A, except for the altered RBS (bold) upstream of the ATG start codon (underlined) in base position 1. (B,C) Induction of ClpB proteins in wild-type (WT) Synechococcus and the complementation strains EcB and EcB1. All 3 strains were shifted from 37 to 48.5°C for 90 minutes, and cellular proteins were isolated at selected times and then separated by polyacrylamide gel electrophoresis on the basis of equal Chl content. (B) Immunoblot detection of SyClpC, SyClpB1–93, and –79 in WT, and SyClpC and EcClpB-93 and -79 in EcB1 using the yeast Hsp104 antibody. (C) Immunoblot detection of EcClpB proteins in the EcB and EcB1 strains using a specific polyclonal antibody. Results are representative of 3 replicates
Synechococcus ΔclpB1 cells were transformed with linearized pEcB or pEcB1 according to the method of Van der Plas et al (1990). Putative transformants were selected on BG-11 plates supplemented with kanamycin and chloramphenicol (5 μg mL−1 of each). Correct insertion of each complementation plasmid into the NSL and complete segregation within the ΔclpB1 genome was verified by Southern blot analysis as described by Eriksson and Clarke (1996).
DNA sequence analysis
Dideoxy sequencing of the PCR-amplified DNA regions within the pEcB and pEcB1 constructs was performed using selected oligonucleotides and a thermal cycle amplification system (DYEnamicTM ET terminator cycle sequencing kit, Amersham Pharmacia). Sequencing reactions were analyzed on an automated sequencer (ABI377, Perkin Elmer) and viewed with the AutoAssembler software (Perkin Elmer).
High temperature experiments
All experiments were performed with 37°C grown cultures in early exponential growth phase at a chlorophyll (Chl) concentration of 2 to 3 μg mL−1. Chl content and cell number were calculated from whole cell spectra according to Myers et al (1980). At time zero, a volume of cells (20 mL) was taken from the 37°C cultures and transferred to an identical growth tube in a 48.5°C water-bath for 90 minutes (heat shock and preconditioning treatment), while another (20 mL) was transferred to a tube at 54°C for 15 minutes (lethal heat treatment). After 90 minutes, the 48.5°C-treated cells were transferred to 54°C for 15 minutes (thermotolerance treatment). For all temperature shifts, the light intensity was maintained at 50-μmol photons m−2 s−1 and the culture continuously bubbled with 5% CO2 in air. At each selected time, a given volume of culture was taken for absorbance measurements (1 mL), protein isolation (4 mL pelleted and frozen in liquid N2), and cell survival determination (100 μL). For cell survival determinations, the 100-μL volumes were serially diluted in fresh, sterile BG-11 medium, and then 5 μL from each dilution was spotted onto predried BG-11 agar plates. Plates were grown for several days under low light (∼5 μmol photons m−2 s−1) at 30°C. Cell survival was determined from the number of colonies counted under a light microscope and multiplied by the appropriate dilution factor. Variations in cell number within replicate cultures were normalized by dividing the calculated number of viable cells from each temperature treatment by the A750 (Eriksson and Clarke 1996). Three independent replicas were carried out for all high temperature experiments.
Protein sample preparation and Western blot analysis
Total proteins were extracted from frozen cell pellets according to the method of Clarke et al (1993). Protein samples containing equal amounts of Chl (0.25 μg) were separated on 3–8% gradient polyacrylamide Tris-acetate NuPAGE gels (Novex, San Diego, CA) and transferred to PVDF membranes (Immobilon Millipore Bedford, MA). Hsp100/Clp protein were detected using the yeast Hsp104 antibodies (antiserum 2-3; Parsell et al 1991). This serum was generated in rabbits to a 16-amino acid synthetic peptide corresponding to the Gly-rich loop of the first ATP-binding site from the yeast Hsp104 protein, a region highly conserved for all Clp/Hsp100 proteins. Because the Synechococcus ClpB1, ClpC, and E coli ClpB proteins have identical amino acid sequences in this region, this serum cross-reacts equally to each protein. A second antibody was also used that specifically recognizes both E coli ClpB proteins (Park et al 1993). All primary antibodies were detected with a horseradish peroxidase–conjugated, anti-rabbit secondary antibody made in donkey (Amersham Pharmacia) and visualized by enhanced chemiluminescence (ECL, Amersham Pharmacia). All immunoblots were performed on samples from at least 3 independent replicate experiments. Density scanning of X-ray films from each replicate immunoblot were carried out with an AlphaImager and associated software (Alpha Innotech Corp, San Leandro, CA).
RESULTS
Construction of E coli clpB complementation strain in Synechococcus ΔclpB1 mutant
The entire clpB gene from E coli was ligated into the pNSL vector along with the chloramphenicol-resistance gene as a selectable marker. A 200-bp fragment, containing the high temperature–inducible promoter of the Synechococcus clpB1 gene, was inserted upstream of the E coli clpB gene (Fig 1A). This promoter fragment confers to selected genes ∼85% of the heat shock inducibility of the native clpB1 gene in Synechococcus (Clarke and Eriksson, unpublished). The resulting pEcB construct was transformed into the Synechococcus ΔclpB1 strain (lacking the form of Synechococcus ClpB protein necessary for acquired thermotolerance) and integrated into the chromosome via recombination at the neutral site locus. This neutral site locus is a region of the Synechococcus chromosome that causes no phenotypic changes upon transformation as previously described (Bustos and Golden 1992). Correct insertion of the pEcB construct into the neutral site and its complete segregation were verified by Southern blotting.
Synthesis of E coli ClpB in Synechococcus
Under standard growth conditions, the strain resulting from transformation with the pEcB construct (ie, EcB) had no obvious phenotypic changes from wild-type Synechococcus and the original ΔclpB1 mutant, as shown by its generation time (7.5 ± 0.5 h), pigment composition, and cell morphology. The heat shock induction of the E coli clpB gene under the control of the Synechococcus clpB1 promoter was analyzed in the EcB strain using the high temperature stress conditions previously defined for Synechococcus (Eriksson and Clarke 1996). Wild-type and EcB cultures were shifted from their standard growth temperature of 37°C to 48.5°C for 90 minutes, and cell protein extracts from each strain were analyzed by Western blotting using a yeast Hsp104 antibody (Fig 1B) or an E coli ClpB-specific antibody (Fig 1C). The first antibody was made to a highly conserved 16-amino acid region within the first ATP-binding domain of the yeast Hsp104 protein and cross-reacts equally to the Synechococcus ClpC (SyClpC) and both ClpB1 (SyClpB1) proteins, as well as E coli ClpB (EcClpB). As shown in Figure 1B, the typical protein profiles for SyClpB1 and SyClpC during heat shock were observed in wild-type Synechococcus (Eriksson and Clarke 1996); ie, a high constitutive level of SyClpC that remained unchanged throughout the heat treatment, and heat shock induction of the normally low constitutive level of full-length SyClpB1 (SyClpB1–93), with the truncated SyClpB1 (SyClpB1–79) induced to a lesser extent. In the EcB strain, the high constitutive level of SyClpC hides the heat shock induction of the EcClpB-93 protein due to their similar molecular masses. In comparison, the level of EcClpB-79 in the EcB strain was significantly higher before and during the high temperature treatment compared to the SyClpB1–79 protein in wild-type Synechococcus.
To avoid the interference of the SyClpC protein, an antibody specific to the EcClpB proteins was used to determine their heat induction profiles in the EcB strain (Fig 1C). Using this antibody, the EcClpB-93 protein in the EcB strain was shown to have a similar induction profile during heat shock as SyClpB1–93 in wild-type Synechococcus, as was expected by their common gene promoters. However, the amount of EcClpB-93 protein synthesized in EcB was considerably lower relative to SyClpB1–93 in wild type, as can be deduced by the amounts of EcClpB-79 detected by both Hsp104 and EcClpB antibodies. Indeed, the level of EcClpB-79 protein in the EcB strain was many times higher than EcClpB-93, opposite to the normal proportion of both types of ClpB protein in both E coli and Synechococcus (Park et al 1993; Eriksson and Clarke 1996).
Restored thermotolerance in Synechococcus ΔclpB1 strain producing E coli ClpB
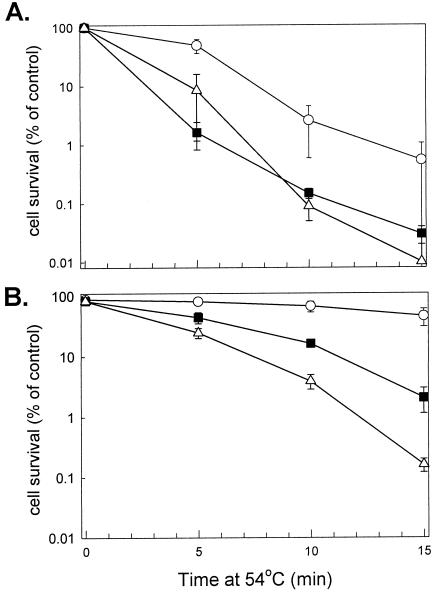
Thermotolerance assays were simultaneously performed on the 3 Synechococcus strains; wild-type, EcB, and ΔclpB1. The assay comprised of shifting each strain from the standard growth temperature of 37°C to either the severe temperature of 54°C for 15 minutes or first pretreating them at 48.5°C for 1.5 hours before the shift to 54°C. The development of thermotolerance was determined by cell survival measurements, with samples taken at selected time points, serially diluted, and then spotted on BG-11 plates (Fig 2). Quantification of the degree of acquired thermotolerance gained by each strain is shown in Figure 3.
Fig 2.
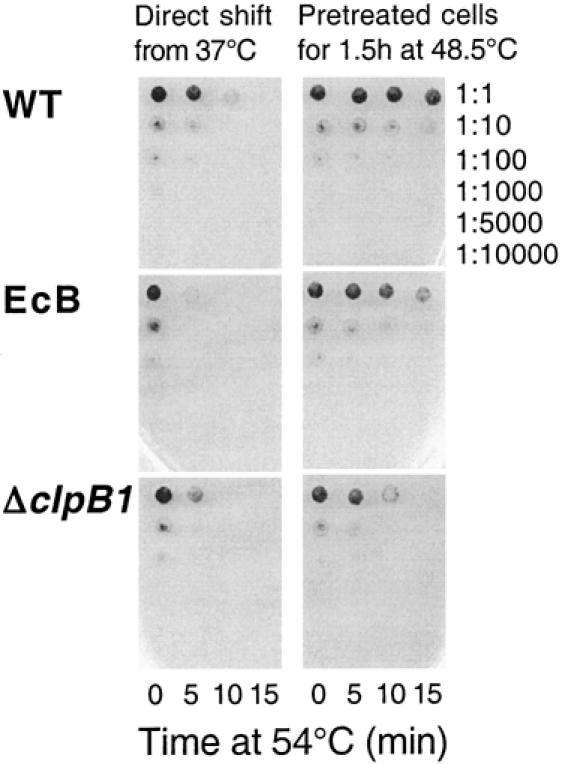
Cell survival assay for acquired thermotolerance in EcB strain. Cells of WT, EcB, and ΔclpB1 grown at 37°C were shifted either directly to 54°C for 15 minutes (left column) or first preconditioned at 48.5°C for 1.5 hours before being exposed to 54°C for 15 minutes (right column). At the indicated time points, cells were removed and serially diluted from 1:1 to 1:10 000 and then spotted on BG-11 plates. Shown is a representative thermotolerance experiment of 3 independent replicas
Fig 3.
Quantification of thermotolerance developed in the EcB strain. Synechococcus WT (○), EcB (▪), and ΔclpB1 (▵) cultures grown at 37°C were either directly shifted to 54°C for 15 minutes (A) or first preheated at 48.5°C for 1.5 hours before the shift to 54°C for 15 minutes (B). Average numbers of viable cells after each temperature treatment are expressed as percentages of the value for the 37°C control (100%). All values are averages ± SE for 3 independent experiments
As shown in Figures 2 and 3A, all 3 strains rapidly lost viability upon the direct shift from 37 to 54°C. Although the wild type was initially more resistant than the ΔclpB1 strain to this severe temperature treatment, this difference became less significant by 15 minutes. More importantly, the EcB strain exhibited the same degree of susceptibility to the 54°C shift as did the ΔclpB1 strain. When cells were pretreated at 48.5°C for 90 minutes prior to the shift to 54°C, the wild type developed higher thermotolerance than the ΔclpB1 mutant (Figs 2 and 3B), similar to that previously observed (Eriksson and Clarke 1996). In comparison, the EcB strain acquired 10 times the thermotolerance of the ΔclpB1 strain, demonstrating that the EcClpB proteins could restore thermotolerance to the Synechococcus ΔclpB1 strain.
Although EcClpB clearly confers increased thermotolerance to the ΔclpB1 mutant, the level of restored thermotolerance in EcB was less than that of the wild type (Fig 3). However, being a heterologous protein, EcClpB would not be expected to complement ClpB1 function completely and produce wild-type levels of thermotolerance in Synechococcus. The 200-bp clpB1 promoter fragment used in the complementation construct also produces ∼85% of the high temperature–induced expression of the native clpB1 gene (data not shown), thus decreasing slightly the level of EcClpB produced in the EcB strain. Another contributing factor could be the relatively small amounts of EcClpB-93 synthesized in the EcB strain relative to the amount of EcClpB-79 (Fig 1C).
Construction of second E coli clpB complementation strain in Synechococcus ΔclpB1 mutant
One likely explanation for the relatively low levels of EcClpB-93 produced in the EcB strain was differential translational efficiency for the EcClpB-93 and -79 proteins. Examination of the 2 translational start domains for both EcClpB proteins revealed sequential and positional differences in the putative RBS. Seven bases upstream of the start ValGTG codon for EcClpB-79 is an RBS with the classical GGAGG motif, whereas the putative RBS for EcClpB-93, originating from the Synechococcus clpB1 gene, is less conserved and further away from the ATG (10 bases, Fig 1A). It is plausible, therefore, that the relatively high constitutive level of EcClpB-79 in the EcB strain is due to increased affinity for available ribosomes, which subsequently interferes with the translation of EcClpB-93 during both normal growth and heat shock.
To overcome the potential problem of differential translation, a second construct was made (ie, pEcB1) in which the classical RBS site was added upstream of, and closer to, the EcClpB-93 start codon (Fig 4A). Therefore, the only difference between the pEcB1 and pEcB constructs was the 13 bases upstream of the start Met. As for pEcB, the new pEcB1 construct was transformed into the Synechococcus ΔclpB1 mutant, and the resulting strain was termed EcB1. Insertion of pEcB1 into the neutral site locus and its complete segregation in EcB1 was confirmed by Southern blotting.
Increased levels of EcClpB-93 protein restored higher acquired thermotolerance
As for EcB, no phenotypic changes were observed under standard growth conditions in the new EcB1 strain relative to the wild-type and ΔclpB1 strains. The levels of EcClpB-93 and -79 were again examined during a high temperature shift with the 2 separate antibodies, Hsp104 and E coli ClpB. In Figure 4B, the increased amount of EcClpB-93 in the EcB1 strain can be seen as a substantial thickening of the double EcClpB-93/SyClpC band. Using the E coli ClpB-specific antibodies, around 10 times more EcClpB-93 protein was detected in EcB1 than in the EcB strain, both before and during the high temperature shift. The EcClpB-79 content in EcB1 was also twice that in the EcB strain (Fig 4C). In comparison to wild-type Synechococcus, the amounts of EcClpB1–93 and -79 in the EcB strain prior to and during the heat shift were considerably higher than SyClpB1–93 and −79, despite being less heat shock inducible (Fig 4B). Despite this, the relative proportion of EcClpB-93 to -79 in EcB1 was similar to what is normally found in wild-type E coli (Park et al 1993) as well as for homologous ClpB proteins in other eubacteria (Eriksson and Clarke 1996; Grandvalet et al 1999).
With the additional EcClpB-93 protein produced in the EcB1 strain, its ability to develop thermotolerance was directly compared to that of the first complementation strain, EcB. The increased constitutive levels of both EcClp-93 and -79 did not significantly increase the basal resistance of the EcB1 strain to the direct shift from 37 to 54°C (data not shown). However, after preconditioning at 48.5°C for 90 minutes, the degree of acquired thermotolerance in EcB1 was 3-fold higher than that of the EcB strain (Fig 5).
Fig 5.
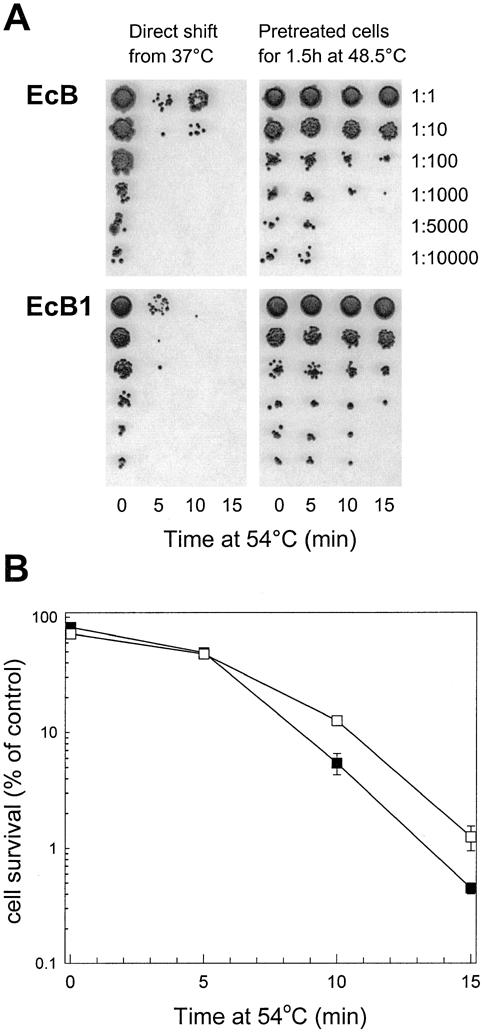
(A) Cell survival assay for acquired thermotolerance in EcB and EcB1 complementation strains. Thermotolerance experiments and cell survival determinations were done as described in Figure 2. Shown are representative experiments from 3 independent replicas for each strain. (B) Quantification of thermotolerance developed in the EcB and EcB1 strains. Synechococcus complementation strains EcB (▪) and EcB1 (□) grown at 37°C were preheated at 48.5°C for 1.5 hours before the shift to 54°C for 15 minutes. Average numbers of viable cells after each temperature treatment are expressed as percentages of the value for the 37°C control (100%). All values are averages ± SE for 3 independent experiments
DISCUSSION
E coli ClpB confers developed thermotolerance to Synechococcus
In an earlier study we have shown that the heat shock protein ClpB1 contributes to the development of thermotolerance in wild-type Synechococcus, the first such example for a bacterial form of ClpB (Eriksson and Clarke 1996). Previously, ClpB involvement in the acquisition of thermotolerance had only been demonstrated in yeast (Sanchez and Lindquist 1990), in which it functions as a molecular chaperone in the resolubilization of protein aggregates that accumulate during severe heat stress (Parsell et al 1994). Plant ClpB homologues were later shown to also confer thermotolerance to yeast (Lee et al 1994; Schirmer et al 1994), suggesting that most eukaryotic ClpB proteins function in a similar fashion. In contrast, ClpB is apparently not involved in the development of thermotolerance in E coli but instead contributes to the basal tolerance of the bacterium to high temperature stress (Squires et al 1991). In this study we examined whether the seemingly different roles of ClpB during high temperature stress in Synechococcus and E coli were due to intrinsic functional differences in the 2 proteins. We showed that the E coli ClpB protein could facilitate the development of thermotolerance in the cyanobacterium Synechococcus, suggesting the E coli protein has similar functional properties as the Synechococcus ClpB1 protein. Indeed, the extent of thermotolerance conferred by the EcClpB proteins to Synechococcus closely matched that conferred by plant ClpB to yeast (Schirmer et al 1994).
Although E coli ClpB complements the function of ClpB1 in Synechococcus, its apparent lack of involvement in acquired thermotolerance in E coli infers that the underlying mechanism for this form of heat tolerance might differ in certain eubacteria. In yeast, ClpB is essential for the acquisition of thermotolerance (Sanchez and Lindquist 1990). It promotes the resolubilization of heat-denatured proteins that form large aggregates, initiating refolding of these proteins in cooperation with the DnaK/DnaJ chaperone system (Glover and Lindquist 1998). Similar cooperation between ClpB and the DnaK system in recovering aggregated proteins has also been observed in E coli (Zolkiewski 1999) and Thermus thermophilus (Motohashi et al 1999). Therefore, the role of ClpB as a molecular chaperone would appear to be similar in both eukaryotes and eubacteria, initiating the reactivation process of heat-aggregated proteins. This is further supported by the observation that increased constitutive levels of ClpB protein in yeast, as a result of ectopic clpB gene expression, enhances cell survival to direct high temperature shifts (Lindquist and Kim 1996), similar to the role of native ClpB in E coli and other bacteria (Squires et al 1991; Allan et al 1998). Overall, these findings suggest that the dissolution of heat-induced protein aggregates by ClpB is critical for the development of thermotolerance in wild-type yeast and cyanobacteria but not in E coli. Instead, this activity would seem to be more important in E coli during direct shifts to extreme high temperatures, during which the loss of ClpB has the greatest impact.
The truncated form of E coli ClpB contributes to developed thermotolerance
Another interesting outcome of this study was the noticeable contribution of EcClpB-79 to the restoration of thermotolerance in the Synechococcus ΔclpB1 mutant. In the first complementation strain EcB, the truncated EcClpB-79 comprised over 90% of the total EcClpB protein produced, and yet the level of developed thermotolerance was significantly higher than the original ΔclpB1 strain. This suggests that the EcClpB-79 protein actively contributed to the acquired thermotolerance restored to the ΔclpB1 strain. Although the exact role of the truncated ClpB-79 remains unclear, its cotranslation with the full-length ClpB-93 is a conserved feature for all known eubacterial clpB mRNAs. Previous studies of the 2 ClpB types from E coli have proposed a regulatory role for EcClpB-79. Although both forms of ClpB possess ATPase activity, the truncated EcClpB-79 protein lacked the protein stimulated increase in ATPase activity exhibited by EcClpB-93. This protein-stimulated activity was increasingly inhibited by incorporation of EcClpB-79 subunits into the EcClpB-93 oligomeric complex (Park et al 1993). It was proposed that EcClpB-79 lacked a necessary protein-binding domain and thus regulated the function of EcClpB-93. This interpretation implies that the truncated form of ClpB may autoregulate the chaperone activity of the full-length ClpB and is thus not involved in the development of thermotolerance in organisms such as Synechococcus. However, the results in this study would suggest that ClpB-79 actively participates in the development of thermotolerance in Synechococcus, a result we have recently confirmed in a Synechococcus strain producing ClpB1–79 but not ClpB1–93 (Clarke and Eriksson, in preparation).
Translational regulation in Synechococcus
Another unexpected result of this study was the apparent translational regulation that occurs in Synechococcus conferred by the sequence and position of the RBS; a form of regulation that to our knowledge has yet to be investigated closely in cyanobacteria. This regulation was first apparent in the EcB strain, in which the amount of EcClpB-79 produced during heat shock was many times higher than that of EcClpB-93, in marked contrast to the normal proportion of both ClpB forms in various eubacteria (Park et al 1993; Eriksson and Clarke 1996; Grandvalet et al 1999). Although the clpB1 promoter fragment included in the pEcB construct produces slightly less heat-inducible gene expression than the native clpB1 promoter (∼85%), the amount of EcClpB-79 was higher in the EcB strain than the corresponding SyClpB1–79 in wild-type Synechococcus. Conversely, the level of EcClpB-93 in EcB was many times lower than the amount of its SyClpB1–93 counterpart in wild-type Synechococcus. Closer examination of the complementation construct revealed the high-affinity RBS (GGAGG) 7 bases upstream of the start ValGTG for EcClpB-79 and the less conserved RBS derived from the Synechococcus clpB1 promoter 10 bases from the ATG start for EcClpB-93. Therefore, the relatively high level of EcClpB-79 was almost certainly due to a more efficient initiation of translation at the GGAGG RBS, which concomitantly reduced the translational efficiency of EcClpB-93. This was confirmed by simply changing the RBS in the clpB1 promoter to that of the native E coli clpB promoter (GGAGG 5 bases from the ATG) in the pEcB1 construct, which resulted in a dramatic rise in EcClpB-93 content along with a slight increase in the amount of EcClpB-79.
One consequence of these findings concerning the RBS is the apparent role this feature has on the regulation of SyClpB in wild Synechococcus. It would appear that the low constitutive level of SyClpB1 proteins is due not only to a low level of clpB1 gene expression, but also to a less efficient translation of the available clpB1 transcripts. The contribution of this translational repression is clearly demonstrated by the high constitutive levels of EcClpB proteins in the 2 complementation strains. Given that the native clpB gene in E coli contains the GGAGG RBS at both translational start codons, then the low constitutive level of ClpB proteins in E coli would appear to be controlled primarily at the level of gene expression. Moreover, the degree of heat shock induction of the EcClpB-93 protein varied in the complementation strains depending on the origin of its RBS. Although the constitutive level of EcClpB-93 was very low in the EcB strain, the extent of its induction during heat shock was similar to that of SyClpB1–93 in wild-type Synechococcus. In comparison, the high constitutive level of EcClpB-79 in EcB rose much less. When the RBS for EcClpB-93 was altered in the EcB1 strain, its degree of heat shock inducibility dropped to that of the EcClpB-79 protein despite its now high constitutive level. When related to the SyClpB1 proteins in wild-type Synechococcus, these results would suggest that translational control also influences the increase in SyClpB1 proteins at high temperatures. It is possible that the level of heat shock inducibility of the EcClpB proteins in the EcB1 strain reflects the increase in expression of the clpB1 gene during heat shock in wild-type Synechococcus. An increased affinity for ribosome binding at the native RBS within the clpB1 transcripts may then contribute to the overall amount of SyClpB1 proteins produced in heat shocked Synechococcus. Although such translational regulation of Hsp synthesis is rare in eubacteria, there are several examples of ribosomal modifications during heat stress in eukaryotes that may alter ribosome binding specificity, including changes in the phosphorylation state of certain ribosomal proteins within the initiation region (Glover 1982; Duncan and Hershey 1989).
Acknowledgments
We thank Professors Catherine L. Squires for the plasmid (pClpB) containing the E coli clpB gene, Susan Lindquist for the yeast Hsp104 antibody, Chin Ha Chung for the E coli ClpB-specific antibody, and C. Peter Wolk for the chloramphenicol-resistance gene. We also thank Dr Vaughan Hurry for a critical reading of the manuscript. This work is supported by grants from the Swedish Natural Science Research Council.
REFERENCES
- Allan E, Mullany P, Tabaqchali S. Construction and characterization of a Helicobacter pylori clpB mutant and role of the gene in the stress response. J Bacteriol. 1998;180:426–429. doi: 10.1128/jb.180.2.426-429.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos SA, Golden SS. Light-regulated expression of the psbD gene family in Synechococcus sp. PCC 7942: evidence for the role of duplicated psbD genes in cyanobacteria. Mol Gen Genet. 1992;232:221–230. doi: 10.1007/BF00280000. [DOI] [PubMed] [Google Scholar]
- Clarke AK, Soitamo A, Gustafsson P, O̊quist G. Rapid interchange between two distinct forms of cyanobacterial photosystem II reaction-center protein D1 in response to photoinhibition. Proc Natl Acad Sci U S A. 1993;90:9973–9977. doi: 10.1073/pnas.90.21.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan RF, Hershey JWB. Protein synthesis and protein phosphorylation during heat stress, recovery, and adaptation. J Cell Biol. 1989;109:1467–1481. doi: 10.1083/jcb.109.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M-J, Clarke AK. The heat shock protein ClpB mediates the development of thermotolerance in the cyanobacterium Synechococcus sp. strain PCC 7942. J Bacteriol. 1996;178:4839–4846. doi: 10.1128/jb.178.16.4839-4846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover CVC. Heat shock induces rapid dephosphorylation of a ribosomal protein in Drosophila. Proc Natl Acad Sci U S A. 1982;79:1781–1785. doi: 10.1073/pnas.79.6.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- Grandvalet C, de Crécy-Lagard V, Mazodier P. The ClpB ATPase of Streptomyces albus G belongs to the HspR heat shock regulon. Mol Microbiol. 1999;31:521–532. doi: 10.1046/j.1365-2958.1999.01193.x. [DOI] [PubMed] [Google Scholar]
- Hoskins JR, Pak M, Maurizi MR, Wickner S. The role of the ClpA chaperone in proteolysis by ClpAP. Proc Natl Acad SciU S A. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama-Fujimura Y, Gottesman S, Maurizi MR. A multiple-component, ATP-dependent protease from Escherichia coli. J Biol Chem. 1987;262:4477–4485. [PubMed] [Google Scholar]
- Lee Y-RJ, Nagao RT, Key JL. A soybean 101-kD heat shock protein complements a yeast HSP104 deletion mutant in acquiring thermotolerance. Plant Cell. 1994;6:1889–1897. doi: 10.1105/tpc.6.12.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt SA, Fearon K, Danese PN, Mason TL. Hsp78 encodes a yeast mitochondrial heat shock protein in the Clp family of ATP-dependent proteases. Mol Cell Biol. 1993;13:6304–6313. doi: 10.1128/mcb.13.10.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Lindquist S, Kim G. Heat-shock protein 104 expression is sufficient for thermotolerance in yeast. Proc Natl Acad Sci U S A. 1996;93:5301–5306. doi: 10.1073/pnas.93.11.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Watanabe Y, Yohda M, Yoshida M. Heat-inactivated proteins are rescued by the DnaK·J-GrpE set and ClpB chaperones. Proc Natl Acad Sci U S A. 1999;96:7184–7189. doi: 10.1073/pnas.96.13.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J, Graham JR, Wang RT. Light harvesting in Anacystis nidulans studied in pigment mutants. Plant Physiol. 1980;66:1144–1149. doi: 10.1104/pp.66.6.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Kim KI, Woo KM, Seol JH, Tanaka K, Ichihara A, Ha DB, Chung CH. Site-directed mutagenesis of the dual translational initiation sites of the clpB gene of Escherichia coli and characterization of its gene products. J Biol Chem. 1993;268:20170–20174. [PubMed] [Google Scholar]
- Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S. The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet. 1993;27:437–496. doi: 10.1146/annurev.ge.27.120193.002253. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Sanchez Y, Stitzel JD, Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- Rippka R, Deruelles J, Waterbury JB, Herdman M, Stainer RY. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- Sanchez Y, Lindquist SL. HSP104 required for induced thermotolerance. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Glover JR, Singer MA, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- Schirmer EC, Lindquist S, Vierling E. An Arabidopsis heat shock protein complements a thermotolerance defect in yeast. Plant Cell. 1994;6:1899–1909. doi: 10.1105/tpc.6.12.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M, Neupert W, Langer T. Hsp78, a Clp homologue within mitochondria, can substitute for chaperone functions of mt-hsp70. EMBO J. 1995;14:3434–3444. doi: 10.1002/j.1460-2075.1995.tb07349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira SK, Chou J, Richaud FV, Casadaban MJ. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983;25:71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- Squires CL, Pedersen S, Ross BM, Squires C. ClpB is the Escherichia coli heat shock protein F84.1. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MW, Maurizi MR. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J Biol Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- Van der Plas J, Hegeman H, De Vrieze G, Tuyl M, Borrias M, Weisbeek P. Genomic integration system based on pBR322 sequences for the cyanobacterium Synechococcus sp. PCC 7942 transfer of genes encoding plastocyanin and ferredoxin. Gene. 1990;95:39–48. doi: 10.1016/0378-1119(90)90411-j. [DOI] [PubMed] [Google Scholar]
- Wang J, Hartling JA, Flanagan JM. The structure of ClpP at 2.3 Å resolution suggests a model for ATP-dependent proteolysis. Cell. 1997;91:447–456. doi: 10.1016/s0092-8674(00)80431-6. [DOI] [PubMed] [Google Scholar]
- Wojtkowiak D, Georgopoulos C, Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- Zolkiewski M. ClpB cooperates with DnaK, DnaJ, and GrpE in suppressing protein aggregation. J Biol Chem. 1999;274:28083–28086. doi: 10.1074/jbc.274.40.28083. [DOI] [PubMed] [Google Scholar]