Abstract
The majority of breast cancer deaths result from metastases rather than from direct effects of the primary tumor itself. Recently, Landemaine and colleagues described a six-gene signature purported to predict lung metastasis risk. They analyzed gene expression in 23 metastases from breast cancer patients (5 lung, 18 non-lung) identifying a 21-gene signature. Expression of 16 of these was analyzed in primary breast tumors from 72 patients with known outcome, and six were selected that were predictive of lung metastases: DSC2, TFCP2L1, UGT8, ITGB8, ANP32E, and FERMT1. Despite the value of such a signature, our analysis indicates that this analysis ignored potentially important confounding factors and that their signature is instead a surrogate for molecular subtype.
Introduction
Breast tumors are heterogeneous, and different subtypes have greater or lesser propensity to metastasize to particular organ sites. Basal-like and luminal B subtypes have the greatest likelihood to metastasize to lung (40% and 36.7%, respectively; ref. 1). In contrast, ERBB2+, luminal A, and normal-like tumors preferentially metastasize to other sites, with rare recurrence in lung (1). Given this difference in metastatic profile, we examined Landemaine's six-gene signature using their training, validation, and other data sets and found it to be more predictive of molecular subtype than of metastasis site independent of subtype.
Materials and Methods
Gene expression data sets
Published data sets were downloaded from the Gene Expression Omnibus4 or ArrayExpress5 database; accession numbers are given in Supplementary Table S1. Data were normalized using robust multiarray averaging (2) using the Bioconductor package affy (R, version 2.1, Bioconductor version 1.8). The EMC-344 data set, available only as MAS5 processed data, was quantile normalized and converted to log 2 expression values. Expression profiles were visualized using average linkage hierarchical clustering using the Bioconductor package made4 (3) using Euclidean distance or 1 – Pearson correlation coefficient distance, as appropriate.
Assigning molecular subtype
Breast cancers can be subdivided into clinically relevant, prognostic molecular subtypes based on expression of characteristic genes. Basal-like breast cancer is typically negative for expression of ESR1/PGR/ERBB2; ERBB2+ tumors have amplification of the ERBB2 gene; and the ESR1+ luminal subtype can be subdivived by grade: luminal A (low grade) and luminal B (high grade). To assign subtypes in the various data sets analyzed, we examined expression of 12 Affymetrix probe sets (Supplementary Table S2) representing estrogen receptor (ER) genes (ESR1, PGR, and GATA3), ERBB2 genes (ERBB2 and GRB7), and the grade signature described by Ma and colleagues (4). Using data from these probe sets, we performed hierarchical clustering analysis and assigned subtype based on overall expression patterns; our assignments were consistent with reported clinical assignments where available. As validation, assignments were compared with those based on the intrinsic breast cancer quantitative reverse transcription-PCR (RT-PCR) signature (5).
Mapping of the six-gene signature to other microarray platforms
The Landemaine signature consists of six probe sets (204751_x_at, 227642_at, 228956_at, 211488_s_at, 208103_s_at, and 60474_at) that were reported to correspond to DSC2, TFCP2L1, UGT8, ITGB8, ANP32E, and FERMT1, respectively (6). The data sets used in our analysis were obtained on two different GeneChip platforms: U133Plus2 (6, 7) and U133A (refs. 8–11; see Supplementary Table S1). Only four of the six probe sets were present on U133A (the “intersection set”); the missing probe sets are 227642_at (TFCPL1) and 228956_at (UGTB8). It should be noted that neither of these two probe sets maps to the coding region of their respective gene sequences in EnsEMBL (release 50). Consequently, we expanded our analysis to include all probe sets mapping to Landemaine's six genes. Fourteen and 10 probe sets map to the six genes on U133Plus2 and U133A arrays, respectively (Supplementary Table S3; the “all-mapped set”). In our analysis, we used Landemaine's six probe sets, the intersection set, and the all-mapped set. Affymetrix arrays often contain multiple probe sets for individual genes, including probe sets with partial matches to other genes and probe sets for alternate splice forms for individual genes; these can sometimes give conflicting results although not always. The decision to use multiple array-based probe sets was motivated by our desire to both replicate Landemaine's analysis and take a more inclusive approach to Landemaine's signature, particularly when comparing across array designs.
Statistical analyses
All analyses were done using the R statistical language (release 2.7.1) and Bioconductor (release 2.2). Associations between clinical and biological covariates and expression of the six-gene signature were analyzed using two methods. First, we applied Goeman's globaltest (Bioconductor package globaltest version 4.10.0), which is based on an empirical Bayesian generalized linear model where the regression coefficients between expression data and clinical outcome are the random variables estimated using a goodness-of-fit test (12). Second, we used an ANCOVA approach (Bioconductor package GlobalAncova version 3.6.0) to test for the association between expression values and clinical covariates (13, 14). GlobalAncova compares linear models via the extra sum of squares principle and thus tests whether the expectation of expression levels differs between clinical covariates for a given group of genes. There was a high level of consensus between these two approaches. R scripts for both analyses are included in Supplementary Materials.
Comparison to published gene expression signatures
Gene symbols for each of the lung metastasis six-gene signature were searched against GenSigDB, a collection of more than 250 breast cancer gene signatures that we manually curated from published studies.6
Results
High-grade basal-like and luminal B tumors have a propensity to metastasize to lung (1). However, studies identifying gene expression signatures predictive of lung metastases have focused on heterogeneous patient groups without considering subtype-specific effects (6, 8). Landemaine's six-gene signature, the focus of our analysis, is claimed to predict lung metastasis risk independent of other factors. Our analysis, using their original training data set (6), their validation data (8, 9), and three additional breast cancer data sets withdefined subtypes (7, 10, 11), finds Landemaine's six genes to be more discriminative in identifying triple-negative basal-like tumors rather than in identifying the site of distant metastasis independent of molecular subtype.
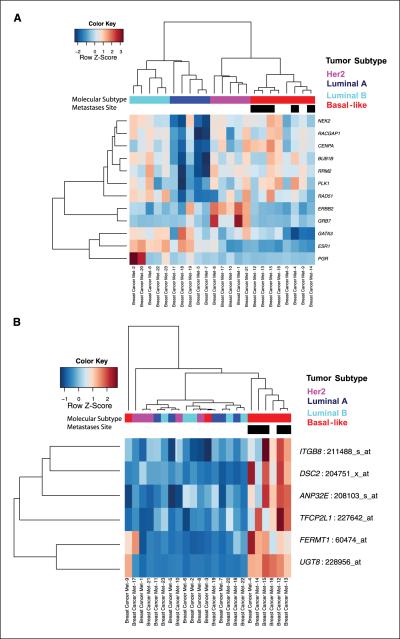
Hierarchical clustering analysis of probe sets that categorize breast cancer molecular subtypes (Supplementary Table S2) was used to assign subtypes in Landemaine's 23 metastasis samples (Fig. 1; Table 1A). Eight were negative for ESR1, PGR, and ERBB2 gene expression and expressed genes associated with high grade, typical of basal-like breast tumors. Five metastasis samples expressed high levels of ERBB2 and GRB7 and were classified as ERBB2+. Of the 10 positive for ESR1 expression, 5 were identified as high-grade (luminal B) and 5 as low-grade (luminal A) tumors (Fig. 1A). All of the lung metastasis (n = 5) samples were classified as basal-like molecular subtype (n = 8) and this relationship was significant (Pearson χ2 test, P < 0.01), indicating a potentially confounding covariate within this data set. This is supported by further hierarchical clustering analysis using Landemaine's six-gene signature, which shows a clear separation between basal-like and non-basal-like tumors (Fig. 1B; globaltest, P < 0.0001; GlobalAncova, P < 0.001).
Figure 1.
Heat maps show results of hierarchical clustering analysis of gene expression profiles of (A) 12 intrinsic molecular subtype genes (Affymetrix probe sets are given in Supplementary Table S2) and (B) the six-gene signature in the Landemaine data set (6).
Table 1.
Cross-tabulation of molecular subtypes and propensity to metastasize to lung
| A. Landemaine data set | ||
|---|---|---|
| Lung metastases tissue | Non-lung metastases tissue | |
| Basal-like | 5 | 3 |
| ERBB2+ | 0 | 5 |
| Luminal A | 0 | 5 |
| Luminal B | 0 | 5 |
| B. MSK data set | ||||
|---|---|---|---|---|
| Metastasis to bone but not to lung | Metastasis to both lung and bone | Metastasis to lung but not to bone | No metastasis to lung or bone | |
| Basal-like | 1 | 4 | 6 | 14 |
| ERBB2+ | 1 | l | 2 | 14 |
| Luminal A | 3 | 0 | 0 | 7 |
| Luminal B | 4 | 0 | 1 | 24 |
Unfortunately, Landemaine's sample annotation lacks information necessary to confirm our subtype assignment: No information is available on the total number of patients profiled (it is possible that there were fewer than 23 patients, some with metastases to multiple sites), relevant clinical and histopathologic data, or gene expression profiles from the primary tumors. Consequently, we validated our observation of confounding effects by analyzing additional published data sets, including those used by Landemaine for confirmation.
We first examined expression profiles of primary breast cancers from patients at Memorial Sloan-Kettering Cancer Center (“MSK” data set) for which metastasis status was known (8); this data set was used by Landemaine for validation. MSK contained profiles from 98 tumors, 82 of which had 3-year follow-up annotation including information on metastasis. These 82 samples were assigned to molecular subtypes: basal-like (n = 25), ERBB2+ (n = 18), luminal A (n = 10), and luminal B (n = 29; Supplementary Figs. S1 and S2). Six of the nine tumors with lung metastases had a basal-like profile (Table 1B), and the association between molecular subtype and metastasis site was significant (χ2 test, P < 0.05).
We then applied global gene set analysis to test whether the six-gene signature is predictive of metastasis site or subtype (Supplementary Table S4). Globaltest (12) and GlobalAncova (13, 14) analyses reported a significant association between expression of the six-gene signature and molecular subtype (intersection or all-mapped probe sets, P < 0.0001; n = 82). Whereas there was a marginally significant association with metastasis site (intersection probe set: globaltest, P = 0.05; GlobalAncova, P < 0.05; all-mapped probe set: globaltest, P = 0.1; GlobalAncova, P < 0.05; n = 82), this was no longer significant when adjusted for molecular subtype (globaltest or GlobalAncova, P > 0.05). By contrast, the association between expression of the six genes and subtype remained highly significant even when corrected for metastasis status (globaltest or GlobalAncova, P < 0.0001; n = 82). When only the basal-like breast samples were considered, Landemaine's six genes were not able to predict propensity to metastasize to lung or non-lung sites using either the intersection (globaltest or GlobalAncova, P > 0.05; n = 26) or all-mapped probe sets (globaltest or GlobalAncova, P > 0.05; n = 26).
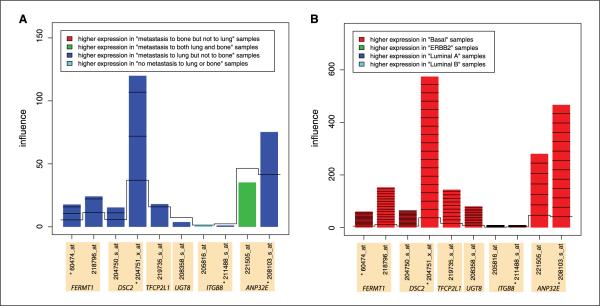
Returning to the full set of 82 MSK samples, we examined the contribution of each of Landemaine's six genes to the association with subtype. The model with four intersection probe sets was influenced most strongly by expression of DSC2 (probe set 204751_x_at) and ANP32E (probe sets 208103_s_at), which were expressed in the basal-like samples. When the 10 all-mapped probe sets are considered, the same genes, DSC2 (probe set 204751_x_at) and ANP32E (probe sets 208103_s_at and 221505_at), most strongly influenced the association (Fig. 2; Supplementary Fig. S3). The association between expression of Landemaine's six genes and metastasis site was not evaluated in luminal B tumors in MSK due to insufficient sample size (Table 1B; n = 1). These analyses indicate that Landemaine's six-gene signature is predictive of subtype, but not metastasis site, in the MSK data set.
Figure 2.
Results of globaltest analysis, which tested the association between gene expression profiles and (A) metastasis status and (B) molecular subtype in the MSK data set (8). The bar height indicates the influence of the respective gene on the test statistic. Note that A and B are very different scales. The color shows in which of the phenotype group the gene has higher expression values. *, four probe sets that intersect with the Landemaine six probe sets.
Landemaine also validated their signature on a larger cohort (n = 344) of early-stage patients (EMC-344, 9), some of whom had lung metastases as the first site of relapse (n = 31) or among cumulative sites of distant relapse (n = 42; see Supplementary Tables S5 and S6 for summaries). We found that although the intersection probe sets (but not the all-mapped probe sets) were significantly associated with first (globaltest, P < 0.001; GlobalAncova, P < 0.05) or all lung metastasis events (globaltest or GlobalAncova, P < 0.01), this effect remains only marginally significant when corrected for subtype (globaltest or GlobalAncova; intersection probe sets, P < 0.05; Supplementary Table S7; Supplementary Figs. S4 and S5). By contrast, subtype is highly significant (globaltest or GlobalAncova; intersection or all-mapped probe sets, P < 0.0001; Supplementary Fig. S6 and Table S7) and remains so even when corrected for first or all lung metastases (globaltest or GlobalAncova; intersection or all-mapped probe sets, P < 0.0001; Supplementary Fig. S5). When we examined specific breast cancer subtypes in the EMC-344 data set, there was no association between first or all lung metastasis events and expression of the Landemaine signature (intersection or all-mapped probe sets) in basal-like tumors (n = 84; P > 0.05). There is a weak association between expression of the intersection probe sets, but not of the all-mapped probe sets, and first recurrence to lung in luminal B breast cancer (globaltest or GlobalAncova, P < 0.05; n = 95). However, there is no association between expression of these probe sets (intersection or all-mapped) and lung metastases in luminal B breast cancer (n = 95). Therefore, assessment of the Landemaine signature indicates that although strongly associated with subtype, there is little evidence to support it as a predictor of lung metastases in two of their validation data sets.
We then tested the association between expression of the six genes and subtype in three additional publicly available breast cancer data sets (7, 10, 11). In each case, molecular subtypes were provided with the sample annotation. In a study of primary breast tumors, Farmer and colleagues (10) defined three tumor subtypes: an ER-positive luminal group (n = 27) and two ER-negative subtypes, basal-like (n = 16) and an androgen receptor-positive group, which they called molecular apocrine (n = 6). The molecular apocrine tumors expressed ERBB2 and shared features with the ERBB2+ subtype. Among Landemaine's six genes (using either 4 intersection probe sets or 10 all-mapping probe sets), we observed a significant association between expression of these and subtype using either globaltest or GlobalAncova analysis (P < 0.0001; Supplementary Fig. S7). In both, ANP32E and DSC2 most influenced the model and were both significantly up-regulated in basal-like tumors relative to the other subtypes.
Basal-like tumors cluster with and are phenotypically similar to BRCA1-deficient breast tumors. Bothare ER negative, display high levels of chromosome abnormalities, and have poorer prognosis compared with other subtypes. In a study of BRCA1 breast cancer and sporadic basal-like breast cancer gene expression using U133Plus2 arrays, Richardson and colleagues (7) provided both BRCA1 status and subtype information for their samples: sporadic basal-like (n = 18), BRCA1-deficient (n = 2), non-basal-like (n = 20), and normal breast (n = 7). Both GlobalAncova and globaltest identified a significant association between expression of Landemaine's six genes and the basal-like phenotype (P < 0.0001; Supplementary Fig. S8). DSC2, ANP32E, and ITGB8 had greatest influence on the test statistic in both analyses.
In an analysis of 51 breast cancer cell lines, Neve and colleagues (11) divided basal-like cell lines into two groups, basal B and basal A, based on morphology and patterns of gene expression. The basal B were less differentiated, displayed a greater mesenchymallike appearance, and were also more invasive in Boyden chamber assays than were basal A or luminal cells (11). We examined the expression of Landemane's six predictive genes in these cell lines to determine their expression profiles in highly invasive basal B and other cell line subtypes. Global gene set analysis found these genes to be associated with expression in both basal A and basal B, but not the luminal, breast cancer cell lines (P < 0.0001; Supplementary Fig. S9). In bothglobaltest and GlobalAncova analyses, expression of DSC2 and FERMT1 was associated with basal A and basal B cell lines, respectively. However, although ANP32E influenced both models, it was associated with basal A by GlobalAncova and with basal B by globaltest. Therefore, although the six-gene signature is associated with expression in basal-like cell lines, it is not specific for basal A or the more aggressive basal B subtype.
These additional analyses further support the association between expression of Landemaine's six genes and basal-like breast cancer. It is interesting to note that expression of DSC2 and ANP32E strongly influences the association between the six-gene signature and subtype, specifically basal-like, in each analysis. Because of this strong association, we investigated whether Landemaine's genes had been previously reported to be important in basal-like breast cancer. We checked the six genes against GenSigDB, a manually curated database of more than 250 published microarray gene expression signatures in breast cancer.6 We found that five of the six genes had been previously identified in breast cancer gene signatures (Table 2; Supplementary Materials); only FERMT1 was novel.
Table 2.
Appearance of six-signature genes in other published predictive gene sets
| Publication in which the gene signature was described |
Genes [present (1) or absent (0)] |
|||||||
|---|---|---|---|---|---|---|---|---|
| Title | Reference | PubMed ID | ANP32E | DSC2 | TFCP2L1 | UGT8 | ITGB8 | FERMT1 |
| Identification of molecular apocrine breast tumours by microarray analysis. | (10) | 15897907 | 1 | 1 | 0 | 1 | 0 | 0 |
| Gene expression profiling predicts clinical outcome of breast cancer. | (20) | 11823860 | 0 | 1 | 0 | 1 | 0 | 0 |
| An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. | (19) | 16141321 | 1 | 1 | 0 | 0 | 0 | 0 |
| Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancers. | (16) | 16651414 | 1 | 0 | 1 | 0 | 0 | 0 |
| Breast cancer classification and prognosis based on gene expression profiles from a population-based study. | (15) | 12917485 | 0 | 1 | 0 | 0 | 0 | 0 |
| Breast cancer molecular subtypes respond differently to preoperative chemotherapy. | (21) | 16115903 | 1 | 0 | 0 | 0 | 0 | 0 |
| Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. | (22) | 17389037 | 0 | 0 | 0 | 0 | 1 | 0 |
| Bromodomain 4 activation predicts breast cancer survival. | (23) | 18427120 | 1 | 0 | 0 | 0 | 0 | 0 |
| Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay. | (5) | 16626501 | 0 | 1 | 0 | 0 | 0 | 0 |
| The molecular portraits of breast tumors are conserved across microarray platforms. | (24) | 16643655 | 0 | 0 | 1 | 0 | 0 | 0 |
| Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. | (17) | 14737219 | 1 | 0 | 0 | 0 | 0 | 0 |
| Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. | (25) | 17910759 | 0 | 0 | 1 | 0 | 0 | 0 |
| Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. | (26) | 18044827 | 0 | 0 | 0 | 0 | 1 | 0 |
NOTE: Further details about the genes and gene signatures are provided in Supplementary Materials.
Whereas roles for DSC2 (a desmocollin) or ANP32E (a member of the leucine-rich acidic nuclear phosphoprotein 32 family) are not well established in breast cancer, these two genes significantly associated with subtype in our analyses and have been previously associated with ER-negative, basal-like breast cancer in multiple studies (Table 2). DSC2 is listed in five microarray breast cancer gene expression signatures, including those that define “intrinsic” molecular subtypes (5, 15). It is a member of the 53-gene set optimized for real-time quantitative RT-PCR subtyping of breast cancer (5). ANP32E was identified in six predictive gene expression signatures. ANP32E was found to be differentially expressed in triple-negative medullary basal-like breast cancer (16) and is a member of the wound response signature (17), which has been found to be predictive of poor prognosis. Whereas it has been reported to regulate the activity of the tumor suppressor protein phosphatase PP2A in cerebellar synatogenesis (18), we know of no study that has reported such a role for it in breast cancer. Expression of both DSC2 and ANP32E is also associated with p53 mutation (19), which is predictive of poor patient outcome and is most frequently observed in patients with basal-like or ERBB2+ breast cancer. The role of ANP32E and DSC2 in basal-like breast cancer warrants further investigation.
In analyzing the NKI and EMC data sets, Landemaine used survival to validate their lung metastasis signature. Basal-like breast cancer, the wound healing signature, p53 mutation, and ER-negative breast cancer are all associated with poor survival. Given these strong associations, it is not unexpected that Landemaine found these genes to be predictive of patient survival.
Discussion
Our analysis of Landemaine's signature indicates that it is confounded by subtype and that it instead predicts basal-like subtype rather than lung metastasis. The true test of the power of Landemaine's six-gene signature to predict lung metastasis would be to show its ability to differentiate between basal-like breast cancer with and without lung metastases, or to selectively identify lung metastases arising in patients with luminal B (highgrade, ER+) breast cancer, but this is not something that can be investigated with the available data. More generally, our work suggests that more comprehensive sample annotation of gene expression data is necessary and that greater care must be taken in analyzing gene expression signatures to account for confounding effects.
Supplementary Material
Acknowledgments
Grant support: National Cancer Institute of the U.S. NIH, grant R01-CA098522-01, and the Dana-Farber Cancer Institute Womens Cancer Program, Boston, Massachusetts and the Kittredge Foundation.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
6A.C. Culhane, T. Schwarzl, K.C. Picard, et al. GenSigDB, a resource of manually curated cancer gene expression signatures, in preparation.
Disclosure of Potential Conflicts of Interest No potential conflicts of interest were disclosed.
References
- 1.Smid M, Wang Y, Zhang Y, et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 2008;68:3108–14. doi: 10.1158/0008-5472.CAN-07-5644. [DOI] [PubMed] [Google Scholar]
- 2.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 3.Culhane AC, Thioulouse J, Perrière G, Higgins DG. MADE4: an R package for multivariate analysis of gene expression data. Bioinformatics. 2005;21:2789–90. doi: 10.1093/bioinformatics/bti394. [DOI] [PubMed] [Google Scholar]
- 4.Ma XJ, Salunga R, Dahiya S, et al. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin Cancer Res. 2008;14:2601–8. doi: 10.1158/1078-0432.CCR-07-5026. [DOI] [PubMed] [Google Scholar]
- 5.Perreard L, Fan C, Quackenbush JF, et al. Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay. Breast Cancer Res. 2006;8:R23. doi: 10.1186/bcr1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Landemaine T, Jackson A, Bellahcene A, et al. A six-gene signature predicting breast cancer lung metastasis. Cancer Res. 2008;68:6092–9. doi: 10.1158/0008-5472.CAN-08-0436. [DOI] [PubMed] [Google Scholar]
- 7.Richardson AL, Wang ZC, De Nicolo A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minn AJ, Gupta GP, Padua D, et al. Lung metastasis genes couple breast tumor size and metastatic spread. Proc Natl Acad Sci U S A. 2007;104:6740–5. doi: 10.1073/pnas.0701138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farmer P, Bonnefoi H, Becette V, et al. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–71. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 11.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goeman JJ, van de Geer SA, de Kort F, van Houwelingen HC. A globaltest for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–9. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- 13.Hummel M, Meister R, Mansmann U. GlobalANCOVA: exploration and assessment of gene group effects. Bioinformatics. 2008;24:78–85. doi: 10.1093/bioinformatics/btm531. [DOI] [PubMed] [Google Scholar]
- 14.Mansmann U, Meister R. Testing differential gene expression in functional groups. Methods Inf Med. 2005;44 [PubMed] [Google Scholar]
- 15.Sotiriou C, Neo SY, McShane LM, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A. 2003;100:10393–98. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bertucci F, Finetti P, Cervera N, et al. Gene expression profiling shows medullary breast cancer is a subgroup of basal breast cancers. Cancer Res. 2006;66:4636–44. doi: 10.1158/0008-5472.CAN-06-0031. [DOI] [PubMed] [Google Scholar]
- 17.Chang HY, Sneddon JB, Alizadeh AA, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2:E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costanzo RV, Vilá-Ortíz GJ, Perandones C, Carminatti H, Matilla A, Radrizzani M. Anp32e/Cpd1 regulates protein phosphatase 2A activity at synapses during synaptogenesis. Eur J Neurosci. 2006;23:309–24. doi: 10.1111/j.1460-9568.2005.04555.x. [DOI] [PubMed] [Google Scholar]
- 19.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102:13550–55. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van 't Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 21.Rouzier R, Perou CM, Symmans WF, et al. Breast cancer molecular subtypes respond differently to preoperative chemotherapy. Clin Cancer Res. 2005;11:5678–85. doi: 10.1158/1078-0432.CCR-04-2421. [DOI] [PubMed] [Google Scholar]
- 22.Turashvili G, Bouchal J, Baumforth K, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crawford NP, Alsarraj J, Lukes L, et al. Bromodomain 4 activation predicts breast cancer survival. Proc Natl Acad Sci U S A. 2008;105:6380–5. doi: 10.1073/pnas.0710331105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kreike B, van Kouwenhove M, Horlings H, et al. Gene expression profiling and histopathological characterization of triple-negative/basal-like breast carcinomas. Breast Cancer Res. 2007;9:R65. doi: 10.1186/bcr1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergamaschi A, Tagliabue E, Sørlie T, et al. Extracellular matrix signature identifies breast cancer subgroups withdifferent clinical outcome. J Pathol. 2008;214:357–67. doi: 10.1002/path.2278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.