Abstract
We have previously proposed that sequence variation of the CD101 gene between NOD and C57BL/6 (B6) mice accounts for the protection from type 1 diabetes (T1D) provided by the Idd10 region, a <1 Mb region on mouse chromosome 3. Here, we provide further support for the hypothesis that Cd101 is Idd10 using haplotype and expression analyses of novel Idd10 congenic strains coupled to the development of a CD101 knockout mouse. Susceptibility to T1D was correlated with genotype-dependent CD101 expression on multiple cell subsets, including FoxP3+ regulatory CD4+ T cells, CD11c+ dendritic cells and Gr1+ myeloid cells. The correlation of CD101 expression on immune cells from four independent Idd10 haplotypes with the development of T1D supports the identity of Cd101 as Idd10. Since CD101 has been associated with T regulatory and antigen presentation cell functions, our results provide a further link between immune regulation and susceptibility to T1D.
Keywords: Rodent, Diabetes, Autoimmunity
Introduction
Type 1 diabetes (T1D) in humans and in an animal model of T1D, the NOD mouse, is genetically complex: variation at a large number of genes influences disease susceptibility. Using a series of congenic strains, we previously mapped one such insulin-dependent diabetes (Idd) gene, Idd10, to a 950 kb region on mouse chromosome 3 (1). Sequencing of Cd101, one of seven genes within the Idd10 region showed multiple variants between the NOD and C57BL/6 (B6) strains, including ten single nucleotide polymorphisms (SNPs) causing amino acid substitutions. Four of the substitutions were nonconservative, with two of the four potentially causing an alteration in N-linked glycosylation (1).
Although the biological functions of CD101 remain unclear and there are no known ligands for CD101, this transmembrane molecule having seven Ig-like domains is expressed by multiple subsets of immune cells including FoxP3+ regulatory T cells, effector CD4 and CD8 T cells, granulocytes, dendritic cells, and monocytes in humans and mice (2–6). There are multiple lines of evidence using human cells suggesting that CD101 modulates T cell activation either directly or indirectly via dendritic cells that express CD101 (5–8). CD101 expression levels on mouse regulatory T cells were demonstrated to be positively correlated with functional suppression (3).
Here, we have examined further the candidacy of CD101 as Idd10. We have performed a haplotype analysis using newly developed Idd10 congenic strains, results from which are consistent with the hypothesis that Cd101 is Idd10. In addition, using a newly developed monoclonal antibody to CD101, we show that expression levels of CD101 on multiple cell subsets are genotype-dependent, including CD101 levels on FoxP3+ regulatory CD4+ T cells, CD11c+ dendritic cells and Gr1+ myeloid cells. Notably, increased CD101 expression on Gr1+ cells in the bone marrow correlates with an increased percentage of Gr1+ cells. Consistent with this positive correlation between increased CD101 expression and Gr1+ cells in the bone marrow of Idd10 congenic mice, the percentage of Gr1+ cells in the bone marrow was reduced in a newly developed CD101 KO strain.
Materials and Methods
Mice
All mice were housed under specific pathogen-free conditions, and the appropriate institutional review committee approved experimental procedures. NOD/MrkTac (NOD) and C57BL/6NTac (B6) mice were purchased from Taconic Inc. (Germantown, NY). The NOD.B6 Idd10 (N16) strain (Taconic line 3538) was developed from the NOD.B6 Idd3 Idd10 (N12) strain (Taconic line 1100) (9), by backcrossing to NOD and genotyping progeny using NOD/B6 polymorphic markers to isolate the Idd10 congenic segment. NOD.A/J Idd10 (N10) and NOD.CAST Idd10 (N9) were developed by backcrossing A/J and CAST/EiJ mice that were obtained from The Jackson Laboratory (Bar Harbor, ME) to the NOD background using polymorphic markers near and in the Idd10 region to define recombination events.
Genotyping
DNA extraction for genotyping and genotyping methods were described previously (9). Primer3 (10) was used to design primers for PCR that were then synthesized by Sigma-Genosys (Haverhill, U.K.). Sequences of D3Nds and D3Mit microsatellite markers are available at http://www-gene.cimr.cam.ac.uk/todd/public_data/mouse/NDS/NDSMicrosTop.html and http://mouse.ensembl.org, respectively. All remaining primers and probes used in this study are available in Supplemental Table 1.
Generation of a CD101 null B6 strain
A 10 kb targeting vector was designed to the B6 CD101 sequence with a 4.6 kb 5’-homology arm and a 2.9 kb 3’-homology arm. The CD101 targeting vector contains one loxP site immediately before the ATG-start codon. This loxP site is preceded by a second ATG-start codon in frame with the CD101 coding sequence; the targeted locus will encode the CD101 protein with a 15 amino acid tag. A second loxP site is inserted distal of the PGK-neo selectable marker cassette. This loxP site is followed by three stop codons (TAATAATAA). Using Cre recombinase, the sequence between the two loxP sites can be removed resulting in the deletion of exon 1 of CD101, but leaving the 15 amino acid tag which is now in frame with the three stop codons. The CD101 gene was targeted in B6-derived Bruce4 cells, chimeras produced and mated with Cre deleter mice, and mice having one CD101 knockout allele backcrossed to B6/Tac mice for 7 generations. Since Bruce4 ES cells have non-B6 regions derived from the strain used to donate the Thy1 congenic region present in the B6 strain from which the Bruce4 ES cells were derived (11) and the Cre deleter mice also have non-B6 regions (12), a 1449 SNP marker panel across 19 autosomes and the X chromosome, averaging a genetic interval of 5 Mbps (1449 marker panel, Taconic) was used to verify that non-B6 regions were not detected in the backcrossed CD101 knockout mice. Sequences of primers used to genotype the WT and targeted CD101 alleles are included in Supplemental Table 1.
Generation of anti-mouse CD101 mAbs
RNA was isolated from B6 splenocytes (Absolutely RNA, Stratagene, La Jolla, CA), and cDNA was transcribed using reverse transcriptase (SuperScript II, Invitrogen, Carlsbad, CA) and CD101-specific primers. The full-length extracellular domain of CD101 was cloned by PCR (Pfu polymerase, Stratagene) from the cDNA and fused in-frame with the hinge and Fc domains of mouse IgG2a (mutated to alter complement and Fc binding epitopes), and fidelity was assured by sequencing. This CD101-Ig construct was sub-cloned into the pEFIRES-P expression vector (Hobbs, Jitrapakdee et al. 1998), and stably transfected in CHO-K1 cells (Lipofectamine, Invitrogen, Carlsbad, CA). The Ig fusion protein was purified by protein A-Sepharose absorption and quantified by detection of mouse IgG2a via ELISA (BD Pharmingen, San Diego, CA).
Purified protein was utilized for immunization and generation of rat hybridomas. Purified CD101-Ig (50 µg) and 80 µg of E. coli DnaK protein were mixed and conjugated to amino-polystyrene beads (Spherotech Inc., Lake Forest, IL). The bead-protein conjugate was combined with RIBI adjuvant and was used for immunization of a Lewis rat. The animal was injected 5 times over a 15-day period with an aliquot of conjugate containing 5 µg CD101-Ig. On day 18, lymphocytes were harvested, fused to the X63-Ag8.653 cell line, and distributed into ten 96-well plates. Seven days later, wells were screened in a direct ELISA for reactivity with CD101-Ig and for lack of reactivity with an irrelevant Fc-fusion protein. Five anti-CD101 mAbs were developed and one (clone 307707), a rat IgG2aκ, is available from R&D Systems (Minneapolis, MN). We have verified that the avidity of clone 307707 is identical for the B6 and NOD allotypes by observing that saturation of CD101 staining was achieved with the same concentration of clone 307707 and that a 30-fold titration of the reagent produced an identical step-wise reduction of staining on both allotypes. These results indicate that clone 307707 recognizes a monomorphic epitope on the CD101 protein, although which of the 7 Ig domains present in the CD101 molecule is recognized by clone 307707 is unknown. Thus, the allele-specific expression differences described in this manuscript are not explained by differential binding of the mAb clone 307707 to the different allotypes.
Variant identification in Idd10
A portion of the Idd10 region has been resequenced using NOD BAC clones selected and sequenced at WTSI, and deposited at EMBL (http://www.ebi.ac.uk/embl/) (AL663099, AL669922, AL669933, AL645760, AL645757, AL669937, AL672281). To identify polymorphisms between NOD and B6 BAC-based sequences in the Idd10 region, NOD-derived sequenced was aligned against the B6 mouse genome sequence as described previously (9). The NOD, A/J and CAST genomes have been sequenced using next generation sequencing technology (http://www.sanger.ac.uk/resources/mouse/genomes/). SNPs were entered into T1DBase (13, 14) and displayed graphically using GBrowse (15). All of the annotation of the Idd10 interval can be viewed at http://www.t1dbase.org.
Flow cytometry and intracellular cytokine staining
Single cell suspensions were prepared from spleen, lymph nodes, thymus and bone marrow. Red blood cells were not removed. Cell surface expression of CD11c, B220, the beta chain of the TCR, CD44, NK1.1, CD4, CD8, and Gr1, was detected using fluorescently-labeled monoclonal antibodies obtained from eBioscience (San Diego, CA). A monoclonal antibody specific for CD11c (clone N418, BioLegend, San Diego, CA) was also used in some experiments. Phycoerythrin-labeled anti-CD101 (clone 307707) was obtained from R&D. Intracellular FoxP3 was detected following permeabilization of the cells (Mouse Regulatory T Cell Staining Kit, eBioscience). Isotype controls were purchased from eBioscience. Cells were analyzed on a LSR II (BD Biosciences, San Jose, CA) or a CyAn (Dako/Beckman Coulter, Miami, FL) flow cytometer; flow cytometry data were assessed using FlowJo software (Tree Star, Ashland, OR).
Diabetes frequency studies
All diabetes cumulative frequency studies were conducted using female mice. The presence of T1D was tested every 14 days beginning at 84 days of age by the detection of urinary glucose >500 mg/dL using Diastix (Miles, Elkhart, IN). Studies were terminated at 196 days of age. Kaplan-Meier survival curves were plotted for each mouse strain, and these were compared using the logrank test (Prism®4 software).
Results
Multiple haplotypes in the Idd10 region
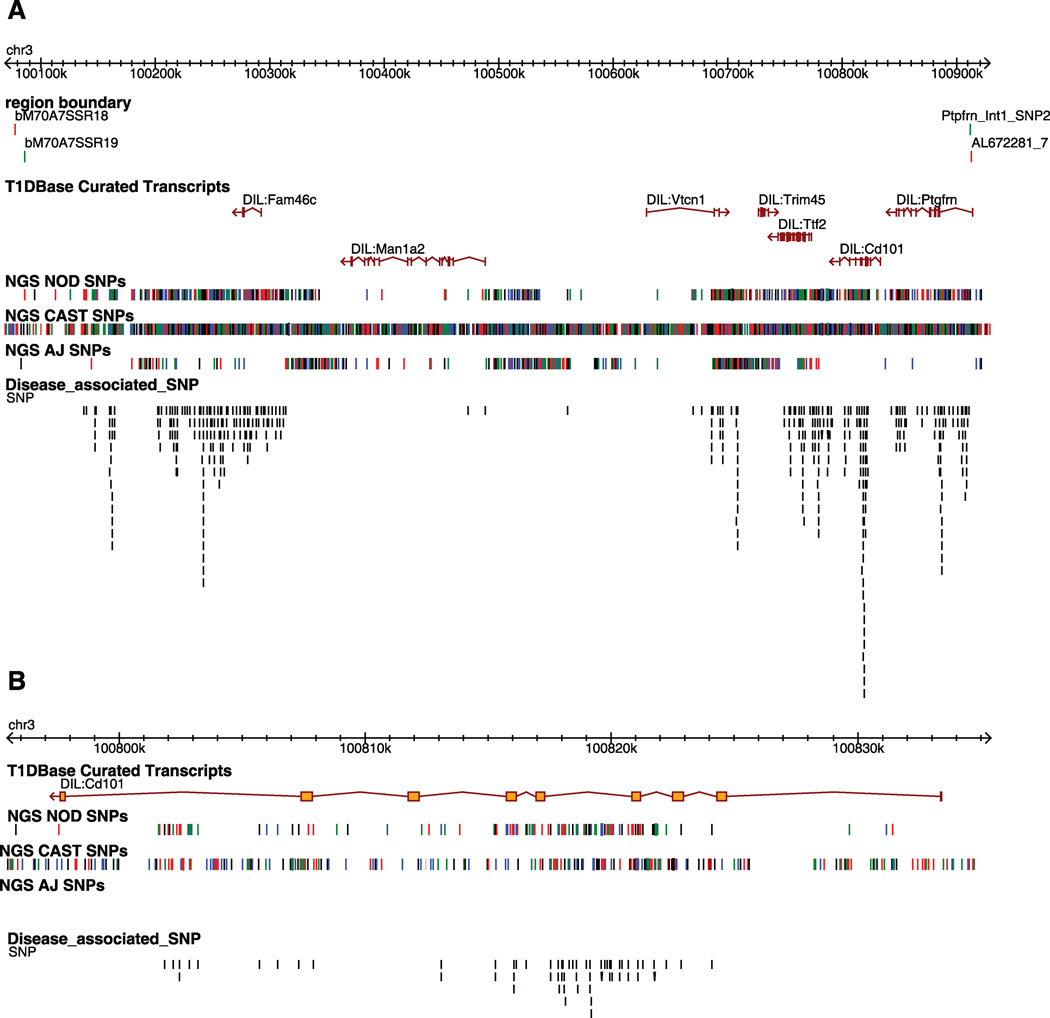
We previously defined the Idd10 region to 950 kb on mouse chromosome 3 using the NOD.B6 congenic strain R323 and R93 (1). Since this publication, additional sequence information has become available (see below) enabling the refinement of the Idd10 region to approximately 836 kb (the B6 haplotype is 835,887 bp). The distal boundary of Idd10 is now located within intron 1 of Ptgfm (Figs. 1 and 2). Seven coding genes are encoded within the Idd10 region (Fig. 1). Although five pseudogenes and one non-coding RNA are notated in Ensembl in the Idd10 region, on inspection of the underlying data, evidence for these genes is inconclusive. Cd101 was highlighted previously as an Idd10 candidate gene (1) because of the expression of its protein CD101 on multiple cell types in the immune system (2–5) and SNPs distinguishing the NOD and B6 Cd101 alleles that cause 10 amino acid substitutions in the CD101 protein, two of which are predicted to alter glycosylation (Supplemental Fig. 1).
Figure 1. Gene content and allelic variants in the Idd10 region on mouse chromosome 3.
A, The in and out boundaries of the Idd10 region are represented as green and red lines, respectively. The T1Dbase curated transcripts track displays the gene content of the Idd10 region. The NGS NOD, NGS CAST and NGS A/J SNP tracks represent the location of sequence polymorphisms relative to B6 identified from Next Generation Sequencing (NGS) performed at the WTSI; black, red, blue, and green lines represent the G, T, C, and A nucleotides, respectively. Where no SNPs are present, the sequence is identical to B6. Note that where multiple SNPs are located close together the lines in the SNPs track may represent more than one SNP. The disease-associated SNP track compares the SNPs present in NOD.B6 Idd10 and NOD.A/J Idd10 mice, which are T1D-resistant, and the NOD and NOD.CAST Idd10 strains that are relatively T1D-susceptible; a black line indicates the position at which one allele of a SNP is shared by the susceptibility haplotypes (NOD and CAST) and the other allele of the SNP is shared by the protective haplotypes (B6 and A/J). Disease associated SNPs are shown vertically in an ‘expanded view’ in order to distinguish SNPs that are close together. B, Zoomed in view of Cd101. The position of the SNPs in this figure is based on NCBI build 37.
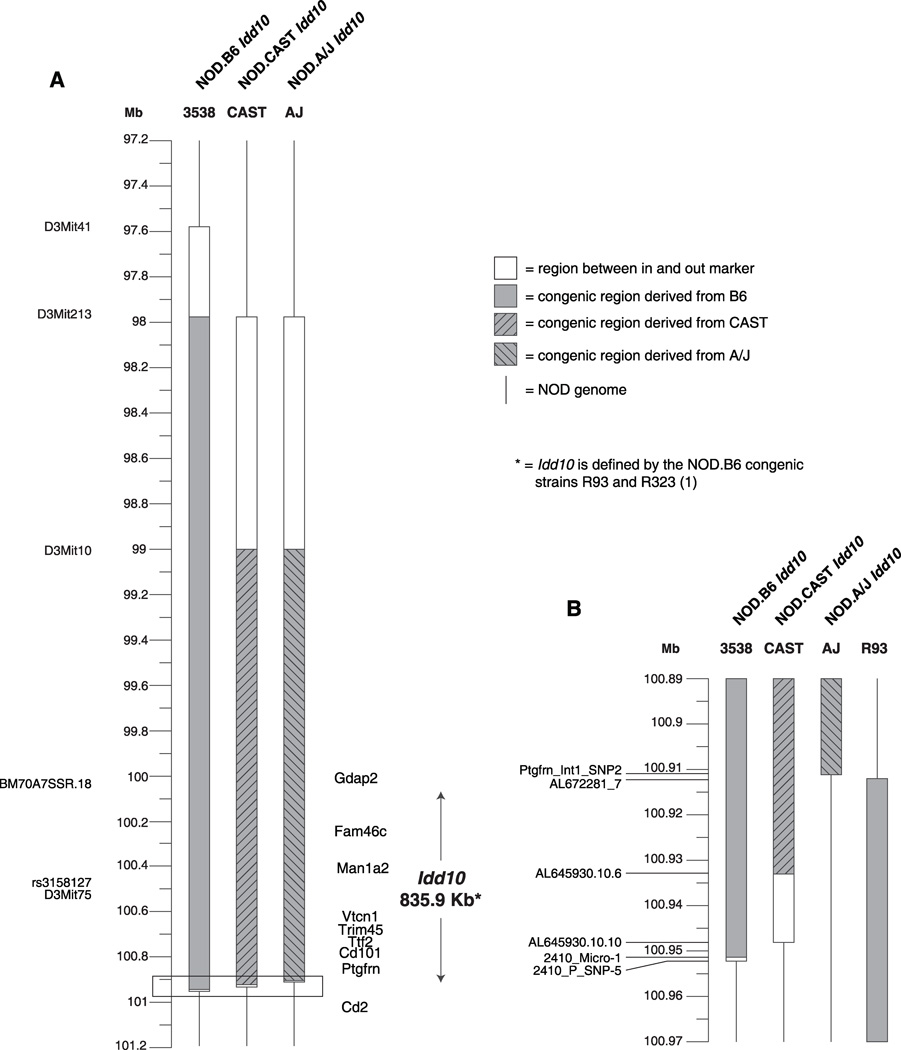
Figure 2. Idd10 congenic mouse strains.
A, The introgressed regions present in the NOD.B6, NOD.CAST and NOD.A/J Idd10 congenic strains are depicted together with the previously defined Idd10 interval and gene content. B, Zoomed in region showing that the distal recombination points of the NOD.B6, NOD.CAST and NOD.A/J Idd10 congenic strains and the proximal recombination point of R93 (a congenic strain that defined the distal boundary of Idd10 (1)), which all map within an approximately 40 Kb region. The distal and proximal boundaries of the NOD.A/J Idd10 and R93 strains, respectively, are defined by the same two markers, between which no additional polymorphisms are present; therefore no region is indicated between the in and out markers for these two boundaries. The position of the markers is based on NCBI build 37.
Next Generation Sequence (NGS) for the entire genomes of 17 inbred strains has become available, including that of the NOD/ShiLtJ, A/J and CAST/EiJ strains (http://www.sanger.ac.uk/resources/mouse/genomes/). In addition, NOD BAC-based sequence from selected regions of the NOD/MrkTac genome (http://www.sanger.ac.uk/Projects/M_musculus-NOD/) including a portion of the Idd10 region is publically available (NOD tile path in Supplemental Fig. 2). The variation determined by NGS (NOD/ShiLtJ) and the BAC-based methods (NOD/MrkTac) are essentially identical in this region (Supplemental Fig. 2) so we have made use of NGS to interrogate NOD/B6 variation through Idd10, which is displayed in the “NGS NOD SNPs” track in Fig. 1A. Inspection of the B6/NOD variation present throughout the Idd10 region shows that only Man1a2 and Vtcn1 have areas of low SNP density when comparing the B6 and NOD haplotypes. Assessment of Idd10 sequence variation amongst inbred strains, originally by targeted hand-sequencing (data not shown) highlighted two Idd10 haplotypes, CAST and A/J that could be potentially useful at defining disease-associated SNPs in the region. A SNP is considered to be disease associated when one allele at the SNP is associated with disease susceptibility and an alternate allele is associated with disease resistance. Although a single SNP can cause alteration in disease susceptibility, usually such SNPs are part of complex haplotypes where many SNPs within a gene are co-inherited. By determining the disease susceptibility phenotype of divergent haplotypes, the potential list of disease-causing SNPs can be reduced, as we did for the Idd3 and Idd5.1 regions (16, 17). SNPs between the B6 and CAST strains are shown on the “NGS CAST SNPs” track. The CAST strain from M. musculus castaneous is one of several inbred mouse strains isolated from the wild; such strains have the attribute of having a greater diversity of sequence variants than inbred laboratory strains (18–20). In the case of Idd3, the disease phenotype of an Idd3 haplotype isolated from the wild (from the CZECH strain) was instrumental in eliminating the hypothesized candidacy of B6/NOD amino acid variants in the IL-2 molecule that alter IL-2 glycosylation as being disease-causing (17). These results were later confirmed when the knock-in of the B6 variants into the NOD Il2 allele was shown to alter glycosylation but did not result in protection from disease (21). In the case of Idd10, the CD101 molecule has 10 amino acid differences between the B6 and NOD alleles; of these 10, CAST shares identity with B6 at 6 and with NOD at 4 (Table 1). The A/J haplotype, which is also observed in the BALB/c, C3H/HeJ, and AKR/J strains (data not shown), is of particular interest because it differs from the B6 haplotype throughout Idd10 except for Cd101, where no SNPs are observed (Fig. 1A, 1B). We therefore hypothesized that if Cd101 variation was responsible for the Idd10-mediated T1D protection provided by the B6 haplotype, the A/J haplotype should also confer resistance from disease. On the other hand, if a NOD.A/J Idd10 congenic strain were to have NOD-like T1D susceptibility, this phenotype would eliminate Cd101 as an Idd10 candidate gene.
Table 1. Disease-associated SNPs in the coding regions of genes in the Idd10 region.
Based on NGS sequence from the B6, A/J, CAST and NOD strains, all synonymous and nonsynonymous SNPs in the coding regions of the genes in the Idd10 region are detailed. A SNP is considered to be disease associated when the T1D-resistant B6 and A/J strains share an allele and the NOD and CAST share a different allele.
| Gene | Chromosome | Position | B6 | A/J | CAST/EiJ | NOD/ShiLtJ | Consequence | Disease Associated? | Domain |
|---|---|---|---|---|---|---|---|---|---|
| Fam46c | 3 | 100277113 | G | G | G | A | Synonymous | No | |
| Vtcn1 | 3 | 100687984 | A | A | G | G | Synonymous | Yes | |
| Vtcn1 | 3 | 100692196 | C | C | C | T | Synonymous | No | |
| Vtcn1 | 3 | 100692211 | T | C | C | C | Synonymous | No | |
| Vtcn1 | 3 | 100696491 | A | G | G | G | Synonymous | No | |
| Vtcn1 | 3 | 100696535 | T | T | C | C | F263S | Yes | Not in a predicted domain |
| Trim45 | 3 | 100732051 | A | G | G | G | I476V | No | Filamin-type immunoglobulin domain |
| Ttf2 | 3 | 100754209 | A | A | A | C | Synonymous | No | |
| Ttf2 | 3 | 100758116 | T | T | T | C | Synonymous | No | |
| Ttf2 | 3 | 100765628 | T | T | A | A | E438D | Yes | Not in a predicted domain |
| Ttf2 | 3 | 100765633 | G | G | G | A | P437T | No | Not in a predicted domain |
| Ttf2 | 3 | 100765736 | A | A | G | G | Synonymous | Yes | |
| Ttf2 | 3 | 100766616 | G | T | G | T | Q355R | No | Not in a predicted domain |
| Ttf2 | 3 | 100766653 | C | C | C | T | Synonymous | No | |
| Ttf2 | 3 | 100766680 | G | G | G | A | Synonymous | No | |
| Ttf2 | 3 | 100766729 | T | T | C | C | E317G | Yes | Not in a predicted domain |
| Ttf2 | 3 | 100766773 | T | T | C | C | Synonymous | Yes | |
| Ttf2 | 3 | 100766804 | C | C | C | T | K292R | No | Not in a predicted domain |
| Ttf2 | 3 | 100766811 | T | T | C | C | T290A | Yes | Not in a predicted domain |
| Ttf2 | 3 | 100766943 | T | T | G | G | T246P | Yes | Not in a predicted domain |
| Ttf2 | 3 | 100767156 | T | T | T | A | S175C | No | Not in a predicted domain |
| Ttf2 | 3 | 100767380 | G | G | G | C | R100P | No | Not in a predicted domain |
| Ttf2 | 3 | 100773436 | G | G | G | A | Synonymous | No | |
| Cd101 | 3 | 100807698 | C | C | C | T | A860T | No | Ig domain 6 |
| Cd101 | 3 | 100815755 | T | T | T | C | Synonymous | No | |
| Cd101 | 3 | 100815776 | T | T | T | C | I648M | No | Ig domain 5 |
| Cd101 | 3 | 100815861 | C | C | C | T | R620H | No | Ig domain 5 |
| Cd101 | 3 | 100816001 | G | G | G | A | Synonymous | No | |
| Cd101 | 3 | 100816004 | C | C | C | T | Synonymous | No | |
| Cd101 | 3 | 100816027 | T | T | C | C | N565D | Yes | Ig domain 5 |
| Cd101 | 3 | 100816040 | A | A | G | G | Synonymous | Yes | |
| Cd101 | 3 | 100816051 | G | G | C | C | H557D | Yes | Ig domain 5 |
| Cd101 | 3 | 100817166 | C | C | C | T | D451N | No | Ig domain 4 |
| Cd101 | 3 | 100820836 | C | C | C | T | Synonymous | No | |
| Cd101 | 3 | 100820871 | C | C | C | T | V392I | No | Ig domain 3 |
| Cd101 | 3 | 100820982 | A | A | A | G | Synonymous | No | |
| Cd101 | 3 | 100821092 | A | A | G | G | Synonymous | Yes | |
| Cd101 | 3 | 100821106 | C | C | T | T | V318A | Yes | Ig domain 3 |
| Cd101 | 3 | 100821159 | C | C | C | T | E296K | No | |
| Cd101 | 3 | 100822813 | A | A | G | G | V175A | Yes | Ig domain 2 |
| Ptgfrn | 3 | 100847330 | A | A | C | C | Synonymous | Yes | |
| Ptgfrn | 3 | 100881063 | A | A | A | G | Synonymous | No | |
| Ptgfrn | 3 | 100884112 | A | A | G | G | Synonymous | Yes |
Protection from T1D in NOD.B6 Idd10 and NOD.A/J Idd10 mice is equivalent thereby supporting the hypothesis that Idd10 is the CD101 gene
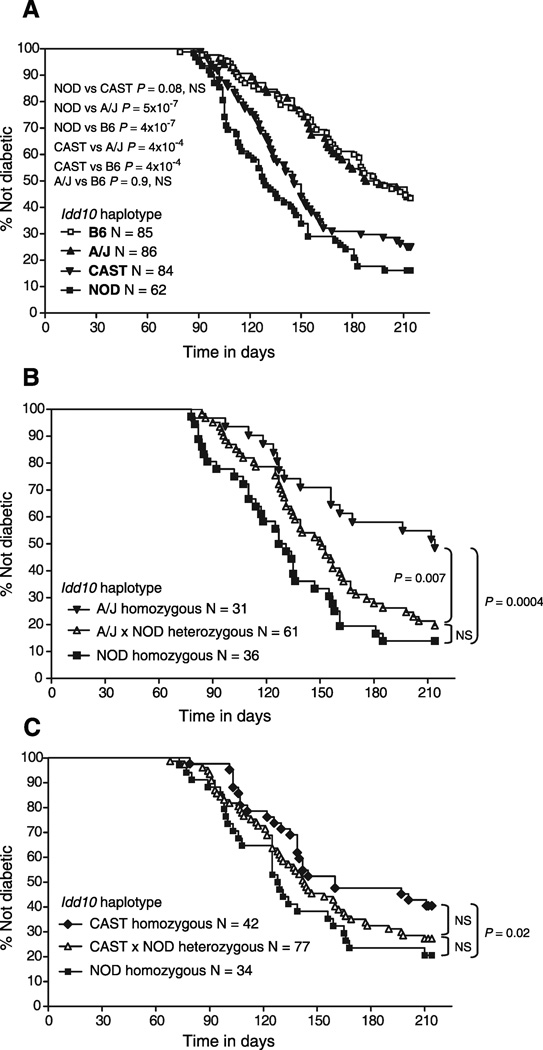
NOD mice congenic for the Idd10 regions of B6, A/J and CAST were generated (Fig. 2) in order to assess their ability to mediate protection from T1D (Fig. 3A). Of note, we found that the distal boundary of the A/J congenic strain occurs in the same 1705 bp region as the proximal boundary of the NOD.B6 R93 congenic strain (between the markers labeled Ptgfm_Int1_SNP2 and AL672281_7 (Fig. 2B), which are both in the first intron of Ptgfm) that, via truncation analysis, defined the distal boundary of Idd10 (22). Since we have identified an additional two independent congenic strains that recombine in this same 1705 bp region (data not shown), the first intron of Ptgfm is likely to be a recombination hotspot (23). Because the Idd10 region was defined previously using congenic interval truncation analyses, this report is the first demonstrating the protection mediated by the isolated B6-derived Idd10 region (Fig. 3A). Consistent with the hypothesis that Cd101 is Idd10, the B6 and A/J haplotypes, which are identical at Cd101, but not at other genes within Idd10, produced equivalent levels of protection from T1D (Fig. 3A and 3B). In Fig. 3B, the A/J haplotype was assessed using littermate progeny from heterozygous breeders to control for any residual A/J-derived gene regions remaining outside of the known congenic interval. Since the mice typed as NOD homozygous at Idd10 developed a NOD-like frequency (Fig. 3B, squares), this verifies the assumption that the protection observed in the NOD.A/J Idd10 congenic strain is due solely to the congenic interval. Although a trend to protection was observed, one dose of the protective A/J-derived Idd10 haplotype did not produce significant disease prevention indicating that protection by Idd10 is recessive (Fig. 3B). In contrast to the potent disease protection mediated by the B6 and A/J haplotypes, the CAST Idd10 haplotype failed to provide significant protection from T1D as compared to the NOD strain in one experiment (Fig. 3A, P = 0.08) and provided only a modest level of protection in another (Fig. 3C, P = 0.02). In a direct comparison, the CAST haplotype was less protective than both the A/J and B6 haplotypes (Fig. 3A, P = 0.0004 for both comparisons). In summary, the CAST haplotype at Idd10 provides a NOD-like susceptibility to T1D while the A/J haplotype provides B6-like protection from disease.
Figure 3. The A/J and B6 haplotypes at Idd10 provide equivalent protection from type 1 diabetes.
The frequency of diabetes was determined in the indicated number (N) of females in each cohort. A, NOD, NOD.B6 Idd10, NOD.CAST Idd10 and NOD.A/J Idd10 congenic mice were bred using breeders homozygous for the NOD, B6, CAST and A/J haplotypes, respectively. B, NOD and NOD.A/J Idd10 congenic mice were intercrossed and the resulting F1 progeny intercrossed to produce an F2 generation. Mice were genotyped at Idd10 following the development of diabetes or at the end of the 196 day observation period. C, NOD and NOD.CAST Idd10 congenic mice were used to generate an F2 cohort for assessing the development of diabetes as described in B.
Disease-associated SNPs in Cd101 and other genes in the Idd10 region
Based on the disease phenotypes determined from the T1D frequency studies, we queried which SNPs held in common by the T1D-protective B6 and A/J Idd10 haplotypes differed from those held in common by the susceptible CAST and NOD haplotypes (disease-associated SNPs). At least 445 disease-associated SNPs are found in the Idd10 region (Fig. 1A), the majority of which are most likely nonfunctional variants that are in linkage disequilibrium with functional ancestral polymorphisms (24). Of the 445 disease-associated SNPs, 16 occur in coding regions and 9 of these 16 are nonsynonymous SNPs. There are disease-associated SNPs in Cd101 that confer non-conservative amino acid changes (H557D and N565D) and potentially alter N-linked glycosylation (N565D) (Table 1, Supplemental Fig. 1). Two additional disease-associated SNPs in Cd101 confer conservative amino acid changes (V175A and V318A). Additional disease-associated SNPs conferring amino acid substitutions are present in two other Idd10 genes, Vtcn1 and Ttf2 (Fig. 1 and Table 1). Disease-associated F263S in the Vtcn1-encoded B7H4 protein occurs in an unstructured portion of the molecule and T246P, T290A, E317G and E438D are located within an unstructured portion of the Ttf2-encoded transcription factor. Disease-associated SNPs are located in the introns of all genes located in Idd10 except for Trim45 and could potentially cause allele-specific expression differences by altering splicing or transcription enhancer elements. In addition, 3’ and 5’ UTR disease-associated SNPs are present in most of the Idd10 region genes. Thus, although Cd101 is implicated as being disease-causing by the fact that the T1D-resistant Idd10 haplotypes of the A/J and B6 strains are identical-by-descent only at the Cd101 gene within the Idd10 region, other genes in the region, with the exception of Trim45 and perhaps Man1a2, which has only two disease-associated SNPs in intron 1 (Fig. 1A), remain viable candidates since disease-associated SNPs are present in each of them. It is also important to note that even if Cd101 is Idd10, the causative SNP or SNPs are amongst 62 disease-associated SNPs in Cd101 (Fig. 1B). It is equally plausible that the causative SNP(s) could alter CD101 function by amino acid replacement or by influencing CD101’s expression, or both.
Development and initial characterization of a CD101 null mouse
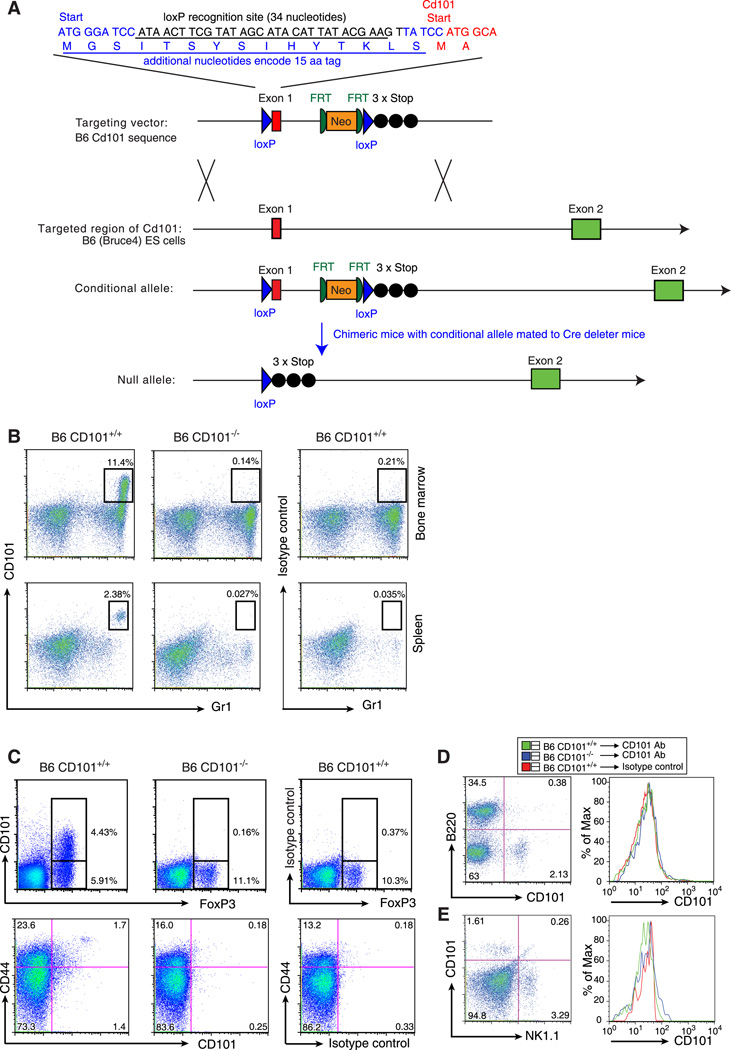
To test further the possibility that the CD101 gene is Idd10, we have targeted the CD101 gene in the context of the B6 haplotype (Fig. 4A). Ultimately we will test this Cd101 null allele for its influence on the frequency of T1D following extensive backcrossing to the NOD background, a long term experiment that is in progress. Here, we have performed an initial characterization of the B6 CD101 null strain, which has B6-like viability until at least one year of age (N = 30 males and 30 females); B6 CD101 null mice aged one year displayed no splenomegaly or lymphadenopathy (N= 20 males and 20 females). Monoclonal antibodies specific for CD101 were developed (see Materials and Methods) to characterize CD101 expression and confirm the lack of expression of the CD101 protein in B6 CD101 null mice (Figure 4B–E). The most prominent expression of CD101 in the tissues tested (bone marrow, spleen, lymph nodes and thymus) is on Gr1+ cells in the bone marrow (Fig. 4B, upper left hand panel). Developing myeloid cells having the highest levels of Gr1 are those that co-express CD101. As expected, expression of CD101 on bone marrow cells from CD101 null mice was not detected (Fig. 4B, upper middle panel); the minimal staining observed was equivalent to that obtained using an isotype control for the directly conjugated anti-CD101 mAb (Fig. 4B, upper right panel). We confirmed the CD101 expression reported recently (3) on a portion of CD4+ FoxP3+ regulatory T cells (Fig. 4C, upper left panel) and CD44+ CD4+ T cells (Fig. 4C, lower left panel) and a small number of CD8+ T cells (data not shown). No expression of CD101 was observed on B or NK cells (Figs. 4D and 4E, respectively). In all cell types examined from CD101 null B6 mice (Fig. 4B–D, data not shown), CD101 expression was not detected, verifying that CD101 null mice do not express CD101 protein.
Figure 4. Development of a B6 mouse having a targeted mutation in the CD101 gene.
A, The strategy used to generate a CD101 null mouse is depicted. Details are included in the Materials and Methods section. B–E, CD101 expression on various cell subsets and lack of expression on cells from B6 CD101 null mice. B, Bone marrow and spleen cells were gated by light scatter (see Supplemental Fig. 3 for examples of light scatter gating) to include all cells except erythrocytes and analyzed for CD101 and Gr1 expression. C, Splenic CD4+ T cells were assessed for CD101, CD44 and FoxP3 expression. See text for further details. Data shown are representative of at least 3 independent experiments.
Genotype-dependent CD101 expression on Gr1+ bone marrow cells
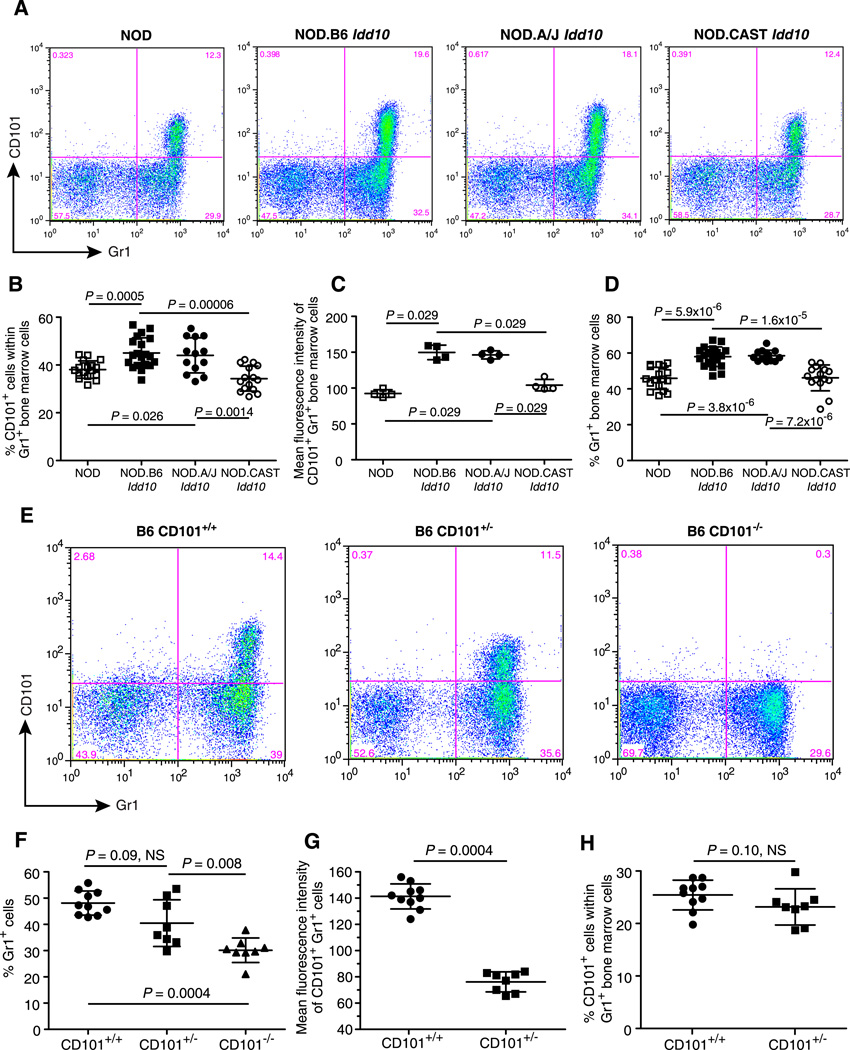
Following the validation of the newly developed anti-CD101 mAb, CD101 expression on cells types expressing CD101 was assessed on cells obtained from the bone marrow, spleen, lymph nodes and thymus of NOD, NOD.B6 Idd10, NOD.A/J Idd10, and NOD.CAST Idd10 congenic mice to determine if genotype-determined CD101 expression could be correlated with disease susceptibility. The most obvious genotype-dependent CD101 expression observed was that amongst cells having the highest levels of CD101, which are Gr1+ cells in the bone marrow and spleen (Fig. 5, Supplemental Fig. 3). Within the Gr1+ cells in the bone marrow, CD101 was expressed on 38% and 34% of the Gr1+ bone marrow cells in the T1D-susceptible NOD and NOD.CAST Idd10 strains, respectively, whereas a higher percentage of cells were CD101 positive in the T1D-resistant NOD.B6 Idd10 and NOD.A/J Idd10 strains, 45% and 44%, respectively (Fig. 5A, 5B, Supplemental Fig. 3A). In addition, the mean fluorescence intensity of the CD101 positive cells was approximately 50% higher for the T1D resistant strains as compared to the susceptible strains (Fig. 5A, 5C). A further correlation that did not involve CD101 expression was observed: there were higher percentages of Gr1+ cells in the bone marrow cells of the T1D-resistant NOD.B6 Idd10 and NOD.A/J Idd10 strains, 58% and 59%, respectively, as compared to NOD and NOD.CAST Idd10 bone marrow cells where 46% were Gr1+ in both strains (Fig. 5A, 5D). No difference was detected in the absolute number of cells recovered from the bone marrow of the congenic strains suggesting that the allelic status of Idd10 influences the likelihood of cells to become Gr1+ (data not shown).
Figure 5. Cd101 genotype-dependent phenotypes in bone marrow cells.
A, Representative dot plots showing the expression of CD101 and Gr1 on bone marrow cells from NOD, NOD.B6 Idd10, NOD.A/J Idd10, and NOD.CAST Idd10 mice; gating strategy is shown in Supplemental figure 3A. B, Data for the percentage of CD101+ cells within the Gr1+ population are compiled from three independent experiments using groups of female mice 11–13 weeks of age. Each point represents an individual mouse (N = 17, 20, 13, and 15 for the NOD, NOD.B6 Idd10, NOD.A/J Idd10, and NOD.CAST Idd10 strains, respectively). C, The MFI of CD101+ Gr1+ cells for one representative experiment of three is shown. D, The percentage of Gr1+ cells in the non-erythroid light scatter gate was compiled from the same mice described in Fig. 5C. E, Representative examples of CD101 and Gr1 expression on bone marrow cells from wild type B6 mice (CD101+/+), B6 mice having one copy of the null allele of Cd101 (CD101+/−), and CD101 null B6 mice (CD101−/−). F, The percentage of Gr1+ cells in the non-erythroid light scatter gate was compiled from 3 independent experiments using groups of female mice 11–14 weeks of age. Each point represents an individual mouse; N = 10 for B6 (CD101+/+), N = 8 for mice expressing only one allele of Cd101 (CD101+/−) and N = 8 for CD101 null B6 mice (CD101−/−). G, The MFIs of CD101+ Gr1+ cells for the CD101+/+ and CD101+/− mice described in Fig. 5F are shown. H, The percentages of CD101+ cells within the Gr1+ population for the CD101+/+ and CD101+/− mice described in Fig. 5F are shown. Comparisons between groups were performed using the Mann-Whitney nonparametric test. Error bars indicate the standard deviation of the mean.
Reduction of Gr1+ cells in the bone marrow of CD101 knockout mice
Because a correlation exists in the NOD Idd10 congenic mice between protective haplotypes and higher CD101 expression on Gr1+ cells as well as an increased percentage of Gr1+ cells in the bone marrow, we tested the hypothesis that the percentage of Gr1+ cells would be altered in B6 mice lacking CD101 expression (Fig. 5E–H ). Strikingly, the percentage of Gr1+ cells in the bone marrow of CD101 null mice, 30%, was substantially less than the 48% Gr1+ cells in the bone marrow of wild type B6 mice (Fig. 5E, 5F). The levels of Gr1+ cells in B6 mice expressing one copy of the wild type CD101 gene (B6 CD101+/− mice) were intermediate (40% Gr1+) to those on the bone marrow cells of B6 and B6 CD101 null mice. As expected, the expression of CD101 on Gr1+, CD101+ bone marrow cells of CD101 hemizygous mice (MFI of 76) was found to be approximately 50% of that observed on this subset in wild type B6 mice (MFI of 141) (Fig. 5E, 5G). No difference in the percentage of Gr1+ cells that are CD101+ was detected between the two strains (Fig. 5E, 5H).
Genotype-dependent CD101 expression in spleen cells
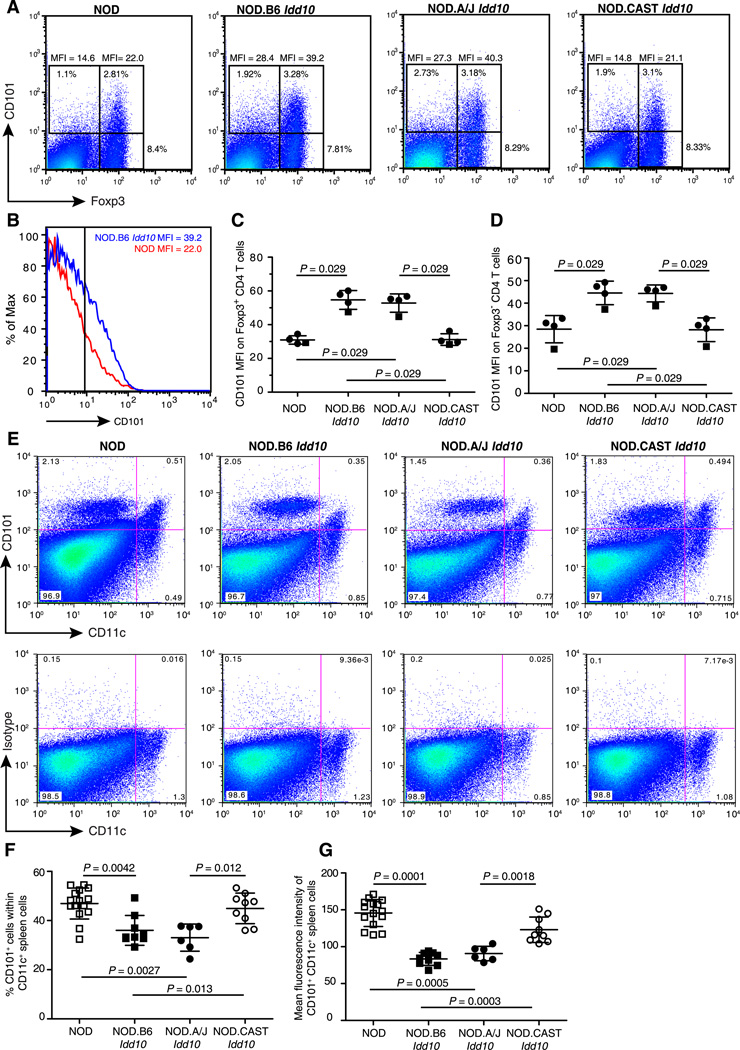
As well as the bone marrow, high levels of CD101 expression on Gr1+ cells was also observed in the spleen, with the CD101 MFI approximately 50% higher on Gr1hi, CD101+ spleen cells obtained from mice having a protective Cd101 genotype (Supplemental Fig. 3B, 3C, 3D). Amongst the Gr1hi cells in the spleen, although the percent of Gr1hi CD101+ did not differ between strains (Supplemental Fig. 3E), there were an increased number of Gr1hi CD101neg cells in mice with a Cd101 susceptible genotype (Supplemental Fig. 3F). Amongst splenic lymphocytes, the Foxp3+ population of CD4 T cells has the most notable expression of CD101 (Fig. 4C, Fig. 6A). Expression of CD101 was increased in FoxP3+ cells from strains having a protected haplotype (NOD.B6 Idd10 and NOD.A/J Idd10 strains) as compared to this regulatory subset in NOD and NOD.CAST Idd10 mice (Fig. 6A–6C). CD101+ CD4 T cells not expressing FoxP3, although a much more rare population than the FoxP3+ CD4 T cells, also had higher expression of the CD101 in cells from mice protected from T1D (Fig. 6A, 6D).
Figure 6. Cd101 genotype-dependent CD101 expression in spleen cells.
A, Splenocytes were gated on cells expressing the beta chain of the TCR and CD4 and assessed for CD101 and FoxP3 expression. Representative examples of the indicated congenic strains are shown. B, Representative examples of CD101 expression on FoxP3+ cells are shown for a NOD female (red histogram) and a NOD.B6 Idd10 female (blue histogram). Genotype-dependent expression of CD101 on FopP3+ CD4 T cells (C) and FoxP3− CD4 T cells (D) from 2 independent experiments are compiled; 3 mice from each strain were tested on one day (closed circles) and one mouse from each strain on a different day (closed squares). A third experiment showed the same genotype-dependent expression differences in both T cell subsets (not shown). Comparisons between groups were performed using the Mann-Whitney nonparametric test. E, Representative dot plots showing expression of CD101 and CD11c on spleen cells from NOD, NOD.B6 Idd10, NOD.A/J Idd10, and NOD.CAST Idd10 mice (top panel) and the isotype control (bottom panel). F, Data for the percentage of CD101+ cells within the CD11c+ population. Each point represents an individual mouse (N = 15, 9, 6, and 9 for the NOD, NOD.B6 Idd10, NOD.A/J Idd10, and NOD.CAST Idd10 strains, respectively). G, The MFI of CD101+ CD11c+ cells was compiled from the same mice described in F. Comparisons between groups were performed using the Mann-Whitney nonparametric test.
Finally, investigation of CD101 expression on CD11c+ cells in the spleen showed that expression was predominant only on CD11chi dendritic cells (Fig. 6E). Most of the CD101-expressing CD11chi cells belong to the myeloid dendritic cell subset since all co-express CD11b and CD4 but less than 5% and 20% express CD8 or B220, respectively (Supplemental Fig. 4). We were surprised to find that CD101 expression was higher on CD11c+ cells from mice having the susceptible NOD and CAST Idd10 haplotypes as compared to the resistant B6 and A/J haplotypes (Fig. 6F, 6G), a genotype-dependent pattern opposite to that observed on Gr1+ cells and CD4+ T cells (Fig. 5 and Fig. 6A–D).
Discussion
Using four independent Idd10 haplotypes, NOD, CAST, B6, and A/J, we have demonstrated correlations between T1D susceptibility and 1) CD101 protein sequence variants and variants that potentially influence the expression of Cd101, 2) decreased CD101 expression on FoxP3+ CD4 T cells, 3) increased expression of CD101 on CD4+ CD11b+ CD11c+ dendritic cells and 4) decreased CD101 expression on Gr1+ myeloid cells. The bone marrow cells of the T1D susceptible NOD and NOD.CAST Idd10 strains also had a lower percentage of Gr1+ cells as compared to the two congenic strains protected from T1D, NOD.B6 Idd10 and NOD.A/J Idd10. Our data are consistent with the hypothesis that genetic variation of Cd101 is responsible for mediating Idd10’s T1D-modifying effects.
CD101 expression differences between susceptible and protective Cd101 haplotypes are modest, which is consistent with most variant genes that contribute to complex traits such as autoimmune disease where a 2-fold difference in expression or function of a disease associated protein is considered large (17, 18, 25–27). It is also notable that the direction of the expression differences is not the same in all tissues and in some cell types no difference in CD101 expression has been observed. This suggests that regulation of CD101 expression is mediated at least in part by tissue-specific transcription factors or that an extrinsic effect of Cd101 haplotypes influences CD101 expression in other cell types in a complex manner. Detailed analyses of mixed bone marrow chimeras will be required to determine if CD101 expression differences are extrinsic or intrinsic to each cell type.
In comparing the Cd101 genomic sequence of the two T1D-susceptible haplotypes, NOD and CAST, with the two T1D-protective haplotypes, B6 and A/J, the number of disease associated SNPs in Cd101 was reduced from 125 (B6/NOD variation only) to 62 disease associated SNPs (Fig. 1A, 1B). Notably, only 4 of the 10 B6/NOD amino acid differences previously reported (1) were shown to be disease associated in the haplotype analysis (Table 1); the highest density of disease-associated SNPs was found in introns 3, 4 and 5 (Fig 1B). Although no disease-associated Cd101 SNPs alter nucleotides known to be critical for splicing (consensus sequences at the donor, acceptor and branch sites), it is possible that the observed disease-associated CD101 expression changes are caused by one or more SNPs altering splicing efficiency and therefore total protein production. Mechanisms that could account for a reduction in splicing efficiency include one or more causal SNPs altering intronic or exonic splicing enhancer or silencer motifs as well as SNPs changing the secondary structure of the pre-mRNA in a manner by which the ordered removal of introns is made less efficient. Alternatively, disease-associated SNPs could alter the efficiency of Cd101 pre-mRNA transcription by influencing regulatory regions in the 5’ and 3’ UTR as well as transcriptional enhancer regions that are present in introns. The additional complexity of genotype-dependent cell type-specific expression seen in this and a related study (6) suggests the possibility that the binding efficiency of tissue-specific transcription factors that influence splicing/transcription are influenced by polymorphisms that define distinct Cd101 haplotypes. Although none have been described, CD101 isoforms created by alternative splicing could be expressed in a genotype-dependent manner. However, the CD101-specific monoclonal antibody used in the current study could fail to recognize one or more of the putative isoforms if the epitope recognized is missing due to splicing out of the sequence encoding the epitope or there is a conformational change induced by the deletion of nearby amino acids. If this scenario is true, we could be underestimating the differential expression of CD101. Although we favor the hypothesis that the disease-causing sequence variation in Cd101 is causing an alteration in gene expression, it is conceivable that the four disease-associated amino acid changes in CD101 (Table 1) are causal, and the functional difference mediated by changing one or more of these amino acids drives the observed expression differences.
Results from B6 CD101 knockout mice support the hypothesis that CD101 expression changes in Idd10 congenics strains are directly functional: in both models reduced CD101 expression on myeloid cells in the bone marrow is correlated with a reduction in the percentage of Gr1+ bone marrow cells. Since in the CD101 KO model a change of amino acids is not required for the phenotypic change in the Gr1+ bone marrow population, this argues that it is the CD101 expression change in the Idd10 congenic mice that influences the generation of Gr1+ cells. The role of CD101 in myeloid cell development could extend beyond Gr1+ cells; myeloid lineage cells in the bone marrow are progenitors of multiple cell subsets in the periphery, including dendritic cells, macrophages, myeloid suppressor cells and granulocytes. The detailed effect of Cd101 genotype on various precursor populations within the bone marrow and peripheral cell subsets awaits further definition, as does the question of how CD101 contributes to the generation of Gr1+ cells in the bone marrow. It is possible that CD101 influences the efficiency of maturation of Gr1+ cells via signaling or through CD101-mediated cell-cell interactions.
Although the data present do not prove the hypothesis that Cd101 is the Idd10 gene influencing the development of T1D, the observed differential expression of CD101 on cell types important in immune regulation invites speculation on potential causal pathways influenced by the Idd10 region. CD11c+ antigen presenting cells and FoxP3+ Tregs are known to be essential for eliciting (28) and regulating (17, 29), respectively, the autoimmune response to islets. It is important to consider that these same cell types may or may not contribute to N. aro-mediated liver autoimmunity described by Mohammed et al (6). As opposed to T1D pathogenesis, there is a likely causal role for differentially-expressed CD101 on Gr1+ cells in a disease process that is partially dependent on the clearance rate of infectious bacteria.
As is the case for CD101, expression of most immune molecules is not restricted to one subset of cells. In regard to increased expression of CD101 on FoxP3+ CD4 T cells found in mice having the disease-protective genotypes, it is notable that the expression of CD101 on FoxP3+ CD4 T cells is correlated with an increased ability to suppress effector T cells (3). Punkosdy et al (30) recently demonstrated that CD101 expression on FoxP3+ Tregs is greatly increased during a chronic viral infection extending the observation that there is dynamic regulation of CD101 on this T cell subset. Multiple studies have demonstrated that FoxP3+ CD4 T cells can suppress T1D in the NOD model (17, 29, 31). It is tempting to speculate that NOD.B6 Idd10 and NOD.A/J Idd10 FoxP3+ CD4 T cells have an increased ability to mediate suppression since they express higher levels of CD101 on this important regulatory cell population as compared to NOD and NOD.CAST Idd10 mice (Fig. 6E–H).
Although it is likely, as argued above, that the genetic variation in CD101 accounts for T1D susceptibility, we have also demonstrated that other genes within the ~800 kb Idd10 region have potentially causal disease-associated sequence differences; thus, the gene responsible for the phenotypic effects of Idd10 must still be considered to be unknown. No genes in the syntenic region on human Chromosome 1p13-1p12 show evidence of association with T1D, including CD101 itself (www.t1dbase.org). We are currently developing a NOD.B6 Idd10 mouse lacking CD101 expression by backcrossing B6 CD101−/− mice to the NOD parental strain. If variants of Cd101 do indeed modify T1D progression by altering CD101 expression or function, a prediction would be that the frequency of T1D in NOD.B6 Idd10 CD101−/− mice would differ from that of NOD.B6 Idd10 mice. Furthermore, if the reduction of CD101 expression on cell types such as Tregs contributes to the susceptibility phenotypes of the NOD and CAST Idd10 haplotypes, we would predict that NOD.B6 Idd10 CD101−/− and NOD.B6 Idd10 CD101+/− mice will be more susceptible to T1D than NOD.B6 Idd10 mice. However, if the increased expression of CD101 on a subset of CD11c+ cells which is observed in mice having a susceptible Idd10 haplotype is causal for T1D susceptibility, an opposite prediction would be made: NOD.B6 Idd10 CD101−/− mice would be expected to be protected from T1D. If CD101 expression on both Tregs and CD11c+ cells contribute to T1D pathogenesis, the disease status of NOD.B6 Idd10 CD101−/− mice would be difficult to predict.
The importance of CD101 in the immune response has been recently demonstrated in a model of infection-induced liver autoimmunity where disease can be induced on both the B6 and NOD backgrounds (6). NOD Idd10 congenic mice having the B6 and A/J T1D-protective haplotypes showed delayed clearance of Novosphingobium aromaticivorans in the liver and subsequently developed more severe liver autoimmunity than NOD and NOD.CAST Idd10 mice. On the B6 background, reduction of CD101 expression decreased bacterial clearance in the liver following infection and increased the autoimmune response.
Further elucidation of the roles of particular cell subsets expressing CD101 in T1D and infection-induced liver autoimmunity will increase our understanding of the cellular functions that are modulated by this molecule. It is likely that the pleiotropic expression and function of CD101 during immune homeostasis and immune challenge contributes to a complex functional balance of innate and adaptive effector and regulatory cells populations, a balance that likely differs depending on the target tissue. Thus, it would not be surprising if the pivotal CD101-expressing cell type in T1D differs from that critical for influencing the progression of infection-induced liver autoimmunity.
Supplementary Material
Acknowledgements
We would like to thank Dr. Jane Rogers for assistance with NOD BAC clone sequencing.
Abbreviations
- Idd
insulin-dependent diabetes
- BAC
bacterial artificial chromosome
- SNP
single nucleotide polymorphism
- T1D
type 1 diabetes
- NOD
non-obese diabetic
- B6
C57BL/6
- WTSI
Wellcome Trust Sanger Institute
- NGS
Next Generation Sequence
- MFI
mean fluorescence intensity
Footnotes
HIF was funded by a Wellcome Trust 4-year studentship. LSW and JAT are supported by a joint grant from the Juvenile Diabetes Research Foundation (JDRF), the Wellcome Trust and the National Institute for Health Research (NIHR) Biomedical Research Centre. Cambridge Institute for Medical Research (CIMR) is in receipt of a Wellcome Trust Strategic Award (079895). This work was also supported by Award Number P01AI039671 (LSW and JT) from the National Institute of Allergy and Infectious Diseases. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy And Infectious Diseases or the National Institute of Health. JM is supported by the Lupus Research Institute, a grant from the UC microbial pathogenesis core center, PHS Grant P30 DK078392 and by Award Number R01DK084054 from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The resequencing of Idd10 using NOD BACs was performed at WTSI and was funded by the Immune Tolerance Network (ITN) contract AI 15416, which was sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), and the Juvenile Diabetes Research Foundation International (JDRF). The availability of NOD congenic mice through the Taconic Emerging Models Program has been supported by grants from the Merck Genome Research Institute, NIAID, and the JDRF.
References
- 1.Penha-Goncalves C, Moule C, Smink LJ, Howson J, Gregory S, Rogers J, Lyons PA, Suttie JJ, Lord CJ, Peterson LB, Todd JA, Wicker LS. Identification of a structurally distinct CD101 molecule encoded in the 950-kb Idd10 region of NOD mice. Diabetes. 2003;52:1551–1556. doi: 10.2337/diabetes.52.6.1551. [DOI] [PubMed] [Google Scholar]
- 2.Bagot M, Martinel I, Charue D, Weill F, Boulland ML, Wechsler J, Freeman GJ, Bensussan A, Boumsell L. CD101 is expressed by skin dendritic cells. Role in T-lymphocyte activation. Tissue Antigens. 1997;50:439–448. doi: 10.1111/j.1399-0039.1997.tb02898.x. [DOI] [PubMed] [Google Scholar]
- 3.Fernandez I, Zeiser R, Karsunky H, Kambham N, Beilhack A, Soderstrom K, Negrin RS, Engleman E. CD101 surface expression discriminates potency among murine FoxP3+ regulatory T cells. J Immunol. 2007;179:2808–2814. doi: 10.4049/jimmunol.179.5.2808. [DOI] [PubMed] [Google Scholar]
- 4.Rivas A, Ruegg CL, Zeitung J, Laus R, Warnke R, Benike C, Engleman EG. V7, a novel leukocyte surface protein that participates in T cell activation. I. Tissue distribution and functional studies. J Immunol. 1995;154:4423–4433. [PubMed] [Google Scholar]
- 5.Russell GJ, Parker CM, Sood A, Mizoguchi E, Ebert EC, Bhan AK, Brenner MB. p126 (CDw101), a costimulatory molecule preferentially expressed on mucosal T lymphocytes. J Immunol. 1996;157:3366–3374. [PubMed] [Google Scholar]
- 6.Mohammed JP, Fusakio ME, Rainbow DB, Moule C, Fraser HI, Clark J, Todd JA, Peterson LB, Savage PB, Bluestone JA, Wills-Karp M, Ridgway WM, Wicker LS, Mattner J. Identification of Cd101 as a suscptibility gene for Novosphingbobium aromaticivorans - induced liver autoimmunity. J Immunol. doi: 10.4049/jimmunol.1003525. co-submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouloc A, Bagot M, Delaire S, Bensussan A, Boumsell L. Triggering CD101 molecule on human cutaneous dendritic cells inhibits T cell proliferation via IL-10 production. Eur J Immunol. 2000;30:3132–3139. doi: 10.1002/1521-4141(200011)30:11<3132::AID-IMMU3132>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 8.Soares LR, Rivas A, Tsavaler L, Engleman EG. Ligation of the V7 molecule on T cells blocks anergy induction through a CD28-independent mechanism. J Immunol. 1997;159:1115–1124. [PubMed] [Google Scholar]
- 9.Fraser HI, Dendrou CA, Healy B, Rainbow DB, Howlett S, Smink LJ, Gregory S, Steward CA, Todd JA, Peterson LB, Wicker LS. Nonobese diabetic congenic strain analysis of autoimmune diabetes reveals genetic complexity of the Idd18 locus and identifies Vav3 as a candidate gene. J Immunol. 2010;184:5075–5084. doi: 10.4049/jimmunol.0903734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 11.Hughes ED, Qu YY, Genik SJ, Lyons RH, Pacheco CD, Lieberman AP, Samuelson LC, Nasonkin IO, Camper SA, Van Keuren ML, Saunders TL. Genetic variation in C57BL/6 ES cell lines and genetic instability in the Bruce4 C57BL/6 ES cell line. Mamm Genome. 2007;18:549–558. doi: 10.1007/s00335-007-9054-0. [DOI] [PubMed] [Google Scholar]
- 12.Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulbert EM, Smink LJ, Adlem EC, Allen JE, Burdick DB, Burren OS, Cavnor CC, Dolman GE, Flamez D, Friery KF, Healy BC, Killcoyne SA, Kutlu B, Schuilenburg H, Walker NM, Mychaleckyj J, Eizirik DL, Wicker LS, Todd JA, Goodman N. T1DBase: integration and presentation of complex data for type 1 diabetes research. Nucleic Acids Res. 2007;35:D742–D746. doi: 10.1093/nar/gkl933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smink LJ, Helton EM, Healy BC, Cavnor CC, Lam AC, Flamez D, Burren OS, Wang Y, Dolman GE, Burdick DB, Everett VH, Glusman G, Laneri D, Rowen L, Schuilenburg H, Walker NM, Mychaleckyj J, Wicker LS, Eizirik DL, Todd JA, Goodman N. T1DBase, a community web-based resource for type 1 diabetes research. Nucleic Acids Res. 2005;33:D544–D549. doi: 10.1093/nar/gki095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, Lewis S. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Araki M, Chung D, Liu S, Rainbow DB, Chamberlain G, Garner V, Hunter KM, Vijayakrishnan L, Peterson LB, Oukka M, Sharpe AH, Sobel R, Kuchroo VK, Wicker LS. Genetic evidence that the differential expression of the ligand-independent isoform of CTLA-4 is the molecular basis of the Idd5.1 type 1 diabetes region in nonobese diabetic mice. J Immunol. 2009;183:5146–5157. doi: 10.4049/jimmunol.0802610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamanouchi J, Rainbow D, Serra P, Howlett S, Hunter K, Garner VE, Gonzalez-Munoz A, Clark J, Veijola R, Cubbon R, Chen SL, Rosa R, Cumiskey AM, Serreze DV, Gregory S, Rogers J, Lyons PA, Healy B, Smink LJ, Todd JA, Peterson LB, Wicker LS, Santamaria P. Interleukin-2 gene variation impairs regulatory T cell function and causes autoimmunity. Nat Genet. 2007;39:329–337. doi: 10.1038/ng1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ridgway WM, Healy B, Smink LJ, Rainbow D, Wicker LS. New tools for defining the 'genetic background' of inbred mouse strains. Nat Immunol. 2007;8:669–673. doi: 10.1038/ni0707-669. [DOI] [PubMed] [Google Scholar]
- 19.Wade CM, Kulbokas EJ, 3rd, Kirby AW, Zody MC, Mullikin JC, Lander ES, Lindblad-Toh K, Daly MJ. The mosaic structure of variation in the laboratory mouse genome. Nature. 2002;420:574–578. doi: 10.1038/nature01252. [DOI] [PubMed] [Google Scholar]
- 20.Wiltshire T, Pletcher MT, Batalov S, Barnes SW, Tarantino LM, Cooke MP, Wu H, Smylie K, Santrosyan A, Copeland NG, Jenkins NA, Kalush F, Mural RJ, Glynne RJ, Kay SA, Adams MD, Fletcher CF. Genome-wide single-nucleotide polymorphism analysis defines haplotype patterns in mouse. Proc Natl Acad Sci U S A. 2003;100:3380–3385. doi: 10.1073/pnas.0130101100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamanaka M, Rainbow D, Schuster-Gossler K, Eynon EE, Chervonsky AV, Wicker LS, Flavell RA. Amino acid polymorphisms altering the glycosylation of IL-2 do not protect from type 1 diabetes in the NOD mouse. Proc Natl Acad Sci U S A. 2009;106:11236–11240. doi: 10.1073/pnas.0904780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyons PA, Armitage N, Lord CJ, Phillips MS, Todd JA, Peterson LB, Wicker LS. Mapping by genetic interaction: high resolution congenic mapping of the type 1 diabetes loci Idd10 and Idd18 in the NOD mouse. Diabetes. 2001 doi: 10.2337/diabetes.50.11.2633. [DOI] [PubMed] [Google Scholar]
- 23.Buard J, de Massy B. Playing hide and seek with mammalian meiotic crossover hotspots. Trends Genet. 2007;23:301–309. doi: 10.1016/j.tig.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Limaye N, Belobrajdic KA, Wandstrat AE, Bonhomme F, Edwards SV, Wakeland EK. Prevalence and evolutionary origins of autoimmune susceptibility alleles in natural mouse populations. Genes Immun. 2008;9:61–68. doi: 10.1038/sj.gene.6364446. [DOI] [PubMed] [Google Scholar]
- 25.Dendrou CA, Wicker LS. The IL-2/CD25 pathway determines susceptibility to T1D in humans and NOD mice. J Clin Immunol. 2008;28:685–696. doi: 10.1007/s10875-008-9237-9. [DOI] [PubMed] [Google Scholar]
- 26.Irie J, Reck B, Wu Y, Wicker LS, Howlett S, Rainbow D, Feingold E, Ridgway WM. Genome-wide microarray expression analysis of CD4+ T Cells from nonobese diabetic congenic mice identifies Cd55 (Daf1) and Acadl as candidate genes for type 1 diabetes. J Immunol. 2008;180:1071–1079. doi: 10.4049/jimmunol.180.2.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Saxena V, Ondr JK, Magnusen AF, Munn DH, Katz JD. The countervailing actions of myeloid and plasmacytoid dendritic cells control autoimmune diabetes in the nonobese diabetic mouse. J Immunol. 2007;179:5041–5053. doi: 10.4049/jimmunol.179.8.5041. [DOI] [PubMed] [Google Scholar]
- 29.Tang Q, Adams JY, Penaranda C, Melli K, Piaggio E, Sgouroudis E, Piccirillo CA, Salomon BL, Bluestone JA. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity. 2008;28:687–697. doi: 10.1016/j.immuni.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Punkosdy GA, Blain M, Glass DD, Lozano MM, O'Mara L, Dudley JP, Ahmed R, Shevach EM. Regulatory T-cell expansion during chronic viral infection is dependent on endogenous retroviral superantigens. Proc Natl Acad Sci U S A. 2011;108:3677–3682. doi: 10.1073/pnas.1100213108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sgouroudis E, Albanese A, Piccirillo CA. Impact of protective IL-2 allelic variants on CD4+ Foxp3+ regulatory T cell function in situ and resistance to autoimmune diabetes in NOD mice. J Immunol. 2008;181:6283–6292. doi: 10.4049/jimmunol.181.9.6283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.