Abstract
Francisella tularensis is a gram negative facultative intracellular bacterium that causes the zoonotic disease tularemia. Free-living amebae, such as Acanthamoeba and Hartmannella, are environmental hosts of several intracellular pathogens. Epidemiology of F. tularensis in various parts of the world is associated with water-borne transmission, which includes mosquitoes and amebae as the potential host reservoirs of the bacteria in water resources. In vitro studies showed intracellular replication of F. tularensis within A. castellanii cells. Whether ameba is a biological reservoir for Francisella in the environment is not known. We used Hartmannella vermiformis as an amebal model system to study the intracellular life of F. novicida. For the first time we show that F. novicida survives and replicates within H. vermiformis. The iglC mutant strain of F. novicida is defective for survival and replication not only within A. castellanii but also in H. vermiformis cells. In contrast to mammalian cells, where bacteria replicate in the cytosol, F. novicida resides and replicates within membrane-bound vacuoles within the trophozoites of H. vermiformis. In contrast to the transient residence of F. novicida within acidic vacuoles prior to escaping to the cytosol of mammalian cells, F. novicida does not reside transiently or permanently in an acidic compartment within H. vermiformis when examined 30 min after initiation of the infection. We conclude that F. tularensis does not replicate within acidified vacuoles and does not escape into the cytosol of H. vermiformis. The Francisella pathogenicity island locus iglC is essential for intra-vacuolar proliferation of F. novicida within H. vermiformis. Our data show a distinct intracellular lifestyle for F. novicida within H. vermiformis compared to mammalian cells.
Keywords: Francisella novicida, Hartmannella vermiformis, vacuolar replication, LysoTracker, iglC
Introduction
Francisella tularensis is a gram negative, facultative intracellular bacterium that causes the zoonotic disease tularemia in humans and animals, and various recent reviews in this special topic issue have discussed various aspects of Francisella (Chong and Celli, 2010; Meibom and Charbit, 2010; Akimana and Abu Kwaik, 2011; Asare and Abu Kwaik, 2011; Bosio, 2011; Bröms et al., 2011; Cremer et al., 2011; Dai et al., 2011; Gavrilin and Wewers, 2011; Jones et al., 2011; Zogaj and Klose, 2011). Tularemia is a zoonotic disease of the northern hemisphere. Humans acquire infection by exposure to infected arthropod vectors, or by handling, ingesting, or inhaling infectious materials. F. tularensis has been isolated from over 250 animal species, including fish, birds, amphibians, rabbits, squirrels, hares, voles, ticks, and flies (Santic et al., 2010; Akimana and Abu Kwaik, 2011). Three closely related subspecies of F. tularensis have been identified: tularensis, holarctica, and mediasiatica (Forsman et al., 1994). Recently F. novicida has been accepted as new species (Sjöstedt, 2005). It has been suggested that holarctica ssp. has a strong association with water-borne disease (Greco et al., 1987; Thelaus et al., 2009; Broman et al., 2011). An in vitro study showed that F. tularensis subsp. holarctica can survive and grow within Acanthamoeba castellanii (Abd et al., 2003). In addition, F. tularensis subsp. holarctica was found within amebal cysts, suggesting potential for long-term survival and an important environmental reservoir for tularemia. The isolation of the bacterium from a water eco-system, as well as from natural spring water (Thelaus et al., 2009; Willke et al., 2009; Broman et al., 2011), supports the hypothesis that protozoa may serve as a reservoir for F. tularensis in nature (Morner, 1992; Thelaus et al., 2009; Broman et al., 2011). Very little is known about the F. tularensis–ameba interaction.
It has been shown that within mammalian and arthropod-derived cells, the Francisella containing phagosome (FCP) transiently matures to an acidified late endosomal stage with limited fusion to lysosomes, followed by rapid bacterial escape into the host cell cytosol (Clemens et al., 2004; Chong et al., 2008; Santic et al., 2008, 2009; Asare and Abu Kwaik, 2011). The FCP is acidified by the vATPase proton pump within 15–30 min of phagosome biogenesis, which is essential for subsequent rapid disruption of the FCP and escape of F. tularensis into the host cell cytosol, where the bacterium replicates (Chong et al., 2008; Santic et al., 2008; Chong and Celli, 2010; Asare and Abu Kwaik, 2011; Bröms et al., 2011; Dai et al., 2011). Inhibition of the vATPase proton pump causes a significant delay in phagosomal escape and blocks bacterial proliferation (Chong et al., 2008; Santic et al., 2008; Chong and Celli, 2010; Asare and Abu Kwaik, 2011; Bröms et al., 2011), indicating a major role for acidification of the FCP in rapid bacterial escape into the cytosol and subsequent replication (Chong et al., 2008; Santic et al., 2008; Chong and Celli, 2010; Asare and Abu Kwaik, 2011; Bröms et al., 2011).
A gene cluster, the Francisella pathogenicity island (FPI), that regulates phagosomal escape and intracellular survival of F. tularensis within macrophages, has been identified (Nano et al., 2004; Nano and Schmerk, 2007; Meibom et al., 2009). It has been suggested to encode a type VI-like secretion system (de Bruin et al., 2007; Nano and Schmerk, 2007; Bingle et al., 2008; Ludu et al., 2008; Barker et al., 2009; Bröms et al., 2011). It has also been shown that IglC is essential for avoiding lysosomal fusion (Santic et al., 2005b; Bonquist et al., 2008) and for bacterial escape into the host cytosol (Lindgren et al., 2004; Santic et al., 2005a) in macrophages. In addition, the iglC mutation diminishes intracellular replication in A. castellanii (Lauriano et al., 2004).
Free-living amebae such as Acanthamoeba and Hartmannella are environmental hosts of several intracellular pathogens such as Legionella, Chlamydia, and Mycobacterium (Amann et al., 1997; Abu Kwaik et al., 1998; Steinert et al., 1998; Molmeret et al., 2005). It has been shown that legionellae interact with their protozoan hosts and mammalian cells in a similar way (Harb et al., 2000). Since the host reservoir of F. tularensis in water systems is not known, we used H. vermiformis, which is the most predominant non-pathogenic ameba in water resources, as an amebal model system to study the intracellular life of F. novicida. Our data indicate that F. novicida survives within H. vermiformis and that the bacteria do not escape into the cytoplasm, which is very distinct from the lifestyle of F. novicida within mammalian cells. The iglC bi-cistronic locus plays an important role in intra-vacuolar replication in H. vermiformis.
Materials and Methods
Bacteria and protozoan strains and media
The wild type (wt) F. novicida strain U112 and it isogenic iglC mutant were grown on buffered-charcoal yeast extract (BCYE) agar plates and have been described previously (Santic et al., 2005b). Construction of ΔiglC::ermC has been described previously (Lauriano et al., 2003). The iglD gene was not affected. The tetracycline-resistant plasmid pKK214, encoding green fluorescent protein (GFP), was introduced into F. novicida (Abd et al., 2003).
Acanthamoeba castellanii and H. vermiformis were obtained from the American Type Culture Collection, 30234 and 1034, respectively. The amebae were grown in medium 30234 and 1034 at 25°C, as described elsewhere (Pedersen et al., 2001; Viswanathan et al., 2002).
Infection and intracellular survival assay in amebal cells
Infection of protozoan strains with F. novicida has been described previously (Abu Kwaik, 1996; El-Etr et al., 2009). Briefly, triplicate cultures of protozoan strains were seeded into 96-well plates at 1 × 105 amebal cells/well and allowed to adhere for a few hours at 25°C. The amebae were washed and infected with F. novicida at a multiplicity of infection (MOI) of 10. After coincubation for 15 min, the cells were washed once and incubated with 100 μg gentamicin/ml for 1 h at 37°C and 5% CO2, followed by gentamicin treatment at the end of all time points examined. The amebae were then washed once to remove gentamicin and lysed with Triton-X100 (0.1%) for 10 min. The number of F. novicida in each well was determined by plating serial dilutions on BCYE agar plates.
Confocal laser scanning and electron microscopy
For confocal microscopy, acidification of the Francisella containing vacuoles (FCVs) was determined using the lysosomotropic agent LysoTracker Red DND-99 (Molecular Probes). H. vermiformis cells were grown on glass cover slips in 24-well plates and then used for subsequent invasion assays with live or heat-killed bacteria. Briefly, triplicate cultures of protozoan strains were seeded into plates at 1 × 105 amebal cells/well and allowed to adhere for a few hours at 25°C. The amebae were washed and infected with F. novicida at a MOI of 10. After coincubation for 15 min, the cells were washed once and incubated with 100 μg gentamicin/ml for 1 h at 37°C and 5% CO2. Thirty minutes prior to the time point, the amebal cells were washed and incubated with 1 ml of 1 μM LysoTracker, washed three times with PBS, fixed with 4% paraformaldehyde, and then mounted on glass slides for confocal microscopy analysis. All confocal microscopy analyses were performed on one hundred infected amebal cells from three different cover slips, for each time point in each experiment, and all experiments were performed three times. The analysis of colocalization has been performed on individual optical sections. The quantification was performed manually on a FV1000 Olympus confocal microscope. The images shown in the figures are stacks of 15 one-micron-thick Z-series sections.
For electron microscopy, triplicate cultures of protozoan strains were seeded into 12-well plates at 1 × 105 amebal cells/well and allowed to adhere for a few hours at 25°C. The amebae were washed and infected with F. novicida at a MOI of 10. After coincubation for 15 min, the cells were washed once and incubated with 100 μg gentamicin/ml for 1 h at 37°C and 5% CO2. At several time intervals, the infected and uninfected monolayers were fixed for transmission electron microscopy with glutaraldehyde, post fixed with OsO4 in Sorenson's buffer (pH 7.4), dehydrated with ethanol, and embedded in Epon (Miller-Stephenson), as described previously (Santic et al., 2005b). The sections were stained with lead citrate and uranyl acetate and examined with a Phillips Morgany transmission electron microscopy.
Results
Intracellular replication of F. novicida within A. castellanii and H. vermiformis
Previous studies have shown that F. tularensis subsp. holarctica and tularensis, and F. novicida survive and replicate in A. castellanii (Abd et al., 2003; Greub and Raoult, 2004; Lauriano et al., 2004; Hazlett et al., 2008; El-Etr et al., 2009). Since H. vermiformis is the most predominant non-pathogenic ameba in water supplies, we determined the ability of F. novicida to replicate within H. vermiformis and compared that to A. castellanii during early and late stages of infection. In addition, it has been shown that iglC is required for growth in macrophages and Acanthamoeba (Abd et al., 2003; Greub and Raoult, 2004; Lauriano et al., 2004; Hazlett et al., 2008; El-Etr et al., 2009), but there is no evidence about the intracellular replication of F. novicida within H. vermiformis.
The amebal cells (1 × 105 amebal cells/well) were infected with F. novicida and/or the iglC mutant at a MOI of 10 for 15 min, followed by treatment with gentamicin for 1 h to kill extracellular bacteria, followed by further incubation. The time at the end of 15 min of infection was considered T0. At different time points after infection (2, 24, 48, and 72 h), the amebal cells were lysed and bacteria were grown on agar plates to determine the number of colony forming units (CFU).
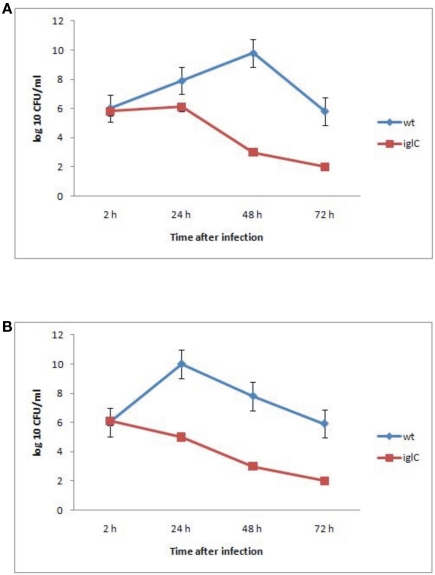
The data showed that F. novicida exhibited robust replication and bacterial numbers increased by 10,000-fold by ∼48 h after infection within A. castellanii cells (Figure 1A). We also determined the ability of F. novicida to replicate within H. vermiformis. F. novicida replicated in H. vermiformis cells at a much faster rate than in A. castellanii cells, and the bacterial number increased by 10,000-fold within 24 h after infection (Figure 1B). The iglC mutant was unable to multiply in A. castellanii or H. vermiformis, and their viability decreased dramatically by 48 h post-infection (Figures 1A,B). Only around 10% of cells were lysed by 48 h after infection (data not shown). The above data showed that F. novicida survives and replicates intracellularly within A. castellanii and H. vermiformis, and that the iglC bi-cistronic locus is necessary for intracellular growth and survival within H. vermiformis.
Figure 1.
Iintracellular growth of F. novicida and its isogenic mutant iglC in A. castellanii (A) and in H. vermiformis (B). The cells were infected for 15 min, followed by gentamicin treatment, and determination of the number of intracellular bacteria at the indicated time points. The error bars represent standard deviations of triplicate samples and results shown are representative of three independent experiments.
F. tularensis replicates in vacuoles within H. vermiformis cells
It has been shown that the FCP matures to a late-endosome-like-phagosome prior to bacterial escape into the cytosol of macrophages, where bacterial proliferation occurs (Clemens et al., 2004; Santic et al., 2005b, 2010; Chong and Celli, 2010; Asare and Abu Kwaik, 2011; Bröms et al., 2011). The process of phagosomal disruption is rapid and occurs within 30 min. of infection in mammalian cells (Chong et al., 2008; Santic et al., 2008; Chong and Celli, 2010; Asare and Abu Kwaik, 2011; Bröms et al., 2011). The iglC locus is essential for bacterial escape into the cytosol of macrophages (Lindgren et al., 2004; Santic et al., 2005b; Asare and Abu Kwaik, 2011). Therefore, we examined at the ultra-structural level the intracellular infection of H. vermiformis with F. novicida.
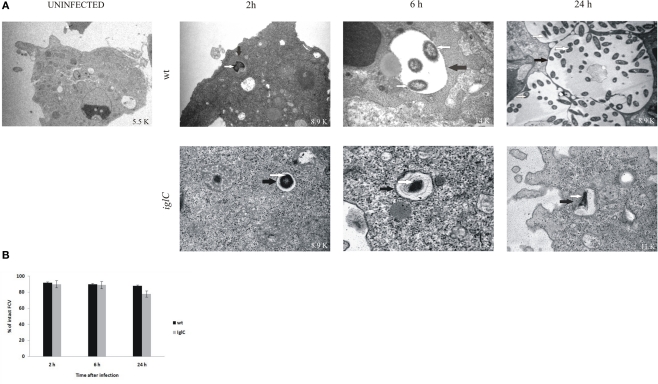
Up to 2 h after infection by F. novicida, the bacteria were localized in intact vacuoles within H. vermiformis (Figures 2A,B), and efficient phagocytosis of bacteria by the amebae was observed. Almost 95% of the vacuoles harboring F. novicida had intact vacuolar membranes. By 6 h after infection, the bacteria were still enclosed in intact vacuoles within amebal cells, and still 90% of H. vermiformis harboring the bacteria had intact vacuolar membranes (Figures 2A,B). There was clear evidence that by 6 h bacterial replication had been initiated, since only one bacterium per vacuole was detected at 2 h while two to six bacteria per vacuole were detected by 6 h post-infection. During all time points examined more than 90% of bacteria were intra-vacuolar. Only 10% of the intracellular bacteria were localized in the cytosol.
Figure 2.
Francisella novicida replicates in vacuoles of H. vermiformis. (A) Representative electron micrographs of H. vermiformis infected with wt F. tularensis subsp. novicida or the iglC mutant at 2, 6, and 24 h after infection. Thin black arrows show intact vacuolar membranes and white arrows show bacteria. (B) Quantitative analysis of the integrity of vacuolar membranes containing wt F. novicida or iglC in H. vermiformis cells. The percentage of disrupted vacuoles harboring bacteria at different time points.
By 24 h after infection ∼90% of the vacuoles were intact with a clear and distinct vacuolar membrane (Figures 2A,B). Similar results to intra-vacuolar localization of the wt strain have been also observed for the iglC mutant. The mutant resides in intact vacuoles within H. vermiformis cells at all time points after infection (Figures 2A,B). However, the iglC mutant failed to replicate within the vacuole. In addition, H. vermiformis did not differentiate into cysts during the time points examined. There have been just a few cysts containing multiplying bacteria.
We conclude that in contrast to mammalian cells, where bacteria do not replicate in the vacuole but escape into the cytosol where they replicate, F. novicida does not escape into the cytosol, but replicates within the vacuoles in H. vermiformis.
F. tularensis does not reside in an acidic compartment within H. vermiformis
Previous data in human macrophages showed that within the first 15 min after infection, ∼90% of the FCPs acquire the lysosomotropic dye LysoTracker, which concentrates in acidic compartments (Chong et al., 2008; Santic et al., 2008; Chong and Celli, 2010; Asare and Abu Kwaik, 2011; Bröms et al., 2011). Colocalization of phagosomes harboring the iglC mutant with the LysoTracker dye was persistent, which is consistent with fusion to the lysosomes and failure of the iglC mutant to escape into the macrophage cytosol (Lindgren et al., 2004; Santic et al., 2005b; Chong and Celli, 2010; Asare and Abu Kwaik, 2011; Bröms et al., 2011). The acquired lysosomotropic dye is gradually lost by 30–60 min post-infection, which coincides with bacterial escape into the cytosol of human monocyte derived macrophages (hMDMs; Chong et al., 2008; Santic et al., 2008; Chong and Celli, 2010; Asare and Abu Kwaik, 2011; Bröms et al., 2011). This transient acidification is essential for subsequent bacterial escape and replication in the macrophage cytosol (Chong et al., 2008; Santic et al., 2008; Chong and Celli, 2010; Asare and Abu Kwaik, 2011; Bröms et al., 2011). Based on our ultra-structural studies, we did not observe dramatic changes between 1 and 2 h after infection and 6 and 12 h after infection; therefore we monitored the acidification of the vacuoles at 30 min, 1, and 12 h by using the lysosomotropic agent LysoTracker Red DND-99, which concentrates in acidified vesicles and compartments.
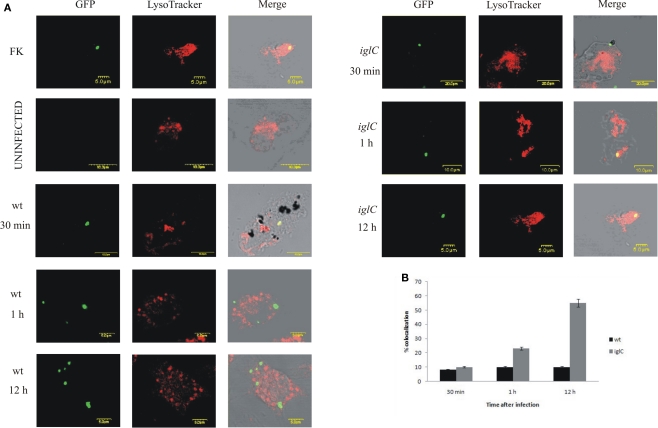
Our results showed that 90% of the wt bacteria in FCVs did not co-localize with the LysoTracker Red DND-99 dye at 30 min, 1, or 12 h post-infection (Figures 3A,B). In contrast, many of the iglC mutant-containing vacuoles acquired the LysoTracker Red DND-99 dye (∼55% colocalization) at 12 h after infection (Figures 3A,B). We conclude that F. tularensis does not reside transiently or permanently in acidic compartments within H. vermiformis, which is distinct from mammalian cells.
Figure 3.
Francisella novicida does not reside in acidic compartments within H. vermiformis cells. (A) Representative confocal microscopy images of colocalization of FCVs with the LysoTracker dye by the GFP-expressing wt F. novicida and iglC at 30 min, 1, and 12 h post-infection is shown. Uninfected cells were used as a negative control, while formalin killed bacteria served as a positive control. The images are representatives of 100 infected cells examined from three different cover slips. The results shown are representative of three independent experiments. (B) Quantification of colocalization of the LysoTracker DND-99 dye with the FCVs of the bacterium at 30 min, 1, and 12 h post-infection is shown. The results shown are representative of three independent experiments, and error bars represent standard deviations of triplicate samples.
Discussion
Epidemiology of F. tularensis in various parts of the world is associated with water-borne transmission, which includes mosquitoes and amebae as the potential host reservoirs of the bacteria in water resources (Thelaus et al., 2009; Chong and Celli, 2010; Akimana and Abu Kwaik, 2011; Asare and Abu Kwaik, 2011; Bosio, 2011; Broman et al., 2011; Bröms et al., 2011; Dai et al., 2011; Gavrilin and Wewers, 2011; Jones et al., 2011; Zogaj and Klose, 2011). However, the main aquatic reservoir of the bacterium is still not known, but likely includes mosquitoes and amebae. Recently, it has been shown that F. novicida and LVS utilize A. castellanii as a natural reservoir (Abd et al., 2003; Hazlett et al., 2008; El-Etr et al., 2009). In addition, F. tularensis LVS and F. novicida survive in A. castellanii for weeks of infection (Abd et al., 2003; Hazlett et al., 2008; El-Etr et al., 2009). Surprisingly, F. novicida multiplied to a much higher degree in H. vermiformis in comparison to what has been found in A. castellanii (Abd et al., 2003; Hazlett et al., 2008; El-Etr et al., 2009). Another explanation is that we used rich 30234 medium compared to the El-Etr et al. (2009) study, where they used High Salt buffer, which does not support F. novicida replication. At 24 h after infection there were 1010 CFU/ml in H. vermiformis. In addition, there was a sudden drop in bacterial numbers, probably due to lysis by amebae. El-Etr et al. (2009) showed that by 30 min post-infection, the bacteria were in spacious vacuoles and continued to replicate until 24 h after infection. They suggested that spacious vacuoles are not lysosomal in nature, and that enclosure within these vacuoles may provide a survival advantage to F. novicida, SCHU S4 and other virulent strains of F. tularensis. However, it was not examined whether the bacteria replicated within vacuoles, and whether formation of vacuoles was necessary for efficient replication (El-Etr et al., 2009). The long-term survival of pathogenic F. tularensis in amebae is dependent on induction of amebal cyst formation. In comparison to A. castellanii, H. vermiformis did not differentiate into cysts. There have been just a few cysts containing multiplying bacteria, which is very different from A. castellanii. The same culture conditions have been used as in the El-Etr et al. (2009) study. It is possible that the encystation process is delayed in H. vermiformis in comparison to A. castellanii infected with F. novicida.
Lauriano et al. (2004) showed that F. tularensis mglA and iglC mutant strains are not only defective for survival and replication within the macrophage-like cell line, but also within A. castellanii. In the present study we examined the interaction of F. novicida with H. vermiformis and the role of the FPI gene iglC in this interaction. The results show, for the first time, that F. novicida can survive and grow within H. vermiformis. The bacteria replicate and grow in vacuolar structures inside the trophozoites of H. vermiformis. Our ultra-structural studies showed that the vacuoles are tight and intact at 2, 6, and 24 h after infection. The iglC locus plays an important role in survival and replication of bacteria within H. vermiformis cells.
Other previous studies have shown that within mammalian cells, F. tularensis resides in acidic vacuoles before escaping to the cytosol where it replicates (Chong et al., 2008; Santic et al., 2008; Chong and Celli, 2010; Asare and Abu Kwaik, 2011; Bröms et al., 2011). Recently El-Etr et al. (2009) have shown that ∼40% of the FCVs harboring F. novicida co-localized with the acidic dye, LysoTracker Red, in A. castellanii vacuoles at 2 h after infection. However, our results in H. vermiformis show that only 10% of the FCVs harboring F. novicida co-localize with the acidic dye at 30 min, 1, and 12 h after infection. The effect of the lysotracker was examined in uninfected cells as well as in H. vermiformis cells with formalin killed bacteria. In both cases H. vermiformis cells were in the trophozoite stage, and the lysotracker did not affect the physiology of the amebae. Whether the lysotracker affects cyst formation or vice versa has not been examined in this study. El-Etr et al. (2009) also showed that there was little difference between colocalization of the FCVs with LysoTracker Red for the different Francisella subsp. (holarctica-derived LVS and novicida). Our data show that many of the iglC mutant-containing FCVs acquired the LysoTracker Red DND-99 dye at a late stage of infection of H. vermiformis, which likely coincided with loss of bacterial viability (Figure 1). Similar observations have been described in macrophages (Santic et al., 2005b). Our results clearly show that F. novicida does not reside transiently or permanently in acidic compartments within H. vermiformis cells after 30 min of initiation of the infection.
Conclusion
There are major differences in the life style of F. tularensis within various protozoa and macrophages. The bacteria multiply to a higher degree in H. vermiformis in comparison to A. castellanii cells (Abd et al., 2003; Hazlett et al., 2008; El-Etr et al., 2009). The formation of cysts is not significant in H. vermiformis cells compared to Acanthamoeba (Abd et al., 2003; Hazlett et al., 2008; El-Etr et al., 2009). Our results showed intra-vacuolar replication of F. novicida within H. vermiformis. This is very different from mammalian cells, where cytosolic location of bacteria is a key component in productive intracellular replication. In H. vermiformis the bacteria do not escape from the vacuole into the cytosol to replicate. In contrast, they are enclosed in vacuoles. Despite the major differences between the intracellular lifestyles within H. vermiformis and macrophages, the IglC protein is essential for bacterial proliferation in both hosts. It is clear that this protein is essential for intra-vacuolar proliferation in amebae, but is also essential for evasion of lysosomal fusion/and or bacterial escape into the cytosol in macrophages. It is likely that these two events in the evolutionarily distant host cells are due to a secondary effect of the biological function of IglC, which still remains to be determined.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work is supported by a grant of Ministry of Science, Education and Sports, Republic of Croatia (grant no. 062-0621273-0950). We thank Elizabeth Hartland for H. vermiformis cells and Snake C. Jones for proofreading the manuscript. Yousef Abu Kwaik is supported by Public Health Service Awards R01AI43965 and R01AI069321 from the National Institute of Allergy and Infectious Diseases (NIAID) and by the Commonwealth of Kentucky Research Challenge Trust Fund.
References
- Abd H., Johansson T., Golovliov I., Sandstrom G., Forsman M. (2003). Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69, 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu Kwaik Y. (1996). The phagosome containing Legionella pneumophila within the protozoan Hartmannella vermiformis is surrounded by the rough endoplasmic reticulum. Appl. Environ. Microbiol. 62, 2022–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu Kwaik Y., Gao L.-Y., Stone B. J., Venkataraman C., Harb O. S. (1998). Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64, 3127–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akimana C., Abu Kwaik Y. (2011). Francisella–arthropod vector interaction and its role in patho-adaptation to infect mammals. Front. Microbio. 2:34. 10.3389/fmicb.2011.00034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R., Springer N., Schonhuber W., Ludwig W., Schmid E. N., Muller K.-D., Michel R. (1997). Obligate intracellular bacterial parasites of Acanthamoebae related to Chlamydia spp. Appl. Environ. Microbiol. 63, 115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asare R., Abu Kwaik Y. (2011). Exploitation of host cell biology and evasion of immunity by Francisella tularensis. Front. Microbio. 1:145 10.3389/fmicb.2010.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J. R., Chong A., Wehrly T. D., Yu J. J., Rodriguez S. A., Liu J. (2009). The Francisella tularensis pathogenicity island encodes a secretion system that is required for phagosome escape and virulence. Mol. Microbiol. 74, 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingle L. E., Bailey C. M., Pallen M. J. (2008). Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11, 3–8 [DOI] [PubMed] [Google Scholar]
- Bonquist L., Lindgren H., Golovliov I., Guina T., Sjostedt A. (2008). MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect. Immun. 76, 3502–3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosio C. (2011). The subversion of the immune system by Francisella tularensis. Front. Microbio. 10.3389/fmicb.2011.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broman T., Thelaus J., Andersson A.-C., Bäckman S., Wikström P., Larsson E., Granberg M., Karlsson L., Bäck E., Eliasson H., Mattsson R., Sjöstedt A., Forsman M. (2011). Molecular detection of persistent Francisella tularensis subspecies holarctica in natural waters. Int. J. Microbiol. 10.1155/2011/851946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms J. E., Sjöstedt A., Lavander M. (2011). The role of the Francisella tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front. Microbio. 1:136 10.3389/fmicb.2010.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A., Celli J. (2010). The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front. Microbio. 1:138 10.3389/fmicb.2010.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong A., Wehrly T. D., Nair V., Fischer E. R., Barker J. R., Klose K. E., Celli J. (2008). The early phagosomal stage of Francisella tularensis determines optimal phagosomal escape and Francisella pathogenicity island protein expression. Infect Immun. 76, 5488–5499 10.1128/IAI.00682-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens D. L., Lee B. Y., Horwitz M. A. (2004). Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect. Immun. 72, 3204–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T. J., Butchar J. P., Tridandapani S. (2011). Francisella subverts innate immune signaling: focus on PI3K/Akt. Front. Microbio. 10.3389/fmicb.2011.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S., Mohapatra N. P., Schlesinger L. S., Gunn J. S. (2011). Regulation of Francisella tularensis virulence. Front. Microbio. 1:144 10.3389/fmicb.2010.00144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin O. M., Ludu J. S., Nano F. E. (2007). The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth. BMC Microbiol. 7, 1. 10.1186/1471-2180-7-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Etr S. H., Margolis J. J., Monack D., Robison R. A., Cohen M., Moore E., Rasley A. (2009). Francisella tularensis type A strains cause the rapid encystment of Acanthamoeba castellanii and survive in amoebal cysts for three weeks postinfection. Appl. Environ. Microbiol. 75, 7488–7500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsman M., Sandstrom G., Sjostedt A. (1994). Analysis of 16S ribosomal DNA sequences of Francisella strains and utilization for determination of the phylogeny of the genus and for identification of strains by PCR. Int. J. Syst. Bacteriol. 44, 38–46 10.1099/00207713-44-1-38 [DOI] [PubMed] [Google Scholar]
- Gavrilin M. A., Wewers M. D. (2011). Francisella recognition by inflammasomes: differences between mice and men. Front. Microbio. 10.3389/fmicb.2011.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco D., Allegrini G., Tizzi T., Ninu E., Lamanna A., Luzi S. (1987). A waterborne tularemia outbreak. Eur. J. Epidemiol. 3, 35–38 [DOI] [PubMed] [Google Scholar]
- Greub G., Raoult D. (2004). Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17, 413–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harb O. S., Gao L.-Y., Abu Kwaik Y. (2000). From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2, 251–265 [DOI] [PubMed] [Google Scholar]
- Hazlett K. R., Caldon S. D., McArthur D. G., Cirillo K. A., Kirimanjeswara G. S., Magguilli M. L., Malik M., Shah A., Broderick S., Golovliov I., Metzger D. W., Rajan K., Sellati T. J., Loegering D. J. (2008). Adaptation of Francisella tularensis to the mammalian environment is governed by cues which can be mimicked in vitro. Infect. Immun. 76, 4479–4488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. W., Broz P., Monack D. M. (2011). Innate immune recognition of Francisella tularensis: activation of type-I interferons and the inflammasome. Front. Microbio. 10.3389/fmicb.2011.00016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauriano C. M., Barker J. R., Nano F. E., Arulanandam B. P., Klose K. E. (2003). Allelic exchange in Francisella tularensis using PCR products. FEMS Microbiol. Lett. 229, 195–202 [DOI] [PubMed] [Google Scholar]
- Lauriano C. M., Barker J. R., Yoon S. S., Nano F. E., Arulanandam B. P., Hassett D. J., Klose K. E. (2004). MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc. Natl. Acad. Sci. U.S.A. 101, 4246–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H., Golovliov I., Baranov V., Ernst R. K., Telepnev M., Sjostedt A. (2004). Factors affecting the escape of Francisella tularensis from the phagolysosome. J. Med. Microbiol. 53, 953–958 [DOI] [PubMed] [Google Scholar]
- Ludu J. S., Nix E. B., Duplantis B. N., de Bruin O. M., Gallagher L. A., Hawley L. M., Nano F. E. (2008). Genetic elements for selection, deletion mutagenesis and complementation in Francisella spp. FEMS Microbiol. Lett. 278, 86–93 [DOI] [PubMed] [Google Scholar]
- Meibom K. L., Barel M., Charbit A. (2009). Loops and networks in control of Francisella tularensis virulence. Future Microbiol. 4, 713–729 10.2217/fmb.09.37 [DOI] [PubMed] [Google Scholar]
- Meibom K. L., Charbit A. (2010). Francisella tularensis metabolism and its relation to virulence. Front. Microbio. 1:140 10.3389/fmicb.2010.00140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M., Horn M., Wagner M., Santic M., Abu Kwaik Y. (2005). Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71, 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morner T. (1992). The ecology of tularaemia. Rev. Sci. Tech. 11, 1123–1130 [PubMed] [Google Scholar]
- Nano F. E., Schmerk C. (2007). The Francisella pathogenicity island. Ann. N. Y. Acad. Sci. 1105, 122–137 [DOI] [PubMed] [Google Scholar]
- Nano F. E., Zhang N., Cowley S. C., Klose K. E., Cheung K. K., Roberts M. J., Ludu J. S., Letendre G. W., Meierovics A. I., Stephens G., Elkins K. L. (2004). A Francisella tularensis pathogenicity island required for intramacrophage growth. J. Bacteriol. 186, 6430–6436 10.1128/JB.186.19.6430-6436.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L. L., Radulic M., Doric M., Abu Kwaik Y. (2001). HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Infect. Immun. 69, 2569–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M., Akimana C., Asare R., Kouokam J. C., Atay S., Kwaik Y. A. (2009). Intracellular fate of Francisella tularensis within arthropod-derived cells. Environ. Microbiol. 11, 1473–1481 [DOI] [PubMed] [Google Scholar]
- Santic M., Al-Khodor S., Abu Kwaik Y. (2010). Cell biology and molecular ecology of Francisella tularensis. Cell. Microbiol. 12, 129–139 [DOI] [PubMed] [Google Scholar]
- Santic M., Asare R., Skrobonja I., Jones S., Abu Kwaik Y. (2008). Acquisition of the vacuolar ATPase proton pump and phagosome acidification are essential for escape of Francisella tularensis into the macrophage cytosol. Infect. Immun. 76, 2671–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santic M., Molmeret M., Abu Kwaik Y. (2005a). Modulation of biogenesis of the Francisella tularensis subsp. novicida-containing phagosome in quiescent human macrophages and its maturation into a phagolysosome upon activation by IFN-gamma. Cell. Microbiol. 7, 957–967 10.1111/j.1462-5822.2005.00529.x [DOI] [PubMed] [Google Scholar]
- Santic M., Molmeret M., Klose K. E., Jones S., Abu Kwaik Y. (2005b). The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm. Cell. Microbiol. 7, 969–979 10.1111/j.1462-5822.2005.00526.x [DOI] [PubMed] [Google Scholar]
- Sjöstedt A. B. (2005). “Genus I. Francisella Dorofe'ev 1947, 176AL,” in Bergey's Manual of Systematic Bacteriology, 2nd Edn. Vol. 2, Part B, eds Brenner D. J., Krieg N. R., Staley J. T., Garrity G. M. (New York: Springer; ), 200–210 [Google Scholar]
- Steinert M., Birkness K., White E., Fields B., Quinn F. (1998). Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl. Environ. Microbiol. 64, 2256–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelaus J., Andersson A., Mathisen P., Forslund A. L., Noppa L., Forsman M. (2009). Influence of nutrient status and grazing pressure on the fate of Francisella tularensis in lakewater. FEMS Microbiol. Ecol. 67, 69–80 [DOI] [PubMed] [Google Scholar]
- Viswanathan V. K., Kurtz S., Pedersen L. L., Abu-Kwaik Y., Krcmarik K., Mody S., Cianciotto N. P. (2002). The cytochrome c maturation locus of Legionella pneumophila promotes iron assimilation and intracellular infection and contains a strain-specific insertion sequence element. Infect. Immun. 70, 1842–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willke A., Meric M., Grunow R., Sayan M., Finke E. J., Splettstosser W., Seibold E., Erdogan S., Ergonul O., Yumuk Z., Gedikoglu S. (2009). An outbreak of oropharyngeal tularaemia linked to natural spring water. J. Med. Microbiol. 58, 112–116 [DOI] [PubMed] [Google Scholar]
- Zogaj X., Klose K. E. (2011). Genetic manipulation of Francisella tularensis. Front. Microbio. 1:142 10.3389/fmicb.2010.00142 [DOI] [PMC free article] [PubMed] [Google Scholar]