Abstract
Soda lakes represent a unique ecosystem with extremely high pH (up to 11) and salinity (up to saturation) due to the presence of high concentrations of sodium carbonate in brines. Despite these double extreme conditions, most of the lakes are highly productive and contain a fully functional microbial system. The microbial sulfur cycle is among the most active in soda lakes. One of the explanations for that is high-energy efficiency of dissimilatory conversions of inorganic sulfur compounds, both oxidative and reductive, sufficient to cope with costly life at double extreme conditions. The oxidative part of the sulfur cycle is driven by chemolithoautotrophic haloalkaliphilic sulfur-oxidizing bacteria (SOB), which are unique for soda lakes. The haloalkaliphilic SOB are present in the surface sediment layer of various soda lakes at high numbers of up to 106 viable cells/cm3. The culturable forms are so far represented by four novel genera within the Gammaproteobacteria, including the genera Thioalkalivibrio, Thioalkalimicrobium, Thioalkalispira, and Thioalkalibacter. The latter two were only found occasionally and each includes a single species, while the former two are widely distributed in various soda lakes over the world. The genus Thioalkalivibrio is the most physiologically diverse and covers the whole spectrum of salt/pH conditions present in soda lakes. Most importantly, the dominant subgroup of this genus is able to grow in saturated soda brines containing 4 M total Na+ – a so far unique property for any known aerobic chemolithoautotroph. Furthermore, some species can use thiocyanate as a sole energy source and three out of nine species can grow anaerobically with nitrogen oxides as electron acceptor. The reductive part of the sulfur cycle is active in the anoxic layers of the sediments of soda lakes. The in situ measurements of sulfate reduction rates and laboratory experiments with sediment slurries using sulfate, thiosulfate, or elemental sulfur as electron acceptors demonstrated relatively high sulfate reduction rates only hampered by salt-saturated conditions. However, the highest rates of sulfidogenesis were observed not with sulfate, but with elemental sulfur followed by thiosulfate. Formate, but not hydrogen, was the most efficient electron donor with all three sulfur electron acceptors, while acetate was only utilized as an electron donor under sulfur-reducing conditions. The native sulfidogenic populations of soda lakes showed a typical obligately alkaliphilic pH response, which corresponded well to the in situ pH conditions. Microbiological analysis indicated a domination of three groups of haloalkaliphilic autotrophic sulfate-reducing bacteria belonging to the order Desulfovibrionales (genera Desulfonatronovibrio, Desulfonatronum, and Desulfonatronospira) with a clear tendency to grow by thiosulfate disproportionation in the absence of external electron donor even at salt-saturating conditions. Few novel representatives of the order Desulfobacterales capable of heterotrophic growth with volatile fatty acids and alcohols at high pH and moderate salinity have also been found, while acetate oxidation was a function of a specialized group of haloalkaliphilic sulfur-reducing bacteria, which belong to the phylum Chrysiogenetes.
Keywords: sulfur-oxidizing bacteria, sulfidogenesis, sulfate-reducing bacteria, sulfur reduction, thiosulfate reduction, soda lakes
Introduction
Sulfur is an important element for life. Apart of its structural importance as a component of various proteins, a complex sulfur cycle exists in the biosphere based on the ability of sulfur atom to change its valence from −2 (sulfide) to +6 (sulfate). Most of the sulfur redox reactions are not spontaneous at ambient conditions and, therefore, are catalyzed by prokaryotes capable of exploiting various inorganic sulfur compounds in their energy-transformation (“-generating”) metabolism, either as electron donors (lithotrophic sulfur-oxidizing prokaryotes, anoxygenic phototrophic bacteria) or as electron acceptors (sulfidogenic prokaryotes). The biological sulfur cycle is an integral part of the whole biogeochemical cycle and connected to many other main element cycles, such as carbon (CO2 fixation, oxidation of organic compounds, formation of organo-sulfur compounds), nitrogen (thiodenitrification), and metal (formation of metal sulfides) cycles. Due to its significance, the microbial sulfur cycle is very well investigated, both in the oxidative and reductive branches. Most of the knowledge, however, was obtained from habitats that are very acidic (e.g., acid mine drainage) or at neutral pH (e.g., marine and fresh waters). Until recently, practically nothing was known about the possibility of dissimilatory sulfur transformations at extremely high pH. This review summarizes 15 years of research on the microbial sulfur cycle in naturally occurring habitats with a stable high pH and a high salinity called “soda lakes.”
Soda Lakes as a Unique Habitat
Soda lakes form a specific type of salt lakes where sodium carbonate/bicarbonate is among the dominant salts. Soda lakes are located in areas with dry climate conditions that facilitate salt accumulation in depressions by evaporative concentration. The earliest scientific record on the natron-depositing salt lakes (Wadi an Natrun, Egypt) already appeared in 1898 (Schweinfurth and Lewin, 1898). The genesis of soda lakes is based on the leaching of sodium-rich rocks by high CO2-containing ground waters concomitant with low Ca and Mg background (Eugster, 1970). The presence of sodium carbonate at high concentrations (soluble alkalinity) creates a uniquely stable, natural alkaline habitat, buffered from the acidifying influence of atmospheric CO2 with pH values of brines usually above 9.5, and salt concentrations varying within a broad range, depending on the local conditions. Sodium is the dominant cation in the brines of soda lakes, while chloride and sulfate are, apart from carbonate/bicarbonate, the dominant anions. So, soda lakes seem to be the only natural environment with appropriate conditions for the stable development of obligately alkaliphilic, salt-tolerant microorganisms, which usually grow optimally at a pH around 10 and need at least 0.1 M Na+ for growth. Most studied examples of typical soda lakes are located in Kenya and Tanzania (Bogoria, Magadi, Natron), in Egypt (Wadi Natrun), and California (Mono Lake). Another area abundant in saline alkaline lakes is Central Asia, where the Transbaikal dry steppe stretches from south Siberia to north-east China. Here the lakes are much smaller, more shallow, and with a relatively unstable water regimen and a freezing winter period causing frequent and substantial fluctuations of the water salinity and temperature.
Our studies were mostly focused on Siberian soda lakes, although sulfur-oxidizing bacteria (SOB) were also actively investigated in samples from Kenyan and American soda lakes. The main characteristics of the lakes studied are given in Table 1.
Table 1.
Main characteristics of soda lakes studied.
| Area | Examples | Total salts, g l−1 | pH | Total carbonate alkalinity (M) |
|---|---|---|---|---|
| Kenya | Magadi, Bogoria, Crater lake, Elmenteita | 20–220 | 9.5–11.0 | 0.12–1.16 |
| Wadi Natrun (Egypt) | Rozita, Beida, Gaar, Fazda, Zugm, Hamra, Umm-Risha, Khadra | 200–380 | 9.5–10.3 | 0.11–0.75 |
| California | Mono lake, Searles lake, Owens lake | 90–220 | 9.7 | 0.5–1.0 |
| Washington State | Soap lake | 20–140 | 9.9 | No data |
| Transbaikal Steppe (Russia) | Hilganta, Hadyn, Ulan-Nor, Gorbunka | 5–40 | 9.5–10.2 | 0.02–0.11 |
| Kulunda Steppe (Altai, Russia) | Tanatar lake system, Bitter lake system, Cock soda lake | 20–380 | 9.3–10.6 | 0.02–5.2 |
| North-eastern Mongolia | Hotontyn, Shar-Burdiin, Dzun-Uldziit, Baga-Nur | 5–360 | 9.2–10.5 | 0.02–1.20 |
The unusually high pH imposes several chemical changes important for the microbial sulfur cycling. First of all, sulfide is present entirely in the ionic form as HS−, which, in contrast to H2S, cannot freely cross the membrane and therefore, is not as toxic as H2S. Accordingly, alkaliphilic SOB and sulfidogens can tolerate much higher substrate/product concentrations than their neutrophilic counterparts. Next, sulfide can react chemically with insoluble sulfur forming soluble polysulfides that are stable at high pH. At aerobic conditions polysulfide is rapidly oxidized to thiosulfate, while at anaerobic conditions it is stable, and hence can accumulate to high concentrations representing a true substrate for sulfur-reducing prokaryotes. High alkalinity would be favorable for SOB, because of a buffering effect of the sulfuric acid produced during sulfur oxidation in the periplasm. On the other hand, domination of carbonate over bicarbonate at pH values close to 10 and above is unfavorable for CO2 fixation, setting a limitation for the growth of autotrophic alkaliphiles at extremely high pH.
Sulfur-Oxidizing Bacteria in Soda Lakes
Distribution and diversity
The first pure cultures of SOB able to grow at pH 10 were isolated from the low salt soda lake Hadyn in Tuva (Russia). Strains AL2 and AL3 became the type strains of type species for the two different genera of haloalkaliphilic SOB, Thioalkalivibrio, and Thioalkalimicrobium, respectively, commonly found in soda lakes all over the world (Sorokin et al., 2000). Subsequent investigation of the samples from different geographic locations (Table 1) resulted in the isolation of more than 100 strains of obligately alkaliphilic chemolithoautrophic SOB. This revealed a previously unrecognized fact that SOB can thrive in extremely high pH/salt habitats (Sorokin and Kuenen, 2005; Sorokin et al., 2006). Apart from that, soda lakes yielded numerous strains of heterotrophic haloalkaliphiles identified as Halomonas spp., capable of sulfide and thiosulfate oxidation to sulfur and tetrathionate, respectively, without obvious energy gain (Sorokin, 2003; this group is out of the scope of this review).
Combining enrichment and serial dilution procedures demonstrated a relatively high density of viable SOB in the surface sediments of various soda lakes (103–108 cells/cm3) capable of chemolithoautotrophic growth at pH 10 and salt concentrations from 0.2 to 4 M total Na+ in the form of carbonates. These bacteria apparently took advantage of the extremely high buffering capacity of the soda media thereby reaching high biomass density which, in the case of neutrophilic SOB, would only have been possible under pH-controlled conditions.
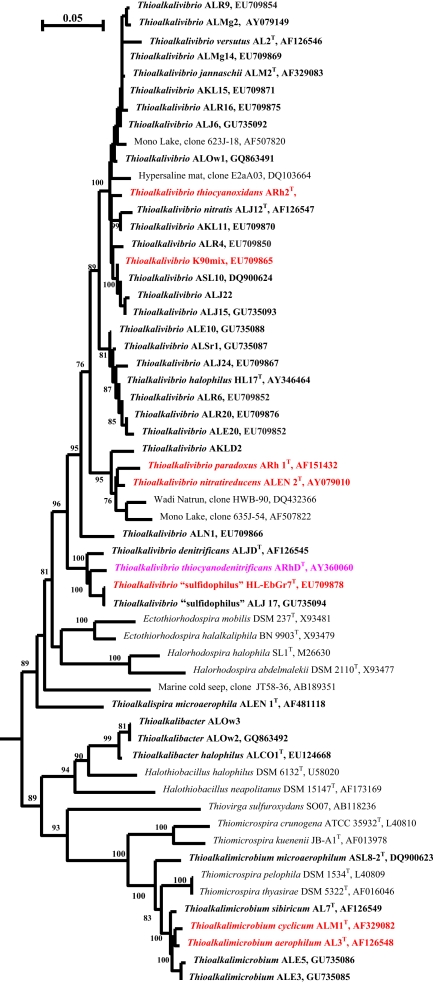
The soda lake SOB isolates formed four distinct phylogenetic groups within the Gammaproteobacteria and were described as four new genera Thioalkalimicrobium, Thioalkalivibrio (Sorokin et al., 2001a, 2002a, 2007a), Thioalkalispira (Sorokin et al., 2002b), and Thioalkalibacter (Banciu et al., 2008; Figure 1). The first three genera include exclusively obligately alkaliphilic salt-tolerant organisms, except for a single facultatively alkaliphilic Thioalkalivibrio halophilus. The genus Thioalkalibacter includes a single species, Thb. halophilus, which is a facultatively alkaliphilic halophile. In general, the Thioalkalimicrobium group dominated enrichment cultures from the low-saline Asian soda lakes. It also could be enriched from the more saline soda lakes, but only from fresh samples. The genus Thioalkalivibrio dominated in enrichments from hypersaline soda lakes and from aged samples, and was always dominant when the enrichment medium contained >1.5 M total Na+. Therefore, the most important selective force influencing survival of alkaliphilic SOB in soda lakes appeared to be the salt concentration. The genus Thioalkalivibrio is apparently the most widely distributed SOB type in soda lakes. Among its more than 100 isolates, several large geographical populations each containing about 20 isolates could be distinguished (Foti et al., 2006). Extremely salt-tolerant isolates from the Kulunda Steppe and Mongolia are clustered together and also resemble strains from Mono Lake and Soap Lake, while most of the Wadi Natrun isolates are clustering with Tv. halophilus. Such clustering correlates with the anionic composition of the brines (see below).
Figure 1.
Phylogenetic position of haloalkaliphilic SOB from soda lakes (in bold) within the Gammaproteobacteria based on 16S rRNA sequence analysis. The strain abbreviations are as follows: AL, strains from Transbaikal region (Russia); AKL, strains from Kulunda Steppe (Altai, Russia); ALMg, strains from north-eastern Mongolia; ALJ, strains from Kenya; ALE, strains from Wadi Natrun in Egypt; ASL, strain from Soap Lake (USA); ASLr, strain from Searles Lake (USA); ALOw, strains from Owens lake (California); ALN1, extremely haloalkaliphilic nitrate-reducing strain from Wadi Natrun; ALR, strains from a lab-scale sulfide-oxidizing bioreactor; HL-EbGr7, a strain from a full-scale sulfide-oxidizing bioreactor. In red are the strains with sequenced genome and in pink are strains which genome sequencing is in progress. Numbers at the nodes indicate the percentage of bootstrap values for the clade of this group in 1000 replications (the values for maximum-likelihood method are given in parentheses). Only values above 70% are shown. Bar, 5 substitutions per 100 nucleotides (nt).
The only other culturable evidences on haloalkaliphilic SOB came from the dry soda lake Texcoco in Mexico (Granada et al., 2009), although none of the isolates were characterized.
The presence of the SOB related to the genus Thioalkalivibrio detected in 16S rRNA gene-based libraries has been reported over the past 10 years in various saline habitats, including marine (AB189351, AB286100), hypersaline mats (DQ103664; Sørensen et al., 2005), and soda lakes (Humayoun et al., 2003; Mesbah et al., 2007). Marine clones, however, had only 90–95% sequence similarity to the species of Thioalkalivibrio, and, therefore, might belong to another SOB genus. Use of a specific functional marker for the key enzyme of the Calvin cycle (RuBisCO gene cbb) to detect autotrophic members of the soda lake microbial communities showed the common presence of the Thioalkalivibrio-related sequences in hypersaline soda lakes and clones related to the Thioalkalimicrobium–Thiomicrospira cluster (Giri et al., 2004; Kovaleva et al., 2011).
Basic physiology of haloalkaliphilic SOB
The comparative properties of soda lake SOB are summarized in Table 2. In their basic physiology, the soda lake SOB represent typical chemolithoautotrophic thiotrophs using the Calvin cycle for inorganic carbon fixation (Tourova et al., 2006, 2007) and growing best with thiosulfate (batch) and sulfide (substrate-limited chemostat). The main difference with their neutrophilic counterparts is not in sulfur metabolism (except for less sulfide toxicity and polysulfide stability at high pH), but in the possibility to realize the lithoautotrophic phenotype in general at extremely high pH and salinity. The potential for this kind of metabolism was not known before. So, the main characteristic of the soda lake SOB is their preference for a sodium carbonate-based environment with optimal growth at pH around 10. The most robust data for the growth/activity pH response were obtained in pH-controlled continuous cultures (Sorokin et al., 2003a; Banciu et al., 2004a). The growth pH maximum reached in this experimental set up was 10.5 for both genera, whilst metabolic activity remained active up to pH 11. Our explanation is that the growth, i.e., CO2 fixation, was limited by inorganic carbon availability (domination of unavailable carbonate over available bicarbonate), whereas sulfur metabolism could still proceed at he higher pH values. The situation is somewhat analogous to that of the xerophytic plants inhabiting cold wetlands – there is enough water present, but it is not possible to take it up, because it is too cold.
Table 2.
Basic properties of haloalkaliphilic SOB from soda lakes.
| Property |
Thioalkalimicrobium spp. |
Thioalkalispira microaerophila |
Thioalkalivibrio spp |
Thioalkalibacter halophilus |
|---|---|---|---|---|
| Number of validated species* | 4 | 1 | 9 | 1 |
| Close relative | Thiomicrospira pelophila | No | Ectothiorhodospira | Halothiobacillus |
| Cell morphology | Rods and spirilla | Spirilla | Rods, vibrios, spirilla, and cocci | Rods |
| Sulfur compounds oxidized | HS−, Sn2−, S2O32− | HS−, Sn2−, S2O32− | HS−, Sn2−, S2O32−, S8, SO32−, S4O62−, SCN− | HS−, Sn2−, S2O32− |
| Electron acceptors | O2 | O2 microaerophile | O2, N2O | O2 |
| pH optimum | 9.5–10.0 | 10.0 | 10.0–10.2 | 8.5 |
| Upper salt limit | 1.5 M Na+ | 1.4 M Na+ | 4.3 M Na+ | 3.5 M Na+ |
| Maximal specific growth rate | 0.33 h−1 | 0.08 h−1 | 0.25 h−1 | 0.22 h−1 |
| Maximal growth yield with | 3.5 mg protein mmol−1 |
5.8 mg protein mmol−1 |
6.5 mg protein mmol−1 |
3.5 mg protein mmol−1 |
| Rate of HS− oxidation | Extremely high | Low | Low | High |
| Dominant compatible solute | Ectoine | nd | Glycine betaine | Ectoine |
| Yellow membrane pigment | – | + | + | – |
| nifH | – | + | – | – |
| Distribution | Asia, Africa, North America | Egypt | Asia, Africa, North America | Asia, North America |
*Up to date.
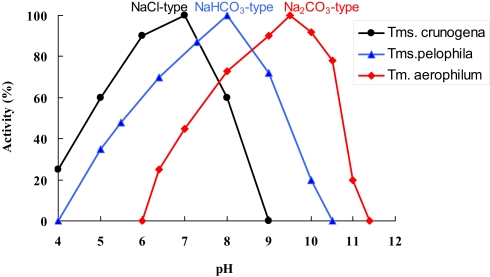
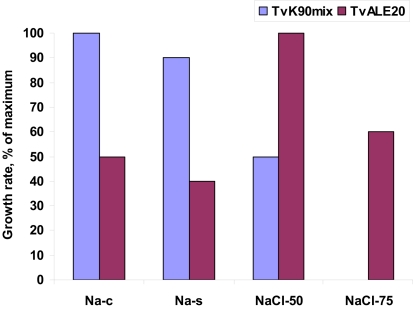
Direct comparison of the growth, c.q. activity of Thioalkalimicrobium aerophilum with its close phylogenetic relatives, Thiomicrospira pelophila and Thiomicrospira crunogena, showed that the three species correspond to three different pH-anion types which may be common also for other physiological groups: the neutrophilic “NaCl type” (marine Tms. crunogena), the alkalitolerant “NaHCO3 type” (littoral Tms. pelophila) and the alkaliphilic “Na2CO3 type” (soda lake Tm. aerophilum; Figure 2). The first two types cannot tolerate the presence of carbonate and pH 10, which are specific optimal conditions for the soda lake organisms. Further work with two facultatively alkaliphilic and extremely salt-tolerant SOB, Thioalkalivibrio halophilus (Banciu et al., 2004b). and Thioalkalibacter halophilus (Banciu et al., 2008) allowed us to identify a previously ignored biological difference between halophiles (NaCl-loving) and natronophiles (soda-loving) organisms and bring forward the concept of natronophily (Table 3). The essence of the latter is that the preference of soda lake microbes for soda might be at least partially explained by its weak electrolytic properties as compared to the strongly electrolytic NaCl. A direct measurement of osmotic parameters of two media used in our experiments with the mentioned SOB species, both containing in total 4 M Na+ at pH 10, but one based on NaCl and another – on sodium carbonate buffer, showed two times higher values for the former, which is also obvious from conductivity measurements. This means that the osmotic burden in saturated soda brines (as well as in sodium sulfate brines) is two times less than in NaCl brines. Therefore, the natronophiles would need two times less specific compatible solutes concentration than the organisms living in NaCl brines. Compatible solutes (or osmolytes) in most of the bacteria are small organic molecules accumulating at high salt to protect the cytoplasm from desiccation (Roeßler and Müller, 2001; Roberts, 2005). The experimental results obtained for the two facultatively alkaliphilic halophilic SOB species grown in NaCl-dominate and in soda-dominated brines confirmed this conclusion (Figure 3). So, from one hand, life at extremely high pH demands extra energy for pH homeostasis, but, on the other hand this is balanced by much less cost of compatible solute synthesis than in neutraphilic halophiles. Such a tendency can also be seen in comparison of the growth dynamic of a typical obligate natronophilic SOB Thioalkalivibrio K90mix from hypersaline soda lakes in Siberia and a haloalkaliphilic Thioalkalivibrio ALE20 from hypersaline alkaline lakes in Wadi Natrun. The former definitely preferred to grow in week electrolytes and was inhibited by NaCl above 2 M, while the latter grew optimally in presence of at least 2 M NaCl (Figure 4). Moreover, Thioalkalivibrio K90mix has another extraordinary difference from halophiles. It can still grow well at pH 10 when 90% of sodium carbonate is replaced by potassium carbonate (i.e., 3.6 M K+/0.4 M Na+ as carbonates), while halophilic SOB from chloride-sulfate lakes could not grow already at 50% replacement (Sorokin, 2008). Obviously, the microorganisms from true soda lakes with preference for sodium carbonates cannot be called (as is usually done in the literature) haloalkaliphiles, but, must be called natronophiles. We also have evidence of the specific requirement for sodium cations for sulfide- and TMPD-dependent respiratory activity of the isolated membrane vesicles from Thioalkalivibrio strains pointing to the involvement of sodium-based elements in the bioenergetic machinery of natronophilic chemolithoautotrophs (Grischuk et al., 2003).
Figure 2.
Illustration of the three different anion-pH types of closely related SOB species from saline habitats. Thiomicrospira crunogena is a deep-see marine halophile, Thiomicrospira pelophila – a bicarbonate-loving alkalitolerant type from an littoral marine habitat, which may have temporal pH increase due to the phototrophic activity, and Thioalkalimicrobium aerophilum is a typical carbonate-loving type from a soda lake. 100% rates were 0.8, 2.2, and 3.8 μmol O2 (mg protein min)−1 for Tm. crunogena, Tm. Pelophila, and Tm aerophilum, respectively.
Table 3.
Influence of anion composition of the medium on accumulation of compatible solutes by facultatively alkaliphilic extremely salt-tolerant SOB from hypersaline alkaline lakes.
| Parameter | Tv. halophilus | Tb. halophilus | ||
|---|---|---|---|---|
| Na-carbonate | NaCl | Na-carbonate | NaCl | |
| Osmotic pressure (osm kg−1) | 5.0 | 9.3 | nd | nd |
| Ectoine, % of dry weight | – | – | 5.2 | 13.5 |
| Glycine betaine, % of dry weight | 12.4 | 19.8 | 1.3 | 0.9 |
| Total compatible solute content, % of dry weight | 12.4 | 19.8 | 6.5 | 14.4 |
Thioalkalivibrio halophilus was grown at 4 M total Na+, Thioalkalibacter halophilus – at 3 M total Na+, both at pH 10.
Figure 3.
Illustration of the concept of natronophily among the extremely salt-tolerant soda lake SOB, showing the influence of counter-anion of sodium on growth of two different types of extremely salt-tolerant Thialkalivibrio isolates. The strains were grown at 4 M total Na+ and pH 10. Sodium carbonate and sodium sulfate represent weak electrolytes in contrast to the strong electrolyte NaCl. Thioalkalivibrio K90mix represent a typical natronophile from soda lakes in the Kulunda Steppe, while Thioalkalivibrio ALE20 represents a haloalkaliphile from hypersaline alkaline lakes of Wadi Natrun. Na-c, medium with 90% carbonates/10% Cl− Na-s, medium with 90% sulfate/10% carbonates; NaCl50, medium with 50% Cl−/50% carbonate; NaCl75, medium with 75% Cl−/25% carbonate.
Figure 4.
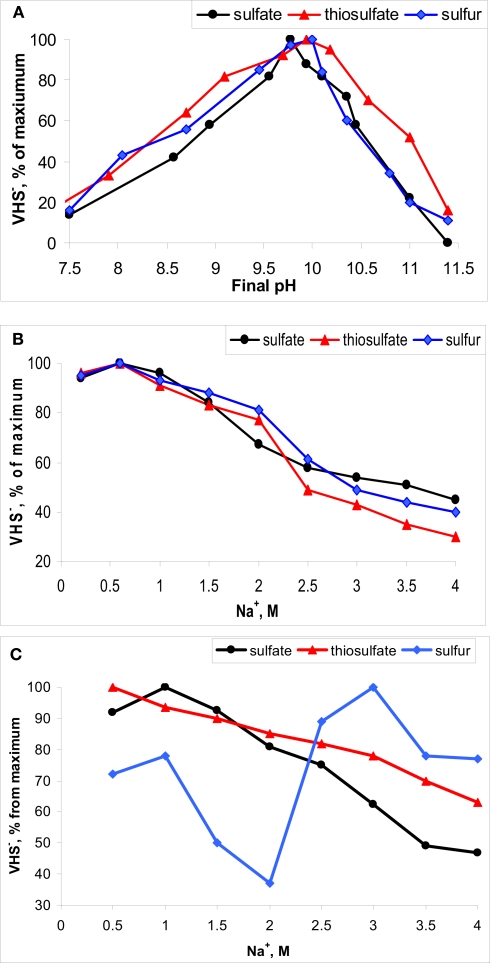
Examples of pH (A) and salt (B,C) on the rate of sulfidogenesis (VHS−) in benthic microbial communities of soda lakes. (A,B), moderately saline lake Tanatar-5; (C), hypersaline lake Tanatar-1 (both in Kulunda Steppe, Altai, Russia). Electron donor – formate.
Specific metabolic properties of members of the genus thioalkalivibrio
Thiodenitrification at extremely high pH
Anoxic enrichments from soda lake sediments with thiosulfate or sulfide as electron donor and nitrate as electron acceptor were positive only at a relatively low salinity (0.5–1.5 M total Na+), but the enrichment culture did not normally yield complete denitrifiers. Instead, partial denitrifiers only reducing nitrate to nitrite with formation of copious sulfur were dominating. One of the enrichment cultures from Wadi Natrun, however, resulted in the complete denitrification of nitrate to nitrogen gas with only intermediate nitrite production. A stable co-culture of two facultatively anaerobic haloalkaliphilic SOB was selected from this enrichment consisting of a nitrate to nitrite reducing and nitrite/N2O-reducing members (Sorokin et al., 2003b). They represented two different species of the genus Thioalkalivibrio, Tv. Nitratireducens, and Tv. denitrificans, respectively. The type strain of the latter has originally been obtained from the enrichment with N2O as electron acceptor (Sorokin et al., 2001b). In pure culture, Tv. nitratireducens could grow anaerobically by nitrate-reduction to nitrite (Sorokin et al., 2003c), while the second organism, Tv. denitrificans, lacking NAR, was capable of further reducing nitrite to N2 via N2O (Sorokin et al., 2001b):

This consortium represented the first example of microbial thiodenitrification at extremely high pH. Moreover, in a chemostat at pH 10 Tv. denitrificans grew well with N2O and polysulfide – a form of autotrophic denitrification unique to alkaliphiles. Apparently, all the denitrifying reductases in these SOB must be adapted to pH 10 or higher, since they are located in the periplasmic space. Indeed, the in vitro assays showed that Nir and Nos activity had a pH optimum above 10, while Nar is less alkalitolerant with an optimum at pH 9.5.
Thiocyanate oxidation at extremely high pH
Thiocyanate (N≡C–S−) is a one-carbon sulfur compound which is much more difficult to utilize as energy source for SOB as compared to other inorganic sulfur species. Only a few neutrophilic Thiobacillus species can use it as an energy source after a primary degradation to sulfide, ammonia and CO2, for which two basic pathways have been proposed, either through the C≡N or the C–S bond hydrolytic cleavage, respectively (Kelly and Baker, 1990):
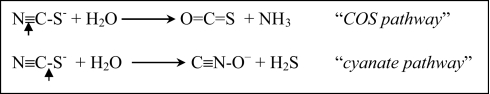
The existence of the “COS pathway” has recently been proven for the neutrophilic SOB Thiobacillus thioparus and halophilic Thiohalophilus thiocyanatoxidans, and the enzyme responsible for the primary degradation of the nitrile bond of thiocyanate was characterized as a specific type of Cobalt-containing nitrile hydratase (Katayama et al., 1998; Bezsudnova et al., 2007). On the other hand, no direct proof of the “cyanate pathway” had been offered until recently.
Aerobic enrichment cultures in medium containing thiocyanate at pH 10 and moderate salinity from various soda lake sediments resulted in the isolation of several strains of haloalkaliphilic, obligately autotrophic SOB growing on thiocyanate as energy and nitrogen source (Sorokin et al., 2001c). The isolates fell into two distinct lineages within the genus Thioalkalivibrio and were described as Tv. thiocyanoxidans and Tv. paradoxus (Sorokin et al., 2002c). At denitrifying conditions another novel species was obtained and described as Tv. thiocyanodenitrificans, which is capable of anaerobic growth with thiocyanate as e-donor and nitrate as e-acceptor at pH 10 (Sorokin et al., 2004a). The aerobic species accumulated large amounts of cyanate in the medium during thiocyanate utilization, which was the first direct confirmation of the involvement of the “cyanate pathway” in the primary thiocyanate degradation in pure bacterial cultures. Interestingly, the cyanase activity (whose presence had been implicated previously as the only confirmation of the “cyanate” pathway in thiobacilli; Youatt, 1954) was completely blocked during growth of the alkaliphilic SOB on thiocyanate. Another peculiar property of the degradation system in alkaliphiles emerged during a detailed study of the mechanism of thiocyanate degradation. The result of a biochemical investigation indicated that the primary degradation reaction resulting in cyanate formation is not hydrolysis (as is the case in the COS pathway), but, rather, a direct oxidation of the sulfane atom to the stage of elemental sulfur according to the following scheme (unpublished results):
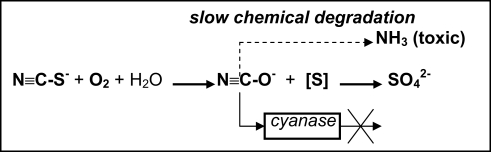
At present, the responsible enzyme has been purified and the gene has been sequenced. The protein has no significant homology to any oxido-reductases in the database. The genomes of all three representatives of the SCN-oxidizing alkaliphilic SOB are in the process of genome sequencing, which must help to better understand their thiocyanate metabolism.
Genome information from two strains of the genus thioalkalivibrio
The complete genome sequencing of two Thioalkalivibrio representatives has just been finished and annotated by the Joint Genome Institute. The low salt-tolerant isolate from a full-scale Thiopaq bioreactor in the Netherlands (see also last section), Thioalkalivibrio “sulfidophilus” HL-EbGr7, belongs to the Thioalkalivibrio denitrificans cluster, and the extremely natronophilic strain Thioalkalivibrio spp. K90mix obtained from a mixed sample of sediments from different Siberian hypersaline soda lakes is a typical representative of the core group of the genus (see Figure 1). The first is microaerophilic and prefers sulfide as the substrate, the latter grows best at fully aerobic conditions with thiosulfate. According to the genome information, the sulfur oxidation pathways in these two SOB strains are different. The Tv. “sulfidophilus” pathway is a combination of the partial “Sox pathway” (lacking soxCD) for oxidation of the sulfane atom of thiosulfate and the reversed sulfate reduction pathway for the oxidation of zero-valent sulfur atom to sulfate, similar to what has been recognized in green and purple anaerobic SOB Chlorobium (Sakurai et al., 2010) and Allochromatium (Sander and Dahl, 2009). Sulfide oxidation is apparently proceeding through the flavocytochrome c (Fcc) sulfide dehydrogenase (Muyzer et al., 2011). A partial “Sox pathway” also operates in Thioalkalivibrio K90mix. However, the genes coding for the reversed Dsr cluster are absent and there is no obvious candidate to fulfill the central function of oxidation of zero-valent sulfur atom to sulfite. On the other hand, there are two possibilities for sulfite oxidation. The aps and sorA genes, present in the chromosome, are coding for the AMP-dependent (APS reductase) and independent sulfite dehydrogenases, respectively. Further work is needed to fully understand the sulfur oxidation mechanism in these SOB species.
Sulfidogenesis in Soda Lakes
Activity of sulfidogenesis in soda lake sediments
Usually, the top sediments in soda lakes have a dark color and a high content of acid labile sulfides (HS− + FeS) indicating active sulfidogenesis. For example, in the sediment pore brines of Kulunda soda lakes we detected up to 5 mM free sulfide. A limited number of direct in situ measurements of sulfate reduction rates with (Gorlenko et al., 1999; Sorokin et al., 2004b; Kulp et al., 2006) and sulfate reduction potential in sediment slurries (Kulp et al., 2007) indicated the presence of active sulfate-reducing bacteria (SRB) populations in various soda lakes with rates comparable to those observed in coastal marine sediments. More detailed recent studies conducted with sediments of Kulunda Steppe soda lakes confirmed active sulfate reduction, but also demonstrated that the most active sulfidogenesis was with elemental sulfur as electron acceptor, followed by thiosulfate, while sulfate was less active (Sorokin et al., 2010a). Sulfidogenesis was stimulated by addition of electron donors of which formate was the most efficient. Hydrogen did not lead to increased sulfide production with any of the inorganic sulfur acceptors, whilst acetate only stimulated sulfur reduction. Furthermore, native benthic sulfidogenic populations in Kulunda soda lakes clearly exhibited an obligately alkaliphilic pH response with optimal activity at pH 9–10 (Figure 4A), while a salinity above 2 M of total Na+ significantly inhibited the activity of sulfate reduction. The same result has also been well documented for hypersaline chloride–sulfate lakes at neutral pH (Brandt et al., 2001; Sørensen et al., 2004). However, thiosulfate and, especially sulfur reduction, in hypersaline soda lakes, such as Bitter-1 and Tanatar-1 in Kulunda Steppe, were much less sensitive to high salt concentrations (Figures 4B,C).
Molecular analysis of diversity of SRB in soda lakes
16S rRNA gene-based molecular analysis of the soda lake prokaryotic communities only rarely detected typical SRB from the Deltaproteobacteria. Particularly, sequences affiliated with the previously described genus Desulfonatronum from soda lakes (Pikuta et al., 1998, 2003; Zhilina et al., 2005) were found in Mono Lake (Scholten et al., 2005), Wadi Natrun lakes (Mesbah et al., 2007), and in Kulunda Steppe lakes (Foti et al., 2008). More specific analysis using the functional marker dsrAB, encoding an essential enzyme in the sulfate reduction pathway, demonstrated the presence of SRB sequences belonging to the orders Desulfovibrionales and Desulfobacterales in Siberian soda lakes (Foti et al., 2007). In the Desulfovibrionales, the sequences were affiliated with the genera Desulfonatronum and Desulfonatronovibrio (Zhilina et al., 1997), previously found in soda lakes, while the detection of dsrAB clones within the order Desulfobacterales indicated the presence of yet unknown heterotrophic SRB in soda lakes.
Culture-dependent analysis of sulfidogens from soda lakes
Lithotrophic SRB
Until recently, only a few SRB's have been cultured from soda lakes. They belong to two genera of the family of the haloalkaliphilic Desulfovibrionales, i.e., Desulfonatronovibrio (a single species), and Desulfonatronum (three species). The former is obligate lithotroph, utilizing only H2 and formate, while the three species of the genus Desulfonatronum also oxidize EtOH and lactate to acetate in addition to H2 and formate. Both are moderately alkaliphilic with a pH optimum of 9–9.5 and moderately salt-tolerance with the upper limit of 1.5 M total Na+ for Desulfonatronum and 2.0 M for Desulfonatronovibrio. The finding of these SRB was a significant discovery, because it was assumed that SRB could not grow under these conditions. However, the question remained, whether there are other SRB in soda lakes, which would be able to grow at much higher salinity occurring in hypersaline soda lakes and whether they would utilize other substrates.
Our enrichment attempts using soda media with 3–4 M Na+ at pH 10 and sulfate as the electron acceptor were negative. However, when sulfate was replaced by sulfite or thiosulfate, positive sulfidogenic enrichments were obtained from the Kulunda Steppe and Wadi Natrun either with formate as electron donor or even without any additional electron donors. The latter was totally unexpected, but soon it became clear, that autotrophic growth by sulfite/thiosulfate disproportionation is a common trait among haloalkaliphilic SRB. Despite a very low energy yield (see comparison below), it allowed the SRB to outcompete the acetogens, which were otherwise more successful at extremely natronophilic conditions.
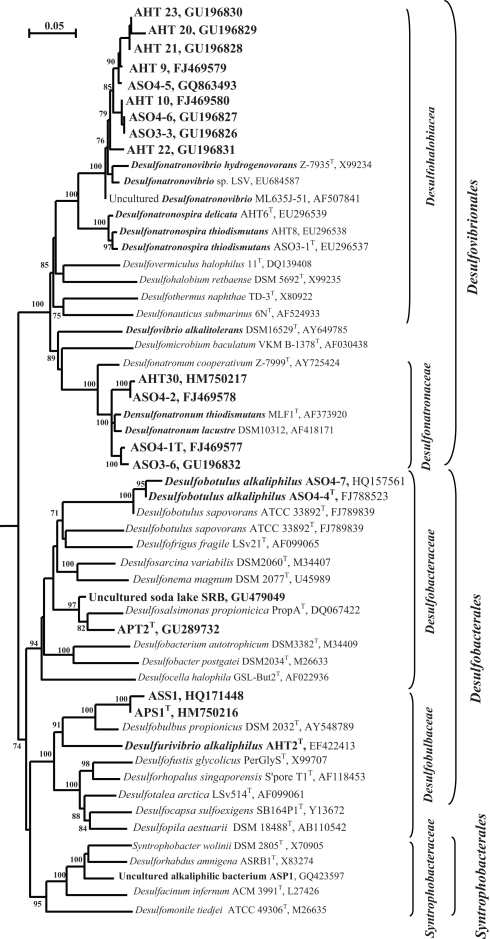
The disproportionation enrichments resulted in the isolation of several extremely natronophilic SRB strains growing optimally at 1.5–2 M total Na+ (and up to 4 M Na+) and pH 10 either by sulfite/thiosulfate disproportionation autotrophically or by “normal” dissimilatory sulfate reduction with H2, formate and lactate as e-donors. The group was described as a new genus Desulfonatronospira within the family Desulfohalobiaceae with Desulfonatronospira thiodismutans as the type species (Figure 5; Sorokin et al., 2008). 16% (w/w) glycine betaine was detected in the cells of Desulfonatronospira thiodismutans grown at 3 M total Na+ accumulated as a compatible solute. The cost of such an “investment” is enormous (Oren, 1999) and, yet, the energy metabolism of this organism is apparently sufficient, although the growth rate is extremely low. The genome of the type strain is currently sequenced by the Joint Genome Institute and its analysis must shed some light on the molecular mechanism of adaptation to extreme natronophilic conditions.
Figure 5.
16S rRNA-based phylogeny of haloalkaliphilic SRB from soda lakes within the Deltaproteobacteria (sequences are in bold). Numbers at the nodes indicate the percentage of bootstrap values for the clade of this group in 1000 replications (the values for maximum-likelihood method are given in parentheses). Only values above 70% are shown. Bar, 5 substitution per 100 nt. E. coli was used as an outgroup.
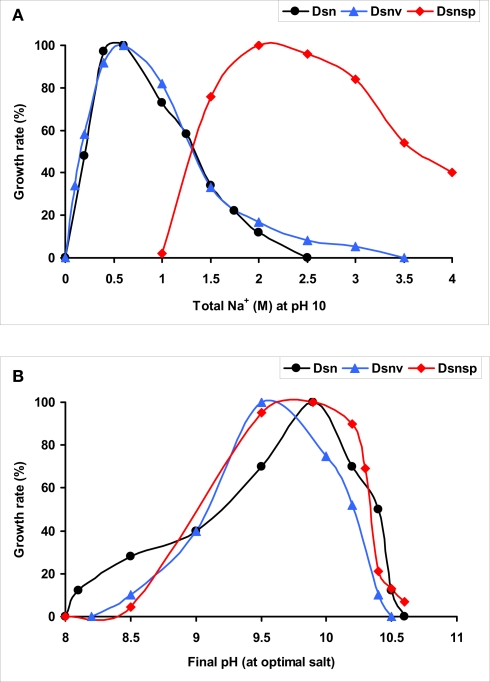
The SRB isolates from Kulunda soda lake sediments, which were enriched at moderate salinity (0.6–2.0 M Na+), pH 10 and sulfate as the electron acceptor (Table 4), could only be enriched and further separated from acetogens by using formate as the electron donor and sulfate as the electron acceptor or at disproportionating conditions. The multiple lithotrophic SRB's isolated from soda lake sediments (Table 4) fell into the two known Desulfovibrionales genera, Desulfonatronum, and Desulfonatronovibrio. However, none of them were closely related (less than 97% of 16S rRNA gene sequence similarity) to the described species (Figure 5). This demonstrated a high diversity of these two lithotrophic haloalkaliphilic SRB genera in soda lakes and corresponded well to the results of culture-independent studies. All isolates were obligate alkaliphiles with a pH optimum around 9.5–10. The upper salt limit was 2 M total Na+ for the Desulfonatronum isolates and 3 M total Na+ for the Desulfonatronovibrio. However, all of them grew optimally at low salt (around 0.5 M). Figure 6 shows the comparative salt/pH profiles for the three groups of lithotrophic soda lake SRB. The most common and ecologically important physiological trait of these SRB was the capacity to grow litho(auto)trophically by thiosulfate disproportionation. It must be mentioned, that, despite the observed high potential (activity) of resting cells for sulfur reduction, none of the SRB isolates were able to use elemental sulfur as an e-acceptor for growth. Production of sulfide in the presence of elemental sulfur at high pH would lead to the formation of polysulfide and, apparently this compound is much more toxic to alkaliphilic SRB than sulfide.
Table 4.
Novel lithotrophic SRB isolates from soda lakes (type strains are in bold).
| Strain number | Source | Enrichment conditions (pH 10) | Key properties | Affiliation | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Lake | Area | e-donor/ C source |
e-acceptor | Total Na+ (M) | Autotrophy | Dismutation | pH range (optimum) | Maximal salt-tolerance M Na+ | μmax h−1 | ||
| ASO4-1 | Tanatar-1 | Kulunda Steppe (Altai, Russia) |
Formate/ | Sulfate | 0.6 | 2 Strains | + | 8.0–10.5 (9.3–10.0) | 1.3–2.3 | 0.042–0.055 | Desulfonatronum |
| ASO4-2 | Picturesque | Pyruvate | Sulfate | ||||||||
| ASO3-6 | Cock soda | EtOH* | Sulfite | ||||||||
| AHT30 | Mixed sample | Formate/ acetate |
Sulfate | ||||||||
| AHT9 | Tanatar-5 | Kulunda Steppe | Formate/ acetate |
Thiosulfate | 0.6 | 4 Strains | + | 8.5–10.5 (9.5–10.0) | 2.0–3.0 | 0.035–0.040 | Desulfonatronovibrio |
| AHT10 | Bitter-1 | Formate/ acetate |
Thiosulfate | 2.0 | |||||||
| ASO4-6 | Cock soda | Formate/ acetate |
Sulfate | 0.6 | |||||||
| ASO3-2 | Tanatar-5 | EtOH* | Thiosulfate | 0.6 | |||||||
| ASO3-3 | Bitter-1 | Formate/ acetate |
Sulfite | 0.6 | |||||||
| AHT20 | Tanatar-5 | Thiosulfate/ acetate |
Thiosulfate | 0.6 | |||||||
| AHT21 | Cock Soda | Thiosulfate/ acetate |
Thiosulfate | 0.6 | |||||||
| AHT22 | Tanatar-5 | Thiosulfate/ acetate |
Thiosulfate | 0.6 | |||||||
| AHT23 | Bitter-1 | Thiosulfate/ acetate |
Thiosulfate | 0.6 | |||||||
| AHT33 | Tanatar-5 | Thiosulfate/ acetate |
Thiosulfate | 0.6 | |||||||
| ASO4-5 | Owens | California | Formate/ acetate |
Sulfate | 2.0 | ||||||
| ASO3-1 | Mixed sediment | Kulunda Steppe | Sulfite | Sulfite | 4.0 | + | + | 8.5–10.6 (9.5–10.0) | 3.5–4.0 | 0.020 | Desulfonatronospira |
| AHT8 | Formate/ acetate |
Thiosulfate | 3.0 | ||||||||
| AHT26 | Thiosulfate/ acetate |
Thiosulfate | 4.0 | ||||||||
| AHT6 | Wadi Natrun | Egypt | Thiosulfate/ acetate |
Thiosulfate | 3.0 | ||||||
*Subsequent isolation of pure cultures was possible after replacement of EtOH by formate.
Figure 6.
Salt [(A), at pH 10] and pH [(B), at optimum salinity] profiles for growth in three subgroups of lithotrophic SRB from soda lakes. Dsn, Desulfonatronum isolates, Dsnv, Desulfonatronovibrio isolates, Dsnsp, Desulfonatronospira thiodismutans. 100% growth rates were 0.05, 0.04, and 0.02 h−1 for Dsn, Dsnv, and Dsnp, respectively.
Heterotrophic SRB
As was mentioned above, the molecular studies indicated the presence of members of the order Desulfobacterales in soda lakes. Most of them, in contrast to Desulfovibrionales, can utilize volatile fatty acids (VFA) as electron donor and carbon source. Our enrichments at pH 10 with sediments from Kulunda Steppe soda lakes resulted in positive sulfidogenic cultures with propionate, butyrate, and benzoate, but only at the lowest salinity used (0.6 M total Na+). From the seven positive enrichments, five pure cultures and two syntrophic associations were eventually obtained. Their basic properties are presented in Table 5. Four pure cultures belonged to three different clusters within the order Desulfobacterales which corresponds to the results of dsr detection (Foti et al., 2007), while the SRB member of the syntrophic propionate-utilizing culture was a member of the order Synthrophobacterales (Figure 5). The SRB member of the benzoate-utilizing association was isolated under autotrophic conditions with H2 and sulfate and identified as a member of the genus Desulfonatronum. This situation was similar to that of the acetate-utilizing syntrophic culture from soda lakes, which yielded Desulfonatronum cooperativum (Zhilina et al., 2005). The organism responsible for benzoate oxidation was identified by sequence analysis as a novel representative of the class Clostridia (89% sequence similarity to Natranaerobius spp.; unpublished results).
Table 5.
Properties of heterotrophic SRB from soda lakes in Kulunda Steppe enriched and isolated at pH 10 and 0.6 M total Na+.
| Strain | Enrichment conditions | Key properties | Affiliationnumber | ||||||
|---|---|---|---|---|---|---|---|---|---|
| e-donor/ C source |
e-acceptor | e-donor/ C source |
e-acceptor | c/i* | pH optimum | Upper salt limit (M Na+) | μmax (h-1) | ||
| ASO4-4 | Butyrate | Sulfate | C4–C9 VFA, | Sulfate, | i | 10.0 | 1.75 | 0.03 | Desulfobotulus alkaliphilus |
| ASO4-7 | BuOH | Sulfate | BuOH | sulfite | |||||
| APT2 | Propionate | C3–C8 VFA, | Sulfate, sulfite | c | 10.0 | 1.5 | 0.019 | Desulfobacteraceae | |
| APS1 | Propionate | Sulfate | Propionate, | Sulfate, | i | 9.4–9.5 | 1.75 | 0.020 | “Desulfobulbus alkaliphilus” |
| ASS1 | Succinate >> propionate | PrOH, BuOH | sulfite | ||||||
| ASP1-syntr | Propionate | Sulfate | Propionate, PrOH | Sulfate | nd | 9.5 | 1.5 | nd | Synthrophobacteraceae |
*c, complete oxidizers; i, incomplete oxidizers; nd, not determined.
From the pure cultures of heterotrophic SRB currently only two similar strains selected with butyrate/BuOH and sulfate are validly described as a new species of the genus Desulfobotulus, Dsb. alkaliphilus (Sorokin et al., 2010b). They oxidize a range of linear VFA incompletely to acetate. Another pair of incomplete oxidizers selected with propionate and sulfate represents a novel haloalkaliphilic member of the genus Desulfobulbus. Finally, strain APT2, a novel member of the family Desulfobacteraceae, is a first cultured example of haloalkaliphilic SRB completely oxidizing VFA to CO2. However, it cannot utilize external acetate as energy source, which is similar to its nearest phylogenetic relative, halophilic Desulfosalsimonas propionicica (Kjeldsen et al., 2010). None of the SRB isolates, listed in Table 5 can grow litho(auto)trophically with H2 or formate or by disproportionation of sulfite/thiosulfate, in contrast to the haloalkaliphilic members of the order Desulfovibrionales.
Gram-positive SRB in soda lakes
The 16S rRNA-based approach showed the presence in Mono Lake of two novel clusters of Clostridiales related to Desulfotomaculum (Scholten et al., 2005), although no dsr sequences belonging to the Gram-positive SRB have been found yet in soda lakes. Our attempt to enrich and isolate endospore-forming SRB from soda lakes in Kulunda Steppe using pasteurized sediments were not successful. Instead, a binary sulfidogenic culture consisting of two endospore-forming haloalkaliphilic clostridia has been selected with pyruvate as the e-donor/C source and thiosulfate as the e-acceptor. Both could grow with thiosulfate, but only one (related to Natronovirga) reduced thiosulfate partially to sulfite and sulfide, while the second member, described as a novel genus and species Desulfitispora alkaliphila, was able to grow with sulfite as an e-acceptor. So, it may be considered as a “crippled” SRB lacking a pathway for sulfate activation. Interestingly, when grown together, the growth rate and final biomass yield of the consortium with thiosulfate was higher, than for Desulfitispora alone, indicating a positive cooperation (Sorokin and Muyzer, 2010a).
Dissimilatory sulfur-reducing bacteria in soda lakes
As was already mentioned above, sulfur reduction was the most active sulfidogenic process in soda lake sediments. Two interesting results were that (1) acetate was apparently utilized as the e-donor only when sulfur acted as the e-acceptor and (2) that in hypersaline soda lakes the activity of lithotrophic sulfur reduction ( and formate-dependent) was not inhibited and, in some cases, even stimulated, at saturated salt conditions, in contrast to sulfate/thiosulfate reduction. Also assuming that alkaliphilic SRB were unable to use elemental sulfur as an e-acceptor, it became clear, that there must be specialized dissimilatory sulfur-reducing microbes in soda lakes different from SRB.
Enrichments with either acetate or propionate as e-donors and sulfur as e-acceptor at moderate salinity from the Kulunda soda lake sediments showed relatively fast growth with copious polysulfide formation and high dominance of a single spiral-shaped morphotype. Two pure cultures obtained from the enrichments shared some common phenotypic traits (Table 6) and were described as a novel genus and species Desulfurispira natronophila, a member of the phylum Chrysiogenetes (Sorokin and Muyzer, 2010b). The most important property of these bacteria was fast growth with and complete oxidation of acetate and propionate, which was lacking when sulfur had been replaced by sulfate. Another important property was the relatively high salt-tolerance (despite the original selection at low salt) of 3 M Na+ at pH 10 for sulfur reduction with acetate. The final amount of reduced sulfur (sulfide/sulfane of polysulfide) in these sulfidogenic alkaliphiles was several times higher that in the SRB cultures, reaching 80 mM. The most obvious question, why sulfur reduction, but not sulfate reduction was active under soda lake conditions, may, at least partially, be answered by the (beneficial) stability of the true electron acceptor (which is not insoluble sulfur itself, but soluble polysulfide) at high pH and by proton formation during the reaction. Both contribute to a favorable change of the available reaction energy in comparison to standard neutral conditions. According to our calculations, those changes make sulfur reduction with acetate [ΔG′ = (−50)–(−70) kJ/mol acetate] more favorable than sulfate reduction (ΔG0′ = −47 kJ/mol acetate; Sorokin et al., 2010a).
Table 6.
Phenotypic comparison of Desulfurispira natronophila from soda lake isolates with related bacteria from the phylum Chrysiogenetes.
| Characteristic | Desulfurispira natronophila | Desulfurispirillum alkaliphilum | Chrysiogenes arsenatis | ||
|---|---|---|---|---|---|
| AHT11T | AHT19 | ||||
| Cell morphology | Spirillum with bipolar flagellum | Vibrio, single polar flagellum | |||
| Autotrophic growth | – | – | – | – | |
| ELECTRON DONORS | |||||
| H2 | + | – | + | – | |
| Formate | + | – | – | – | |
| Acetate, pyruvate, lactate, malate, succinate, fumarate | + | + | + | + | |
| Propionate | + | + | + | – | |
| Citrate | – | – | + | – | |
| Glycerol | + | – | – | – | |
| ELECTRON ACCEPTORS | |||||
| O2 | – | – | – | – | |
| Sulfur (polysulfide) | + | + | + | – | |
| ( to NH3) | – | – | + | + | |
| N2O | – | – | – | – | |
| + | + | – | + | ||
| Fe3+, | – | – | – | – | |
| Oxidation of HS− with nitrate | – | – | + | nd | |
| pH range (optimum) | 8.5–10.9 (10.2) | 8.2–10.5 (9.8) | 8.0–10.2 (9.0) | Neutrophilic | |
| Salt range (M Na+) | 0.2–3.0 | 0.2–3.0 | 0.1–2.5 | Na-independent | |
| Habitat | Soda lakes | Bioreactor | Gold mine | ||
The most essential properties are highlighted in bold.
Further exploration of potential of dissimilatory sulfur reduction in soda lakes at extremely high salt and pH 10 (saturated soda brines) gave unexpected microbiological results. With H2, formate, and acetate as the electron donors three extremely natronophilic bacterial strains identical in morphology and very close to each other phylogenetically were enriched and isolated from hypersaline soda lakes in the Kulunda Steppe and from Wadi Natrun (Table 7). They had 96–97% 16S rRNA gene sequence similarity to Natroniella acetigena – a heterotrophic homoacetogenic natronophile from soda lake Magadi (Zhilina et al., 1995). In contrast to the type species, the novel isolates were obligate sulfur respirers and not capable of acetogenic growth with organic compounds, but, instead, could grow lithoautotrophically with formate and hydrogen. One of the isolates was even able to utilize acetate as an e-donor, which is supposed to be a final metabolic product in this genus.
Table 7.
Properties of extremely natronophilic, obligately sulfur-reducing isolates from hypersaline soda lakes in comparison with their closest phylogenetic relatives.
| Property | Natroniella “sulfidigena” (3 strains) | Natroniella acetigena | Natronolimno biusstrain AHT32 | Natronolimnobius innermongolicus |
|---|---|---|---|---|
| Taxonomy | Bacteria, Halanaerobiales | Euryarchaeota, Halobacteriales | ||
| Acetogenesis | – | EtOH, lactate, pyruvate, PrOH | – | – |
| Anaerobic growth with sulfur as e-acceptor | + | – | + | –* |
| Aerobic growth | – | – | C2–C8 VFA, yeast extract | C6-sugars, propionate, citrate, lactate, malate, pyruvate, fumarate |
| Use of H2 and formate as e-donors | + | – | + | – |
| Acetate ase-donor | + (In one strain) | – | + | + |
| Autotrophy | + | – | +** | – |
| pH optimum | 9.8–10.0 | 10.0 | 10.0 | 9.5 |
| Minimal salt requirement (M Na+) | 1.5–2.0 | 1.7 | 2.5 | 1.7 |
| Habitat | Hypersaline soda lakes |
The most essential properties are highlighted in bold.
*Sulfide was produced aerobically; **needs to be confirmed.
Even more unexpected results came from enrichments under the same conditions but with C4–C6 VFA as the e-donors. In all of them the dominant morphotype was represented by irregular coccid cells that looked flat – a typical characteristic of the haloarchaea. The isolation of a pure culture, was possible only when the VFA were replaced by formate. The isolated strain grew by sulfur respiration utilizing only H2, formate, and pyruvate as the e-donors in pure culture. It could gradually be adapted for aerobic heterotrophic growth with C2–C8 VFA through microoxic conditions and in the presence of oxygen the biomass acquired the typical red pigmentation of haloarchaea. Phylogenetic analysis put this isolate into the cluster of natronarchaea with an uncharacterized isolate from Little Lake Magadi in Kenya (Jones et al., 1998) and the members of recently described genus Natronolimnobius (Itoh et al., 2007) as closest relatives. As far as we know, none of the so far described members of the order Halobacteriales have the type of metabolism as possessed by strain AHT32, which makes it truly unique.
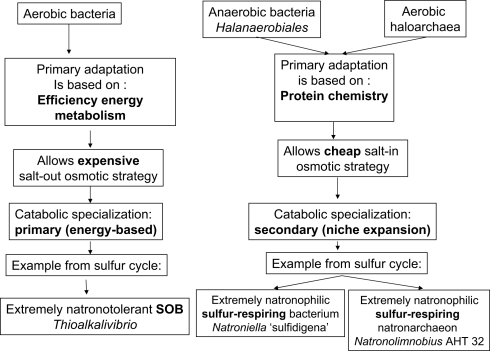
These two examples of extremely natronophilic obligatory sulfur-respiring prokaryotes from hypersaline soda lakes with unexpected phylogenetic affiliation to completely different metabolic groups are important for the general understanding of mechanisms of adaptation of prokaryotic life to extreme salinity. If one accepts the hypothesis of A. Oren about the existence of a direct relation between the ability of prokaryotes to grow at high salt and their efficiency of catabolism (Oren, 1999, 2011), an attempt can be made to rationally explain different cases of the extreme natronophilism observed in sulfur cycle prokaryotes from hypersaline soda lakes (Figure 8). The essence of concept is that there may be two different ways of adaptation to cope with such extreme conditions. One is based on a very efficient energy metabolism, such as in the aerobic SOB of the genus Thioalkalivibrio, providing a possibility to generate sufficient energy required for the synthesis of high concentrations of organic compatible solutes. In that case the metabolic specialization is primary to the secondary adaptation to high salt. In the second strategy it is opposite, i.e., existing structural adaptation to high salt is primary to the low energy-yielding metabolic specialization. The second strategy is represented by the two examples of extremely natronophilic sulfur respirers – the bacterium Natroniella “sulfidigena” and the archaeon Natronolimnobius AHT32. Both have in common the energy-inexpensive (“cheap”) osmotic strategy based on the import of K+, giving them an advantage over the competitors with an “organic” osmotic strategy in exploiting the low energy catabolic reactions at extremely high salt. Of course, this is only part of the story, there must be other factors involved in such “fitness.” For example, several strains of the genus Thioalkalivibrio and all members of the genus Thioalkalimicrobium can not grow at extreme salinity, having the same type of energy metabolism as the extremely salt-tolerant representatives of the genus Thioalkalivibrio. Another exception – Desulfonatronospira with an organic compatible solute strategy, growing chemolithoautotrophically in saturated soda brines (albeit very slowly) using energy-poor catabolic reactions (disproportionation). Comparative analysis of genomes from low and high salt-tolerant Thioalkalivibrio strains might shed a light on those additional properties.
Figure 8.
A generalized microbial sulfidogenic system detected in soda lakes at pH 9.5–11.
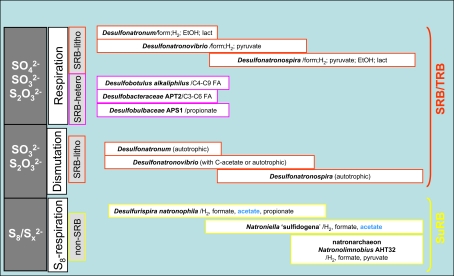
A generalized scheme showing various sulfidogenic processes and microbes involved, which have been found so far in soda lakes, is given in Figure 7. The main factor influencing the soda lake SRB responsible for sulfate reduction and thiosulfate reduction/disproportionation is salinity. Sulfur reduction is a function of specialized groups, whose most important properties include the potential to utilize acetate as the e-donor and to grow in saturated soda brines.
Figure 7.
Possible adaptation scenario of prokaryotes to functioning in saturated soda brines: case of sulfur cycling.
Application of haloalkaliphilic SOB
Full-scale bioreactors (Thiopaq reactors, Paques B.V., The Netherlands) offer an attractive alternative to chemical processes for efficient H2S removal from gases. Briefly, H2S is stripped from the gas phase into alkaline solution at pH 9. Since the gases also contain CO2, the resulting spent liquid is essentially sulfide solution in sodium bicarbonate, which is then transferred to a second reactor containing haloalkaliphilic SOB. The reactor is operating at extreme oxygen-limited conditions and very low redox potential to limit the oxidation of sulfide to elemental sulfur (Janssen et al., 2009). At such conditions both aerobic and anaerobic part of the microbial sulfur cycle can coexist. Recent analysis of the microbial communities in several of these reactors using DGGE and cultivation approach showed that the dominant population was represented by a single microaerophilic sulfide-specialized haloalkaliphilic SOB species belonging to the genus Thioalkalivibrio (Tv. “sulfidophilus”). Several other alkaliphilic and alkalitolerant SOB species belonging to the genera Thioalkalivibrio, Thioalkalimicrobium, and Thiomicrospira (Tm. pelophila group) could be isolated from the reactors as well, but they represented minor populations. Although molecular analysis did not detect any significant presence of the anaerobic sulfur cycle bacteria, alkalitolerant SRB, and sulfur-reducing bacteria could be enriched from the reactor biomass. The SRB belonged to Desulfonatronum spp. and Desulfovibrio alkalitolerans (Abildgaard et al., 2006), whilst the sulfur reducers were represented by a novel genus and species Desulfurispirillum alkaliphilum related to Desulfurispira natronophila from soda lakes (Sorokin et al., 2007b).
General Conclusions
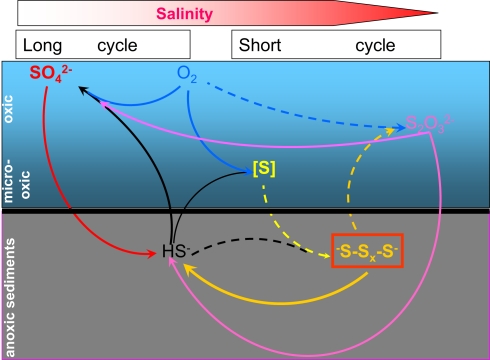
Fifteen years of research on the microbial sulfur cycle in extremely alkaline and saline (soda) lakes showed that the cycle is complete and active at native pH up to salt-saturating conditions. The major (cultured) players include four different groups of aerobic and denitrifying chemolithoautotrophic SOB within the Gammaproteobacteria, two major groups of SRB within the Deltaproteobacteria and three different groups of sulfur-reducing prokaryotes. The culture-independent molecular approach also indicated the presence of several groups of SOB and SRB, which so far lack cultured representatives. The inherent properties of these sulfur-catabolizing prokaryotes from soda lakes are their obligate alkaliphily and preference for sodium carbonate as salt over sodium chloride. Being a weak electrolyte, sodium carbonate brines impose two times less osmotic pressure than strongly electrolytic NaCl brines with the same sodium content and, thus, are beneficial for the soda lake organisms, most of which are rather natrono(soda)philic than halo(sodium chloride)alkaliphilic. Nevertheless, even sodium carbonates at saturating concentrations, as present in hypersaline soda lakes, impose severe limitations on possible sulfur cycle catabolic reactions. In particular, only a few SOB and a single SRB have been found capable of growing in saturated soda brines. On the other hand, sulfur-reducing extreme natronophiles, possessing an “inorganic” osmotic strategy, may drive sulfidogenesis at soda-saturated conditions with simple electron donors, such as H2, formate, and acetate. In fact, it may be speculated that two types of sulfur cycling exist at the soda lake sediment interface depending primarily on salinity (Figure 9). At low-to-moderate salinity the “long” sulfur cycle is possible with complete oxidation of sulfide/thiosulfate to sulfate by moderately natronophilic SOB and sulfate/thiosulfate reduction by natronophilic SRB. In saturated soda brines only a limited number of players can operate, such as the extremely natronotolerant SOB Thioalkalivibrio and the extremely natronophilic sulfur reducers, such as very specialized representatives of the Halanaerobiales and of the natronoarchaea. Also assuming less oxygen availability, the “short” sulfur cycle is most probable at salt-saturating conditions, with a specific reduced sulfur compound stable at high pH, i.e., polysulfide, as the central intermediate. In this cycle, sulfide is incompletely oxidized to elemental sulfur that in turn, chemically reacts with sulfide to form polysulfide. Polysulfide is a true substrate for sulfur reducers, but can also easily be converted to thiosulfate abiotically.
Figure 9.
Proposed scheme of microbial sulfur cycling in soda lakes. Dashed lines indicate abiotic reactions and solid lines show microbial conversions. The horizontal color gradient shows increase in salinity from left to right.
Future research in this field will obviously benefit from focused work on the bioenergetics of natronophiles with different metabolism and from whole genome analysis to understand the adaptation mechanisms to extreme natron-alkaline conditions. Recently, a metagenomic project on Kulunda Steppe soda lakes has been approved in the Community Sequencing Program of the Joint Genome Institute of the US Department of Energy, which should further advance research in this direction.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by RFBR grant 10-04-00152 to Dimitry Y. Sorokin and by the Dutch Foundations NWO and STW over the previous years of research. We are grateful to our colleagues Brian Jones, William Grant, George Zavarzin, Vladimir Gorlenko, and Holly Pinkart for providing samples from soda lakes in Kenya and the USA.
References
- Abildgaard L., Nielsen M. B., Kjeldsen K. U., Ingvorsen K. (2006). Desulfovibrio alkalitolerans sp. nov., a novel alkalitolerant, sulfate-reducing bacterium isolated from district heating water. Int. J. Syst. Evol. Microbiol. 56, 1019–1024 [DOI] [PubMed] [Google Scholar]
- Banciu H., Sorokin D. Y., Kleerebezem R., Muyzer G., Galinski E. A., Kuenen J. G. (2004a). Influence of sodium on the growth of haloalkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio versutus strain ALJ 15 in continuous culture. Extremophiles 8, 185–192 10.1007/s00792-004-0376-5 [DOI] [PubMed] [Google Scholar]
- Banciu H., Sorokin D. Y., Galinski E. A., Muyzer G., Kleerebezem R., Kuenen J. G. (2004b). Thialkavibrio halophilus sp. nov., a novel obligately chemolithoautotrophic facultatively alkaliphilic and extremely salt-tolerant sulfur-oxidizing bacterium from a hypersaline alkaline lake. Extremophiles 8, 325–334 10.1007/s00792-004-0391-6 [DOI] [PubMed] [Google Scholar]
- Banciu H., Sorokin D. Y., Tourova T. P., Galinski E. A., Muntyan M. S., Kuenen J. G., Muyzer G. (2008). Influence of salts and pH on growth and activity of a novel facultatively alkaliphilic, extremely salt-tolerant, obligately chemolithoautotrophic sufur-oxidizing gammaproteobacterium Thioalkalibacter halophilus gen. nov., sp. nov. from south-western Siberian soda lakes. Extremophiles 12, 391–404 10.1007/s00792-008-0142-1 [DOI] [PubMed] [Google Scholar]
- Bezsudnova E. Y., Sorokin D. Y., Tichonova T. V., Popov V. O. (2007). System of primary thiocyanate degradation in a novel halophilic sulfur-oxidizing bacterium Thiohalophilus thiocyanoxidans. Biochim. Biophys. Acta 1774, 1563–1570 [DOI] [PubMed] [Google Scholar]
- Brandt K. K., Vester F., Jensen A. N., Ingvorsen K. (2001). Sulfate reduction dynamics and enumeration of sulfate-reducing bacteria in hypersaline sediments of the Great Salt Lake (Utah, USA). Microb. Ecol. 41, 1–11 [DOI] [PubMed] [Google Scholar]
- Eugster H. P. (1970). Chemistry and origins of the brines of Lake Magadi. Mineral. Soc. Am. Spec. Publ. 3, 215–235 [Google Scholar]
- Foti M., Ma S., Sorokin D. Y., Rademaker J. L. W., Kuenen J. G., Muyzer G. (2006). Genetic diversity and biogeography of haloalkaliphilic sulfur-oxidizing bacteria belonging to the genus Thioalkalivibrio. FEMS Microbiol. Ecol. 56, 95–101 [DOI] [PubMed] [Google Scholar]
- Foti M., Sorokin D. Y., Lomans B., Mussman M., Zakharova E. E., Pimenov N. V., Kuenen J. G., Muyzer G. (2007). Diversity, activity and abundance of sulfate-reducing bacteria in saline and hypersaline soda lakes. Appl. Environ. Microbiol. 73, 2093–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti M., Sorokin D. Y., Zakharova E. E., Pimenov N. V., Kuenen J. G., Muyzer G. (2008). Bacterial diversity and activity along a salinity gradient in soda lakes of the Kulunda Steppe (Altai, Russia). Extremophiles 12, 133–145 10.1007/s00792-007-0117-7 [DOI] [PubMed] [Google Scholar]
- Giri B. J., Bano N., Hollibaugh J. T. (2004). Distribution of RuBisCO genotypes along a redox gradient in Mono Lake, California. Appl. Environ. Microbiol. 70, 3443–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlenko V. M., Namsaraev B. B., Kulyrova A. V., Zavarzina D. G., Zhilina T. N. (1999). Activity of sulfate-reducing bacteria in the sediments of the soda lakes in south-east Transbaikal area. Microbiology 68, 580–586 [Google Scholar]
- Granada C., Revah S., Le Borgne S. (2009). Diversity of culturable bacteria in an alkaliphilic sulfur-oxidizing microbial consortium. Adv. Mat. Res. 71–73, 137–140 10.4028/www.scientific.net/AMR.71-73.137 [DOI] [Google Scholar]
- Grischuk Y. V., Muntyan M. S., Popova I. V., Sorokin D. Y. (2003). Ion transport coupled to terminal oxidase functioning in the extremely alkaliphilic halotolerant bacterium Thioalkalivibrio. Biochemistry 68, 385–390 [DOI] [PubMed] [Google Scholar]
- Humayoun S. B., Bano N., Hollibaugh J. T. (2003). Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69, 1030–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T., Yamaguchi T., Zhou P., Takashina T. (2007). Natronolimnobius baerhuensis gen. nov., sp. nov. and Natronolimnobius innermongolicus sp. nov., novel haloalkaliphilic archaea isolated from soda lakes in Inner Mongolia, China. Extremophiles 9, 111–116 10.1007/s00792-004-0426-z [DOI] [PubMed] [Google Scholar]
- Janssen A. J. H., Lens P., Stams A. J. M., Plugge C. M., Sorokin D. Y., Muyzer G., Dijkman H., van Zessen E., Luimes P., Buisman C. J. N. (2009). Application of bacteria involved in the biological sulfur cycle for paper mill effluent purification. Sci. Total Environ. 407, 1333–1343 [DOI] [PubMed] [Google Scholar]
- Jones B. E., Grant W. D., Duckworth A. W., Owenson G. G. (1998). Microbial diversity of soda lakes. Extremophiles 2, 191–200 10.1007/s007920050060 [DOI] [PubMed] [Google Scholar]
- Katayama Y., Matsushita Y., Kaneko M., Kondo M., Mizuno T., Nyunoya H. (1998). Cloning of genes coding for the subunits of thiocyanate hydrolase of Thiobacillus thioparus THI 115 and their evolutionary relationships to nitrile hydratase. J. Bacteriol. 180, 2583–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D. P., Baker S. C. (1990). The organosulfur cycle: aerobic and anaerobic processes leading to turnover of C1-sulfur compounds. FEMS Microbiol. Rev. 87, 241–246 [Google Scholar]
- Kjeldsen K. U., Jakobsen T. F., Glastrup J., Ingvorsen K. (2010). Desulfosalsimonas propionicica gen. nov. sp. nov., a novel halophilic sulfate-reducing member of the family Desulfobacteraceae isolated from sediment of Great Salt Lake (Utah). Int. J. Syst. Evol. Microbiol. 60, 1060–1065 [DOI] [PubMed] [Google Scholar]
- Kovaleva O. L., Tourova T. P., Muyzer G., Kolganova T. V., Sorokin D. Y. (2011). Diversity of RuBisCO and ATP citrate lyase genes in soda lake sediments. FEMS Microbiol. Ecol. 35, 37–47 [DOI] [PubMed] [Google Scholar]
- Kulp T. R., Han S., Saltikov C. V., Lanoil B. D., Zargar K., Oremland R. S. (2007). Effects of imposed salinity gradients on dissimilatory arsenate reduction, sulfate reduction, and other microbial processes in sediments from two California soda lakes. Appl. Environ. Microbiol. 73, 5130–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp T. R., Hoeft S. E., Miller L. G., Saltikov C., Murphy J. N., Han S., Lanoil B., Oremland R. S. (2006). Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles Lakes, California. Appl. Environ. Microbiol. 72, 6514–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesbah N. M., Abou-El-Ela S. H., Wiegel J. (2007). Novel and unexpected prokaryote diversity in water and sediments of the alkaline hypersaline lakes of the Wadi An Natrun, Egypt. Microb. Ecol. 54, 598–617 [DOI] [PubMed] [Google Scholar]
- Muyzer G., Sorokin D. Y., Mavromatis K., Lapidus A., Clum A., Ivanova N., Pati A., d'Haeseleer P., Woyke T., Kyrpides N. C. (2011). Complete genome sequence of Thioalkalivibrio “sulfidophilus” HL-EbGr7. Stand. Genomic Sci. 4, 10.4056/sigs.1483693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. (1999). Bioenergetic aspects of halophilism. Microbiol. Mol. Biol. Rev. 63, 34–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. (2011). Thermodynamic limits to microbial life at high salt concentrations. Environ. Microbiol. [Epub ahead of print]. 10.1111/j.1462-2920.2010.02365.x [DOI] [PubMed] [Google Scholar]
- Pikuta E. V., Hoover R. B., Bej A. K., Marsic D., Whitman W. B., Cleland D., Krader P. (2003). Desulfonatronum thiodismutans sp. nov., a novel alkaliphilic, sulfate-reducing bacterium capable of lithoautotrophic growth. Int. J. Syst. Evol. Microbiol. 53, 1327–1332 [DOI] [PubMed] [Google Scholar]
- Pikuta E. V., Zhilina T. N., Zavarzin G. A., Kostrikina N. A., Osipov G. A., Rainey F. A. (1998). Desulfonatronum lacustre gen. nov., sp. nov.: a new alkaliphilic sulfate-reducing bacterium utilizing ethanol. Microbiology 67, 105–113 [Google Scholar]
- Roberts M. F. (2005). Organic compatible solutes of halotolerant and halophilic microorganisms. Saline Syst. 1, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeßler M., Müller V. (2001). Osmoadaptation in bacteria and archaea: common principles and differences. Environ. Microbiol. 3, 743–754 [DOI] [PubMed] [Google Scholar]
- Sakurai H., Ogawa T., Shiga M., Inoue K. (2010). Inorganic sulfur-oxidizing system in green sulfur bacteria. Photosyn. Res. 104, 163–176 [DOI] [PubMed] [Google Scholar]
- Sander J., Dahl C. (2009). “Metabolism of inorganic sulfur compounds in purple bacteria,” in The Purple Sulfur Bacteria, eds Hunter N., Daldal F., Thurnauer M. C., Beatty J. T. (Dordrecht: Springer Science + Business Media B.V.), 595–692 [Google Scholar]
- Scholten J. C. M., Joye S. B., Hollibaugh J. T., Murrell J. C. (2005). Molecular analysis of the sulfate reducing and archaeal community in a meromictic soda lake (Mono Lake, California) by targeting 16S rRNA, mcrA, apsA, and dsrAB genes. Microb. Ecol. 50, 29–39 [DOI] [PubMed] [Google Scholar]
- Schweinfurth G., Lewin L. (1898). Beiträge zur Topographie und Geochemie des ägyptischen Natron-Thals. Z. Gesamte Erdk. 33, 1–25 [Google Scholar]
- Sørensen K. B., Canfield D. E., Oren A. (2004). Salinity responses of benthic microbial communities in a solar saltern (Eilat, Israel). Appl. Environ. Microbiol. 70, 1608–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen K. B., Canfield D. E., Teske A. P., Oren A. (2005). Community composition of a hypersaline endoevaporitic microbial mat. Appl. Environ. Microbiol. 71, 7352–7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin D. Y. (2003). Oxidation of inorganic sulfur compounds by obligately organotrophic bacteria. Microbiology 72, 725–739 10.1023/B:MICI.0000008363.24128.e5 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y. (2008). “Diversity of halophilic sulfur-oxidizing bacteria in hypersaline habitats,” in Microbial Sulfur Metabolism, Proceedings of the International Symposium on Microbial Sulfur Metabolism, 29.06.2006 to 02.07.2006, eds Dahl C., Friedrich C. G. (Münster: Springer-Berlin; ), 225–237 [Google Scholar]
- Sorokin D. Y., Banciu H., Robertson L. A., Kuenen J. G. (2006). “Haloalkaliphilic sulfur-oxidizing bacteria,” in The Prokaryotes: A Handbook on the Biology of Bacteria, 3rd Edn, Vol. 2, eds Dworkin M., Falkow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (New York: Springer-Verlag; ), 969–984 [Google Scholar]
- Sorokin D. Y., Banciu H., van Loosdrecht M., Kuenen J. G. (2003a). Growth physiology and competitive interaction of obligately chemolithoautotrophic, haloalkaliphilic, sulfur-oxidizing bacteria from soda lakes. Extremophiles 7, 195–203 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Antipov A. N., Kuenen J. G. (2003b). Complete denitrification in coculture of obligately chemolithoautotrophic haloalkaliphilic sulfur-oxidizing bacteria from a hypersaline soda lake. Arch. Microbiol. 180, 127–133 10.1007/s00203-003-0567-y [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Tourova T. P., Sjollema K. A., Kuenen G. J. (2003c). Thioalkalivibrio nitratireducens sp. nov., a nitrate-reducing member of an autotrophic denitrifying consortium from a soda lake. Int. J. Syst. Evol. Microbiol. 53, 1779–1783 10.1099/ijs.0.02615-0 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Foti M., Tindall B. J., Muyzer G. (2007a). Desulfurispirillum alkaliphilum gen. nov. sp. nov., a novel obligately anaerobic sulfur- and dissimilatory nitrate-reducing bacterium from a full-scale sulfide-removing bioreactor. Extremophiles 11, 363–370 10.1007/s00792-006-0048-8 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Foti M., Pinkart H. C., Muyzer G. (2007b). Sulfur-oxidizing bacteria in Soap Lake (Washington, USA), a meromictic, haloalkaline lake with an unprecedented high sulfide content. Appl. Environ. Microbiol. 73, 451–455 10.1128/AEM.02087-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin D. Y., Gorlenko V. M., Tourova T. P., Kolganova T. V., Tsapin A. I., Nealson K. H., Kuenen J. G. (2002a). Thioalkalimicrobium cyclicum sp. nov. and Thioalkalivibrio jannaschii sp. nov., new species of alkaliphilic, obligately chemolithoautotrophic sulfur-oxidizing bacteria from a hypersaline alkaline Mono Lake (California). Int. J. Syst. Evol. Microbiol. 52, 913–920 10.1099/ijs.0.02034-0 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Tourova T. P., Kolganova T. V., Sjollema K. A., Kuenen J. G. (2002b). Thioalkalispira microaerophila gen. nov., sp. nov., a novel lithoautotrophic, sulfur-oxidizing bacterium from a soda lake. Int. J. Syst. Evol. Microbiol. 52, 2175–2182 10.1099/ijs.0.02339-0 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Tourova T. P., Lysenko A. M., Mityushina L. L., Kuenen J. G. (2002c). Thioalkalivibrio thiocyanooxidans sp. nov. and Thioalkalivibrio paradoxus sp. nov., novel alkaliphilic, obligately autotrophic, sulfur-oxidizing bacteria from the soda lakes able to grow with thiocyanate. Int. J. Syst. Evol. Microbiol. 52, 657–664 10.1099/ijs.0.02034-0 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Kuenen J. G. (2005). Haloalkaliphilic sulfur-oxidizing bacteria in soda lakes. FEMS Microbiol. Rev. 29, 685–702 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Lysenko A. M., Mityushina L. L., Tourova T. P., Jones B. E., Rainey F. A., Robertson L. A., Kuenen J. G. (2001a). Thioalkalimicrobium aerophilum gen. nov., sp. nov. and Thioalkalimicrobium sibericum sp. nov., and Thioalkalivibrio versutus gen. nov., sp. nov., Thioalkalivibrio nitratis sp. nov. and Thioalkalivibrio denitrificans sp. nov., novel obligately alkaliphilic and obligately chemolithoautotrophic sulfur-oxidizing bacteria from soda lakes Int. J. Syst. Evol. Microbiol. 51, 565–580 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Kuenen J. G., Jetten M. (2001b). Denitrification at extremely alkaline conditions in obligately autotrophic alkaliphilic sulfur-oxidizing bacterium Thioalkalivibrio denitrificans. Arch. Microbiol. 175, 94–101 10.1007/s002030000210 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Tourova T. P., Lysenko A. M., Kuenen J. G. (2001c). Microbial thiocyanate utilization under highly alkaline conditions. Appl. Environ. Microbiol. 67, 528–538 10.1128/AEM.67.2.528-538.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin D. Y., Muyzer G. (2010a). Haloalkaliphilic spore-forming sulfidogens from soda lake sediments and description of Desulfitispora alkaliphila gen. nov., sp. nov. Extremophiles 14, 313–320 10.1007/s00792-010-0310-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin D. Y., Muyzer G. (2010b). Desulfurispira natronophila gen. nov. sp. nov.: an obligately anaerobic dissimilatory sulfur-reducing bacterium from soda lakes. Extremophiles 14, 349–355 10.1007/s00792-010-0314-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin D. Y., Robertson L. A., Kuenen J. G. (2000). Isolation and characterization of obligately chemolithoautotrophic alkaliphilic sulfur-oxidizing bacteria. Ant. V. Leeuwenhoek 77, 251–260 10.1023/A:1002445704444 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Rusanov I. I., Pimenov N. V., Tourova T. P., Abbas B., Muyzer G. (2010a). Sulfidogenesis at extremely haloalkaline conditions in soda lakes of Kulunda Steppe (Altai, Russia). FEMS Microbiol. Ecol. 73, 278–290 10.1111/j.1574-6941.2010.00901.x [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Detkova E. N., Muyzer G. (2010b). Propionate and butyrate dependent bacterial sulfate reduction at extremely haloalkaline conditions and description of Desulfobotulus alkaliphilus sp. nov. Extremophiles 14, 71–77 10.1007/s00792-009-0288-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin D. Y., Tourova T. P., Antipov A. N., Muyzer G., Kuenen J. G. (2004a). Anaerobic growth of the haloalkaliphilic denitrifying sulphur-oxidising bacterium Thialkalivibrio thiocyanodenitrificans sp. nov. with thiocyanate. Microbiology 150, 2435–2442 10.1099/mic.0.27015-0 [DOI] [PubMed] [Google Scholar]
- Sorokin D. Y., Gorlenko V. M., Namsaraev B. B., Namsaraev Z. B., Lysenko A. M., Eshinimaev B. T., Khmelenina V. N., Trotsenko Y. A., Kuenen J. G. (2004b). Prokaryotic communities of the north-eastern Mongolian soda lakes. Hydrobiologia 522, 235–248 10.1023/B:HYDR.0000029989.73279.e4 [DOI] [Google Scholar]
- Sorokin D. Y., Tourova T. P., Henstra A. M., Stams A. J. M., Galinski E. A., Muyzer G. (2008). Sulfidogenesis at extremely haloalkaline conditions by Desulfonatronospira thiodismutans gen. nov., sp. nov., and Desulfonatronospira delicata sp. nov. – a novel lineage of Deltaproteobacteria from hypersaline soda lakes. Microbiology 154, 1444–1453 10.1099/mic.0.2007/015628-0 [DOI] [PubMed] [Google Scholar]
- Tourova T. P., Spiridonova E. M., Berg I. A., Kuznetsov B. B., Sorokin D. Y. (2006). Occurrence, phylogeny and evolution of ribulose-1,5-bisphosphate carboxylase/oxygenase genes in obligately chemolithoautotrophic sulfur-oxidizing bacteria of the genera Thiomicrospira and Thioalkalimicrobium. Microbiology 152, 2159–2169 10.1099/mic.0.28699-0 [DOI] [PubMed] [Google Scholar]
- Tourova T. P., Spiridonova E. M., Berg I. A., Slobodova N. V., Boulygina E. S., Sorokin D. Y. (2007). Phylogeny and evolution of the family Ectothiorhodospiraceae based on comparison of 16S rRNA, cbbL and nifH gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2387–2398 [DOI] [PubMed] [Google Scholar]
- Youatt J. B. (1954). Studies on the metabolism of Thiobacillus thiocyanooxidans. J. Gen. Microbiol. 11, 139–149 10.1099/00221287-11-2-139 [DOI] [PubMed] [Google Scholar]
- Zhilina T. N., Zavarzin G. A., Detkova E. N., Rainey F. A. (1995). Natroniella acetigena gen. nov. sp. nov., an extremely haloalkaliphilic, homoacetic bacterium: a new member of Haloanaerobiales. Curr. Microbiol. 32, 320–326 [DOI] [PubMed] [Google Scholar]
- Zhilina T. N., Zavarzin G. A., Rainey F. A., Pikuta E. N., Osipov G. A., Kostrikina N. A. (1997). Desulfonatronovibrio hydrogenovorans gen. nov., sp. nov., an alkaliphilic, sulfate-reducing bacterium. Int. J. Syst. Bacteriol. 47, 144–149 10.1099/00207713-47-1-144 [DOI] [PubMed] [Google Scholar]
- Zhilina T. N., Zavarzina D. G., Kuever J., Lysenko A. M., Zavarzin G. A. (2005). Desulfonatronum cooperativum sp. nov., a novel hydrogenotrophic, alkaliphilic, sulfate-reducing bacterium, from a syntrophic culture growing on acetate. Int. J. Syst. Evol. Microbiol. 55, 1001–1006 [DOI] [PubMed] [Google Scholar]