Abstract
Microbial communities are at the very basis of life on earth, catalyzing biogeochemical reactions driving global nutrient cycles. However, unlike for plants and animals, microbial diversity is not on the biodiversity–conservation agenda. The latter, however, would imply that microbial diversity is not under any threat by anthropogenic disturbance or climate change. This maybe a misconception caused by the rudimentary knowledge we have concerning microbial diversity and its role in ecosystem functioning. This perspective paper identifies major areas with knowledge gaps within the field of environmental microbiology that preclude a comprehension of microbial ecosystems on the level we have for plants and animals. Opportunities and challenges are pointed out to open the microbial black box and to go from descriptive to predictive microbial ecology.
Keywords: microbial diversity, biodiversity–ecosystem functioning, resistance, resilience, redundancy
Microbial Diversity and Conservation?
Ecosystems collectively determine biogeochemical processes that regulate the Earth System. Loss of biodiversity is generally regarded as detrimental to ecosystems and ecosystem functioning and therefore has been a central issue for environmental scientists during the last decades (Hooper et al., 2005; Balvanera et al., 2006; Cardinale et al., 2006). Microorganisms (i.e., bacteria, archaea, protozoa, and fungi) comprise a major part of the total biomass of organisms inhabiting on earth (Whitman et al., 1998) and represent the largest source of biodiversity (Gans et al., 2005; Sogin et al., 2006; Rusch et al., 2007). They play critical roles in natural biogeochemical processes, sustaining ecosystem functioning (Falkowski et al., 2008). Many of the reactions catalyzed by microorganisms are fundamental to functioning of engineered or managed systems like wastewater treatment plants, agricultural fields, and industrial bioreactors providing many services to our societies (see Table 1). Considering the challenges we are facing with overexploitation of the planet, climate change, pandemics, increasing demands in food production, need for renewable energy and resources (Verstraete et al., 2007), it is remarkable that microbes and their diversity are absent in the ongoing debates about global biodiversity loss and conservations policy, despite various pleas to do so (Colwell, 1997; Cockell, 2005; Cockell and Jones, 2009; Sutherland et al., 2010). The biodiversity–ecosystem function (BEF) research inherently requires the investigation of the relationship between species-assemblies and ecosystem processes. High diversity, rapid generation times, high adaptability due to genome rearrangements, and ubiquitous distribution, have led to the notion that microbial communities are highly redundant and omnipresent and therefore inextinguishable. However, the latter may be a misconception driven by a number of gaps in our assessment and understanding of the functioning of microbial communities and the relevance of microbial diversity in ecosystem functioning being (i) inadequate diversity assessment, (ii) no clear biogeographical patterns, (iii) inability to link diversity to function, (iv) lack of knowledge on functional redundancy, resistance, and resilience of microbial communities.
Table 1.
Major groups of microbes and ecosystem services they provide.
| Microbial group | Process | Ecosystem service | Ecosystem service category |
|---|---|---|---|
| Heterotrophic bacteria/archaea | Organic matter breakdown, mineralization | Decomposition, nutrient recycling, climate regulation, water purification | Supporting and regulating |
| Photoautotrophic bacteria | Photosynthesis | Primary production, carbon sequestration | Supporting and regulating |
| Chemo(litho)autotrophic | Specific elemental transformations (e.g., CH4 oxidation) | Nutrient recycling, climate regulation, water purification | Supporting and regulating |
| Unicellular phytoplankton | Photosynthesis | Primary production, carbon sequestration | Supporting and regulating |
| Archaea | Specific elemental transformation (e.g., metals, CH4 formation, oxidation), often in extreme habitats. | Nutrient recycling, climate regulation, carbon sequestration | Supporting and regulating |
| Protozoa | Mineralization of other microbes | Decomposition, nutrient recycling, soil formation | Supporting |
| Fungi | Organic matter breakdown and mineralization | Decomposition, nutrient recycling, soil formation, primary production (i.e., mycorrhizal fungi) | Supporting |
| Viruses | Lysis of hosts | Nutrient recycling | Supporting |
| All | Production of metabolites (e.g., antibiotics, polymers), degradation of xenobiotics, genetic transformation, and rearrangement | Production of precursors to industrial and pharmaceutical products | Provisional |
| All | Huge diversity, versatility, environmental and biotechnological applications | Educational purposes, getting students interested in science | Cultural |
Inadequate microbial diversity assessment
Diversity of plant and animal communities is traditionally assessed by counting taxonomically described species and calculating diversity indices composed of richness (i.e., number of species) and evenness (relative abundance of species). This approach is also the routine way to assess microbial diversity despite the fact that there are some fundamental discrepancies between macrobial and microbial ecology in the way to define taxonomic units. The first problem is that approx. only 1% of the actual microbial biodiversity is represented as cultured organisms while the characteristics and functions of the remaining 99% are unknown. There are only 7000 microbial species (Achtman and Wagner, 2008) described geno- as well as phenotypically whereas DNA based-methods have identified more than 100 prokaryotic phyla to be present in ecosystems (Alain and Querellou, 2009; Pace, 2009). Ribosomal RNA sequence divergence and DNA–DNA hybridization (DDH) are the main classifiers of microbial taxonomy where less than 70% DDH and less than 98.7% similarity of the 16s rRNA demarcates microbes to belong to different species (Schleifer, 2009). These thresholds are congruent with 95% average nucleotide identity on the whole genome level (Konstantinidis and Tiedje, 2005; Raes and Bork, 2008; Wilmes and Bond, 2009).
However, below the species level the sequence-identity cut-off value used to demarcate species have led to “species” that are still enormously diverse in their genome content, physiology, and ecology (Staley, 2006). Hence, the application of richness and evenness, based on these taxonomic criteria will not be very helpful in studying BEF relationships in microbial communities because there is no direct link between these units and the functions catalyzed by the community. For example, soil contains up to 106 bacterial genotypes g−1 DW (Gans et al., 2005), but the phylotypic diversity estimates are one order of magnitude lower (Roesch et al., 2007). It has been demonstrated recently, that environmental parameters effect (e.g., pH, cattle grazing) microbial communities at taxonomic levels higher than species, hence, indicating a degree of ecological coherence at these higher taxonomic levels. (Lauber et al., 2009; Philippot et al., 2010). However, these relationships are at the level of factors determining the distribution but do not tell anything about the functions catalyzed by these ecological coherent groups and the consequences for ecosystem functioning.
Hence, approaching the BEF question in a meaningful way in microbial communities requires an approach beyond the classical richness and evenness approach of arbitrarily demarcated taxonomic units. There is a need for novel concepts in microbial BEF research regarding ecological relevant units for microbes to by-pass the species hang-up in order to develop generic concepts and theories in microbial ecology. This required paradigm shift away from taxa oriented concepts of community analysis toward functional biodiversity is also starting to be developed for microbial communities. Genomic and proteomic techniques enable high-throughput inventories of microbial functional units (protein coding genes, enzymes, metabolites) which can be regarded as microbial traits being the key components of the overall ecosystem (Green et al., 2008).
Microbial biogeography
The conventional view of microbial distribution of species through space and time has been dominated for decades by the “Baas-Becking” hypothesis (see de Wit and Bouvier, 2006) “everything is everywhere, but the environment selects.” The lack of dispersal limitations of microorganisms would ensure a global distribution, but that local deterministic factors would determine the relative abundance of “latent” and “flourishing” species. This view is in sharp contrast with plants and animals which show clear taxa–area relationships and biogeography. The Baas-Becking legacy is likely the main reason why microbial diversity is not on the biodiversity–conservation agenda. However, the last decade there are a number of studies demonstrating species–area relationships, biogeography, and spatial patterns at various scales for microbes (Horner-Devine et al., 2004; Bell et al., 2005a; Fierer and Jackson, 2006) and more recently even using a functional gene-based approach (Zhou et al., 2008). Next to this, microbial endemism has been reported as well (Cho and Tiedje, 2000; Pommier et al., 2007). Using high-throughput sequencing technology a number of studies clearly demonstrated the presence of habitat specific communities shaped by edaphic factors and historical contingencies (Fierer et al., 2007; von Mering et al., 2007; Lauber et al., 2009). A meta analysis of all currently available 16S rRNA gene sequences revealed clear environmental distributions on the genus or species level with soil and freshwater as least selective habitats while marine, animal, and thermal habitats were most selective (Tamames et al., 2010). Similarly, a meta analysis of all currently available denitrification genes showed that similarities between globally distributed communities corresponded to similarities in habitat salinity and the majority of communities were phylogenetically clustered more than expected by chance, indicating habitat filtering (Jones and Hallin, 2010).
Linking function to identity
Assessing BEF relationships in the classical macro-ecological context would require the connecting of individual microbial species to the biogeochemical processes they catalyze. However, considering the problems in defining relevant ecological units, the metabolic versatility, the large number of unknown species, and the scale issue involved, this is the central problem in the field of environmental microbiology. The majority of studies in the literature have relied on correlating changes in activity to changes in community composition or diversity, and only a few articles can actually show a causal relationship. A myriad of techniques have been developed for linking diversity and function (see Wagner, 2009). However, many of these techniques were based on the analyses of ribosomal RNA or mRNA transcripts of functional genes, indicating only the potential to be involved in specific processes. The use of stable isotope probing (SIP) has evoked a major breakthrough in environmental microbiology (see Chen and Murrell, 2010). Stable isotopes (13C/15N) are incorporated following assimilation of a specific substrate into taxonomically relevant molecules [RNA/DNA (Whiteley et al., 2006; Rasche et al., 2009); PLFA; phospholipids, see (Bodelier et al., 2009b) or proteins (Jehmlich et al., 2009)]. Only the microbes which have actively been incorporating the stable isotopes are detected when analyzing RNA/DNA, after density centrifugation, or PLFA using GC–IRMS or proteins using GC–MS or LC–MS. The major disadvantage of SIP is the use of unnaturally high substrate concentrations, the different label uptake rates per species, and cross feeding. Recent work brought improvements in the shortcomings of traditional SIP studies. By combining SIP with magnetic bead capturing of mRNA (Miyatake et al., 2009), Raman spectroscopy, NanoSIMS (Secondary Ion Beam Mass Spectroscopy; Wagner, 2009) and metagenomics, active species of which no cultured representatives are available or unknown pathways or genes involved in biogeochemical processes were detected, avoiding high substrate concentrations and long-incubation times (see Chen and Murrell, 2010). The challenge in applying SIP-based techniques will be in BEF experiments, where experimental designs allowing for causal and mechanistic conclusions require high sample throughput.
Functional redundancy, resistance, and resilience in microbial ecosystems
Another paradigm that led to the absence of microbial diversity in BEF debate, conservation issues, and global biogeochemical process models is the idea of microbial omnipresence, high adaptability, and functional redundancy. Hence, many species carrying the same function ensures stability of the process when species are lost or temporarily inactive. But is this the rule or are the current methods and approaches used studying redundancy, resistance, and resilience inadequate to answers these questions? Well, the latter is definitely a part of the story. Functional redundancy sensu stricto is difficult to assess in microbial communities, since it requires the contribution of individual community members to processes and separation between diversity and environmental factors, which is a difficult task as outlined in the previous sections. Another part of the story probably lies in natural complexity, making generalizations regarding redundancy using the current approaches rather useless. This becomes apparent in recent overviews and meta-analyses on BEF in microbial communities were equal numbers of studies are found that report on positive, negative, or inconclusive responses of increased diversity on ecosystem processes (Ducklow, 2008). Recently, a comprehensive meta-analyses demonstrated that out of 110 studies more than 70% demonstrate that microbial community composition was not resistant (i.e., the degree to which community composition remains unchanged when disturbed) against disturbances (fertilization; CO2 increase; temperature; carbon amendment; Allison and Martiny, 2008). This held true for broad taxonomic groups (fungi, bacteria, archaea) as well as narrow functional guilds catalyzing specific functions (e.g., methane oxidation, nitrification). The same study demonstrated that the resilience (i.e., the rate at which microbial community composition returns to its original composition after being disturbed) can be in the order of years. An example is the suppression of atmospheric methane consumption in soils by nitrogenous fertilizers which can last for decades after fertilizer application has ceased because there no other microbes to take over (see Bodelier and Laanbroek, 2004). The latter example represents functional guilds which catalyze a specific function and which do not harbor many species and, hence, are less redundant (e.g., methane production and oxidation, ammonia oxidation). Similar “vulnerability “one would expect in symbiotic relationship (e.g., mycorrhiza, N2 fixers) where it has been shown that reduced diversity has led to lower biomass production of the host–plants (see van der Heijden et al., 2008) indicating that species cannot be replaced without consequences for ecosystem functioning. Considering this it is tempting to focus on these narrow functional guilds and these specific symbiont–host relationships when thinking about vulnerable microbial communities. However, quite some studies have demonstrated a direct relationship between diversity and ecosystem process rate for general broad functions, like respiration or denitrification (Setala and McLean, 2004; Wohl et al., 2004; Balser and Firestone, 2005; Bell et al., 2005b; Salles et al., 2009). Hence, reducing the diversity also for these broad range, presumably redundant functions will have consequences for ecosystem functioning. By growing plants on soils inoculated with a microbial community of reduced diversity it was demonstrated that plant biomass, being the outcome of many interacting processes, was reduced (Hol et al., 2010). The absence of “rare” microbes led to higher nutrient content of the plants and made them more susceptible to pest organisms. These indirect ecosystem effects due to reduced microbial diversity will often go unnoticed when only measuring direct connections between diversity and single function catalyzed by the microbes, as has been done most frequently.
The degree of redundancy can also depend very much on the environmental conditions. Under nutrient poor conditions many species may not perform well leading to diversity effects on ecosystem functions which are not detected under optimal conditions (see van der Heijden et al., 2008). Also heavy perturbations of environments with chemicals or heavy metals may lead to such a situation of lowered redundancy (Gans et al., 2005).
The strong focus on species richness, being synonymous for redundancy can also impair our understanding of BEF in microbial communities. Recovery of denitrification after salt stress in laboratory microcosms strongly depended on the initial evenness of the community (Wittebolle et al., 2009). The relative abundance of species before perturbation was of greater importance than the number of species present, all capable of performing denitrification. Hence, even when an ecosystem function is supported by a highly redundant community, the resilience against disturbance can be low when the relative abundance is very uneven. This may be caused by non-resistant dominant community members or density dependent mutualism between species, like was shown for the process of nitrification (Graham et al., 2007) or syntrophic consortia (Kato and Watanabe, 2010).
Maybe one of the most important gaps in our understanding and assessment of redundancy and the BEF in microbial communities is the focus on diversity in relation to one particular, isolated function. However, overall ecosystem functioning is the collective action of many species carrying out various functions, often in interaction. It was shown for a number of datasets that diversity is more important for multifunctionality than it is for single functions (Gamfeldt et al., 2008). Hence, species can contribute to more than one function which introduces another level of redundancy. Hence, when looking at overall ecosystem functions (e.g., primary productivity) it will always take more species than the number of single functions contributing to overall ecosystem function due to the fact that not all species can perform all functions. The latter was also demonstrated for microbial systems where a number of soil functions were measured and the most parsimonious set of species (OTU's on DGGE) was determined which positively influenced these functions (He et al., 2009). By backward-elimination boosted tree analyses it was shown that reducing microbial diversity will eventually lead to loss of function. The positive relationship between diversity and ecosystem-multifunctionality has been explained by complementarity between species, positive selection, or niche differentiation (Loreau and Hector, 2001; Cardinale et al., 2006; Salles et al., 2009). Hence, for overall ecosystem functioning but also for the performance of single functions multiple traits of species have to be taking into account. It is the overall geno- plus phenotype of a microbe which makes it survive or function in a particular habitat.
Another aspect we have to consider in the classical view on microbial communities being functionally highly redundant is the scale issue. Populations or communities of interacting microbes on microbial relevant scales may not consist of many different species due to spatial arrangement or isolation, e.g., along roots, soil pores, plant leaves, biofilms, or microbial flocs in sewage treatment (Lindow and Leveau, 2002; Daims et al., 2006; Battin et al., 2007; Gross et al., 2010). Hence, for example at the soil aggregate scale the number of species present may be very low as will also be redundancy. Whether the role of diversity for microbial ecosystem functioning on the landscape scale is a simple addition of what happens on the micro scale is one of the biggest challenges in microbial BEF research. Nevertheless, it is evident that in heterogeneous, spatially separated micro-habitats colonized by low numbers of cells, functional redundancy is an irrelevant concept.
The growing body of experimental evidence suggest that microbial communities can be sensitive to disturbances and that resilience is linked to diversity. Next to this, functional redundancy is certainly not a general characteristic of microbial communities. However, the issues put forward in the preceding paragraphs preclude firm conclusions regarding sensitivity and vulnerability of microbial communities and what the role of community composition is in this matter. When thinking about conservation, a fundamental problem is that the great majority of microbial diversity is unknown in terms of species identity and associated functional characteristics. Hence, we do not know what to conserve but we also do not know what we have lost.
From Descriptive to Predictive; Toward Understanding and Protection
From species richness to functional biodiversity
It is obvious that the omission of microbial communities in management and conservation of ecosystems is due to a lack of understanding of the functioning and composition of environmental microbial communities leading to the assumption that microbial communities are insensitive, resilient, and redundant. However, getting microbial diversity under the attention of policy and management will require incorporation of microbial diversity into predictive ecosystem process models.
However, this will require a different approach as has been followed till now. Evidently, using richness of taxonomically described species is not the way to follow in microbial BEF research. In parallel to plant–sciences (McGill et al., 2006), a functional biodiversity approach using species, genus, or community functional traits would make much more sense as a framework to be used in microbial BEF research, especially, considering the swapping of genes across microbial taxonomic units (McDaniel et al., 2010). Descriptions of such trait-centered approaches for microbial communities have already been initiated (Allison and Martiny, 2008; Green et al., 2008) awaiting testing and application.
Based on the functional trait approach Allison and Martiny (2008) proposed a conceptual model that incorporates microbial diversity via functional traits into a process model. This model assumes a number of taxa in a functional group. The community process rate is the sum of the products of the abundances and the biomass-specific physiological process rate of all taxa. To predict community response to environmental disturbance, taxa specific physiological response rates to disturbance were introduced which principally have to be determined for every individual taxon. However, the model assumes that functional traits and physiological responses are related to phylogeny and hence can be estimated from phylogenetic information. This would result in aggregating taxa which have similar traits and responses in this model. The challenge remains, however, to establish basic relationships between traits, responses, and phylogeny. This could be achieved using artificial communities of microbes in which abundance and individual physiological responses can be measured for every single taxon in these communities. Methane oxidizing proteobacteria would be an ideal functional group to test this model considering the methodological possibilities to link their function to their phylogeny (Bodelier et al., 2009a,b). A project testing the Allison and Martiny (2008) model using methanotrophs is under way1.
A helpful tool to facilitate the aggregation of phylogenetic groups with common traits may the “ecotype” systematics, where evolutionary and ecological information demarcates groups of organism that have a history of coexistence as distinct lineages and a prognosis of future coexistence, leading to units of bacterial ecology and evolution below the conventional species level-cut-off (Koeppel et al., 2008). This approach may point out phylogenetic units putatively sharing a common eco-physiology, even when no cultured representatives are available, thereby focusing microbial BEF research.
The functional biodiversity approach would also offer the incorporation of interactions multifunctionality and diversity effects across trophic levels, as been proposed recently (de Bello et al., 2010). These authors propose the concept of “trait–ecosystem service clusters” as traits of various organisms that together influence ecosystem services over trophic levels. Ultimately this will be the goal to understand the role of microbial diversity as an integral part of ecosystems. Using gene-expression as analogs of microbial traits, microbial functional molecular ecological networks can be established using functional microarrays (GeoChip) and subsequent statistical analyses to find highly correlated clusters of genes (Zhou et al., 2010) as response to environmental change or perturbation. The identified clusters of genes, designated as ecological network modules, can be linked to ecosystem processes or properties and would fit as such in the trait-service cluster concept. High-throughput genomic and proteomic techniques offer the potential to monitor genes, transcripts, and proteins of microbial communities in spatial as well as temporal high resolution and thereby identify correlated microbial functional traits. However, to use these relationships in microbial BEF studies we have to know what the biology is underlying these correlations and whether they can be used as ecosystem process proxies, possibly linked to phylogenetic categories. This information still has to be retrieved from culture or single cell studies.
Single cell and community eco-physiology
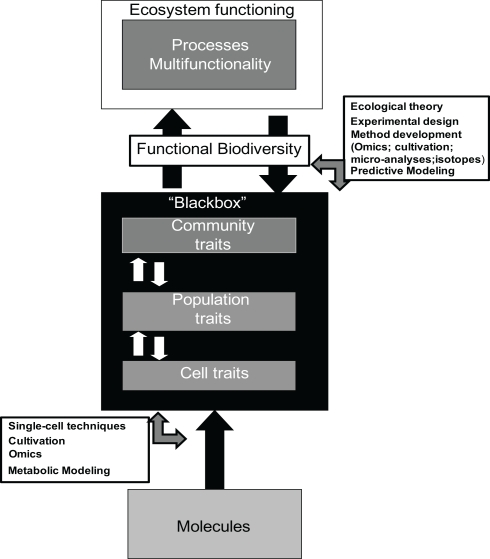
Besides obtaining empirical results demonstrating biodiversity–function relationships for microbial communities, it will be inevitable to understand the mechanistic basis for these observations. These mechanisms are to be found at the cellular level and in the interaction between cells, communities with the environment, and higher trophic levels. However, in microbial BEF these levels of organization have largely been treated as “black boxes” combined with being largely of descriptive nature and disconnected to ecological concepts. Approaches in microbial BEF have been primarily “Top-down” manipulating the “black box” and measuring the effect on singular ecosystem processes (see Figure 1). However, understanding the structure and functioning of microbial communities at the cell and community level, is necessary to understand mechanisms and to generate the knowledge needed to move toward predictions on ecosystem process level. The rapid methodological developments of the last decades are narrowing down the limitations which kept the discipline at the descriptive level. The “omics” techniques enable studying community ecology as well as physiology (Konopka, 2009) of known as well as unknown microbial species. The genome of single microbial cells derived directly from the environment can be sequenced (Woyke et al., 2010) and physiological responses can be recorded (Musat et al., 2008; Konopka et al., 2011). Moreover, in situ adaptation of community members (Denef et al., 2009; Belnap et al., 2010) or in situ profiling of whole genome transcripts and proteins of individual species (Kalyuzhnaya et al., 2010) is feasible. Methodology and concepts are emerging enabling individual-based physiology and ecology (Leveau, 2009; Wagner, 2009; Gross et al., 2010) and even interactions on microbial relevant scale (Kim et al., 2008). Theoretical and conceptual approaches from macro ecology are being applied to understand microbial community structure and to link it to ecosystem processes (Prosser et al., 2007; Allison and Martiny, 2008; Green et al., 2008; Salles et al., 2009). These novel avenues at the level of environmental molecules, cells, and communities will lead us to causal mechanistic understanding of environmental microbial communities, eventually leading to the opening of the “Black box” (see Figure 1). It will take the field of environmental microbiology a step closer toward moving from descriptive to predictive.
Figure 1.
Schematic representation of important elements in elucidating the role of microbial diversity in ecosystem functioning. Crucial element is the application of a Functional Biodiversity concept to link microbial diversity to ecosystem functioning. This approach will facilitate predictive ecosystem modeling and will be fostered by omics techniques. However, to make this conceptual step the mechanistic insight into what is going on in the “black box” being the structure and functioning of microbial communities and underlying populations an cells, needs to be elucidated. Application of ecological theory, conceptual experimental design, novel methodology, and mathematical modeling will be the key to gain access to the knowledge in the “Black box.”
Toward conservation and management
Preservation and management of our ecosystems and ecosystem services has the highest research priority globally (Millennium Ecosystem Assessment, 2005). Biodiversity is a crucial component in preservation of ecosystem services globally and there are many coordinated activities to preserve biodiversity and habitats. In the previous sections it became evident that it is not a matter of a lack of vulnerability but mainly a lack of understanding of microbial ecosystems, which causes the absence of microbial diversity from conservation initiatives. I would say that the lack of knowledge in combination with the crucial services they provide would be the best argument for the protection of microbial diversity and ecosystems. However, for the time being it seems that we have to take the level of knowledge on microbial communities to the same level we have with plants and animals, as outlined in the previous sections, before microbial diversity will be taken seriously in conservation policy. However, this will require multidisciplinarity and input from various scientific disciplines (microbiology, genetics, genomics, ecology, physics, engineering, chemistry, mathematical modeling; biogeochemistry, theoretical biology, systems biology). It will also require the coordinated application of novel technologies embedded in well-designed ecological experimentation that will allow for causal and mechanistic information to emerge. This will also include long-term experiments (Eisenhauer et al., 2010) which are not common in microbial BEF research. Research consortia are needed to channel the explosive developments toward microbial ecological studies and subjects. Achieving this will require intense cooperation and exchange of researchers between scientific disciplines and intense training efforts to educate the researchers which will work in this multidisciplinary field. This cross fertilization between “omics” techniques and experimental microbial ecology is the first step to reach an ecosystem biology approach, incorporating microbial diversity into ecosystem process models. However, we are not yet there. What to do in the mean time. Recently, a “roadmap” toward microbial conservation has been put forward (Cockell and Jones, 2009). Important elements in this plea were habitat conservation, legislation, education, and ethical considerations. Since habitat conservation is already integrated in the EU-legislation2 it would be most efficient to consider taking habitat characteristics into account which are important for microbial communities. In the EU-Soil Framework Directive3 in development, microbes are at least mentioned and taking into consideration (Gardi et al., 2009). Great efforts are under way to make inventories of soil microbial diversity using metagenomics techniques, attempting to catalog what is out there, including associated metadata (Costello et al., 2006; Gardi et al., 2009; Ranjard et al., 2010). A recently started initiative is aiming at sequencing many relevant microbiomes distributed over the globe4. Mining of these data and associated environmental metadata will yield relationships between microbial diversity and important environmental characteristics which can be used in guidelines for habitat preservation in conservation policy. It will also facilitate the establishment of so called “normal operating ranges” for microbial habitats, indicating what the level of microbial diversity and functioning is to be expected in for example “normally” functioning soils. Deviations from this “normal operating range” may be interpreted as disturbances of the soils under investigation. Such an exercise has been carried out in a very intensive soil monitoring network in the Netherlands which included 300 soil sampling sites covering various land-use types and soil types. Monitoring complete foodwebs in these sites for 6 years identified most important life-support functions in these soils including the abundance and diversity of the associated biota. This database led to guidelines for the range of diversity and numbers of organisms that should be present in a specific soil type in order to function normally (Rutgers et al., 2009).
Hence, the policy mindset is going in the right direction, theoretical frameworks are in place, and methodology is developing in a way that in the near future global biodiversity–conservation will embrace all three domains of life on our planet. The incorporation of functional microbial traits into predictive ecosystem models will also facilitate the management of habitats taking environmental effects on microbes into account. Large scale sequencing efforts will provide information on what microbes are out there in our ecosystems. Clever, conceptually, and theoretical supported experimentation will reveal what these microbes are capable of increasing the appreciation for this supporting and regulating domain of life promoting its conservation.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The basis of paper was part of the outcome of the workshop “EuroMicrobe”; exploring the rationale for conservation of environmental microbial communities and associated habitats, held from December 8–10, 2009, Hilversum, the Netherlands. This workshop was part of the ESF EUROCORE Eurodiversity (ERAS-CT-2003-98049, 6th EU-framework programme). This work was also part of the European Science Foundation EUROCORES Programme EuroEEFG as supported by funds from the Netherlands Organisation for Scientific Research (NWO). The author wants to thank Prof. Nico Boon, Dr. Holger Daims, Dr. Ciro Gardi, Dr. Sara Hallin, Dr. Graeme Nicol, Dr. Jakob Pernthaler, Dr. Joana Salles, Prof. Michael Schloter for their constructive comments to the contents of this manuscript. This publication is publication no. 5024 of the Netherlands Institute of Ecology.
Footnotes
References
- Achtman M., Wagner M. (2008). Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6, 431–440 [DOI] [PubMed] [Google Scholar]
- Alain K., Querellou J. (2009). Cultivating the uncultured: limits, advances and future challenges. Extremophiles 13, 583–594 10.1007/s00792-009-0261-3 [DOI] [PubMed] [Google Scholar]
- Allison S. D., Martiny J. B. H. (2008). Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 105, 11512–11519 10.1073/pnas.0801925105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balser T. C., Firestone M. K. (2005). Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry 73, 395–415 10.1007/s10533-004-0372-y [DOI] [Google Scholar]
- Balvanera P., Pfisterer A. B., Buchmann N., He J. S., Nakashizuka T., Raffaelli D., Schmid B. (2006). Quantifying the evidence for biodiversity effects on ecosystem functioning and services. Ecol. Lett. 9, 1146–1156 10.1111/j.1461-0248.2006.00963.x [DOI] [PubMed] [Google Scholar]
- Battin T. J., Sloan W. T., Kjelleberg S., Daims H., Head I. M., Curtis T. P., Eberl L. (2007). Microbial landscapes: new paths to biofilm research. Nat. Rev. Microbiol. 5, 76–81 10.1038/nrmicro1556 [DOI] [PubMed] [Google Scholar]
- Bell T., Ager D., Song J. I., Newman J. A., Thompson I. P., Lilley A. K., van der Gast C. J. (2005a). Larger islands house more bacterial taxa. Science 308, 1884–1884 10.1126/science.1111318 [DOI] [PubMed] [Google Scholar]
- Bell T., Newman J. A., Silverman B. W., Turner S. L., Lilley A. K. (2005b). The contribution of species richness and composition to bacterial services. Nature 436, 1157–1160 10.1038/nature03891 [DOI] [PubMed] [Google Scholar]
- Belnap C. P., Pan C., VerBerkmoes N. C., Power M. E., Samatova N. F., Carver R. L., Hettich R. L., Banfield J. F. (2010). Cultivation and quantitative proteomic analyses of acidophilic microbial communities. ISME J. 4, 520–530 10.1038/ismej.2009.139 [DOI] [PubMed] [Google Scholar]
- Bodelier P. L. E., Bar-Gillisen M.-J., Hordijk K., Damsté J. S., Rijpstra W. I., Geenevasen J. A., Dunfield P. F. (2009a). A reanalysis of phospholipid fatty acids as ecological biomarkers for methanotrophic bacteria. ISME J. 3, 606–617 10.1038/ismej.2009.6 [DOI] [PubMed] [Google Scholar]
- Bodelier P. L. E., Kamst M., Meima-Franke M., Stralis-Pavese N., Bodrossy L. (2009b). Whole-community genome amplification (WCGA) leads to compositional bias in methane-oxidizing communities as assessed by pmoA-based microarray and QPCR. Environ. Microbiol. Rep. 1, 434–441 10.1111/j.1758-2229.2009.00066.x [DOI] [PubMed] [Google Scholar]
- Bodelier P. L. E., Laanbroek H. J. (2004). Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol. Ecol. 47, 265–277 10.1016/S0168-6496(03)00304-0 [DOI] [PubMed] [Google Scholar]
- Cardinale B. J., Srivastava D. S., Duffy J. E., Wright J. P., Downing A. L., Sankaran M., Jouseau C. (2006). Effects of biodiversity on the functioning of trophic groups and ecosystems. Nature 443, 989–992 10.1038/nature05202 [DOI] [PubMed] [Google Scholar]
- Chen Y., Murrell J. C. (2010). When metagenomics meets stable-isotope probing: progress and perspectives. Trends Microbiol. 18, 157–163 10.1016/j.tim.2010.02.002 [DOI] [PubMed] [Google Scholar]
- Cho J. C., Tiedje J. M. (2000). Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66, 5448–5456 10.1128/AEM.66.12.5448-5456.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockell C. S. (2005). The value of microorganisms. Environ. Ethics 27, 375–390 [Google Scholar]
- Cockell C. S., Jones H. L. (2009). Advancing the case for microbial conservation. Oryx 43, 520–526 10.1017/S0030605309990111 [DOI] [Google Scholar]
- Colwell R. R. (1997). Microbial diversity: the importance of exploration and conservation. J. Ind. Microbiol. Biotechnol. 18, 302–307 10.1038/sj.jim.2900390 [DOI] [PubMed] [Google Scholar]
- Costello M. J., Bouchet P., Emblow C. S., Legakis A. (2006). European marine biodiversity inventory and taxonomic resources: state of the art and gaps in knowledge. Mar. Ecol. Prog. Ser. 316, 257–268 10.3354/meps316257 [DOI] [Google Scholar]
- Daims H., Lucker S., Wagner M. (2006). Daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8, 200–213 10.1111/j.1462-2920.2005.00880.x [DOI] [PubMed] [Google Scholar]
- de Bello F., Lavorel S., Diaz S., Harrington R., Cornelissen J. H. C., Bardgett R. D., Berg M. P., Cipriotti P., Feld C. K., Hering D., da Silva P. M., Potts S. G., Sandin L., Sousa J. P., Storkey J., Wardle D. A., Harrison P. A. (2010). Towards an assessment of multiple ecosystem processes and services via functional traits. Biodivers. Conserv. 19, 2873–2893 10.1007/s10531-010-9850-9 [DOI] [Google Scholar]
- de Wit R., Bouvier T. (2006). “Everything is everywhere, but, the environment selects”; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 8, 755–758 10.1111/j.1462-2920.2006.01017.x [DOI] [PubMed] [Google Scholar]
- Denef V. J., VerBerkmoes N. C., Shah M. B., Abraham P., Lefsrud M., Hettich R. L., Banfield J. F. (2009). Proteomics-inferred genome typing (PIGT) demonstrates inter-population recombination as a strategy for environmental adaptation. Environ. Microbiol. 11, 313–325 10.1111/j.1462-2920.2008.01769.x [DOI] [PubMed] [Google Scholar]
- Ducklow H. (2008). Microbial services: challenges for microbial ecologists in a changing world. Aquat. Microb. Ecol. 53, 13–19 10.3354/ame01220 [DOI] [Google Scholar]
- Eisenhauer N., Bessler H., Engels C., Gleixner G., Habekost M., Milcu A., Partsch S., Sabais A. C. W., Scherber C., Steinbeiss S., Weigelt A., Weisser W. W., Scheu S. (2010). Plant diversity effects on soil microorganisms support the singular hypothesis. Ecology 91, 485–496 10.1890/08-2338.1 [DOI] [PubMed] [Google Scholar]
- Falkowski P. G., Fenchel T., Delong E. F. (2008). The microbial engines that drive earth's biogeochemical cycles. Science 320, 1034–1039 10.1126/science.1153213 [DOI] [PubMed] [Google Scholar]
- Fierer N., Jackson R. B. (2006). The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. U.S.A. 103, 626–631 10.1073/pnas.0507535103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N., Morse J. L., Berthrong S. T., Bernhardt E. S., Jackson R. B. (2007). Environmental controls on the landscape-scale biogeography of stream bacterial communities. Ecology 88, 2162–2173 10.1890/06-1746.1 [DOI] [PubMed] [Google Scholar]
- Gamfeldt L., Hillebrand H., Jonsson P. R. (2008). Multiple functions increase the importance of biodiversity for overall ecosystem functioning. Ecology 89, 1223–1231 10.1890/06-2091.1 [DOI] [PubMed] [Google Scholar]
- Gans J., Wolinsky M., Dunbar J. (2005). Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309, 1387–1390 10.1126/science.1112665 [DOI] [PubMed] [Google Scholar]
- Gardi C., Montanarella L., Arrouays D., Bispo A., Lemanceau P., Jolivet C., Mulder C., Ranjard L., Rombke J., Rutgers M., Menta C. (2009). Soil biodiversity monitoring in Europe: ongoing activities and challenges. Eur. J. Soil Sci. 60, 807–819 10.1111/j.1365-2389.2009.01177.x [DOI] [Google Scholar]
- Graham D. W., Knapp C. W., Van Vleck E. S., Bloor K., Lane T. B., Graham C. E. (2007). Experimental demonstration of chaotic instability in biological nitrification. ISME J. 1, 385–393 10.1038/ismej.2007.45 [DOI] [PubMed] [Google Scholar]
- Green J. L., Bohannan B. J. M., Whitaker R. J. (2008). Microbial biogeography: from taxonomy to traits. Science 320, 1039–1043 10.1126/science.1153475 [DOI] [PubMed] [Google Scholar]
- Gross C. A., Reddy C. K., Dazzo F. B. (2010). CMEIAS color segmentation: an improved computing technology to process color images for quantitative microbial ecology studies at single-cell resolution. Microb. Ecol. 59, 400–414 10.1007/s00248-009-9616-7 [DOI] [PubMed] [Google Scholar]
- He J. Z., Ge Y., Xu Z. H., Chen C. R. (2009). Linking soil bacterial diversity to ecosystem multifunctionality using backward-elimination boosted trees analysis. J. Soils Sediments 9, 547–554 10.1007/s11368-009-0120-y [DOI] [Google Scholar]
- Hol W. H. G., de Boer W., Termorshuizen A. J., Meyer K. M., Schneider J. H. M., van Dam N. M., van Veen J. A., van der Putten W. H. (2010). Reduction of rare soil microbes modifies plant-herbivore interactions. Ecol. Lett. 13, 292–301 10.1111/j.1461-0248.2009.01424.x [DOI] [PubMed] [Google Scholar]
- Hooper D. U., Chapin F. S., Ewel J. J., Hector A., Inchausti P., Lavorel S., Lawton J. H., Lodge D. M., Loreau M., Naeem S., Schmid B., Setala H., Symstad A. J., Vandermeer J., Wardle D. A. (2005). Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol. Monogr. 75, 3–35 10.1890/04-0922 [DOI] [Google Scholar]
- Horner-Devine M. C., Lage M., Hughes J. B., Bohannan B. J. M. (2004). A taxa-area relationship for bacteria. Nature 432, 750–753 10.1038/nature03073 [DOI] [PubMed] [Google Scholar]
- Jehmlich N., Schmidt F., Taubert M., Seifert J., von Bergen M., Richnow H. H., Vogt C. (2009). Comparison of methods for simultaneous identification of bacterial species and determination of metabolic activity by protein-based stable isotope probing (protein-SIP) experiments. Rapid Commun. Mass Spectrom. 23, 1871–1878 10.1002/rcm.4084 [DOI] [PubMed] [Google Scholar]
- Jones C. M., Hallin S. (2010). Ecological and evolutionary factors underlying global and local assembly of denitrifier communities. ISME J. 4, 633–641 10.1038/ismej.2009.152 [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya M. G., Beck D. A. C., Suciu D., Pozhitkov A., Lidstrom M. E., Chistoserdova L. (2010). Functioning in situ: gene expression in Methylotenera mobilis in its native environment as assessed through transcriptomics. ISME J. 4, 388–398 10.1038/ismej.2009.117 [DOI] [PubMed] [Google Scholar]
- Kato S., Watanabe K. (2010). Ecological and evolutionary interactions in syntrophic Methanogenic consortia. Microbes Environ. 25, 145–151 10.1264/jsme2.ME10122 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Boedicker J. Q., Choi J. W., Ismagilov R. F. (2008). Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc. Natl. Acad. Sci. U.S.A. 105, 18188–18193 10.1073/pnas.0807935105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeppel A., Perry E. B., Sikorski J., Krizanc D., Warner A., Ward D. M., Rooney A. P., Brambilla E., Connor N., Ratcliff R. M., Nevo E., Cohan F. M. (2008). Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc. Natl. Acad. Sci. U.S.A. 105, 2504–2509 10.1073/pnas.0712205105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka A. (2009). What is microbial community ecology? ISME J. 3, 1223–1230 10.1038/ismej.2009.88 [DOI] [PubMed] [Google Scholar]
- Konopka M. C., Strovas T. J., Ojala D. S., Chistoserdova L., Lidstrom M. E., Kalyuzhnaya M. G. (2011). Respiration response imaging for real-time detection of microbial function at the single-cell level. Appl. Environ. Microbiol. 77, 67–72 10.1128/AEM.01166-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K. T., Tiedje J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 2567–2572 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber C. L., Hamady M., Knight R., Fierer N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120 10.1128/AEM.00335-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveau J. H. J. (2009). Individual-based ecology of plant-associated bacteria. Phytopathology 99, S155–S155 [Google Scholar]
- Lindow S. E., Leveau J. H. (2002). Phyllosphere microbiology. Curr. Opin. Biotechnol. 13, 238–243 10.1016/S0958-1669(02)00313-0 [DOI] [PubMed] [Google Scholar]
- Loreau M., Hector A. (2001). Partitioning selection and complementarity in biodiversity experiments. Nature 412, 72–76 10.1038/35083573 [DOI] [PubMed] [Google Scholar]
- McDaniel L. D., Young E., Delaney J., Ruhnau F., Ritchie K. B., Paul J. H. (2010). High frequency of horizontal gene transfer in the oceans. Science 330, 50–50 10.1126/science.1192243 [DOI] [PubMed] [Google Scholar]
- McGill B. J., Enquist B. J., Weiher E., Westoby M. (2006). Rebuilding community ecology from functional traits. Trends Ecol. Evol. 21, 178–185 10.1016/j.tree.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Millennium Ecosystem Assessment. (2005). Ecosystems and Human Well-being: General Synthesis. United Nations. Available at: www.millenniumassessment.org/en/synthesis.aspx
- Miyatake T., MacGregor B. J., Boschker H. T. S. (2009). Linking microbial community function to phylogeny of sulfate-reducing Deltaproteobacteria in marine sediments by combining stable isotope probing with magnetic-bead capture hybridization of 16S rRNA. Appl. Environ. Microbiol. 75, 4927–4935 10.1128/AEM.00652-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musat N., Halm H., Winterholler B., Hoppe P., Peduzzi S., Hillion F., Horreard F., Amann R., Jorgensen B. B., Kuypers M. M. M. (2008). A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc. Natl. Acad. Sci. U.S.A. 105, 17861–17866 10.1073/pnas.0809329105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R. (2009). Mapping the tree of life: progress and prospects. Microbiol. Mol. Biol. Rev. 73, 565–576 10.1128/MMBR.00033-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L., Andersson S. G. E., Battin T. J., Prosser J. I., Schimel J. P., Whitman W. B., Hallin S. (2010). The ecological coherence of higher bacterial taxonomic ranks. Nat. Rev. Microbiol. 8, 523–529 10.1038/nrmicro2367 [DOI] [PubMed] [Google Scholar]
- Pommier T., Canback B., Riemann L., Bostrom K. H., Simu K., Lundberg P., Tunlid A., Hagstrom A. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol. Ecol. 16, 867–880 10.1111/j.1365-294X.2006.03189.x [DOI] [PubMed] [Google Scholar]
- Prosser J. I., Bohannan B. J. M., Curtis T. P., Ellis R. J., Firestone M. K., Freckleton R. P., Green J. L., Green L. E., Killham K., Lennon J. J., Osborn A. M., Solan M., van der Gast C. J., Young J. P. W. (2007). Essay – the role of ecological theory in microbial ecology. Nat. Rev. Microbiol. 5, 384–392 10.1038/nrmicro1643 [DOI] [PubMed] [Google Scholar]
- Raes J., Bork P. (2008). Systems microbiology – timeline – molecular eco-systems biology: towards an understanding of community function. Nat. Rev. Microbiol. 6, 693–699 10.1038/nrmicro1935 [DOI] [PubMed] [Google Scholar]
- Ranjard L., Dequiedt S., Jolivet C., Saby N. P. A., Thioulouse J., Harmand J., Loisel P., Rapaport A., Fall S., Simonet P., Joffre R., Boure N. C. P., Maron P. A., Mougel C., Martin M. P., Toutain B., Arrouays D., Lemanceau P. (2010). Biogeography of soil microbial communities: a review and a description of the ongoing French national initiative. Agron. Sustain. Dev. 30, 359–365 10.1051/agro/2009033 [DOI] [Google Scholar]
- Rasche F., Lueders T., Schloter M., Schaefer S., Buegger F., Gattinger A., Hood-Nowotny R. C., Sessitsch A. (2009). DNA-based stable isotope probing enables the identification of active bacterial endophytes in potatoes. New Phytol. 181, 802–807 10.1111/j.1469-8137.2008.02744.x [DOI] [PubMed] [Google Scholar]
- Roesch L. F., Fulthorpe R. R., Riva A., Casella G., Hadwin A. K. M., Kent A. D., Daroub S. H., Camargo F. A. O., Farmerie W. G., Triplett E. W. (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 1, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch D. B., Halpern A. L., Sutton G., Heidelberg K. B., Williamson S., Yooseph S., Wu D. Y., Eisen J. A., Hoffman J. M., Remington K., Beeson K., Tran B., Smith H., Baden-Tillson H., Stewart C., Thorpe J., Freeman J., Andrews-Pfannkoch C., Venter J. E., Li K., Kravitz S., Heidelberg J. F., Utterback T., Rogers Y. H., Falcon L. I., Souza V., Bonilla-Rosso G., Eguiarte L. E., Karl D. M., Sathyendranath S., Platt T., Bermingham E., Gallardo V., Tamayo-Castillo G., Ferrari M. R., Strausberg R. L., Nealson K., Friedman R., Frazier M., Venter J. C. (2007). The sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5, 398–431 10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutgers M., Schouten A. J., Bloem J., van Eekeren N., de Goede R. G. M., Akkerhuis G., van der Wal A., Mulder C., Brussaard L., Breure A. M. (2009). Biological measurements in a nationwide soil monitoring network. Eur. J. Soil Sci. 60, 820–832 10.1111/j.1365-2389.2009.01163.x [DOI] [Google Scholar]
- Salles J. F., Poly F., Schmid B., Le Roux X. (2009). Community niche predicts the functioning of denitrifying bacterial assemblages. Ecology 90, 3324–3332 10.1890/09-0188.1 [DOI] [PubMed] [Google Scholar]
- Schleifer K. H. (2009). Classification of bacteria and archaea: past, present and future. Syst. Appl. Microbiol. 32, 533–542 10.1016/j.syapm.2009.09.002 [DOI] [PubMed] [Google Scholar]
- Setala H., McLean M. A. (2004). Decomposition rate of organic substrates in relation to the species diversity of soil saprophytic fungi. Oecologia 139, 98–107 10.1007/s00442-003-1478-y [DOI] [PubMed] [Google Scholar]
- Sogin M. L., Morrison H. G., Huber J. A., Mark Welch D., Huse S. M., Neal P. R., Arrieta J. M., Herndl G. J. (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120 10.1073/pnas.0605127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J. T. (2006). The bacterial species dilemma and the genomic-phylogenetic species concept. Philos. Trans. R. Soc. B Biol. Sci. 361, 1899–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland W. J., Clout M., Cote I. M., Daszak P., Depledge M. H., Fellman L., Fleishman E., Garthwaite R., Gibbons D. W., De Lurio J., Impey A. J., Lickorish F., Lindenmayer D., Madgwick J., Margerison C., Maynard T., Peck L. S., Pretty J., Prior S., Redford K. H., Scharlemann J. P. W., Spalding M., Watkinson A. R. (2010). A horizon scan of global conservation issues for 2010. Trends Ecol. Evol. (Amst.) 25, 1–7 10.1016/j.tree.2009.10.003 [DOI] [PubMed] [Google Scholar]
- Tamames J., Abellan J. J., Pignatelli M., Camacho A., Moya A. (2010). Environmental distribution of prokaryotic taxa. BMC Microbiol. 10, 85. 10.1186/1471-2180-10-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden M. G. A., Bardgett R. D., van Straalen N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310 10.1111/j.1461-0248.2007.01139.x [DOI] [PubMed] [Google Scholar]
- Verstraete W., Wittelbolle L., Heylen K., Vanparys B., de Vos P., van de Wiele T., Boon N. (2007). Microbial resource management: the road to go for environmental biotechnology. Eng. Life Sci. 7, 117–126 10.1002/elsc.200620176 [DOI] [Google Scholar]
- von Mering C., Hugenholtz P., Raes J., Tringe S. G., Doerks T., Jensen L. J., Ward N., Bork P. (2007). Quantitative phylogenetic assessment of microbial communities in diverse environments. Science 315, 1126–1130 10.1126/science.1133420 [DOI] [PubMed] [Google Scholar]
- Wagner M. (2009). Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu. Rev. Microbiol. 63, 411–429 10.1146/annurev.micro.091208.073233 [DOI] [PubMed] [Google Scholar]
- Whiteley A. S., Manefield M., Lueders T. (2006). Unlocking the “microbial black box” using RNA-based stable isotope probing technologies. Curr. Opin. Biotechnol. 17, 67–71 10.1016/j.copbio.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Whitman W. B., Coleman D. C., Wiebe W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583 10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmes P., Bond P. L. (2009). Microbial community proteomics: elucidating the catalysts and metabolic mechanisms that drive the earth's biogeochemical cycles. Curr. Opin. Microbiol. 12, 310–317 10.1016/j.mib.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Wittebolle L., Marzorati M., Clement L., Balloi A., Daffonchio D., Heylen K., De Vos P., Verstraete W., Boon N. (2009). Initial community evenness favours functionality under selective stress. Nature 458, 623–626 10.1038/nature07840 [DOI] [PubMed] [Google Scholar]
- Wohl D. L., Arora S., Gladstone J. R. (2004). Functional redundancy supports biodiversity and ecosystem function in a closed and constant environment. Ecology 85, 1534–1540 10.1890/03-3050 [DOI] [Google Scholar]
- Woyke T., Tighe D., Mavromatis K., Clum A., Copeland A., Schackwitz W., Lapidus A., Wu D. Y., McCutcheon J. P., McDonald B. R., Moran N. A., Bristow J., Cheng J. F. (2010). One bacterial cell, one complete genome. PLoS ONE 5, e10314. 10.1371/journal.pone.0010314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. Z., Deng Y., Luo F., He Z. L., Tu Q. C., Zhi X. Y. (2010). Functional molecular ecological networks. MBio 1, e00169–10 10.1128/mBio.00169-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J. Z., Kang S., Schadt C. W., Garten C. T. (2008). Spatial scaling of functional gene diversity across various microbial taxa. Proc. Natl. Acad. Sci. U.S.A. 105, 7768–7773 10.1073/pnas.0709016105 [DOI] [PMC free article] [PubMed] [Google Scholar]