Abstract
Targeting of messenger RNAs (mRNAs) in neuron processes relies on cis-acting regulatory elements, the nature of which is poorly understood. Here, we report that approximately 30% of the best-known dendritic mRNAs contain a guanine (G)–quadruplex consensus in their 3′-untranslated region. Among these mRNAs, we show by using RNA structure probing that a G–quadruplex is present in the mRNAs of two key postsynaptic proteins: PSD-95 and CaMKIIa. The G–quadruplex structure is necessary and sufficient for the potent and fast localization of mRNAs in cortical neurites and this occurs in a metabotropic glutamate receptor-responsive manner. Thus, G–quadruplex seems to be a common neurite localization signal.
Keywords: mRNA trafficking, Guanine–quadruplex, PSD-95, CaMKIIa, FMRP
Introduction
The transport of messenger RNAs (mRNAs) within cells is important for the establishment and conservation of cell polarity (Hirokawa et al, 2009; Holt & Bullock, 2009). In neurons, the transport of specific mRNAs away from the soma to the dendrites or the axons has been shown to occur for several genes translated locally at synapses or at axonal growth cones, respectively (Besse & Ephrussi, 2008; Andreassi & Riccio, 2009; Martin & Ephrussi, 2009). Transport and localization of mRNAs to the dendrites is one preliminary step in the control of local protein synthesis, a key process for the lasting activity-dependent changes that take place at synapses (Sutton & Schuman, 2006; Bramham & Wells, 2007; Costa-Mattioli et al, 2009). The targeting of mRNAs in neurites is proposed to rely on protein motors (for example, kinesins) complexed with diverse trans-acting factors and RNA-binding proteins that interact with the RNA to be transported through cis-element, usually located in the 3′-untranslated region (UTR) of the mRNAs (Hirokawa et al, 2009; Mili & Macara, 2009). However, the mechanism by which this process occurs is poorly understood. In a few cases, a small sequence could be defined as the cis-acting element involved in mRNA localization. Among the best-known examples are the ‘zipcode’ of β-actin mRNA—a 54-nucleotide-long AC-rich element bound by zip code binding protein 1 (Huttelmaier et al, 2005)—and the cytoplasmic polyadenylation element (CPE), a UUUUAU sequence or similar, found in CaMKIIa and other mRNAs that is bound by CPEB1 (Huang et al, 2003). However, these elements have been shown to be important for localization in few mRNAs; thus, for most mRNAs localized in dendrites, the cis-acting signal involved remains unknown.
Fragile-X mental retardation protein (FMRP) was recently proposed to be a trans-acting factor in the activity-dependent localization of several neuronal mRNAs (Dictenberg et al, 2008). The number of mRNAs affected by this localization is unknown and the way in which FMRP interacts with these localized mRNAs has also not been investigated. Previously, we and others have shown that FMRP binds to intramolecular guanine (G)–quadruplex (or G-quartet) RNA structures (Darnell et al, 2001; Schaeffer et al, 2001). Intramolecular G–quadruplexes are self-assembling, stable nucleic-acid structures that are proposed to occur in a number of genes, both at the DNA and RNA levels, and are involved in several regulatory processes including telomere stabilization, promoter control, recombination and translation control (Lipps & Rhodes, 2009). We tested whether the G–quadruplex structure, the canonical binding site of FMRP, could represent a cis-acting signal for localization of mRNAs in the processes of mouse cortical neurons. We first report the presence of G–quadruplex consensus in a high proportion of dendritic mRNAs and provide experimental evidence of the presence of G–quadruplex in the 3′-UTR of two well-known, dendritically localized mRNAs (PSD-95 and CaMKIIa) to show that G–quadruplex are neurite localization elements.
Results and Discussion
G–quadruplex structures in PSD-95 and CaMKIIa mRNAs
We have previously shown that the intramolecular G–quadruplex consensus (G⩾3N0−6)4 is a potent binding site for the FMRP protein (Schaeffer et al, 2001; Didiot et al, 2008). By using the bioinformatic tool QGRS-mapper (Kikin et al, 2008) and visual inspection, we found that about one-third of the most well-characterized dendritic RNAs (11/34) have a (G⩾3N0−6)4 consensus in their 3′-UTR that is conserved between humans and rodents (Table 1; supplementary Table S1 online). This ratio seems to be significantly higher than that found in the general mRNA population, in which less than 5% of mRNAs from a 500-member random pool have this consensus in their 3′-UTR (supplementary Table S2 online). From the short list of dendritic RNAs, two mRNAs—PSD-95 and CaMKIIa—that have been proposed to be controlled by FMRP (Todd et al, 2003; Zalfa et al, 2007; Dictenberg et al, 2008) were retained for further investigation.
Table 1. Presence of G–quadruplex consensus (G⩾3N0−6)4 in the 3′-UTR of known dendritic RNAs.
| Gene | G–quadruplex in 3′-UTR * | G–quadruplex sequence |
|---|---|---|
| β-Actin | − | |
| APP | + | GGGGCGGGTGGGGAGGGG |
| Arc (Arg3.1) | − | |
| BDNF | + | GGGGATGGGGGATGGGGGG |
| Calmodulin | − | |
| α-CaMKII | + | GGGGGGGCGGGTGGGATGGGAAGAAGGGG |
| CREB | − | |
| Dendrin | + | GGAGGGCAGGGTAGGGTAGGG |
| FMRP | − | |
| G protein gamma 7 sub. | + | GGGAGGGCTGGGGCTTCGGG |
| GABA-A-R-g | − | |
| GLRA1 | + | GGGGGAGGCTGGGAGAGGGGAACGTGGG |
| InsP3R1 | − | |
| Ligatin | − | |
| MAP1b | − | |
| MAP2 | − | |
| MRG15 (MORF4L1) | − | |
| Neurogranin | − | |
| Neurofilament protein 68 | − | |
| NEURL | + | GGGATGGGCCAGGGCCCTGGGTGGG |
| NMDAR1 | − | |
| Pcp2(L7) | − | |
| PEP19 | − | |
| PSD95 (DLG4) | + | GGGGAAAAGGGAGGGATGGGTCTAGGGAGTGGGAAATGCGGGAGGGAGGGTGGGGGGCAGGGGTCGGG |
| RGS5 | − | |
| SAPAP4 (DLGAP4) | + | GGGCGGGGTAGGGGAGGGCAGGGG |
| Shank1 | + | GGGAGGGTCACGGGAGGGGGGAGGGG…GGGGTTGGGGAGGGTGTAGGGGGTGGGGGTGGGGGTGGAAGGAGAGGGGAGAGGGAAGGGGGAGGG |
| Shank3 | + | GGGGCGGGAGGTGCCGGGGGTGGGG…GGGGGGAGGGGGGAGACATTGGG…GGGGTGGGGGGCCCTGGG |
| TrkB | − | |
| Vasopressin (AVP) | − | |
| Oxytocin (OXT) | − | |
| BC1 | − | |
| Ribosomal RNAs | − | |
| tRNAs | − | |
| *G–quadruplex fulfilling consensus (G⩾3N0−6)4 predicted with QGRS-mapper (Kikin et al, 2008) and visual inspection are conserved between human and rodents. References and additional data are provided in supplementary Table S1 online. | ||
| UTR, untranslated region. | ||
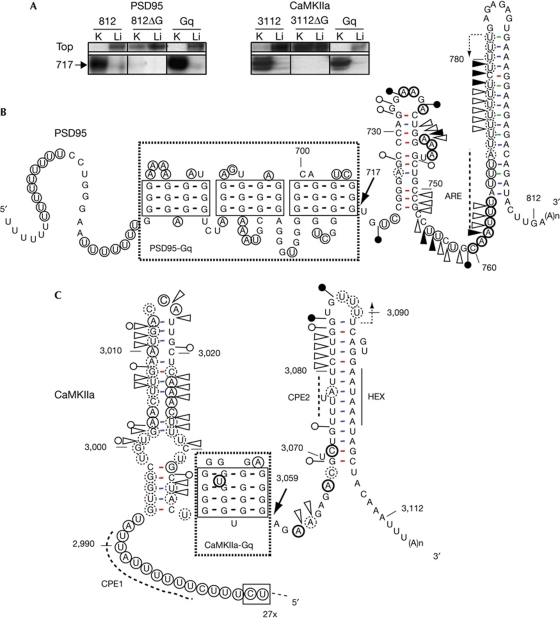
The presence of G–quadruplex structures in PSD-95 and CaMKIIa 3′-UTRs was indicated by a reverse transcription (RT) assay (Fig 1A; supplementary Fig S1A online) and confirmed by a T1 RNase protection assay (supplementary Fig S1B online). The presence of G–quadruplex structures also agrees with the reactivity pattern of the RNAs submitted to enzymatic and chemical probes that enabled the establishment of secondary structure models of the 3′-UTRs (Fig 1B,C). In both mRNAs, the G–quadruplex motif occurred within 100 nucleotides of the 3′ end. Apart from the G–quadruplex, a single-stranded U-rich region upstream and a paired U-rich region downstream (forming a hairpin with polyadenylation signal) were the only features in common between the secondary structures of the 3′ ends of the two mRNAs. The most noticeable difference between the mRNAs is that three independent G–quadruplex can occur simultaneously in PSD-95 instead of only one in CaMKIIa. The fact that the G–quadruplex-forming sequences of the two mRNAs are highly conserved among mammals (supplementary Fig S2 online) suggests that they are biologically important.
Figure 1.
G–quadruplex structures are present in the 3′-UTR of PSD-95 and CaMKIIa mRNAs. (A) K+-dependent RT arrest in PSD-95 (human) and CaMKIIa (rat) 3′-UTR full-length transcripts (PSD95-812 and CaMKIIa-3112, respectively). Numbering from the first 3′-UTR nucleotide. K, 150 mM KCl; Li, 150 mM LiCl. Deletion of Gq-forming sequences eliminates the RT arrests (PSD95-812ΔG, CaMKIIa-3112ΔG), whereas Gq-forming sequences only (PSD95–Gq, CaMKIIa–Gq) are sufficient to arrest RT. Secondary structure models of the 3′ end of PSD95 (B) and CaMKIIa (C) mRNAs deduced from enzymatic (RNase T1 and V1) and chemical probing (CMCT and DMS) as described in the supplementary Methods online. Triangles and circles connected to the line are RNase V1 and T1 cleavages, respectively. Circles around nucleotides are CMCT (G,U) or DMS (A,C) modifications. Symbol thickness is proportional to the degree of reactivity (strong, moderate or weak). Nucleotides without symbol are unreactive or undetermined. Dotted arrows indicate the start position of RT performed to analyse the RNA structure. The thick arrows indicate the strong stops of RT at the 3′ end of Gq structure, as in (A). The regions framed with thick dotted lines are the minimal Gq-forming sequence. Polyadenylation site (HEX), CPE as defined in Huang et al (2003), AU-rich sites (ARE) as proposed in Zalfa et al (2007), are indicated. ARE, AU-rich element; CMCT, 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulphonate; CPE, cytoplasmic polyadenylation element; DMS, dimethyl sulphate; Gq, guanine–quadruplex; HEX, hexamer motif; mRNA, messenger RNA; RT, reverse transcription; UTR, untranslated region.
G–quadruplex sequences are neurite-targeting elements
Both PSD-95 and CaMKIIa mRNAs have been shown to localize to dendrites (Burgin et al, 1990; Zalfa et al, 2007) and the 3′-UTR of CaMKIIa has been shown to be involved in the localization process (Mayford et al, 1996; Mori et al, 2000; Huang et al, 2003). To assess the potential role of the G–quadruplex motif in dendritic RNA localization, we used the λN-GFP system (Daigle & Ellenberg, 2007), which allows visualization of the subcellular localization of a reporter RNA in fixed or live cells by GFP labelling (Fig 2A). By using this system, we visualized the localization of a reporter mRNA bearing the 54-nucleotide dendritic targeting element ‘zipcode’ of the β-actin mRNA (Kislauskis et al, 1994) in the neurites of living (supplementary Fig S3 online) or fixed (data not shown) neurons. DsRed gene encoding reporter mRNAs bearing the full-length 3′-UTR of PSD-95 or CaMKIIa were transfected into primary cortical neuron cultures. Both reporter mRNAs localized in the processes of neurons (Fig 2B, PSD95-812, CaMKIIa-3112) as efficiently as the zipcode-bearing mRNA, confirming the role of CaMKIIa 3′-UTR and demonstrating that of PSD-95 3′-UTR. This localization was confirmed in parallel by in situ hybridization of a multilabelled fluorescent DNA probe on the same mRNAs (Fig 2C).
Figure 2.
Involvement of PSD95 and CaMKIIa G–quadruplex in neurite targeting. (A) λN-GFP system principle and RNA-reporter constructs scheme tested with their neurite localization quantification (percentage of cells showing GFP signal along the neurites among those expressing GFP (n=100, ±standard error). (B) Visualization of reporter mRNAs in cortical neurons with the λN-GFP system (green). DsRed protein expressed from pDsRed-Mono-4BB accumulates in the cytoplasm and acted as a transfection control (red). Scale bar, 20 μm. Images are projections of 10 × 1.5 μm confocal sections. The deletion of the Gq sequences eliminates neurite localization (ΔG constructs), whereas the Gq sequence only (Gq) restores it. (C) Visualization of PSD95 and CaMKIIa 3′-UTRs-containing reporter mRNAs in cortical neurons with fluorescent in situ hybridization using a multilabelled Alexa-488 oligodeoxynucleotide probe (green). DsRed protein is expressed from pDsRed-Mono-4BB as a transfection control (red). Cells were treated as described in (C), but without using p4lambda-N22-3mEGFP-M9. Scale bar, 20 μm. DAPI and merged images are presented in supplementary Fig S4 online. CPE, cytoplasmic polyadenylation element; DAPI, 4,6-diamidino-2-phenylindole; GFP, green fluorescent protein; Gq, guanine–quadruplex; mRNA, messenger RNA; UTR, untranslated region.
To assess their role in neurite RNA localization, the G–quadruplex-forming sequences of PSD-95 and CaMKIIa 3′-UTRs were deleted (boxed sequences in Fig 1B,C). The absence of G–quadruplex structure in the T7 transcripts of 3′-UTRs of both mRNAs was indicated by the disappearance of the K+-dependent RT arrests (Fig 1A, PSD95-812ΔG, CaMKIIa-3112G). The G–quadruplex deletions caused a loss of more than 90%Δ of the signal in neurites (Fig 2), suggesting that the deleted sequences are necessary for the localization process. The localization defect was apparently not due to a change in RNA stability, as the levels of RNA with or without G–quadruplex were unchanged (supplementary Fig S5A online). As the amount of mRNA present in neurites represents only a fraction of the total amount of neuronal mRNA, one cannot totally exclude the fact that those mRNAs being transported in neurites are also protected by the presence of the G-quadruplex at their 3′ end. In fact, active transport and degradation protection mechanisms could be coupled as indicated by the colocalization of mRNA decay components, such as Dcp1, with RNA transport particles (Cougot et al, 2008). In addition, PSD-95 mRNA has been proposed to be protected from degradation by FMRP in hippocampal neurons through AU-rich elements (AREs; Zalfa et al, 2007). Interestingly, these AREs are located close to the G–quadruplex motifs, and could explain how FMRP contributes to RNA stabilization in the hippocampus (Fig 1B); however, this stabilization does not take place in cortical neurons (Zalfa et al, 2007) in which the present study was conducted. Furthermore, neither the length of the poly-A tail, nor the overall translation level of mRNAs bearing the same 3′-UTRs were affected by the deletions (supplementary Fig S5B,C online).
We then aimed to demonstrate their role as neurite-targeting elements (NTEs), and to evaluate the contribution of additional nearby cis-acting elements such as the CPE element, two copies of which are close to the G–quadruplex structure in CaMKIIa mRNA (Fig 1C) and have been proposed to be involved in dendritic localization (Huang et al, 2003). The G–quadruplex-forming sequences alone (Fig 1B,C) were tested out of their natural 3′-UTR context in the heterologous 3′-UTR sequence of pDsRed-Mono-4BB in front of the SV40 polyadenylation signal. The presence of a G–quadruplex structure was confirmed in these mRNAs (Fig 1A, PSD95-Gq, CaMKIIa-Gq) and their neurite-targeting was comparable to that of the full-length UTRs (Fig 2), indicating that the G–quadruplex structures are sufficient for the neurite localization. For CaMKIIa mRNA, this could suggest a small contribution of CPEs to transport in the processes of cortical neurons (this study) compared with hippocampal ones (Huang et al, 2003). Alternatively, G–quadruplex and CPEs could function together in CaMKIIa mRNA, as the efficiency of localization is not fully recovered in CaMKIIa-G–quadruplex (55% compared with 82% in full-length CamKIIa 3′ UTR), and the G–quadruplex deletion in CaMKIIa-3112ΔG might have perturbed the folding of CPEs, masking their contribution to mRNA localization.
The λN-GFP system allowed the visualization of fast anterograde movement (⩾1 μm/s) of λN-GFP-labelled PSD-95 3′-UTR-RNA particles along neurites, similar to what was previously described for active mRNA transport (Fig 3A; supplementary movie S1 online).
Figure 3.
Transport of mRNAs bearing a Gq structure is a fast, dynamic process that is sensitive to group 1 glutamate receptor activation, and a ‘generic’ Gq is sufficient for localization. (A) Time-lapse confocal visualization of the movement of PSD-95 3′-UTRs-containing reporter mRNAs in cortical neurons with the λN-GFP system. Scale bar, 20 μm. Recording at 1 frame/s is presented in supplementary Movie 1 online. (B) The neurite targeting of PSD-95 reporter mRNA is stimulated by 50-μM DHPG treatment for 20 min. PSD95-812ΔG RNA is not sensitive to such treatment. Cells were treated as described in Fig 2, except that p4λN22-3mEGFP-M9 plasmid was absent and reporter RNA was visualized by in situ hybridization of a fluorescent DNA probe; DAPI staining is shown. Histogram shows quantification of neurite fluorescence of FISH signal (arbitrary units), n=15; paired Student's t-test (unstimulated compared with stimulated) *P=0.00014 (PSD95-812), P=0.35 (PSD95-812ΔG). (C) Gq detection by RT in the 3′-UTR of pDsRBB-‘variants’, as described in Fig 1A. The sequence of transcripts is given in supplementary Fig S6 online. (D) Visualization of variant 3′-UTR-containing reporter mRNAs in cortical neurons with λN-GFP. Neurons were transfected with mRNA reporter (pDsRBB-variants) and fluorescence reporter (p4λN22-3mEGFP-M9) plasmids and imaged as shown in Fig 2. The secondary structure model of the variant mRNA reporters (variable region) is shown with quantifications as in Fig 2A. DAPI, 4,6-diamidino-2-phenylindole; DHPG, 3,5-dihydroxyphenylglycine hydrate; FISH, fluorescence in situ hybridization; Gq, guanine–quadruplex; mRNA, messenger RNA; RT, reverse transcription; UTR, untranslated region.
The dendritic localization of CaMKIIa mRNAs had been shown to be enhanced by group 1 metabotropic glutamate receptor agonist 3,5-dihydroxyphenylglycine hydrate (DHPG; Dictenberg et al, 2008). We reproduced this effect with CaMKIIa 3′-UTR (CaMKIIa-3112 +DHPG, supplementary Fig S4 online) and we demonstrated it with PSD-95 3′-UTR (Fig 3B). Remarkably, the deletion of the G–quadruplex in both mRNAs eliminated this effect. Taken together, these data support the idea that G–quadruplex-forming regions in the 3′-UTR of PSD-95 and CaMKIIa are true NTEs that contribute to the activity-dependent transport of mRNAs in neurites.
The G–quadruplex structure is essential for NTE function
To demonstrate that the G–quadruplex structure itself (rather than just its G-rich content) is important for NTE function, structural variants of G–quadruplexes and G-rich-non-G–quadruplex sequences were tested. A positive correlation was established between the ability of a given sequence to adopt a G–quadruplex structure, as defined by its efficiency in arresting RT elongation (Fig 3C), and its efficiency as a NTE (Fig 3D). Thus, all G–quadruplex-forming sequences are efficient NTEs (with efficiencies between 35 and 80%), whereas G-rich sequences that do not promote G–quadruplex formation have no or little NTE activity (Fig 3D, ‘G-rich’ and ‘no-Gq’). A two-layer Gq motif is not detected in vitro and does not have NTE activity. Among the Gq variants, the three-layer G–quadruplex G3A is a better NTE (80%±5%) than the four-layer G–quadruplex G4A (60%±5%), although it is more stable (compare the intensity of RT stops in Fig 3C, top), suggesting that G–quadruplex stability is not the sole determinant of NTE efficiency. Similarly, a G–quadruplex with intervening adenines has relatively higher NTE efficiency than one with uridines. In all examples, the G–quadruplexes are present within a hundred nucleotides of the 3′ end. It remains to be determined whether specific rules apply regarding the position of the G–quadruplex motif within a given mRNA. Together, these data allow us to propose that a G–quadruplex structure is a NTE independently of its surrounding sequences, and to add the G–quadruplex structure to the list of RNA structures with NTE function. The finding that about one-third of the best-known dendritic mRNAs contain a putative G–quadruplex in their 3′-UTR suggests that G–quadruplexes would be among the most prevalent neuronal NTEs.
NTEs are proposed to affect transport by their interaction with trans-acting RNA-binding proteins. Both PSD-95 and CaMKIIa mRNAs have been shown to be regulated by, and to interact with, FMRP (Todd et al, 2003; Muddashetty et al, 2007; Zalfa et al, 2007; Dictenberg et al, 2008). Given this, FMRP seemed to be a good candidate for the G–quadruplex-dependent transport. We verified that FMRP binds directly to the G–quadruplex motif of both PSD-95 and CaMKIIa mRNAs in vitro, and that binding is impaired by deletion of the G–quadruplex and enhanced by K+, compared with Li+ (supplementary Fig S7A,B online). These data support a direct role for FMRP in the G–quadruplex-dependent localization of mRNAs. However, whereas the deletion of G-quartet motif within an mRNA 3′-UTR had a strong effect on its dendritic localization, the lack of FMRP had a more subtle effect. Indeed, the basal transport of reporter mRNAs bearing CaMKIIa or PSD-95 3′-UTRs was unaffected by the absence of FMRP (data not shown) and only the mGluR-triggered transport of the RNA bearing CaMKIIa was diminished (supplementary Fig S7C online), as reported previously (Dictenberg et al, 2008; Kao et al, 2010), whereas the RNA bearing PSD-95 3′-UTR was not significantly changed. Thus, FMRP is not the main trans-acting factor of the G–quadruplex-dependent transport, but it would contribute to their activity-dependent transport. Although FMRP has similar binding affinities in vitro for CaMKIIa and PSD-95 3′-UTRs, our data also suggest that it might have distinct roles in the transport of its different target mRNAs. Whether the intrinsic nature of the G-quartet (for example, a triple three-tetrad structure in PSD-95 as opposed to a single four-tetrad one in CaMKIIa) influences this efficiency remains to be determined.
Finally, because of their cation sensitivity, G–quadruplex structures might have a unique role in activity-dependent transport by ‘sensing’ the cationic intracellular content in relation to neuronal activity status. The modulation of G–quadruplex stability by cation variations could regulate the binding of trans-acting factors, which could then affect the transport level of the mRNAs (such as that observed after mGluR activation). FMRP—the binding of which to G–quadruplexes is modulated by the concentration of monovalent cations—could have a role in this regulatory process. Future work will be required to test this hypothesis and to identify the trans-acting factors that, along with FMRP, contribute to the formation of the G–quadruplex-transport particles, and to determine the other neuronal mRNAs that are subjected to this transport.
Methods
Plasmid constructs. PSD-95 3′-UTR (812-nucleotide human) was amplified from placental DNA by RT-PCR. pCDNA 3.1 vector containing CaMKIIa 3′-UTR (3112-nucleotide rat) was a gift from Y. Mori (Osaka Medical College). p4λN22-3mEGFP-M9-NLS and pmRFP-4-boxB-βactin-zipcode were obtained from J. Ellenberg (EMBL). Details on plasmid constructions, mutagenesis and primers can be found in the supplementary Methods online.
Neuron culture, transfection and stimulation. Primary cortical neurons were prepared from C57Bl/6 mouse embryos at embryonic day 17 and grown on polylysine-coated 24-well plates or 35-mm glass-bottom dishes (Iwaki) in Neurobasal Medium (NBM) supplemented with 1 × B27, 0.5 mM L-glutamine and 100 IU/ml penicillin/streptomycin at 37 °C with 5% CO2. Neurons were transfected at day 7 with 0.1 μg p4λN-3mGFP and 0.25 μg pDsRBB, and 1.2 μl Lipofectamine 2000 (Invitrogen) in 400 μl NBM. The medium was replaced after 3 h with a 1:1 (v:v) mixture of conditioned and fresh NBM. After 20 h, the neurons were fixed for 20 min in 4% paraformaldehyde 1 × PBS (in situ hybridization), imaged directly live (spinning disk confocal) or lysed (luciferase assays, RNA preparation). Where indicated, treatment with 50 μM (S)-DHPG (Tocris) was carried out for 20 min before fixation.
RT assays and RNA structure probing. Primer extensions with RT were performed as described in Schaeffer et al (2001) with 5 pmol of T7 RNA transcripts renatured for 1 min at 90 °C and 5 min at 20 °C in 20 μl of 50 mM Hepes, pH 7.5, 5 mM MgCl2, 150 mM KCl, NaCl or LiCl and 2 μg tRNA. RNA structure probing was performed as described in Moine et al (1998), see details in the supplementary information online.
Fluorescence in situ hybridization and imaging. A DNA probe against DsRed coding region was produced by RT with amine-modified adenines and labelled with AlexaFluor-488 succinimidyl-ester (Invitrogen) as recommended by the manufacturer. Fluorescence in situ hybridization was performed as described in Didiot et al (2008). Fluorescence intensity was measured with Image J on three transfections of cortical neurons for each RNA construct tested, and with three mice each (supplementary information online).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank M.-C. Didiot and S. Pannetier for technical assistance; B. Bardoni, Y. Trottier, N. Daigle, E. Bertrand and L. Lacroix for helpful suggestions; and M. Koch, M. Boeglin and P. Kessler for help with microscopy analyses. M.S. was supported by a grant from Fondation de la Recherche Médicale to J.L.M. This project benefited from ANR and Fondation Jérôme Lejeune grants to H.M.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andreassi C, Riccio A (2009) To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol 19: 465–474 [DOI] [PubMed] [Google Scholar]
- Besse F, Ephrussi A (2008) Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol 9: 971–980 [DOI] [PubMed] [Google Scholar]
- Bramham CR, Wells DG (2007) Dendritic mRNA: transport, translation and function. Nat Rev Neurosci 8: 776–789 [DOI] [PubMed] [Google Scholar]
- Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, Kelly PT (1990) In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J Neurosci 10: 1788–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N (2009) Translational control of long-lasting synaptic plasticity and memory. Neuron 61: 10–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N, Bhattacharyya SN, Tapia-Arancibia L, Bordonne R, Filipowicz W, Bertrand E, Rage F (2008) Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J Neurosci 28: 13793–13804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N, Ellenberg J (2007) LambdaN-GFP: an RNA reporter system for live-cell imaging. Nat Methods 4: 633–636 [DOI] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB (2001) Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell 107: 489–499 [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ (2008) A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell 14: 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiot MC, Tian Z, Schaeffer C, Subramanian M, Mandel JL, Moine H (2008) The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res 36: 4902–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S (2009) Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 10: 682–696 [DOI] [PubMed] [Google Scholar]
- Holt CE, Bullock SL (2009) Subcellular mRNA localization in animal cells and why it matters. Science 326: 1212–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YS, Carson JH, Barbarese E, Richter JD (2003) Facilitation of dendritic mRNA transport by CPEB. Genes Dev 17: 638–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH (2005) Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature 438: 512–515 [DOI] [PubMed] [Google Scholar]
- Kao DI, Aldridge GM, Weiler IJ, Greenough WT (2010) Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc Natl Acad Sci USA 107: 15601–15606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikin O, Zappala Z, D’Antonio L, Bagga PS (2008) GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res 36: D141–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislauskis EH, Zhu X, Singer RH (1994) Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol 127: 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipps HJ, Rhodes D (2009) G-quadruplex structures: in vivo evidence and function. Trends Cell Biol 19: 414–422 [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A (2009) mRNA localization: gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Baranes D, Podsypanina K, Kandel ER (1996) The 3′-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc Natl Acad Sci USA 93: 13250–13255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mili S, Macara IG (2009) RNA localization and polarity: from A(PC) to Z(BP). Trends Cell Biol 19: 156–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moine H, Ehresmann B, Ehresmann C, Romby P (1998) In RNA Structure and Function (Simons, R.W., Grunberg-Manago, M. eds), pp 77–115. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M (2000) Two cis-acting elements in the 3′ untranslated region of alpha-CaMKII regulate its dendritic targeting. Nat Neurosci 3: 1079–1084 [DOI] [PubMed] [Google Scholar]
- Muddashetty RS, Kelic S, Gross C, Xu M, Bassell GJ (2007) Dysregulated metabotropic glutamate receptor-dependent translation of AMPA receptor and postsynaptic density-95 mRNAs at synapses in a mouse model of fragile X syndrome. J Neurosci 27: 5338–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H (2001) The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J 20: 4803–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM (2006) Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127: 49–58 [DOI] [PubMed] [Google Scholar]
- Todd PK, Mack KJ, Malter JS (2003) The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95. Proc Natl Acad Sci USA 100: 14374–14378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalfa F et al. (2007) A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat Neurosci 10: 578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.