Abstract
We describe a new mechanism by which CTG tract expansion affects myotonic dystrophy (DM1). Changes to the levels of a panel of RNAs involved in muscle development and function that are downregulated in DM1 are due to aberrant localization of the transcription factor SHARP (SMART/HDAC1-associated repressor protein). Mislocalization of SHARP in DM1 is consistent with increased CRM1-mediated export of SHARP to the cytoplasm. A direct link between CTG repeat expression and SHARP mislocalization is demonstrated as expression of expanded CTG repeats in normal cells recapitulates cytoplasmic SHARP localization. These results demonstrate a role for the inactivation of SHARP transcription in DM1 biology.
Keywords: MINT, myotonic dystrophy, SHARP, Spen, transcription
Introduction
Myotonic dystrophy (DM1) is an autosomal dominant, multisystem disorder resulting from the expansion of a CTG repeat sequence located in the 3′ untranslated region of DMPK. Expression of expanded CTG tracts has been shown to result in the aberrant splicing of a set of physiologically important RNAs in DM1 (Ranum & Cooper, 2006). Mouse models that recreate DM1-specific defects in key splice regulators do not recapitulate all features of the disease, suggesting that other events have roles in DM1 pathophysiology (Kanadia et al, 2003; Ho et al, 2005). Evidence for the ectopic expression of NKX2–5, a transcriptional regulator, in DM1 muscle supports this observation (Yadava et al, 2008).
SMART/HDAC1-associated repressor protein (SHARP) is a human transcription factor and a component of a multiprotein complex that is known to function as both an activator and a repressor of transcription (Shi et al, 2001; Sierra et al, 2004; Feng et al, 2007). The Drosophila homologue of SHARP, SPEN, has been shown to enhance the neurodegenerative phenotype resulting from the expression of expanded CUG-repeat-encoding RNAs (Mutsuddi et al, 2004). In agreement with the Drosophila studies, we demonstrate here that SHARP is an important factor that mediates CUG toxicity in DM1.
Results And Discussion
Altered steady-state RNA levels in DM1
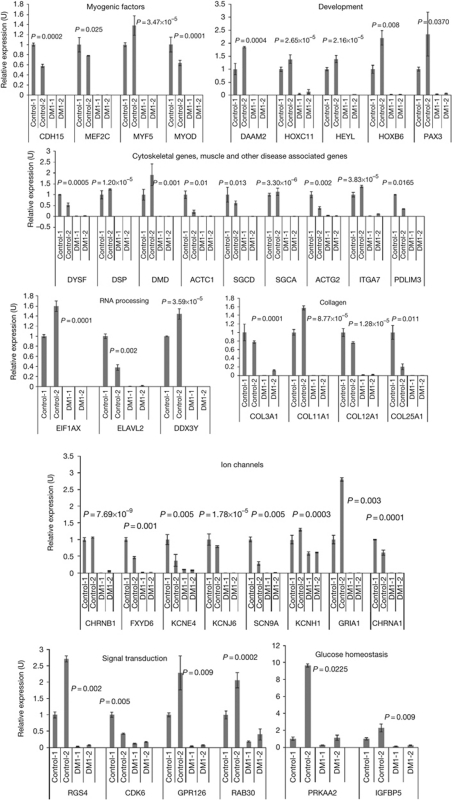
To examine the differences in RNA steady-state levels between normal and DM1 myoblasts, total RNA from two normal and two DM1 myoblast lines that show characteristics of DM1—including expanded CTG tracts, CUG-RNA foci, aberrant splicing and activation of protein kinase-Cα (PKCα; supplementary Fig S1 online)—were analysed in duplicate using Affymetrix human exon 1.0 ST (sense target) arrays. Differential expression analysis (analysis of variance P<0.01; fold change >5, <−5) showed that the majority (76%) of the changes in DM1 myoblasts reflect a reduction in steady-state RNA levels, compared with normal myoblasts. Gene ontology enrichment analyses of the modulated RNAs that show decreased levels in DM1 myoblasts demonstrate that several of these RNAs are involved in muscle development and function (P<1.1 × 10−6). Of these, 65 RNAs encoding myogenic factors, muscle structural proteins, ion channels and proteins involved in glucose homeostasis, development, RNA processing and signal transduction were chosen for validation. Of the 65 RNAs tested, changes in RNA steady-state levels of 39 RNAs were confirmed by real-time PCR analysis (Fig 1) in both DM1 myoblast lines. These spectra of changes are specific to DM1, as they are not observed in facioscapulohumeral muscular dystrophy (FSHD) myoblasts (supplementary Table S1 online). These results demonstrate that DM1 myoblasts have different steady-state levels of several RNAs that participate in muscle development and function, compared with normal myoblasts.
Figure 1.
Quantification of steady-state RNA levels in normal (control) and DM1 myoblasts. Synthesized complementary DNAs were subjected to real-time PCR analysis to measure the expression levels of RNAs in normal and DM1 myoblasts, with GAPDH RNA as an internal control. Error bars (±) represent standard deviation (n=3). P-values were determined by paired Student's t-test. DM1, myotonic dystrophy.
Previous studies have demonstrated that either functional inactivation of the alternative splice factors MBNL1 and MBNL2 or elevated levels of CUG-BP1 or hnRNP H can lead to splice defects in DM1 myoblasts (Paul et al, 2006). As these changes could affect gene expression, we tested whether the transcript levels of the genes that demonstrate reduced steady-state levels in DM1 myoblasts occur as a consequence of altered expression of the splice regulators implicated in DM1. Thus, we depleted MBNL1 or MBNL2 by using a short-interfering RNA (siRNA)-mediated approach or overexpressed CUG-BP1 and hnRNP H in normal myoblasts—both of which result in aberrant splicing of IR—and performed reverse transcription–polymerase chain reaction (RT–PCR) analyses to measure transcript levels of the genes downregulated in DM1 (supplementary Fig S2 online). This analysis demonstrates that the changes in steady-state levels of this set of RNA transcripts are not a result of the altered levels of the four splice factors implicated in DM1 pathophysiology, suggesting that other mechanisms might have roles in altering RNA levels in DM1 myoblasts.
SHARP inactivation results in DM1 RNA defects
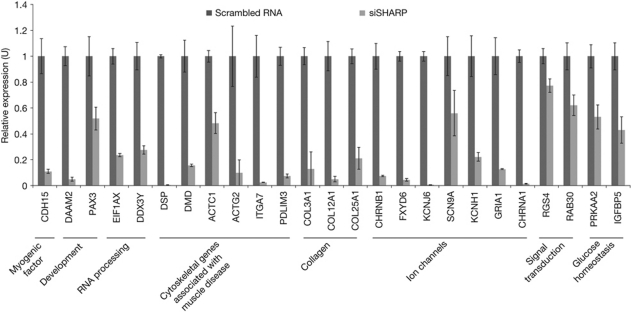
Loss-of-function alleles of MBL (a Drosophila homologue of muscleblind) and the transcription factor SPEN (a Drosophila homologue of SHARP) enhance the Drosophila eye phenotype on expression of expanded CUG repeat tracts (Mutsuddi et al, 2004). We therefore hypothesized that SHARP contributes to the changes in RNA steady-state levels in DM1. To test this hypothesis, SHARP was depleted (approximately 60–80%) in normal myoblasts using the cognate siRNAs, and transcript levels of the 39 RNAs downregulated in DM1 were measured by real-time PCR analysis (Fig 2). These analyses showed downregulation of 25 of the 39 transcripts examined (approximately 64%) in SHARP-depleted myoblasts. Statistical analysis of these data predicts with a 95% confidence that 49–77% of the genes that show decreased levels in DM1 myoblasts would also be reduced in normal myoblasts in which SHARP is inactivated. By contrast to the RNAs that show decreased steady-state levels, nine randomly chosen RNAs that demonstrate increased steady-state levels in DM1 myoblasts were not found to be regulated by SHARP (data not shown).
Figure 2.
SHARP depletion in normal myoblasts recapitulates DM1 RNA steady-state changes. Real-time PCR analysis shows that decreased levels of SHARP in normal myoblasts results in reduced steady-state levels in 25 of the 39 RNAs examined. Error bars (±) represent standard deviation (n=3). P-values determined by paired Student's t-test are listed in supplementary Table S5 online. DM1, myotonic dystrophy; SHARP, SMART/HDAC1-associated repressor protein.
SHARP was identified as a transcriptional co-repressor for steroid hormone receptors; however, the mouse homologue of SHARP (MINT) has been shown to function as a co-activator by stimulating Runx2-dependent activation of the osteocalcin promoter (Sierra et al, 2004). As our analyses show that functional inactivation of SHARP results in downregulation of several transcripts, it is likely that, similar to the osteoblast system, SHARP functions as a transcriptional activator by forming a complex with yet-to-be-identified activators in human myoblasts.
SHARP is normally spliced in DM1 myoblasts
As aberrant RNA splicing has a role in the development of DM1, we tested whether SHARP RNA was abnormally spliced in DM1 myoblasts. The splice pattern of SHARP RNAs in normal and DM1 myoblasts demonstrated no significant differences when examined by RT–PCR analysis. Subsequently, we measured RNA transcript levels of SHARP RNAs and found no difference in the levels of SHARP RNA isoforms, which encode nuclear localization signals, in normal and DM1 myoblasts (supplementary Fig S3 online). Therefore, inactivation of SHARP transcription in DM1 does not seem to be a consequence of transcript downregulation or aberrant splicing.
SHARP is aberrantly localized in DM1 muscle
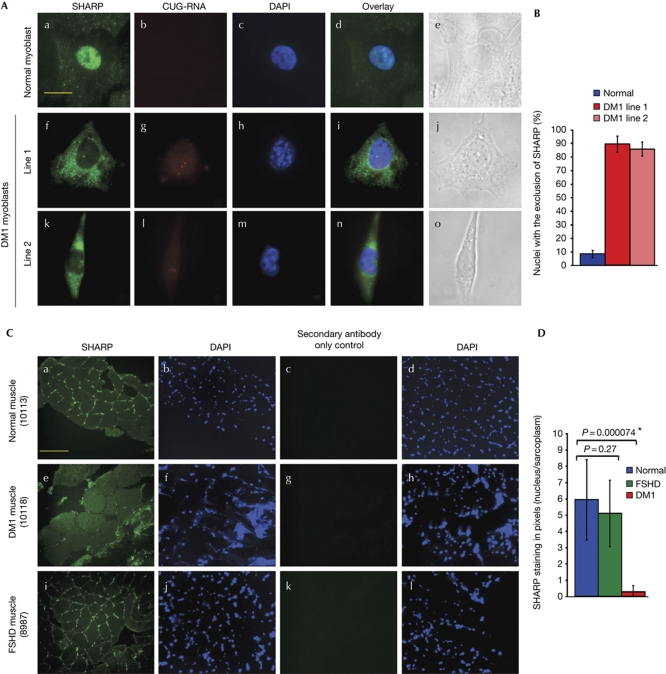
Previous studies have shown that SHARP binds to stem–loop-structured RNA hairpins (Hatchell et al, 2006). Therefore, we examined whether SHARP inactivation occurs as a consequence of aberrant sequestration in CUG foci. To determine whether SHARP is recruited to CUG-RNA foci in DM1 myoblasts, we performed fluorescence in situ hybridization (FISH) using a Cy3-conjugated (CAG)10 oligonucleotide probe to detect CUG-RNA foci, followed by immunofluorescence analysis using polyclonal SHARP antibodies (a gift from Dr Eric Fearon; Feng et al, 2007). In normal human myoblasts, SHARP is diffusely localized, almost exclusively within the nucleus (Fig 3A). In DM1 myoblasts, we found that SHARP co-localized with a small percentage of the foci (approximately 10%; data not shown). Thus, sequestration in CUG foci is not the primary mechanism for SHARP inactivation in DM1 cells. Unexpectedly, we observed that SHARP was predominantly found in the cytoplasm in approximately 90% of DM1 myoblasts (Fig 3A). Aberrant SHARP localization in DM1 myoblasts was confirmed with both polyclonal SHARP antibodies developed by Fearon and colleagues and with a commercially available polyclonal SHARP antibody (Bethyl). The specificity of SHARP antibodies has been shown previously (Feng et al, 2007), and established for the antibodies obtained from Bethyl by using cells in which SHARP was depleted by its cognate siRNAs (supplementary Fig S4A online). These results demonstrate that although a small fraction of SHARP sequesters in CUG foci, functional inactivation of SHARP is primarily a result of mislocalization to the cytoplasm in DM1 myoblasts. By contrast, SHARP localizes exclusively in the nuclei of FSHD myoblast cultures (supplementary Fig S4B online).
Figure 3.
SHARP is abnormally distributed in DM1. (A) Endogenous SHARP is shown in green (a,f,k). Expanded CUG tracts were detected by fluorescence in situ hybridization (red; g,l). Nuclear DAPI staining of normal and DM1 myoblasts (c,h,m) are shown. Merged images demonstrate that endogenous SHARP localizes primarily in the nucleus in normal myoblasts (d) and predominantly in the cytoplasm in the two DM1 myoblast lines (i,n). Bright-field images (e,j,o) are shown. Scale bar, 15 μM. (B) Graphical representation of nuclei with exclusion of SHARP in 90 normal and DM1 cells. Error bars (±) represent standard deviation (n=3). (C) Cross-sections of normal human (10113), DM1 (10118) and FSHD (8987) muscles were immunostained with SHARP antibodies followed by DAPI staining and visualized using × 20 magnification. Endogenous SHARP is shown in green (a,e,i). A staining control in which only the secondary antibody was used is shown in panels c,g,k. Nuclei are stained with DAPI (b,f,j,d,h,l). Scale bar, 175 μm. (D) Graphical representation of nuclear/sarcoplasmic SHARP staining in pixels of normal, FSHD and DM1 muscle is shown. Error bars (±) represent standard deviation (n=8). P-values were determined by paired Student's t-test. Quantification method is described in supplementary Fig S4C online. DAPI, 4,6-diamidino-2-phenylindole; DM1, myotonic dystrophy; FSHD, facioscapulohumeral muscular dystrophy; SHARP, SMART/HDAC1-associated repressor protein.
Notably, myoblast and fibroblast cell lines, which showed small CUG foci, did not show DM1 splice defects or SHARP mislocalization. Thus, these results demonstrate that significant accumulation of toxic CUG-RNA may be required for the DM1 phenotype to manifest. To test whether the mislocalization of SHARP is observed in vivo, we used normal, severely affected muscle biopsies from patients with DM1 and FSHD. Immunohistochemical analysis of muscle cross-sections demonstrate that SHARP localizes in the nuclei of normal and FSHD sections, whereas diminished nuclear signals in conjunction with increased sarcoplasmic staining for SHARP is observed in DM1 sections (Fig 3B, supplementary Fig S4C online).
Although functional inactivation of MBNL1 and MBNL2 or overexpression of CUG-BP1 and hnRNP H did not recapitulate the defects in RNA steady-state levels observed in DM1 myoblasts, we tested whether these changes could alter the localization of SHARP in DM1. Thus, we depleted either MBNL1 or MBNL2, or overexpressed CUG-BP1 or hnRNP H, in normal myoblasts and examined SHARP localization by immunofluorescence. Results of these experiments indicate that mislocalization of SHARP is not a consequence of altered levels of MBNL1, MBNL2, CUG-BP1 and hnRNP H (supplementary Fig S5A,B online). Aberrant activation of PKCα has been considered to stabilize CUG-BP1 in DM1 (Kuyumcu-Martinez et al, 2007). To test whether PKCα has a role in the mislocalization of SHARP in DM1, we functionally inactivated PKCα in DM1 myoblasts using the cognate siRNAs. However, depletion of PKCα did not rescue the aberrant localization of SHARP in DM1 myoblasts, suggesting that other mechanisms are involved in this process (supplementary Fig S5C online).
Leptomycin B reverses SHARP mislocalization in DM1
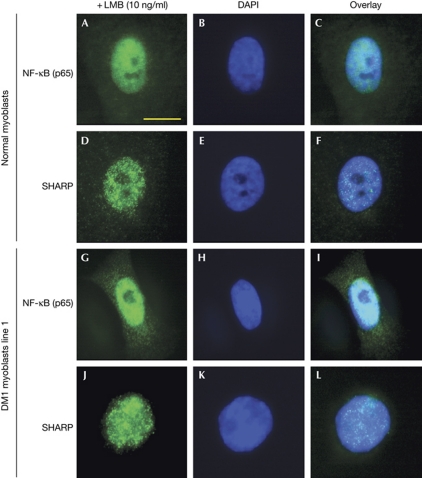
To understand the mechanism for nuclear exclusion of SHARP in DM1 myoblasts, we determined whether this phenomenon reflects global defects in nuclear–cytoplasmic transport or nuclear retention. We therefore examined by immunofluorescence the localization of two shuttling proteins, nuclear factor-κB and ACTIN, and one nuclear transcription factor, SP1, in normal and DM1 myoblasts. All three proteins showed normal localization in DM1 myoblasts, suggesting that the aberrant localization of SHARP was specific to this protein (supplementary Fig S6A,B online). Next, to examine the mechanism of SHARP transport between the nucleus and the cytoplasm, we treated normal and DM1 myoblasts with a CRM1-mediated nuclear export inhibitor, leptomycin B (LMB). Myoblasts were treated with LMB at a concentration of 10–100 ng/ml, and SHARP localization was determined after 24 h by immunofluorescence microscopy using SHARP antibodies. Nuclear retention of SHARP is observed in DM1 myoblasts at a concentration of LMB as low as 10 ng/ml, demonstrating that inhibition of chromosome maintenance region 1 (CRM1)-mediated nucleo-cytoplasmic transport blocks relocalization of SHARP to the cytoplasm in DM1 cells (Fig 4). These results indicate that mislocalization of SHARP to the cytoplasm is not caused by failure to transport newly synthesized SHARP into the nucleus, but is consistent with an increased CRM1-mediated nuclear export of SHARP. Notably, CRM1-mediated transport is not altered per se, as nucleo-cytoplasmic shuttling of nuclear factor-κB, which is mediated by CRM1, is not affected in DM1 cells (supplementary Fig S6A,B online). Thus, expression of expanded CUG repeats may result in the modification of SHARP, which could alter its interaction with CRM1 to allow SHARP mislocalization to the cytoplasm.
Figure 4.
Nuclear retention of SHARP in DM1 myoblasts treated with leptomycin B. Localization of endogenous NF-κB and SHARP proteins in normal and DM1 myoblasts treated with leptomycin B (10 ng/ml for 24 h) is shown in green. NF-κB (A–C, G–I) and SHARP (D–F, J–L) localize predominantly in the nucleus of normal and DM1 myoblasts after leptomycin B treatment. Scale bar, 10 μM. DAPI, 4,6-diamidino-2-phenylindole; DM1, myotonic dystrophy; LMB, leptomycin B; NF-κB, nuclear factor-κB; SHARP, SMART/HDAC1-associated repressor protein.
Nuclear SHARP does not rescue DM1 RNA defects
To determine whether forced relocalization of SHARP to the nucleus is sufficient to rescue DM1 transcription defects, DM1 myoblasts were treated with LMB (10 ng/ml). Although LMB treatment allowed nuclear localization of SHARP in DM1 cells, robust rescue of the expression of five randomly chosen transcripts at 24 h post-LMB treatment was not observed (supplementary Fig S7A,B online). As forced retention of SHARP in the nucleus does not rescue RNA steady-state defects, the expression of CTG tracts might facilitate modification of SHARP, which in turn might inhibit its interaction with other factors that have a role in its normal transcription. Consistent with this interpretation, overexpression of SHARP did not rescue the RNA steady-state defects in DM1 myoblasts (supplementary Fig S7C,E online). The nature and potential modification of the SHARP complex in response to CTG tract expression remains to be determined and is the focus of current investigations.
CTG tract expression results in SHARP mislocalization
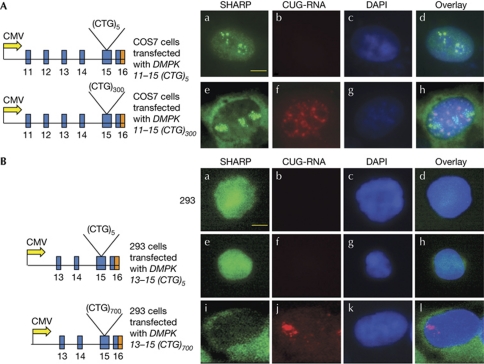
To test whether aberrant localization of SHARP in DM1 is directly linked to the expression of RNA with expanded CUG repeats, we expressed constructs encoding either DMPK exons 11–15 or DMPK exons 13–15 with 5, 300 or 700 CTG repeats (CTG5, 300 or 700) in COS7 and HEK293 cells. Analysis of transfected cells by immunofluorescence microscopy shows that nuclear localization of SHARP is not altered by the expression of constructs encoding five CTG repeats (DMPK-(CTG)5; Fig 5A,B). By contrast, expression of 300 and 700 CTG repeats (DMPK-(CTG)300 and DMPK-(CTG)700), resulted in significant relocalization of SHARP to the cytoplasm in 80–90% of cells (Fig 5A,B). Thus, in accordance with the Drosophila studies, our results demonstrate that SHARP is an important factor that is controlled by the expression of the toxic CUG-RNA and is responsible for maintaining the steady-state levels of several RNAs related to muscle development and function.
Figure 5.
Expression of expanded CTG repeats results in mislocalization of SHARP in COS7 and HEK293 cells. (A) The constructs DMPK 11–15(CTG)5 and DMPK 11–15(CTG)300 were expressed in COS7 cells. Endogenous SHARP (green, a,e) and CUG foci (red, b,f) were visualized 48 h post-transfection. Nuclei were stained with DAPI (c,g). Merged images (d,h) demonstrate that SHARP localizes primarily within the nucleus of cells expressing the DMPK 11–15(CTG)5 and predominantly in the cytoplasm of cells expressing expanded (CTG)300 tracts. (B) Similar results were obtained in HEK293 cells transfected with DMPK 13–15(CTG)5 and DMPK 13–15(CTG)700. Endogenous SHARP is shown in green (a,e,i), CUG foci are shown in red (b,f,j), nuclei stained with DAPI (c,g,k) and merged images (d,h,l) are shown. Scale bar, 20 μM. CMV, cytomegalovirus; DAPI, 4,6-diamidino-2-phenylindole; DM1, myotonic dystrophy; SHARP, SMART/HDAC1 associated repressor protein.
SIX5 and MBNL3 depletion recapitulates DM1 RNA defects
The expansion of CTG repeat tracts in DM1 leads to condensed chromatin and the transcriptional repression of an adjacent homeobox gene, SIX5, in DM1 (Otten & Tapscott, 1995; Thornton et al, 1997). In addition, MBNL3, a member of the muscleblind-protein family, has been previously shown to sequester in CUG-RNA foci (Fardaei et al, 2002). We therefore tested whether MBNL3 and SIX5 depletion recapitulate the RNA steady-state defects observed in DM1, by knocking down the expression of MBNL3 and SIX5 in normal myoblasts (supplementary Fig S8A,C online). The levels of MBNL3 and SIX5 downregulation achieved in our experiments (approximately 90 and 60%, respectively) are probably within the range of the levels of these proteins in DM1 cells, as MBNL3 is expressed at low levels and strongly sequestered in CUGexp RNA aggregates, and only the SIX5 allele adjacent to the expanded CTG tract is epigenetically silenced. In MBNL3-depleted myoblasts, 15 of the 39 transcripts tested (approximately 38%) were downregulated, whereas 28 of the 39 transcripts tested (approximately 72%) were downregulated in SIX5-depleted myoblasts (supplementary Fig 8B,D online). In contrast to SHARP inactivation, MBNL3 and SIX5 depletion resulted in an increase in steady-state levels of 13 of the 39 (33%) and 3 of the 39 (8%) transcripts that are downregulated in DM1 myoblasts, respectively (supplementary Fig 8B,D online). As inactivation of MBNL3 and SIX5 does not alter SHARP localization (supplementary Fig 8E online), these results demonstrate that in addition to SHARP, MBNL3 and SIX5 could either function in conjunction or independently to modulate the expression of genes that are altered in DM1.
In summary, our study demonstrates that SHARP, MBNL3 and SIX5 are important factors that are responsible for maintaining the steady-state level of several RNAs related to muscle development and function in myoblasts. Thus, deregulation of SHARP/MBNL3/SIX5-dependent transcription contributes to the pathology of DM1, in conjunction with altered splice-site selection. As the results detailed in this study elucidate a new mechanism by which DM1 biology is regulated, our data demonstrate that in addition to the reversal of splice alterations, correction of transcriptional defects will be required in order to fully rescue DM1 pathology in vivo.
Methods
DNA constructs, siRNAs and microarray analysis. hnRNP H (L22009) and CUG-BP1 (NM_006560) expression plasmids, siRNAs against MBNL1, MBNL2 and the scrambled siRNA are described in Paul et al (2006). SHARP was amplified by PCR and cloned into Flag-pcDNA3.1. SHARP complementary DNA was a gift from Dr R. Evans. siRNAs for SHARP (Feng et al, 2007), MBNL3 (catalogue number M-017983-01-0005) and SIX5 (catalogue number M-015373-00-0005) were purchased from Dharmacon. Microarray results have been deposited in the Gene Expression Omnibus database (accession number: GSE22498).
Cell culture. Normal (line 2) and DM1 myoblasts were a gift from Dr Charles Thornton. Normal myoblasts (line 1) and FSHD myoblasts (GM17869) were purchased from Lonza and Coriell Cell Repository, respectively. Normal and DM1 myoblasts were immortalized by SV-40. Normal (10113), FSHD (8987) and DM1 (10118) muscle cross-sections were obtained from the EuroBioBank and Telethon network of Genetic Biobanks. Cell culture and transfections were carried out as described by Paul et al (2006).
RNA extraction, RT–PCR and real-time PCR. RNA extraction, RT–PCR and real-time PCR analyses were carried out as described previously (Dansithong et al, 2008). Sequences of primers are shown in supplementary Tables S2,S4 online.
Western blot, FISH and immunofluorescence. Western blot, FISH and immunofluorescence analyses were carried out as described in Paul et al (2006) and Dansithong et al (2008).
Immunohistochemistry. For immunostaining, cryostat cross-sections were fixed with ice-cold methanol for 5 min and permeabilized by using 0.3% Triton X-100/3% bovine serum albumin (BSA) in PBS for 30 min. The tissues were rinsed with PBS and preblocked with 1%, 2% and 3% BSA in PBS for 5 min each followed by incubation with SHARP primary antibody (Bethyl) at 4 °C overnight. Sections were rinsed with PBST and washed with 1%, 2% and 3% BSA in PBS for 5 min each and then incubated in secondary antibody for 1 h at room temperature. Sections were washed as described above and mounted using mounting solution with DAPI (Vector Labs).
LMB treatment. Normal and DM1 myoblasts were treated with LMB (Sigma-Aldrich) at a concentration of 10 ng/ml in media, as well as vehicle control for 24 h at 37 °C with 5% CO2. Cells were fixed using 4% paraformaldehyde before immunofluorescence analysis, as described previously (Dansithong et al, 2008).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 NS50861-04 & R01 NS060839-03) to S.R. and L.C. We thank Dr C. Thornton for the normal and DM1 myoblasts; Drs I. Holt and G.E. Morris for MBNL1 and MBNL2 antibodies; and the EurobioBank and the Telethon Network of Genetic Biobanks (GTB07001F) for muscle sections. Cell and Tissue Imaging Core, USC Norris Comprehensive Cancer Center was used for confocal microscopy.
Footnotes
The authors declare that they have no conflict of interest.
References
- Dansithong W et al. (2008) Cytoplasmic CUG RNA foci are insufficient to elicit key DM1 features. PLoS ONE 3: e3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD (2002) Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet 11: 805–814 [DOI] [PubMed] [Google Scholar]
- Feng Y, Bommer GT, Zhai Y, Akyol A, Hinoi T, Winer I, Lin HV, Cadigan KM, Cho KR, Fearon ER (2007) Drosophila split ends homologue SHARP functions as a positive regulator of Wnt/β-catenin/T-cell factor signaling in neoplastic transformation. Cancer Res 67: 482–491 [DOI] [PubMed] [Google Scholar]
- Hatchell EC et al. (2006) SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol Cell 22: 657–668 [DOI] [PubMed] [Google Scholar]
- Ho TH, Bundman D, Armstrong DL, Cooper TA (2005) Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet 14: 1539–1547 [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS (2003) A muscleblind knockout model for myotonic dystrophy. Science 302: 1978–1980 [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA (2007) Increased steady-state levels of CUG-BP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell 28: 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsuddi M, Marshall CM, Benzow KA, Koob MD, Rebay I (2004) The spinocerebellar ataxia 8 noncoding RNA causes neurodegeneration and associates with staufen in Drosophila. Curr Biol 14: 302–308 [DOI] [PubMed] [Google Scholar]
- Otten AD, Tapscott SJ (1995) Triplet repeat expansion in myotonic dystrophy alters the adjacent chromatin structure. Proc Natl Acad Sci USA 92: 5465–5469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Dansithong W, Kim D, Rossi J, Webster NJ, Comai L, Reddy S (2006) Interaction of muscleblind, CUG-BP1 and hnRNP H proteins in DM1-associated aberrant IR splicing. EMBO J 25: 4271–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA (2006) RNA-mediated neuromuscular disorders. Annu Rev Neurosci 29: 259–277 [DOI] [PubMed] [Google Scholar]
- Shi Y, Downes M, Xie W, Kao HY, Ordentlich P, Tsai CC, Hon M, Evans RM (2001) Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev 15: 1140–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra OL, Cheng SL, Loewy AP, Charlton-Kachigian N, Towler DA (2004) MINT, the Msx2 interacting nuclear matrix target, enhances Runx2-dependent activation of the osteocalcin fibroblast growth factor response element. J Biol Chem 279: 32913–32923 [DOI] [PubMed] [Google Scholar]
- Thornton CA, Wymer JP, Simmons Z, McClain C, Moxley RT III (1997) Expansion of the myotonic dystrophy CTG repeat reduces expression of the flanking DMAHP gene. Nat Genet 16: 407–409 [DOI] [PubMed] [Google Scholar]
- Yadava RS, Frenzel-McCardell CD, Yu Q, Srinivasan V, Tucker AL, Puymirat J, Thornton CA, Prall OW, Harvey RP, Mahadevan MS (2008) RNA toxicity in myotonic muscular dystrophy induces NKX2-5 expression. Nat Genet 40: 61–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.