Abstract
The cellular response to double-strand breaks (DSBs) in DNA is a complex signalling network, mobilized by the nuclear protein kinase ataxia-telangiectasia mutated (ATM), which phosphorylates many factors in the various branches of this network. A main question is how ATM regulates DSB repair. Here, we identify the DNA repair enzyme polynucleotide kinase/phosphatase (PNKP) as an ATM target. PNKP phosphorylates 5′-OH and dephosphorylates 3′-phosphate DNA ends that are formed at DSB termini caused by DNA-damaging agents, thereby regenerating legitimate ends for further processing. We establish that the ATM phosphorylation targets on human PNKP—Ser 114 and Ser 126—are crucial for cellular survival following DSB induction and for effective DSB repair, being essential for damage-induced enhancement of the activity of PNKP and its proper accumulation at the sites of DNA damage. These findings show a direct functional link between ATM and the DSB-repair machinery.
Keywords: ATM, DNA damage response, double strand break repair, polynucleotide kinase/phosphatase, protein phosphorylation
Introduction
The cellular response to DNA double-strand breaks (DSBs) is an extensive signalling network that activates DNA repair and special cell-cycle checkpoints, as well as modulating several metabolic processes (Ciccia & Elledge, 2010). The key activator of this network is the ataxia-telangiectasia mutated (ATM) protein (Derheimer & Kastan, 2010). ATM is missing or inactive in patients with the genetic disorder ataxia-telangiectasia, which is characterized by cerebellar degeneration, immunodeficiency, radiation sensitivity, chromosomal instability and cancer predisposition (Lavin, 2008). In response to DSBs, ATM is activated and phosphorylates a range of substrates in various branches of the DNA-damage response (DDR; Derheimer & Kastan, 2010). ATM is a member of the phosphoinositide-3-kinase-like protein kinase (PIKK) family (Lempiainen & Halazonetis, 2009), which includes two other main factors in the DDR: the catalytic subunit of DNA-dependent protein kinase (DNA-PK) and ATM- and Rad3-related protein (ATR). These protein kinases preferentially phosphorylate serine or threonine residues that are followed by glutamine (S/TQ).
Mammalian polynucleotide kinase/phosphatase (PNKP) is a 57-kDa protein that has a key role in single-strand breaks (SSBs) and DSB repair by non-homologous end joining (reviewed in Weinfeld et al, 2011). PNKP is a DNA-end processor and harbours 5′ DNA kinase and 3′ DNA phosphatase activities. PNKP dephosphorylates ‘illegitimate’ 3′-P ends and phosphorylates 5′-OH ends formed during DNA breakage, converting them back to legitimate substrates for further processing.
Here, we show that upon treatment with DSB-inducing agents, human PNKP (hPNKP) is rapidly phosphorylated on two sites in an ATM-dependent and DNA-PK-independent manner. Furthermore, these phosphorylations are important for DNA-damage-induced enhancement of the activity of PNKP and its proper recruitment to the damaged sites, and subsequently for timely DSB repair and optimal cellular survival following induction of DNA damage.
Results
Identification of PNKP as a potential PIKK substrate
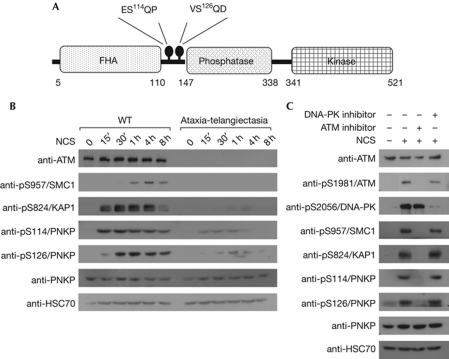
ATM, DNA-PK and ATR share substrate specificity, primarily recognizing S/TQ motifs. Antibodies against phospho-S/TQ sites are usually not specific for a single substrate and can therefore potentially immunoprecipitate various proteins that are phosphorylated on these sites. In such a proteomic screen for new PIKK substrates, an antibody against a phosphopeptide (GSPQE) was used to immunoprecipitate phosphorylated proteins from mouse tissues after whole-body X-irradiation. The immune complexes were resolved on polyacrylamide gels and protein bands—the presence of which seemed to be enhanced in the irradiated tissues—were excised and subjected to mass-spectrometric analysis. Mouse PNKP was identified in one of these bands (supplementary Fig S1A online). Mouse PNKP contains two SQ motifs, one of which, at S114, is contained within a potential antigenic epitope (GSPQ) corresponding to the antibody that was used in the screen. Alignment of the mouse and human PNKP amino-acid sequences showed that although hPNKP contains two SQ motifs at S114 and S126, only the one at S114 is conserved between mice and humans (supplementary Fig S1B online). Both serine residues are located at the ‘intradomain’ between the forkhead-associated and the phosphatase domains of the protein (Fig 1A). DNA-damage-induced phosphorylation of S114 and S126 of hPNKP was also suggested by a high-throughput proteomic screen for ATM/ATR substrates (Matsuoka et al, 2007). Therefore, polyclonal antibodies were raised against the presumed phosphorylated forms of S114 and S126 of hPNKP.
Figure 1.
ATM-mediated phosphorylation of human polynucleotide kinase/phosphatase in response to DNA-damage induction. (A) Scheme of human PNKP domains. The protein contains a forkhead-associated (FHA) domain and phosphatase and kinase domains. ATM targets, S114 and S126, are located between the FHA and phosphatase domains. (B) Kinetics and ATM-dependence of phosphorylation of S114 and S126 of hPNKP in wild-type (WT) and ataxia-telangiectasia lymphoblasts treated with 100 ng/ml neocarzinostatin (NCS) and harvested at the indicated time points. Cellular extracts were subjected to western blotting analysis with the indicated antibodies. (C) Effect of the ATM inhibitor KU-55933 and DNA-PK inhibitor NU-7441 at 10 μM on hPNKP phosphorylation in U2-OS cells. One hour after addition of the inhibitor, cells were treated with 100 ng/ml of NCS for 1 h. Cellular extracts were subjected to western blotting analysis with the indicated antibodies. ATM, ataxia-telangiectasia mutated; DNA-PK, DNA-dependent protein kinase; PNKP, polynucleotide kinase/phosphatase.
DNA damage-induced phosphorylation of hPNKP in cells
To examine the specificity of the antibodies and the possible phosphorylation of hPNKP in response to DNA-damage induction, vectors expressing carboxy-terminal Flag-tagged hPNKP were generated in wild-type and mutant forms. In the mutant proteins, the serine residues at positions 114 and 126 were substituted by alanines, thereby eliminating the corresponding phosphorylation sites. These constructs were expressed in HEK293T cells that were subsequently treated with the radiomimetic drug neocarzinostatin (NCS). The ectopic proteins were immunoprecipitated using a Flag antibody. Western blotting analysis of the immune complexes indicated that, following DNA damage induction, the ectopic hPNKP was phosphorylated in cells on both serine residues and that the antibodies recognized these phosphorylations (supplementary Fig S2 online). While this work was in progress, two high-throughput proteomic screens for DNA damage-induced phosphorylations carried out in our and another lab identified S114 of hPNKP as a target of such phosphorylation (Bennetzen et al, 2010; Bensimon et al, 2010).
hPNKP is an ATM target
We examined the DNA-damage-induced phosphorylation of endogenous PNKP in human wild-type and ataxia-telangiectasia lymphoblastoid cell lines. Both S114 and S126 were rapidly phosphorylated after NCS treatment, and these phosphorylations began to decline within several hours (Fig 1B), a time-course that is typical for many ATM substrates. Importantly, in ATM-deficient ataxia-telangiectasia cells, PNKP phosphorylations were markedly diminished, suggesting that this process is ATM-dependent (Fig 1B). Moreover, pretreatment with the ATM inhibitor KU-55933 (Hickson et al, 2004) prevented these phosphorylations, whereas the DNA-PK inhibitor NU-7441 (Leahy et al, 2004) had no detectable effect (Fig 1C), further suggesting that under these conditions both phosphorylations are ATM dependent.
hPNKP phosphorylation in response to genotoxic stress was further studied by monitoring U2-OS cells for 8 h after treatment with NCS, ionizing radiation, ultraviolet radiation, hydrogen peroxide, the topoisomerase-I inhibitor camptothecin and the replication inhibitor hydroxyurea. PNKP phosphorylation was observed by following all of these treatments (supplementary Fig S3 online), but the variation in its kinetics and intensity suggested that the primary inducers of PNKP phosphorylation were DSBs rather than SSBs. We concluded that PNKP is an ATM target, activated in response to DSB induction.
PNKP phosphorylation sites are important for DSB repair
To investigate the functional significance of DNA-damage-induced PNKP phosphorylation, endogenous PNKP was replaced by ectopic Flag-tagged PNKP in wild-type or phospho-mutant versions. Briefly, endogenous PNKP was stably knocked down in A549 cells using stable expression of a short-hairpin RNA (shRNA), and the cells were complemented by stable expression of wild-type or phospho-mutant Flag-tagged PNKPs encoded by small-interfering RNA-resistant complementary DNAs (cDNAs; Fig 2A). Further analysis indicated that successful protein replacement had been achieved and the expected pattern of phosphorylation was obtained (Fig 2B).
Figure 2.
Loss of polynucleotide kinase/phosphatase or its ataxia-telangiectasia mutated phosphorylation sites is associated with increased sensitivity to double-strand-break-inducing agents. (A) Replacement of endogenous human polynucleotide kinase/phosphatase (hPNKP) in A549 cells by ectopic Flag-tagged wild type (WT) or phosphorylation mutants of hPNKP: S114A, S126A and S114A/S126A. Shown is a western blotting analysis demonstrating the PNKP levels in cell lines with different versions of PNKP. (B) Western blotting analysis of the same cell lines was carried out after treatment with 0.5 μg/ml neocarzinostatin (NCS) for 30 min, with the indicated antibodies. (C) Clonogenic survival of A549 cells transfected with small-interfering RNAs against hPNKP, ATM or green fluorescent protein (GFP) after treatment with varying doses of NCS. The experiment was carried out in triplicate. Bars represent s.e.m. The western blotting analysis on the right shows the extent of protein depletion. (D) Similar experiments with cells in which endogenous hPNKP was replaced by ectopic WT or phospho-mutants of hPNKP, and cells in which ATM had been knocked down. The cells were treated with varying doses of NCS (left) or ionizing radiation (right). ATM, ataxia-telangiectasia mutated; DSB, double-strand break.
The sensitivity of these cells to the cytotoxic effect of DSB-inducing agents was measured using clonogenic survival assays after treatment with NCS or ionizing radiation. Loss of PNKP made the cells hypersensitive to both agents, with intermediate sensitivity between that of wild-type and ATM-deficient cells (Fig 2C,D); complementation of cells devoid of endogenous PNKP with ectopic, wild-type protein restored normal sensitivity (Fig 2D). Importantly, cells expressing non-phosphorylatable PNKP in three versions (S114A, S126A and S114A/S126A) showed variable sensitivities in the range between cells lacking PNKP and cells expressing the wild-type protein (Fig 2D), indicating that all three phospho-mutant protein combinations are not fully able to function in the DSB response.
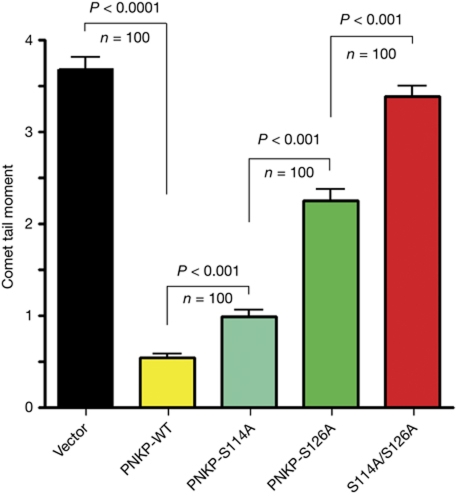
Cellular hypersensitivity to DSB-inducing agents is suggestive of defective DSB repair. A preliminary indication of DSB repair deficiency in cells expressing phospho-mutant PNKP was obtained by following the disappearance of nuclear foci of γH2AX or 53BP1 protein after ionizing radiation treatment (not shown). We then used pulsed-field gel electrophoresis, as described previously (Lundin et al, 2005), as a direct assay of DSBs. The results (supplementary Fig S4 online) reflected the delay in DSB repair in PNKP-deficient cells, but the resolution of this method was not sufficiently fine to discern the differences in DSB repair efficiency associated with the different phospho-mutant versions of PNKP. We therefore used the neutral comet assay, which monitors the presence of DSBs in single cells by microscopic detection of DNA migration in gel (Dhawan et al, 2009). This sensitive assay clearly showed the differences in DSB repair efficiency among cells with different PNKP constitutions (Fig 3, supplementary Fig S5 online). We concluded that ATM-dependent phosphorylation of PNKP represents a signalling pathway that is required for optimal DSB repair.
Figure 3.
Dynamics of double-strand break repair in human A549 cells with different versions of ectopic hPNKP. Direct observation of DNA damage 2 h after treatment with 200 ng/ml of neocarzinostatin (NCS) was carried out using a neutral comet assay (Dhawan et al, 2009). The length and intensity of SYBG green-stained DNA tails relative to heads is shown as the relative comet tail moment (n=100). P-values indicating the statistical difference between samples (Student's t-test) are indicated. Bars represent s.e.m. on the basis of three independent experiments. ATM, ataxia-telangiectasia mutated; DSB, double-strand break; hPNKP, human polynucleotide kinase/phosphatase.
PNKP phosphorylation is required for activity enhancement
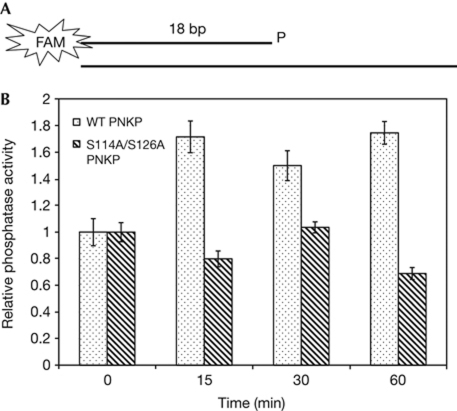
The mechanism of PNKP phosphorylation in DSB repair was studied by analysing the enzymatic activity of PNKP. We assayed its phosphatase activity in extracts of cells in which endogenous PNKP was replaced by ectopic protein in wild-type or phospho-mutants. A duplex oligonucleotide substrate containing a 3′-phosphate and a 5′ overhang (Fig 4A) was incubated with extracts of cells expressing different versions of PNKP (supplementary Fig S6A online) at various time points after NCS treatment. The activity of PNKP was assayed by monitoring the conversion of the 3′-phosphate oligonucleotide to the non-phosphorylated form (supplementary Fig S6B online). Importantly, although the activity in extracts of cells expressing wild-type PNKP was enhanced after NCS treatment, a marked reduction or complete abrogation of this enhancement was detected in cells harbouring the various phospho-mutant versions of the enzyme (Fig 4B, supplementary Fig S6C online). We concluded that ATM-dependent phosphorylation of PNKP in response to DNA-damage induction is important for enhancing the activity of the enzyme in response to excessive damage.
Figure 4.
Enhancement of human polynucleotide kinase/phosphatase activity by DNA-damage induction is dependent on the presence of ataxia-telangiectasia mutated phosphorylation sites. (A) Scheme of the substrate used in the assay. The 3′-phosphate end of the oligonucleotide is labelled with the fluorescent dye fluorescein amidite (FAM), which allows its detection in gels (see Methods section for details). (B) Diagram showing the relative activity of wild-type (WT) and S114A/S126A mutant hPNKP at different time points following neocarzinostatin treatment. Data are the mean of three experiments. Error bars represent s.e.m. hPNKP, human polynucleotide kinase/phosphatase.
The association of PNKP with the scaffold protein XRCC1, which is important for the role of PNKP in SSB repair (Whitehouse et al, 2001; Weinfeld et al, 2011), was not affected by DSB induction and was similar for wild-type and phospho-mutant PNKPs (supplementary Fig S7A online). This observation further suggests that the ATM-mediated pathway leading to enhancement of the activity of PNKP is associated with DSB, rather than SSB, repair. The XRCC4 protein was recently reported to link PNKP with the non-homologous end-joining pathway of DSB repair (Mani et al, 2010). Similar analysis of the PNKP–XRCC4 interaction (supplementary Fig S7B online) indicated that, although the interaction between the proteins was slightly enhanced in response to DSB induction, this enhancement was similar for wild-type and mutant versions of PNKP, implying that it was not mediated by the damage-induced phosphorylation of PNKP.
PNKP phosphorylation in recruitment to damaged sites
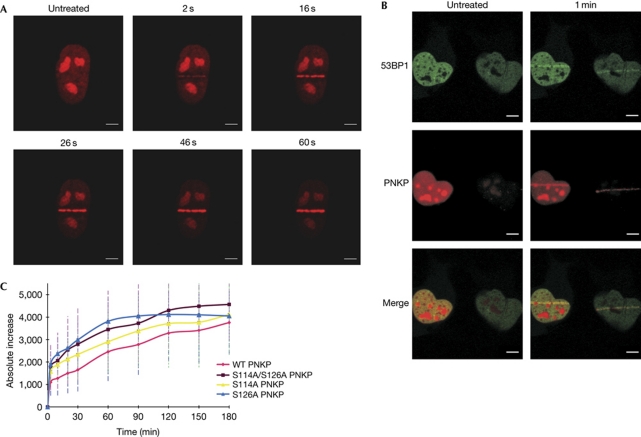
After DSB induction, biochemical fractionation experiments showed that PNKP is recruited to the chromatin fraction (data not shown), probably because of its recruitment to the damaged DNA sites. We explored the possibility that PNKP phosphorylation is involved in this process by following the recruitment of PNKP tagged with a fluorophore to localized DNA damage, induced by a focused laser beam. This system allows the generation of DNA damage including DSBs in small, defined regions within the nucleus, and subsequent monitoring of the spatio-temporal dynamics of proteins recruited to damaged sites (Bekker-Jensen et al, 2006). Cells in which endogenous PNKP had been stably replaced by red fluorescent protein (dsRED)-tagged PNKP in wild-type or phospho-mutants were used in these experiments. Notably, the dsRED-tagged protein was active and showed the DNA damage-induced activation (supplementary Fig S8 online). Recruitment of PNKP was detected seconds after irradiation (Fig 5A). The recruited protein co-localized with the well-documented DDR protein 53BP1 (Fig 5B). The recruitment kinetics of wild-type and mutant PNKPs were similar; however, higher amounts of the mutant proteins were detected at the damaged sites (Fig 5C), suggesting that turnover of the mutant protein at those sites was slower.
Figure 5.
Human polynucleotide kinase/phosphatase is recruited to DNA sites damaged within seconds after damage induction. (A) Rapid recruitment of dsRED–PNKP to DNA sites damaged by a focused laser microbeam. (B) Human PNKP recruited to the damaged site and co-localized with 53BP1, a central factor in the early phase of the DNA-damage response. Scale bars, 10 μM. (C) Recruitment kinetics of dsRED-tagged human PNKP in wild type (WT) and phospho-mutants during the first 3 h after DNA damage induction. The cell nuclei were exposed to a single burst of laser irradiation, images were captured at the indicated time the points and the amounts of dsRED–PNKP were quantified (Yano et al, 2008; Moyal et al, 2011). To compensate for nonspecific photobleaching, the background fluorescence was subtracted from the accumulation spot. Each data point is the average of 10 independent measurements. Error bars represent s.d. PNKP, polynucleotide kinase/phosphatase.
Discussion
Although much of DSB repair seems to be ATM independent, a varying fraction (10–25%) of DSBs are not repaired quickly in the absence of ATM activity (Riballo et al, 2004). One of the mechanisms by which ATM facilitates DSB repair was recently shown to be chromatin decondensation, mediated by various ATM substrates (Ziv et al, 2006; Goodarzi et al, 2008; Beucher et al, 2009; Moyal et al, 2011). However, ATM typically controls downstream processes through several parallel signalling pathways. Here, we show another simple mechanism by which ATM boosts cellular repair capacity—phosphorylation-dependent enhancement of the activity of a repair enzyme—possibly through conformational changes of the protein.
Although both the wild-type and phospho-mutant forms of PNKP are recruited to DSBs with similar kinetics, higher amounts of the non-phosphorylatable protein were detected at damaged sites. This could be due to higher retention or slower turnover of the mutant protein bound to damaged sites because of its reduced repair activity; that is, the mutant protein is recruited to these sites at a normal pace, but is retained there because of its reduced ability to carry out the enzymatic reaction under its responsibility. Conversely, higher amount of DNA damage resulting from insufficient PNKP activity might contribute to enhanced accumulation of the enzyme at the damaged sites.
Our observations agree with studies in which depletion of PNKP resulted in cellular hypersensitivity to ionizing radiation and camptothecin—a topoisomerase I inhibitor (Meijer et al, 2002). PNKP has been considered to be a main factor in SSB repair (Weinfeld et al, 2011). Collectively, our and other data (Koch et al, 2004; Karimi-Busheri et al, 2007; Weinfeld et al, 2011) indicate a role for this enzyme in DSB repair. Proteomic analysis carried out in our lab (not shown) recently indicated that hPNKP is constitutively associated with factors in both SSB repair and the non-homologous end-joining pathway of DSB repair, suggesting that PNKP is involved in both pathways, in agreement with previous evidence (Karimi-Busheri et al, 1998; Whitehouse et al, 2001; Chappell et al, 2002). Another example of direct targeting by ATM of a DNA-damage-processing enzyme is ATM-mediated phosphorylation of tyrosyl-DNA phosphodiesterase (TDP1) in response to DSBs (Das et al, 2009).
The role of PNKP in DSB repair–downstream of ATM—links it to an intricate network, the loss of which leads to, among others, the severe neurological phenotype in ataxia-telangiectasia (Lavin, 2008). Our initial finding of the phosphorylation of PNKP in mouse cerebellum, which led to this work, further supports this notion.
Methods
Vectors and constructs. Full-length PNKP cDNA cloned in pCMV–SPORT6 was obtained from RZPD (IRATp970B0454D; IMAGE ID 5218400). A Flag tag was introduced at the amino-terminus of the protein expressed from this clone by subcloning into pCMV-Tag 2B vector (Stratagene, La Jolla, CA, USA) using BamHI and EcoRI restriction sites. Flag–PNKP was cloned into the pLNCX2 retroviral vector (Clontech Laboratories, Palo Alto, CA, USA) using HindIII and SalI. Flag–PNKP pLNCX2 was also cloned into pDsRed1-C1 vector (Clontech Laboratories) using SalI and BglII.
RNA-interference-mediated silencing and protein replacement. Cell lines stably depleted for various proteins were obtained by expressing the corresponding shRNAs in them, using the retroviral vector pRetroSuper (a gift from R. Agami, Netherlands Cancer Institute, Amsterdam, The Netherlands). ATM expression was knocked down using an shRNA corresponding to positions 912–932 in ATM messenger RNA (Biton et al, 2006). hPNKP depletion was obtained using an shRNA corresponding to positions 517–535 in hPNKP messenger RNA. Cells were infected with retroviral particles according to standard protocols and underwent selection with 5 μg/ml puromycin for 6 days. These cells were subsequently complemented with the retroviral vector pLNCX2 (Clontech Laboratories) expressing Flag-tagged hPNKP, and underwent selection with 500 ng/ml neomycin. To allow for ectopic expression while the shRNA is stably expressed, a silent mutation, T523C, was introduced into the cDNA using the QuickChange Site-Directed Mutagenesis kit (Stratagene).
DNA-damage-response assays. Clonogenic survival, detection and quantification of 53BP1 foci and the comet assay were carried out as described previously (Moyal et al, 2011).
Laser microirradiation and imaging. Localized DNA damage was induced by microirradiation with a pulsed nitrogen laser (Spectra-Physics; 365 nm, 30 Hz pulse). Time-lapse image acquisition and kinetics analysis were carried out as described previously (Yano et al, 2008; Moyal et al, 2011).
PNKP phosphatase assay. The substrate was prepared by annealing a fluorescently labelled oligonucleotide (5′-FAM-TAGCATCGATCAGTCCTC-p) and an unlabelled complementary strand (TCGCTTGACTAACGATGCTAGACCTCGAGGACTGATCGATGCTA; Sigma). The fluorescent dye that we used, fluorescein amidite (FAM), is the most commonly used fluorescent dye for attachment to oligonucleotides with absorbance maximum at 495 nm and emission maximum at 520 nm. The double-stranded oligonucleotide was purified from the gel using Microcon ultracel YM-10 (Millipore). After treating the cells with varying doses of NCS for different times, cellular extracts were prepared by lysing them in Flag lysis buffer (50 mM Tris, pH 7.8, 137 mM NaCl, 1 mM NaF, 1 mM NaVO3, 1% Triton X-100, 0.2% sarcosyl) containing complete EDTA-Free Protease Inhibitor Cocktail (Roche) for 30 min at 4°C. Double-stranded oligonucleotide substrate (12 nM) was incubated with 5 μg of cellular extracts for 1 min at 37°C in a T4 buffer. The reaction was stopped by adding formamide loading buffer and heating at 95°C before it was loaded onto a 14% denaturating polyacrylamide gel. Gels were visualized using Radioisotopic Imaging FLA5100 (FUJIFILM).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank A. Admon and A. Shianskaya for the proteomic analysis, T. Halazonetis for the 53BP1 antibody and R. Agami for the pRetroSuper vector. This work was supported by research grants from the Ataxia-Telangiectasia Medical Research Foundation, the Ataxia-Telangiectasion Ease Foundation and the Israel Cancer Research Fund (Y.S.), The National Institutes of Health of the USA (CA050519, to D.J.C.), The National Cancer Institute (CA113859) and the Avon Foundation for Women (02-2010-063) to M.C.-T.H and The Swedish Cancer Foundation (T.H.). Y.S. is a Research Professor of the Israel Cancer Research Fund.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J (2006) Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol 173: 195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen MV, Larsen DH, Bunkenborg J, Bartek J, Lukas J, Andersen JS (2010) Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response. Mol Cell Proteomics 9: 1314–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensimon A, Schmidt A, Ziv Y, Elkon R, Wang SY, Chen DJ, Aebersold R, Shiloh Y (2010) ATM-dependent and -independent dynamics of the nuclear phosphoproteome after DNA damage. Sci Signal 3: rs3. [DOI] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M (2009) ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J 28: 3413–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biton S, Dar I, Mittelman L, Pereg Y, Barzilai A, Shiloh Y (2006) Nuclear ataxia-telangiectasia mutated (ATM) mediates the cellular response to DNA double strand breaks in human neuron-like cells. J Biol Chem 281: 17482–17491 [DOI] [PubMed] [Google Scholar]
- Chappell C, Hanakahi LA, Karimi-Busheri F, Weinfeld M, West SC (2002) Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. EMBO J 21: 2827–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ (2010) The DNA damage response: making it safe to play with knives. Mol Cell 40: 179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das BB, Antony S, Gupta S, Dexheimer TS, Redon CE, Garfield S, Shiloh Y, Pommier Y (2009) Optimal function of the DNA repair enzyme TDP1 requires its phosphorylation by ATM and/or DNA-PK. EMBO J 28: 3667–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derheimer FA, Kastan MB (2010) Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS Lett 584: 3675–3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan A, Bajpayee M, Parmar D (2009) Comet assay: a reliable tool for the assessment of DNA damage in different models. Cell Biol Toxicol 25: 5–32 [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA (2008) ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell 31: 167–177 [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC (2004) Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 64: 9152–9159 [DOI] [PubMed] [Google Scholar]
- Karimi-Busheri F, Lee J, Tomkinson AE, Weinfeld M (1998) Repair of DNA strand gaps and nicks containing 3′-phosphate and 5′-hydroxyl termini by purified mammalian enzymes. Nucleic Acids Res 26: 4395–4400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi-Busheri F, Rasouli-Nia A, Allalunis-Turner J, Weinfeld M (2007) Human polynucleotide kinase participates in repair of DNA double-strand breaks by nonhomologous end joining but not homologous recombination. Cancer Res 67: 6619–6625 [DOI] [PubMed] [Google Scholar]
- Koch CA, Agyei R, Galicia S, Metalnikov P, O’Donnell P, Starostine A, Weinfeld M, Durocher D (2004) Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J 23: 3874–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF (2008) Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol 9: 759–769 [DOI] [PubMed] [Google Scholar]
- Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC (2004) Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett 14: 6083–6087 [DOI] [PubMed] [Google Scholar]
- Lempiainen H, Halazonetis TD (2009) Emerging common themes in regulation of PIKKs and PI3Ks. EMBO J 28: 3067–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T (2005) Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res 33: 3799–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani RS et al. (2010) Dual modes of interaction between XRCC4 and polynucleotide kinase/phosphatase: implications for nonhomologous end joining. J Biol Chem 285: 37619–37629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S et al. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Meijer M, Karimi-Busheri F, Huang TY, Weinfeld M, Young D (2002) Pnk1, a DNA kinase/phosphatase required for normal response to DNA damage by gamma-radiation or camptothecin in Schizosaccharomyces pombe. J Biol Chem 277: 4050–4055 [DOI] [PubMed] [Google Scholar]
- Moyal L et al. (2011) Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell 41: 529–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E et al. (2004) A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell 16: 715–724 [DOI] [PubMed] [Google Scholar]
- Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN (2011) Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci 36: 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, Weinfeld M, Caldecott KW (2001) XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104: 107–117 [DOI] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ (2008) Ku recruits XLF to DNA double-strand breaks. EMBO Rep 9: 91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y (2006) Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 8: 870–876 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.