Abstract
Human malignant melanoma has poor prognosis because of resistance to apoptosis and therapy. We describe identification of the expression profile of 1,037 mitochondria-focused genes and 84 survival-apoptosis genes in 21 malignant melanoma cell lines and 3 normal melanocyte controls using recently developed hMitChip3 cDNA micro-arrays. Unsupervised hierarchical clustering analysis of 1,037 informative genes, and 84 survival-apoptosis genes, classified these malignant melanoma cell lines into type A (n = 12) and type B (n = 9). Three hundred fifty-five of 1,037 (34.2%) genes displayed significant (P ≤ 0.030; false discovery rate ≤ 3.68%) differences (±≥2.0-fold) in average expression, with 197 genes higher and 158 genes lower in type A than in type B. Of 84 genes with known survival-apoptosis functions, 38 (45.2%) displayed the significant (P < 0.001; false discovery rate < 0.15%) difference. Antiapoptotic (BCL2, BCL2A1, PPARD, and RAF1), antioxidant (MT3, PRDX5, PRDX3, GPX4, GLRX2, and GSR), and proapoptotic (BAD, BNIP1, APAF1, BNIP3L, CASP7, CYCS, CASP1, and VDAC1) genes expressed at higher levels in type A than in type B, whereas the different set of antiapoptotic (PSEN1, PPP2CA, API5, PPP2R1B, PPP2R1A, and FIS1), antioxidant (HSPD1, GSS, SOD1, ATOX1, and CAT), and proapoptotic (ENDOG, BAK1, CASP2, CASP4, PDCD5, HTRA2, SEPT4, TNFSF10, and PRODH) genes expressed at lower levels in type A than in type B. Microarray data were validated by quantitative reverse transcription-PCR. These results showed the presence of two types of malignant melanoma, each with a specific set of dysregulated survival-apoptosis genes, which may prove useful for development of new molecular targets for therapeutic intervention and novel diagnostic biomarkers for treatment and prognosis of malignant melanoma.
Introduction
Human cutaneous malignant melanoma remains as the most deadly form of skin cancer, responsible for 75.2% of skin cancer–related deaths (1). Furthermore, the incidence of a melanoma has been increasing in white populations worldwide, largely due to an increase in thin rather than thick melanomas (2, 3). Mortality rates vary by country, gender, and ethnic origin; in the United States in 2008, there were an estimated 62,480 new cases and 8,420 (13.5%) deaths (1). The primary tumors in early stages are curable by surgical excision, whereas the prognosis of patients with distant metastases not amenable to surgery remains poor (1, 4, 5); the poor outcome has not improved in response to current therapeutic regimens (6–8). The median survival reported for patients with metastatic melanoma in recent phase III clinical trials is only 7 to 9 months (9, 10). Highly resistant to apoptosis and therapy is a unique hallmark of malignant melanoma, although the mechanisms by which melanoma cells protect themselves from drug-induced apoptosis remains largely unknown (11). Tumor cells bypass the apoptotic machinery through mechanisms that may involve dysregulation of antiapoptotic and proapoptotic genes (12–14). Thus, our understanding of the survival-apoptosis biology of malignant melanoma cells is a prerequisite for breakthroughs in the therapy of this malignant disease.
To study survival, antiapoptotic, antioxidant, and proapoptotic genes, and their signaling pathways in malignant cells, we recently developed a third-generation human mitochondria-focused cDNA microarray (hMitChip3) and new bioinformatic tools (15). hMitChip3 contains 37 mitochondrial DNA–encoded genes, 1,098 nuclear DNA–encoded and mitochondria-related genes, and 225 controls, each printed in triplicate. A total of 1,135 mitochondria-related genes include 946 genes associated with 645 molecular functions, 930 genes with 612 biological processes, 476 genes with biological chemistry pathways, 227 genes with 23 reactome events, 237 genes with 320 genetic disorders, and 55 genes with 87 drugs targets (15). Approximately 100 genes are involved in cell survival, antioxidant, antiapoptosis, and proapoptosis. Using the hMitChip3 and bioinformatic tools, we have successfully identified molecular mechanisms underlying survival and apoptosis of the cutaneous malignant melanoma cell lines UACC903 (resistance to apoptosis) and UACC903(+6) (sensitive to apoptosis; refs. 13, 14, 16). These findings show the presence of differentially expressed genes that govern survival and apoptosis of malignant melanoma cells.
In this report, we describe the identification of two types of characteristic expression patterns of 1,037 mitochondria-focused genes, including 84 survival-apoptosis genes, in 21 malignant melanoma cell lines compared with 3 normal melanocytes derived originally from lightly, moderately, and darkly pigmented neonatal foreskin tissues. Applying hMitChip3, we identify expression profiles of 1,037 informative genes and 355 significantly [P ≤ 0.030; false discovery rate (FDR) ≤ 3.68%] differentially (≥2.0-fold) expressed genes that classified 21 malignant melanoma cell lines into two distinct types (type A, n = 12; type B, n = 9). The different sets of survival, anti-apoptotic, antioxidant, and proapoptotic genes displayed significant (P ≤ 0.001; FDR ≤ 0.15%) differences in expression between the type A and the type B malignant melanoma cell lines. These results facilitate our understanding of the different molecular basis for survival of malignant melanoma cells and may also prove useful for the development of new biomarkers for therapeutic intervention as well as potential diagnostic biomarkers for treatment and prognosis for a better clinical outcome of patients with malignant melanoma.
Materials and Methods
Cell Culture
Most malignant melanoma cell lines were purchased from either American Type Culture Collection (ATCC) or University of Arizona Cancer Center (UACC; Tucson, AZ; Table 1). C8161 and MUM2C cell lines were gift generously provided by Dr. Brian Nickoloff (Loyola University Medical Center, Maywood, IL). UACC903(+6) and SRS3 cell lines were developed in Dr. Jeffrey Trent's laboratory (National Human Genome Research Institute, Bethesda, MD). These cell lines were cultured in medium recommended by either ATCC or UACC. UACC903(+6) cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mmol/L l-glutamine, and 600 μg/mL G418 to select for the pSV2neo-tagged chromosome 6 (17). SRS3 was cultured in the same medium in addition of 8 mmol/L l-histidinol dihydrochloride to select for integrated retroviral vector (16). Cell culture medium (RPMI 1640, DMEM, Eagle's MEM, and McCoy's 5A), fetal bovine serum, and trypsin-EDTA were purchased from Mediatech, Inc., and Leibovitz's L-15 medium was purchased from Invitrogen (Life Technologies Cell Culture). Normal human neonatal epidermal melanocytes were purchased from Cascade Biologics (Invitrogen). The melanocytes were cultured in Medium 254 plus phorbol 12-myristate 13-acetate–free human melanocyte growth supplement-2 (Cascade Biologics). One hundred units of penicillin G sodium and 100 μg of streptomycin sulfate (penicillin-streptomycin; MP Biomedicals, LLC) were added into each milliliter of cell culture medium as antibiotics.
Table 1.
Twenty-one human malignant melanoma cell lines and 3 normal control melanocytes
| Malignant melanoma cell lines | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | Name* | Disease | Biopsy site | Xenograft animal models |
Age | Gender | Source and reference | This study |
|
| Tumorigenicity | Metastasis | Type | |||||||
| 1 | A2058 (5) (A) | MM | Lymph node | Yes | Yes | 43 | Male | ATCC (30) | A |
| 2 | A375 (5) (B) | MM | Skin | Yes | Yes | 54 | Female | ATCC (30, 31) | A |
| 3 | C32 (5) (B) | MM | Skin | Yes | Unknown | 53 | Male | ATCC | A |
| 4 | COLO829 (5) (E) | MM | Skin | Unknown | Unknown | 45 | Male | ATCC | A |
| 5 | G361 (5) (C) | MM | Skin | Yes | Yes | 31 | Male | ATCC (30) | A |
| 6 | HT144 (5) (C) | MM | Skin | Yes | Yes | 29 | Male | ATCC (32) | A |
| 7 | HTB65 (5) (B) | MM | Lymph node | Yes | Yes | 78 | Male | ATCC (31, 33) | A |
| 8 | RPMI7951 (5) (B) | MM | Lymph node | Yes | Unknown | 18 | Female | ATCC | A |
| 9 | SKMEL2 (5) (B) | MM | Skin of thigh | Yes | Unknown | 60 | Male | ATCC | A |
| 10 | SRS3 (9) (E) | UACC903(+6) variant | Yes | Unknown | (16) | A | |||
| 11 | UACC3074 (5) (E) | MM | Unknown | Unknown | Unknown | Unknown | Unknown | UACC | A |
| 12 | WM266-4 (5) (b) | MM | Skin | Yes | Yes | 58 | Female | ATCC (34) | A |
| 13 | C8161 (4) (E) | MM | Unknown | Yes | Yes | Unknown | Unknown | (31) | B |
| 14 | MUM2C (4) (E) | MM (Uveal) | Liver | Yes | Yes | Unknown | Male | (23) | B |
| 15 | UACC1227 (5) (F) | MM | Lymph node | Unknown | Unknown | 27 | Male | UACC | B |
| 16 | UACC2565 (5) (F) | MM | Lymph node | Unknown | Unknown | 63 | Male | UACC | B |
| 17 | UACC457 (5) (F) | MM | Abdomen | Unknown | Unknown | 41 | Male | UACC | B |
| 18 | UACC827 (5) (F) | MM | Right breast | Unknown | Unknown | 32 | Female | UACC | B |
| 19 | UACC903 (9) (E) | MM | Back | Yes | Unknown | 35 | Female | UACC (17) | B |
| 20 | UACC903(+6) (11) (E) | UACC903 variant | Yes | Unknown | (17, 35) | B | |||
| 21 | UACC929 (5) (F) | MM | Lymph node | Unknown | Unknown | 31 | Male | UACC | B |
| Normal control melanocytes | ||||||
|---|---|---|---|---|---|---|
| No. | Name | Description | Biopsy site | Age | Gender | Source and reference |
| 1 | HEMn-LP (9) (G) | Normal human epidermal melanocytes, lightly pigmented | Foreskin | Neonatal | Male | Cascade Biologics |
| 2 | HEMn-MP (9) (G) | Normal human epidermal melanocytes, moderately pigmented | Foreskin | Neonatal | Male | Cascade Biologics |
| 3 | HEMn-DP (9) (G) | Normal human epidermal melanocytes, darkly pigmented | Foreskin | Neonatal | Male | Cascade Biologics |
Abbreviation: MM, malignant melanoma.
All cell lines were cultured in this laboratory to 90% confluence for harvesting of cells at the passages (the numbers in parentheses) after receiving from the resources indicated and analysis. The letters in parentheses indicate culture medium used: (A) DMEM, (B) Eagle's MEM, (C) McCoy's 5A, (E) RPMI 1640, (F) L-15, and (G) Medium 254. The cell lines cultured in L-15 medium were under 100% air, that is, without the supplement of 5% carbon dioxide. All other 19 cell lines were cultured in the presence of 5% carbon dioxide.
All cell lines were cultured in the same laboratory under the standard cell culture conditions with exceptions where indicated. Cultured cells in Petri dishes (Greiner Bio-One) were routinely viewed under inverted microscope to assess the degree of confluence and to confirm the absence of any contamination. When cells reached at ~90% confluence under microscope, culture media were removed by aspiration, and the cells were rinsed once with 1× PBS (Mediatech) and then lysed directly by adding Trizol Reagent (Invitrogen). Five cell lines (UACC1227, UACC2565, UACC457, UACC827, and UACC929) were cultured in Leibovitz's L-15 medium that requires no supplement of 5% carbon dioxide. Other 19 cell lines were cultured in the medium indicated in Table 1 with standard supplement of 5% carbon dioxide.
RNA Extraction and Purification
Total RNA was extracted from freshly cultured cells using Trizol Reagent and purified using RNeasy kit (Qiagen), following the commercial instructions, in the same laboratory.
hMitChip3 Microarray
A third-generation human mitochondria-focused cDNA microarray (hMitChip3) containing 37 mitochondrial DNA–encoded genes, 1,098 nuclear DNA–encoded and mitochondria-related genes, and 225 controls were printed as described previously (15). For examples, hMitChip3 contains 84 genes with known involvement in cellular survival, antiapoptosis, antioxidative protection, and proapoptosis. Total RNA (5 μg) per sample was used for microarray labeling and hybridization as previously described (15). Slides were scanned using the ScanArray Express Microarray Scanner (Perkin-Elmer) as described previously (15). Each and every gene on hMitChip3 gene chip was printed in triplicate and triplicate microarray experiments were done for each and every RNA samples.
Microarray Database and Data Analysis
Gene expression database was constructed using FileMaker software (FileMaker Pro, Inc.). Database construction, data filtering, and selection were done as described previously (15). The quantile normalization method (18) in software R version 2.7.1 (The R Foundation for Statistical Computing) was used to normalize microarray data. The normalized expression data were used to cluster and visualize genes and cell lines by using software Cluster version 3.0 (19) and heat map was visualized by using software MapleTree.6
Gene Information Analysis
ID, symbols, and names of genes on hMitChip3 were updated to human UniGene Build 2147 based on IMAGE8 clone ID. Ontology, pathways, and phenotypes of genes were compiled from Entrez9 and DAVID10 Bioinformatics Resources 2008.
Quantitative Reverse Transcription-PCR
Total RNA (2 μg) was reverse transcribed into cDNA by using SuperScript First-Strand Synthesis System (Invitrogen). cDNA (30 ng) was used for quantitative PCRs with the Universal PCR Master Mix (No AmpErase UNG) on an Applied Biosystems 7300 Real-Time PCR System following the manufacturer's instructions. After 40 cycles, expression data were obtained with the 7300 PCR Software. Triplicate quantitative reverse transcription-PCR (qRT-PCR) experiments were done for each gene. Relative RNA levels were calculated using the published methods (20, 21). Taqman probes and primers for quantitative PCR were purchased from Applied Biosystems and include those of glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 4352934E), B-cell CLL/lymphoma 2 (BCL2; Hs00236808_s1), BCL2-related protein A1 (BCL2A1; Hs00187845_m1), catalase (CAT; Hs00156308_m1), glutathione peroxidase 4 (GPX4; Hs00989766_g1), glutathione reductase (GSR; Hs00167317_m1), glutathione synthetase (GSS; Hs00609286_m1), optic atrophy 1 (OPA1; Hs01047019_m1), prohibitin (PHB; Hs00855044_g1), peroxiredoxin 3 (PRDX3; Hs00428953_g1), peroxiredoxin 5 (PRDX5; Hs00738905_g1), protein phosphatase 2 catalytic subunit α isoform (PPP2CA; Hs00427259_m1), and regulatory subunit A β isoform (PPP2R1B; Hs00988483_m1).
Statistics
Statistical calculations were done on triplicate array experiments using XLSTAT 2006 (XLSTAT). Differentially expressed genes were identified arbitrarily by >1.24-fold change in the average expression of the background-subtracted mean intensity ratios of a gene between comparisons. Using the LIMMA package in software R/Bioconductor (version 2.7.1; The R Foundation for Statistical Computing), we calculated the moderated t statistic, raw P values, and log2 fold changes in gene expression and FDR for multiple statistical testing with Benjamini and Hochberg methods (22). The level of statistical significance was set at a P value of <0.05 and FDR of <5%.
Results
Twenty-One Human Malignant Melanoma Cell Lines and 3 Normal Melanocytes
Table 1 lists 21 human malignant melanoma cell lines and 3 normal melanocytes as controls that are frequently used in melanoma research. Of 21 malignant melanoma cell lines, 18 (A2058, A375, C32, COLO829, G361, HT144, HTB65, RPMI7951, SKMEL2, UACC3074, WM266-4, C8161, UACC1227, UACC2565, UACC457, UACC827, UACC903, and UACC929) were derived from malignant melanoma, 2 [UACC903(+6) and SRS3] were variants of UACC903 (16, 17), and 1 (MUM2C) was from liver metastasis of uveal malignant melanoma (23). Eight cell lines (A375, C32, COLO829, G361, HT144, SKMEL2, UACC903, and WM266-4) were derived from skin biopsies, whereas 9 from metastatic sites, including lymph note (A2058, HTB65, RPMI7951, UACC1227, UACC2565, and UACC929), liver (MUM2C), abdomen (UACC457), and breast (UACC827). Fourteen cell lines [A2058, A375, C32, C8161, G361, HT144, HTB65, MUM2C, RPMI7951, SKMEL2, SRS3, UACC903, UACC903(+6), and WM266-4] were tumorigenic in animal models and 8 (A2058, A375, G361, HT144, HTB65, WM266-4, C8161, and MUM2C) were metastatic. Twelve cell lines (A2058, C32, COLO829, G361, HT144, HTB65, SKMEL2, MUM2C, UACC1227, UACC2565, UACC457, and UACC929) were derived from male patients, whereas 5 (A375, RPMI7951, WM266-4, UACC827, and UACC903) from females. The normal melanocytes were originally derived from lightly, moderately, and darkly pigmented neonatal foreskin tissues.
Clusters and Subclusters of 21 Malignant Melanoma Cell Lines Revealed by Unsupervised Hierarchical Clustering Analysis of 1,037 Informative Genes from 81 Microarray Experiments
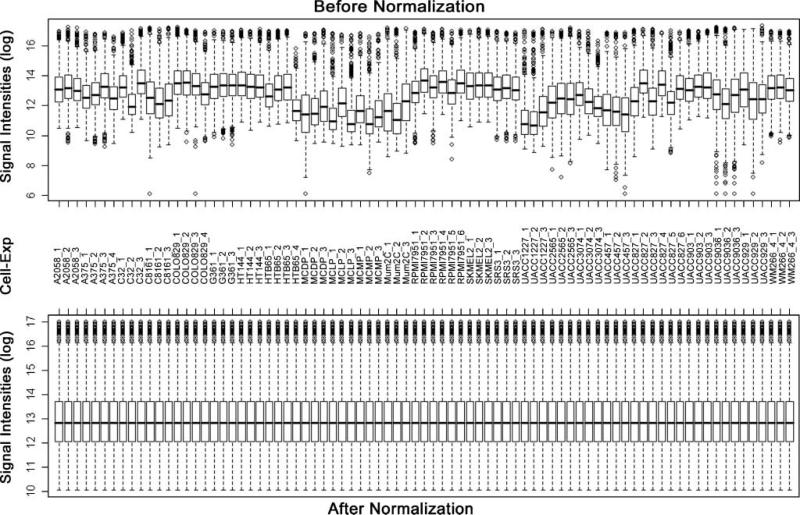
Total RNA samples were extracted from 21 malignant melanoma cell lines and 3 controls and labeled for microarray analysis using hMitChip3. Because use of three replicates in microarray analysis reduces misclassification (24), triplicate microarray experiments were done for all malignant melanoma and control cell lines, except for A375, COLO829, and HTB65 with four replicates and RPMI7951 and UACC827 with six replicates (Fig. 1). Thus, 1,135 genes and control elements on hMitChip3 microarrays were all measured at least nine times (three identical probes per microarray and at least three microarray experiments per cell line), which generated reproducible data for statistical analysis. Microarray data of 4,080 spots across all 81 gene chips used for 24 RNA samples were filtered by uniform criteria within microarray database as described previously (15), which yield 1,037 genes with informative background-subtracted signal intensities for data normalization. Figure 1 shows the box plots of mRNA levels of 1,037 genes in 24 cell lines and all 81 microarray experiments before (Supplementary Table S1)11 and after (Supplementary Table S2)11 data normalization. The mean values of normalized data were used to calculate ratios in gene expression between each malignant melanoma cell line and each control, resulting in a total of 63 comparisons across all 1,037 genes (Supplementary Table S3).11 These data were used for unsupervised clustering analysis, visualization, and identification of differentially expressed genes. The resultant double dendrograms and heat map classified 21 malignant melanoma cell lines into two major groups. Group A consisted of three subclusters (a1, a2, and a3) and group B had two subclusters (b1 and b2). The a1 subcluster contained C32, COLO829, A2058, RPMI7951, and HT144; a2 had G3G1, A375, WM266-4, and SRS-3; and a3 consisted of HTB65, SKMEL2, and UACC3074. The b1 subcluster contained MUM2C, UACC1227, C8161, UACC2565, UACC457, UACC929, UACC903(+6), and UACC903, whereas b2 had only one cell line, UACC827. The multiple cell lines within a subcluster were further classified into smaller groups. Whereas a high percentage of 1,037 genes displayed apparently differential expression patterns between these two groups, a cluster of 28 genes (EHD4, HNRNPA1, CIDEB, DDX18, FDX1, HRBL, CHD6, BDH1, AKAP1, ABCD1, MT1A, MRPL45, PHB2, MCL1, THEM2, MCCC2, TXNRD2, CYC1, ODC1, SDHB, STIP1, PTGES2, MRPS9, MRPL10, PPIB, UCP3, MRPL54, and TAGLN2) was down-regulated in four malignant melanoma cell lines (G361, A375, WM266-4, and SRS3) but up-regulated in the others (Supplementary Fig. S1).11
Figure 1.
Box plots of expression data before and after normalization. The quantile normalization algorithms were used to adjust the values of the background-subtracted mean pixel intensities of triplicate measurements per microarray for each and every set of 1,037 genes. Microarray experiments for most samples were conducted in triplicate, except for A375, COLO829, and HTB65 in four times and for RPMI7951 and UACC827 in six times. In contrast to the prenormalization box plots (top), the postnormalized box plots distribute in the same intervals with the same density center, indicating successful adjustment of data. The postnormalized data were used for further analysis. Cell, cell lines; Exp, independent microarray experiments; log, log2.
Three Hundred Fifty-Five Genes Were Significantly Differentially Expressed between the Type A and Type B Malignant Melanoma Cell Lines
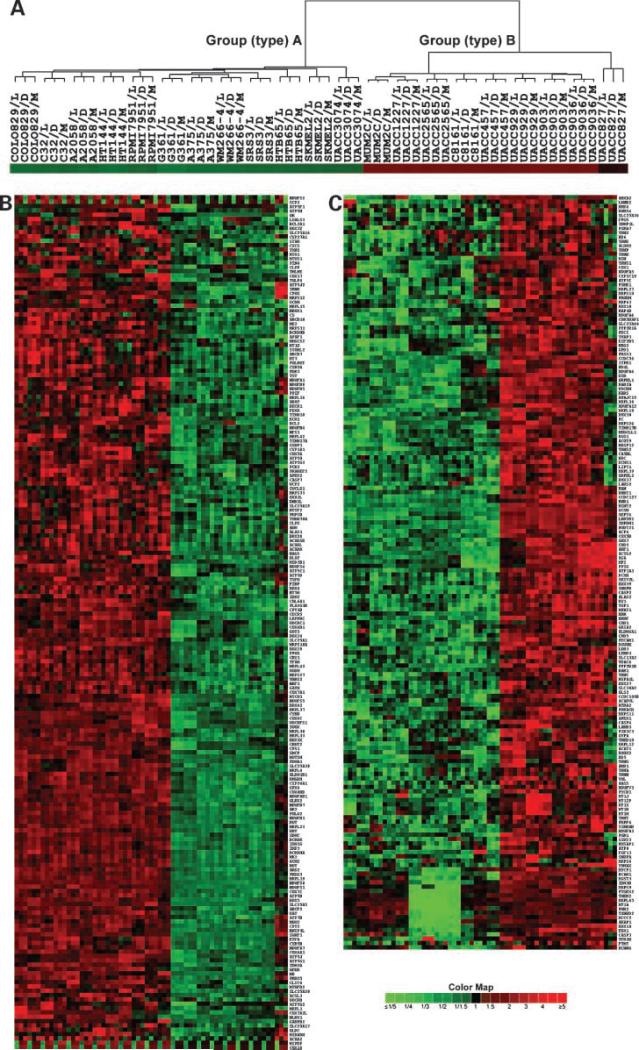
Based on unsupervised cluster results, we calculated average expression levels, ratios, the moderated t statistic, P values, and FDR of genes between the group A and group B malignant melanoma cell lines. The results revealed that 371 of 1,037 genes (40.4%) had ±≥2.0-fold changes in average expression between these two types with a P value of ≤0.030 and FDR of ≤3.68%. Unsupervised cluster analysis of these 371 genes revealed the same group A and group B clusters as expected, and subclusters similar to those derived by using all 1,037 genes. In addition, 16 genes, including MTCP1, BCAR1, MGST3, IDH3B, MRPS9, PTGES2, THEM2, MRPL45, MT1A, PHB2, TXNRD2, MCCC2, AKAP1, DDX18, FDX1, and CASP3, were consistently at lower levels of expression in four malignant melanoma cell lines (G361, A375, WM266-4, and SRS3) than in the others, in comparison with three normal control melanocytes (Fig. 2). Thus, 355 significantly differentially expressed genes defined the group A (type A) and group B (type B) malignant melanoma (Supplementary Tables S4 and S5).11
Figure 2.
Dendrogram and heat map of genes differentially expressed between two types of human malignant melanoma cell lines. A, cluster tree of 63 comparisons among 21 human malignant melanoma cell lines and 3 normal melanocytes, including dark (D), moderate (M), and light (L) pigmented melanocytes. B and C, heat maps of 371 genes with >2-fold changes in average intensities between type A and type B (Supplementary Table S1). Color map indicates fold changes in down-expression (green) and up-expression (red) of 371 genes, with P < 0.03 (t test) and FDR < 4% (Supplementary Table S2). The multiple statistical testing for FDR was done by using the Benjamini and Hochberg method (22).
Identification of 84 Genes with Known Antiapoptosis/ Proapoptosis Functions and Their Expression Patterns
To identify genes involved in cell survival and apoptosis, we analyzed biological information of all 1,037 informative genes. The results revealed that 84 genes (8.1%) had the known roles in survival, antiapoptosis, antioxidation, and proapoptosis (Table 2). Of 84 genes, 73 (86.9%) displayed significant (P ≤ 0.039 and FDR ≤ 4.7%) differences (≥1.24-fold changes) in average expression between the type A and type B malignant melanoma cell lines. The relative expression ratios of 84 genes in all 63 comparisons between each malignant melanoma cell line and each normal control were subjected to clustering analysis. The sample clusters indicated the same type A and type B of malignant melanoma cell lines, but subclusters were slightly different from those based on expression profiles of 1,037 genes. The gene clusters revealed four clusters (GC-1 through GC-4). GC-1 had 17 genes (CASP1, CYCS, BNIP1, BCL2, APAF1, BCL2A1, CASP7, BNIP3L, GLRX2, GPX4, PRDX3, PRDX5, MT3, PPARD, GSR, RAF1, and VDAC1) with a consistently higher level of expression in the type A malignant melanoma cell lines than in type B; GC-2 had 4 genes (CASP10, FAS, GZMH, and PDIA2) with expression higher in the moderately pigmented normal melanocytes than in the lightly and darkly pigmented ones; GC-3 had 21 genes (FIS1, ATOX1, PHB, SOD1, ENDOG, BAK1, API5, PPP2CA, PSEN1, CASP2, CAT, HTRA2, SEPT4, CASP4, PDCD5, PPP2R1B, PPP2R1A, SYK, TNFSF10, PRODH, and VHL) with expression lower in type A than in type B; and GC-4 had 5 genes (CASP3, CIDEB, MCL1, TXNRD2, and PTGES2) with expression lower in 4 malignant melanoma cell lines (G361, A375, WM266-4, and SRS3) than in the rest (except for CASP3 also lower in UACC3074; Supplementary Fig. S2).11
Table 2.
Functions and statistics of 84 genes between the types A and B of human malignant melanoma cell lines
| Role | Symbol | A/B* | P* | FDR* | Gene name |
|---|---|---|---|---|---|
| Antioxidation | |||||
| GLRX2 | 4.75 | 0.000 | 0.000 | Glutaredoxin 2 | |
| PRDX3 | 3.62 | 0.000 | 0.000 | Peroxiredoxin 3 | |
| GPX4 | 3.11 | 0.000 | 0.000 | Glutathione peroxidase 4 | |
| MT3 | 2.82 | 0.000 | 0.000 | Metallothionein 3 | |
| PRDX5 | 2.67 | 0.000 | 0.000 | Peroxiredoxin 5 | |
| GSR | 1.02 | 0.000 | 0.000 | Glutathione reductase | |
| GSS | –0.35 | 0.002 | 0.003 | Glutathione synthetase | |
| GPX7 | –0.35 | 0.032 | 0.039 | Glutathione peroxidase 7 | |
| MPO | –0.40 | 0.001 | 0.001 | Myeloperoxidase | |
| NOS2A | –0.43 | 0.000 | 0.000 | Nitric oxide synthase 2A | |
| GPX1 | –0.55 | 0.017 | 0.022 | Glutathione peroxidase 1 | |
| CAT | –0.87 | 0.000 | 0.000 | Catalase | |
| SOD1 | –1.00 | 0.000 | 0.000 | Superoxide dismutase 1, soluble | |
| HSPD1 | –1.01 | 0.000 | 0.000 | Heat shock 60 kDa protein 1 | |
| TXNRD2 | –1.05 | 0.000 | 0.001 | Thioredoxin reductase 2 | |
| ATOX1 | –1.16 | 0.000 | 0.000 | ATX1 antioxidant protein 1 homologue | |
| PTGES2 | –2.58 | 0.000 | 0.000 | Prostaglandin E synthase 2 | |
| PINK1 | –0.23 | 0.104 | 0.120 | PTEN induced putative kinase 1 | |
| GLRX | –0.13 | 0.146 | 0.166 | Glutaredoxin (thioltransferase) | |
| PRDX2 | 0.22 | 0.148 | 0.169 | Peroxiredoxin 2 | |
| Antiapoptosis | |||||
| BCL2 | 2.57 | 0.000 | 0.000 | B-cell CLL/lymphoma 2 | |
| BCL2A1 | 1.94 | 0.000 | 0.000 | BCL2-related protein A1 | |
| PPARD | 0.78 | 0.000 | 0.000 | Peroxisome proliferative-activated receptor, δ | |
| RAF1 | 0.53 | 0.000 | 0.000 | v-raf-1 murine leukemia viral oncogene homologue 1 | |
| OPA1 | –0.34 | 0.000 | 0.001 | Optic atrophy 1 | |
| BCL2L1 | –0.35 | 0.000 | 0.000 | BCL2-like 1 | |
| HDAC3 | –0.39 | 0.001 | 0.002 | Histone deacetylase 3 | |
| MTP18 | –0.44 | 0.001 | 0.001 | Mitochondrial protein 18 kDa | |
| FIS1 | –0.50 | 0.000 | 0.000 | Fission 1 (mitochondrial outer membrane) homologue | |
| CLN3 | –0.58 | 0.000 | 0.000 | Ceroid lipofuscinosis, neuronal 3, juvenile | |
| MCL1 | –0.75 | 0.014 | 0.018 | Myeloid cell leukemia sequence 1 | |
| BCL2L10 | –0.77 | 0.000 | 0.000 | BCL2-like 10 | |
| PPP2CA | –0.77 | 0.000 | 0.000 | Protein phosphatase 2, catalytic subunit, α isoform | |
| PHB | –0.82 | 0.000 | 0.000 | Prohibitin | |
| API5 | –0.94 | 0.000 | 0.000 | Apoptosis inhibitor 5 | |
| SYK | –0.94 | 0.000 | 0.000 | Spleen tyrosine kinase | |
| PDIA2 | –0.97 | 0.000 | 0.001 | Protein disulfide isomerase family A, member 2 | |
| PSEN1 | –0.98 | 0.000 | 0.000 | Presenilin 1 | |
| VHL | –0.99 | 0.000 | 0.000 | von Hippel-Lindau tumor suppressor | |
| PPP2R1B | –1.08 | 0.000 | 0.000 | Protein phosphatase 2, regulatory subunit A, β isoform | |
| PPP2R1A | –1.69 | 0.000 | 0.000 | Protein phosphatase 2, regulatory subunit A, α isoform | |
| ANXA1 | –0.13 | 0.398 | 0.431 | Annexin A1 | |
| HSPA5 | 0.17 | 0.418 | 0.451 | Heat shock 70 kDa protein 5 | |
| SIRT1 | 0.01 | 0.925 | 0.936 | Sirtuin (silent mating type information regulation 2 homologue) 1 | |
| Proapoptosis | |||||
| BNIP3L | 3.54 | 0.000 | 0.000 | BCL2/adenovirus E1B 19 kDa interacting protein 3-like | |
| CASP7 | 2.27 | 0.000 | 0.000 | Caspase-7, apoptosis-related cysteine peptidase | |
| APAF1 | 1.85 | 0.000 | 0.000 | Apoptotic peptidase activating factor | |
| CYCS | 1.14 | 0.000 | 0.000 | Cytochrome c, somatic | |
| VDAC1 | 0.98 | 0.000 | 0.000 | Voltage-dependent anion channel 1 | |
| CASP1 | 0.87 | 0.000 | 0.000 | Caspase-1, apoptosis-related cysteine peptidase | |
| SLC25A4 | 0.54 | 0.007 | 0.010 | Solute carrier family 25, member 4 | |
| SLC25A6 | 0.53 | 0.000 | 0.000 | Solute carrier family 25, member 6 | |
| BNIP1 | 0.48 | 0.000 | 0.000 | BCL2/adenovirus E1B 19 kDa interacting protein 1 | |
| BID | 0.38 | 0.002 | 0.003 | BH3 interacting domain death agonist | |
| BAD | 0.37 | 0.000 | 0.000 | BCL2-antagonist of cell death | |
| FADD | 0.37 | 0.001 | 0.002 | Fas-associated via death domain | |
| NDUFA13 | 0.29 | 0.018 | 0.023 | NADH dehydrogenase 1α subcomplex, 13 | |
| GZMB | –0.29 | 0.005 | 0.006 | Granzyme B | |
| HRK | –0.40 | 0.001 | 0.001 | Harakiri, BCL2 interacting protein | |
| NFKBIA | –0.44 | 0.001 | 0.001 | Nuclear factor of κ light polypeptide gene enhancer in B cells inhibitor, α | |
| CASP5 | –0.45 | 0.011 | 0.015 | Caspase-5, apoptosis-related cysteine peptidase | |
| APP | –0.45 | 0.000 | 0.000 | Amyloid β (A4) precursor protein | |
| CASP10 | –0.51 | 0.000 | 0.000 | Caspase-10, apoptosis-related cysteine peptidase | |
| ERCC3 | –0.56 | 0.000 | 0.000 | Excision repair cross-complementing rodent repair deficiency, complementation group 3 | |
| GZMH | –0.61 | 0.002 | 0.003 | Granzyme H | |
| DAP3 | –0.67 | 0.000 | 0.000 | Death-associated protein 3 | |
| TP53 | –0.74 | 0.001 | 0.001 | Tumor protein p53 | |
| ENDOG | –0.77 | 0.000 | 0.000 | Endonuclease G | |
| TNFSF10 | –0.81 | 0.000 | 0.000 | Tumor necrosis factor superfamily, member 10 | |
| PDCD5 | –0.95 | 0.000 | 0.000 | Programmed cell death 5 | |
| HTRA2 | –0.98 | 0.000 | 0.000 | HtrA serine peptidase 2 | |
| BAK1 | –1.03 | 0.000 | 0.000 | BCL2-antagonist/killer 1 | |
| CASP4 | –1.27 | 0.000 | 0.000 | Caspase-4, apoptosis-related cysteine peptidase | |
| CASP3 | –1.30 | 0.000 | 0.000 | Caspase-3, apoptosis-related cysteine peptidase | |
| PRODH | –1.74 | 0.000 | 0.000 | Proline dehydrogenase 1 | |
| CASP2 | –1.93 | 0.000 | 0.000 | Caspase-2, apoptosis-related cysteine peptidase | |
| SEPT4 | –3.18 | 0.000 | 0.000 | Septin 4 | |
| BCAP31 | –0.36 | 0.058 | 0.069 | B-cell receptor-associated protein 31 | |
| FAS | –0.68 | 0.067 | 0.079 | Fas (TNF receptor superfamily, member 6) | |
| CASP6 | –0.25 | 0.074 | 0.087 | Caspase-6, apoptosis-related cysteine peptidase | |
| CIDEB | –0.22 | 0.339 | 0.373 | Cell death–inducing DFFA-like effector b | |
| BAX | 0.12 | 0.405 | 0.438 | BCL2-associated X protein | |
| BBC3 | –0.07 | 0.574 | 0.606 | BCL2 binding component 3 | |
| BCL2L13 | 0.03 | 0.789 | 0.812 | BCL2-like 13 |
A/B: fold changes (log2) of average expressions of genes in type A versus those in type B malignant melanoma cell lines. Genes with P > 0.05 are in bold. FDR is by Benjamini and Hochberg method.
Biological Process, Molecular Function, Expression Pattern, and Statistics of 38 Survival-Apoptosis Genes
Focusing on biological process, molecular function, expression pattern, fold change, P value, and FDR of 84 genes, we identified 38 differentially expressed genes between the type A and type B malignant melanoma cell lines with known biological process in cell survival or death (Fig. 3). The survival genes consisted of antiapoptotic (n = 10) and antioxidant (n = 11) genes, whereas the cell death genes (n = 17) had a known proapoptotic role. The average changes of four up-regulated antiapoptotic genes ranged from 0.51-fold to 2.59-fold (log2, the same below) higher in the type A malignant melanoma cell lines than in type B, with P ≤ 2.8 × 10−6 and FDR ≤ 5.4 × 10−6. The average changes of six down-regulated antiapoptotic genes ranged from −0.53-fold to −1.69-fold lower in the type A malignant melanoma cell lines than in type B, with P ≤ 5.1 × 10−8 and FDR ≤ 1.1 × 10−7. Eleven antioxidant genes included 6 genes (MT3, PRDX5, PRDX3, GPX4, GLRX2, and GSR) having ≥0.90-fold higher expression in type A than in type B, with P ≤ 2.9 × 10−6 and FDR ≤ 5.6 × 10−6, and 5 antioxidant genes (HSPD1, GSS, SOD1, ATOX1, and CAT) having ≤−0.36-fold lower expression in type A than in type B, with P ≤ 1.1 × 10−3 and FDR ≤ 1.5 × 10−3 (Fig. 3). Of 17 proapoptotic genes, 9 (ENDOG, BAK1, CASP2, CASP4, PDCD5, HTRA2, SEPT4, TNFSF10, and PRODH) were at ≤−0.75-fold lower expression in the type A malignant melanoma cell lines than in type B, with P ≤ 2.1 × 10−12 and FDR ≤ 7.2 × 10−12. Eight proapoptotic genes (BAD, BNIP1, APAF1, BNIP3L, CASP7, CYCS, CASP1, and VDAC1) were at ≥0.31-fold higher expression in type A than in type B, with P ≤ 2.4 × 10−4 and FDR ≤ 3.7 × 10−4 (Fig. 3).
Figure 3.
Biological process, molecular function, expression pattern, and statistics of 38 survival-apoptosis genes.
qRT-PCR Validation of the Microarray Data
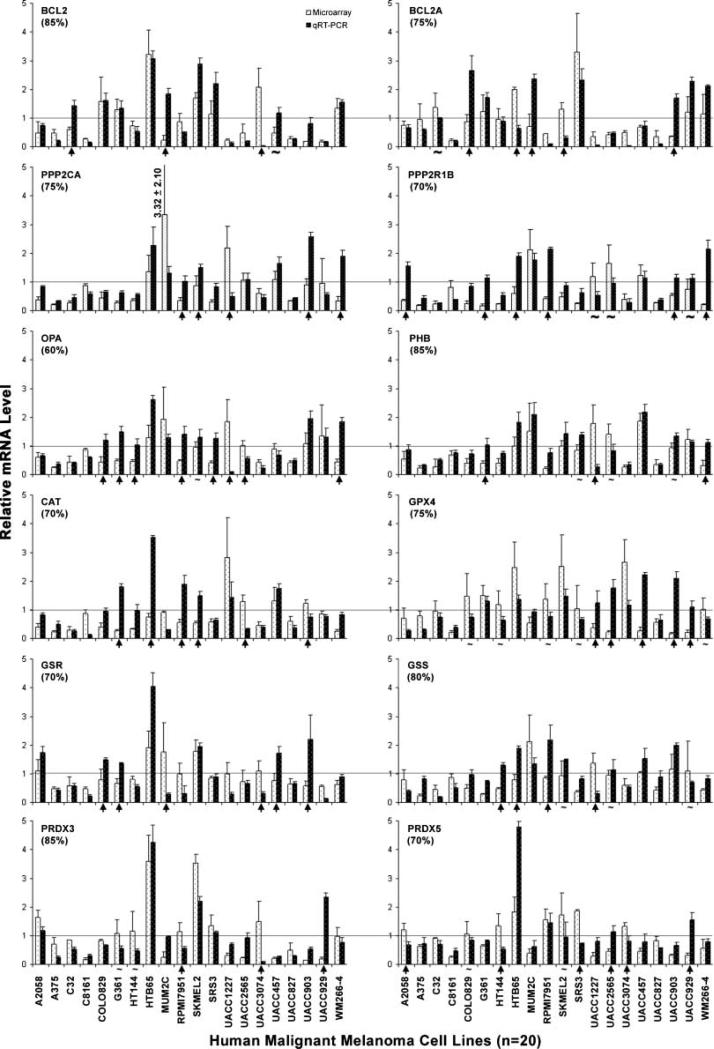
To validate the microarray results, we conducted qRT-PCR analysis on 13 genes, including 6 antiapoptotic genes (BCL2, BCL2A1, PPP2CA, PPP2R1B, OPA1, and PHB) and 6 antioxidant genes (CAT, GPX4, GSR, GSS, PRDX3, and PRDX5) and GAPDH as control for normalization of RNA loading and relative expression in 20 malignant melanoma cell lines (A2058, A375, C32, C8161, COLO829, G361, HT144, HTB65, MUM2C, RPMI7951, SKMEL2, SRS3, UACC1227, UACC2565, UACC3074, UACC457, UACC827, UACC903, UACC929, and WM266-4). The results indicated that of 240 data points tested (12 genes in 20 malignant melanoma cell lines, n = 240), 180 (75%) had essentially similar mRNA changes detected by both methods (Fig. 4). However, 60 (25%) data points displayed a significant (P < 0.05) difference but randomly distributed among 12 genes and 20 cell lines tested. These results showed overall agreement between microarray data and qRT-PCR results.
Figure 4.
The consistency in mRNA levels measured by microarray and qRT-PCR. Bar graphs indicate mRNA levels of 12 genes relative to GAPDH that were measured and analyzed by both microarray and qRT-PCR experiments. The bar with a line indicates the mean and SD of a mRNA level. The horizontal line shows the relative mRNA level of 1 that serves as an indicator for up-expression (above the line) and down-expression (below the line) of a gene. The arrow indicates a significant (P < 0.05) difference between up-expression and down-expression measured by these two methods. The waveline “~” means the difference is not significant (P > 0.05). Of these 240 comparisons (12 genes in 20 malignant melanoma cell lines), 200 (75%) display the consistency.
Discussion
Human malignant melanoma has a poor prognosis (25); differences in survival-apoptosis genes may be promising targets for therapy (26, 27). Our previous study, using mitochondria-focused cDNA microarray hMitChip3 and its related bioinformatic tools, led to identification of differentially expressed survival-apoptosis genes, their signaling pathways, and molecular mechanisms by which malignant melanoma cell lines were either resistant or sensitive to treatment-induced apoptosis (14). In this report, we identified expression patterns of 1,037 mitochondria-focused genes and significantly differentially expressed genes, resulting in the classification of 21 frequently used malignant melanoma cell lines into the type A (n = 12) and type B (n = 9). The microarray data were validated by qRT-PCR analysis of 13 genes in 20 malignant melanoma cell lines. Of 1,037 informative genes, 355 (34.2%) expressed in the completely opposite direction in up-regulation or down-regulation in these two types of malignant melanoma cell lines, indicating two different sets of molecules regardless of cause-effect relationships between differentially expressed genes and malignant melanoma cell malignancy. Of importance in cell survival, the up-regulated antiapoptotic and antioxidant genes and the down-regulated proapoptotic genes in the type A malignant melanoma cell lines were completely different from those in type B. These findings suggest the presence of at least two different survival pathways for these malignant melanoma cell lines. Drugs and methods targeting either pathway need to be designed and tested for development of new therapeutic intervention. Moreover, the patterns of gene expression provide potential biomarkers for development of novel molecular diagnosis and prognosis of malignant melanoma.
Human malignant melanoma cell lines used in this study were derived from biopsy of malignant primary or meta-static melanoma (Table 1). More than a dozen of these malignant melanoma cell lines are tumorigenic and metastatic in xenograft animal models. The fact that both tumorigenic and metastatic melanomas were clustered into either type A or type B indicated highly similar in vivo and in vitro cellular malignancy between type A and type B malignant melanoma cell lines. Although 21 malignant melanoma cell lines were cultured in five different media and all were harvested at ~90% confluence at the indicated passage numbers in the same laboratory after receiving from suppliers, the different culture media and cell passage numbers seem to have no obvious effects on the classification of cells into either type A or type B. For example, the cell lines cultured in RPMI 1640 were grouped in both type A and type B. Furthermore, there were no significant differences in age and gender distributions between these two types of malignant melanoma. Apparently, the clinical information, the xeno-graft tumorigenic and metastatic assays, and in vitro culture conditions were unable to differentiate these two types of malignant melanoma cell lines, suggesting that the underlying genomic diversities determine the molecular differences. Although these results were generated from in vitro cultured cells, our observations are very similar to those reported by Lin et al. (28) that the genomic alterations were strikingly similar between cultured melanoma cells and primary tumors. Therefore, the molecular diagnosis of heterogeneous malignant melanoma for clinical primary tumor tissues is needed for accurate diagnosis and targeted treatment.
One third of 1,037 mitochondria-focused genes (n = 355, 34.2%) displayed significant (P ≤ 0.030 and FDR ≤ 3.68%) differences (±≥2.0-fold) in expression between type A and type B. The known gene ontology of these 355 genes covers a broad biological spectrum, including 322 (91%) molecular functions, 316 (89%) cellular components, 302 (85%) biological processes, 193 (54%) enzymes, and 171 (48%) canonical biochemical and molecular pathways (data not shown). That is, these two types of malignant melanoma cell lines may live in the microenvironments with highly significant different molecules in quantity, quality, and machinery that must be treated with the different drugs aimed at the different molecular targets.
Homeostasis of cell survival and death are controlled by antiapoptotic, antioxidant, and proapoptotic genes. Up-regulation of antiapoptotic and antioxidant genes and down-regulation of proapoptotic genes confer a survival advantage and have been frequently reported to be involved in anticancer therapy. The resultant 38 survival-apoptosis genes displayed highly significant and common changes between the type A and type B malignant melanoma cell lines. Four up-regulated antiapoptotic genes (BCL2, BCL2A1, PPARD, and RAF1), six up-regulated antioxidant genes (MT3, PRDX5, PRDX3, GPX4, GLRX2, and GSR), and nine down-regulated proapoptotic genes (ENDOG, BAK1, CASP2, CASP4, PDCD5, HTRA2, SEPT4, TNFSF10, and PRODH) may lead to corresponding protein changes that confer survival and growth advantage to the type A malignant melanoma cell lines. In contrast, the completely different set of genes may mediate the survival and resistance to apoptosis of the type B malignant melanoma cell lines. These genes included six up-regulated antiapoptotic genes (PSEN1, API5, PPP2CA, PPP2A1A, PPP2R1B, and FIS1), five up-regulated antioxidant genes (HSPD1, GSS, SOD1, ATOX1, and CAT), and eight down-regulated proapoptotic genes (BAD, BNIP1, APAF1, BNIP3L, CASP7, CYCS, CASP1, and VDAC1). All these genes and expression changes seem consistent with phenotypic characteristics of malignant cells. Because these malignant melanoma cell lines are frequently used in melanoma research, our findings of the specific gene products in specific types of malignant melanoma cell lines facilitate study of drug-induced apoptosis of malignant melanoma cells. We do not know the consequence of up-regulation of eight proapoptotic genes (BAD, BNIP1, APAF1, BNIP3L, CASP7, CYCS, CASP1, and VDAC1) and nine proapoptotic genes (ENDOG, BAK1, CASP2, CASP4, PDCD5, HTRA2, SEPT4, TNFSF10, and PRODH) in the type A malignant melanoma and type B malignant melanoma cell lines, respectively. A plausible explanation is that a proapoptotic role of these genes might be recessive due to the up-regulation of so many antiapoptotic and antioxidant genes and the down-regulation of other proapoptotic genes in the same cell lines.
These findings have significant implications for translational research and clinical trials of malignant melanoma therapy. For example, the proapoptotic agent oblimersen has been used as an antisense drug targeted to mitochondrial bcl2 in a randomized phase III clinical trial comparing its combination with dacarbazine against dacarbazine alone in treating 771 patients with metastatic melanoma (29). These clinical trials revealed no statistical difference in overall survival (9.0 months versus 7.8 months; P = 0.077) probably because of a failure to measure bcl2 expression in malignant melanoma tumors (10, 29). Our results showed that 61.9% of malignant melanoma cell lines had up-regulation of bcl2 but 38.1% had the down-regulation (Fig. 3). Thus, the treatment with oblimersen should be administered only to the patients with bcl2 up-regulation, which requires preselection of an appropriate patient population before a clinical trial. In addition, our study revealed that several of 13 malignant melanoma cell lines with bcl2 up-regulation displayed the increase in expression of lactate dehydrogenase (LDH; Supplementary Table S3). If an elevated serum LDH interferes with the oblimersen activity (29), whether the up-regulated cellular LDH has the same effects is interesting and deserves further study for further enhancement of efficacy of oblimersen.
Thirty-eight of 84 (45.2%) antiapoptotic, proapoptotic, and antioxidant genes were identified. Other genes with relatively consistent expression patterns include four genes (CASP10, FAS, GZMH, and PDIA2) expressed at higher levels in the moderately pigmented normal melanocytes than in the lightly and darkly pigmented normal melanocytes and five genes (CASP3, CIDEB, MCL1, TXNRD2, and PTGES2) expressed at lower levels in four malignant melanoma cell lines (G361, A375, WM266-4, and SRS3) than in the other malignant melanoma cell lines (except for CASP3 also lower in UACC3074). Altogether, these genes account for only 56% (47 of 84) of antiapoptotic, proapoptotic, and antioxidant genes, two groups of malignant melanoma cell lines and two sets of survival-apoptosis genes reported may facilitate our understanding of molecular differences of malignant melanoma pathogenesis and may prove useful for the development of new molecular targets for therapeutic intervention, diagnostic markers for treatment indications, and prognostic markers for the clinical outcomes of patients with malignant melanoma.
Acknowledgments
We thank Drs. Paul Meltzer and Brian Nickoloff for generously providing several melanoma cell lines, John Lueders and Jian-Zhong Qin for excellent assistance in preparing and sending cell lines, and Dr. Yidong Chen for helpful discussion on calculation of a FDR using Benjamini and Hochberg method.
Grant support: Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH grant NIH-NIDDK-06-925 (Y.A. Su) and U.S. Department of Defense Congressionally Directed Medical Research Programs grant PT075745 (Y.A. Su). D.M. Su was from Walt Whitman High School (Bethesda, MD) as a special volunteer working on his science project. X. Wang and J. Zhao were supported by Lanzhou University School of Medical Sciences (Lanzhou, Gansu, China). P. He was supported by Division of Hematology, Center for Biological Evaluation and Research, Food and Drug Administration (Bethesda, MD). Y.J. Zhu was supported by Advanced Biomedical Computing Center, National Cancer Institute, NIH (Frederick, MD). O.M. Rennert was supported by Program in Reproductive and Adult Endocrinology, Laboratory of Clinical Genomics, Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH (Bethesda, MD).
Footnotes
ftp://ftp.ncbi.nih.gov/repository/UniGene/, August 4, 2008.
Supplementary material for this article is available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.American Cancer Society . Cancer facts and figures 2008. American Cancer Society; Atlanta: 2008. pp. 1–70. Available from: http://www.cancer.org. [Google Scholar]
- 2.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–7. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 3.MacKie RM, Bray C, Vestey J, et al. Melanoma incidence and mortality in Scotland 1979-2003. Br J Cancer. 2007;96:1772–7. doi: 10.1038/sj.bjc.6603801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garbe C, Blum A. Epidemiology of cutaneous melanoma in Germany and worldwide. Skin Pharmacol Appl Skin Physiol. 2001;14:280–90. doi: 10.1159/000056358. [DOI] [PubMed] [Google Scholar]
- 5.Lens MB, Dawes M. Global perspectives of contemporary epidemiological trends of cutaneous malignant melanoma. Br J Dermatol. 2004;150:179–85. doi: 10.1111/j.1365-2133.2004.05708.x. [DOI] [PubMed] [Google Scholar]
- 6.Eigentler TK, Caroli UM, Radny P, Garbe C. Palliative therapy of disseminated malignant melanoma: a systematic review of 41 randomised clinical trials. Lancet Oncol. 2003;4:748–59. doi: 10.1016/s1470-2045(03)01280-4. [DOI] [PubMed] [Google Scholar]
- 7.Schadendorf D, Ugurel S, Schuler-Thurner B, et al. Dacarbazine (DTIC) versus vaccination with autologous peptide-pulsed dendritic cells (DC) in first-line treatment of patients with metastatic melanoma: a randomized phase III trial of the DC study group of the DeCOG. Ann Oncol. 2006;17:563–70. doi: 10.1093/annonc/mdj138. [DOI] [PubMed] [Google Scholar]
- 8.Tuettenberg A, Schmitt E, Knop J, Jonuleit H. Dendritic cell-based immunotherapy of malignant melanoma: success and limitations. J Dtsch Dermatol Ges. 2007;5:190–6. doi: 10.1111/j.1610-0387.2007.06179.x. [DOI] [PubMed] [Google Scholar]
- 9.Middleton MR, Grob JJ, Aaronson N, et al. Randomized phase III study of temozolomide versus dacarbazine in the treatment of patients with advanced metastatic malignant melanoma. J Clin Oncol. 2000;18:158–66. doi: 10.1200/JCO.2000.18.1.158. [DOI] [PubMed] [Google Scholar]
- 10.Bedikian AY, Millward M, Pehamberger H, et al. Bcl-2 antisense (oblimersen sodium) plus dacarbazine in patients with advanced melanoma: the Oblimersen Melanoma Study Group. J Clin Oncol. 2006;24:4738–45. doi: 10.1200/JCO.2006.06.0483. [DOI] [PubMed] [Google Scholar]
- 11.Meier F, Satyamoorthy K, Nesbit M, et al. Molecular events in melanoma development and progression. Front Biosci. 1998;3:D1005–10. doi: 10.2741/a341. [DOI] [PubMed] [Google Scholar]
- 12.Kim R, Emi M, Tanabe K. The role of apoptosis in cancer cell survival and therapeutic outcome. Cancer Biol Ther. 2006;5:1429–42. doi: 10.4161/cbt.5.11.3456. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Chen Y, Wang BD, He P, Su YA. Differences in apoptosis and cell cycle distribution between human melanoma cell lines UACC903 and UACC903(+6), before and after UV irradiation. Int J Biol Sci. 2007;3:342–8. doi: 10.7150/ijbs.3.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q, Wu J, Nguyen A, et al. Molecular mechanism underlying differential apoptosis between human melanoma cell lines UACC903 and UACC903(+6) revealed by mitochondria-focused cDNA microarrays. Apoptosis. 2008;13:993–1004. doi: 10.1007/s10495-008-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai X, Wu J, Zhang Q, et al. Third-generation human mitochondria-focused cDNA microarray and its bioinformatic tools for analysis of gene expression. Biotechniques. 2007;42:365–75. doi: 10.2144/000112388. [DOI] [PubMed] [Google Scholar]
- 16.Su YA, Ray ME, Lin T, et al. Reversion of monochromosome-mediated suppression of tumorigenicity in malignant melanoma by retroviral transduction. Cancer Res. 1996;56:3186–91. [PubMed] [Google Scholar]
- 17.Trent JM, Stanbridge EJ, McBride HL, et al. Tumorigenicity in human melanoma cell lines controlled by introduction of human chromosome 6. Science. 1990;247:568–71. doi: 10.1126/science.2300817. [DOI] [PubMed] [Google Scholar]
- 18.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 19.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manoli I, Le H, Alesci S, et al. Monoamine oxidase-A is a major target gene for glucocorticoids in human skeletal muscle cells. FASEB J. 2005;19:1359–61. doi: 10.1096/fj.04-3660fje. [DOI] [PubMed] [Google Scholar]
- 21.Holland PM, Abramson RD, Watson R, Gelfand DH. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci U S A. 1991;88:7276–80. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 23.Seftor EA, Meltzer PS, Kirschmann DA, et al. Molecular determinants of human uveal melanoma invasion and metastasis. Clin Exp Metastasis. 2002;19:233–46. doi: 10.1023/a:1015591624171. [DOI] [PubMed] [Google Scholar]
- 24.Lee ML, Kuo FC, Whitmore GA, Sklar J. Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridizations. Proc Natl Acad Sci U S A. 2000;97:9834–9. doi: 10.1073/pnas.97.18.9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soengas MS, Lowe SW. Apoptosis and melanoma chemoresistance. Oncogene. 2003;22:3138–51. doi: 10.1038/sj.onc.1206454. [DOI] [PubMed] [Google Scholar]
- 26.Eberle J, Kurbanov BM, Hossini AM, Trefzer U, Fecker LF. Overcoming apoptosis deficiency of melanoma—hope for new therapeutic approaches. Drug Resist Updat. 2007;10:218–34. doi: 10.1016/j.drup.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Eberle J, Fecker LF, Forschner T, Ulrich C, Rowert-Huber J, Stockfleth E. Apoptosis pathways as promising targets for skin cancer therapy. Br J Dermatol. 2007;156(Suppl 3):18–24. doi: 10.1111/j.1365-2133.2007.07855.x. [DOI] [PubMed] [Google Scholar]
- 28.Lin WM, Baker AC, Beroukhim R, et al. Modeling genomic diversity and tumor dependency in malignant melanoma. Cancer Res. 2008;68:664–73. doi: 10.1158/0008-5472.CAN-07-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorigan P, Eisen T, Hauschild A. Systemic therapy for metastatic malignant melanoma—from deeply disappointing to bright future? Exp Dermatol. 2008;17:383–94. doi: 10.1111/j.1600-0625.2007.00673.x. [DOI] [PubMed] [Google Scholar]
- 30.Ikoma N, Yamazaki H, Abe Y, et al. S100A4 expression with reduced E-cadherin expression predicts distant metastasis of human malignant melanoma cell lines in the NOD/SCID/γCnull (NOG) mouse model. Oncol Rep. 2005;14:633–7. [PubMed] [Google Scholar]
- 31.Welch DR, Bisi JE, Miller BE, et al. Characterization of a highly invasive and spontaneously metastatic human malignant melanoma cell line. Int J Cancer. 1991;47:227–37. doi: 10.1002/ijc.2910470211. [DOI] [PubMed] [Google Scholar]
- 32.Gouon V, Tucker GC, Kraus-Berthier L, Atassi G, Kieffer N. Up-regulated expression of the β3 integrin and the 92-kDa gelatinase in human HT-144 melanoma cell tumors grown in nude mice. Int J Cancer. 1996;68:650–62. doi: 10.1002/(SICI)1097-0215(19961127)68:5<650::AID-IJC16>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 33.Thies A, Mauer S, Fodstad O, Schumacher U. Clinically proven markers of metastasis predict metastatic spread of human melanoma cells engrafted in scid mice. Br J Cancer. 2007;96:609–16. doi: 10.1038/sj.bjc.6603594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luca M, Xie S, Gutman M, Huang S, Bar-Eli M. Abnormalities in the CDKN2 (p16INK4/MTS-1) gene in human melanoma cells: relevance to tumor growth and metastasis. Oncogene. 1995;11:1399–402. [PubMed] [Google Scholar]
- 35.Su YA, Bittner ML, Chen Y, et al. Identification of tumor-suppressor genes using human melanoma cell lines UACC903, UACC903(+6), and SRS3 by comparison of expression profiles. Mol Carcinog. 2000;28:119–27. doi: 10.1002/1098-2744(200006)28:2<119::aid-mc8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]