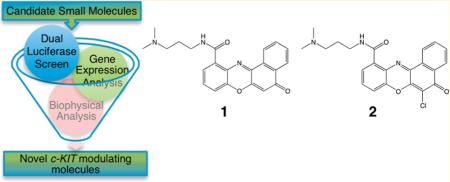
Abstract
There is considerable interest in the structure and function of G-quadruplex nucleic acid secondary structures, their cellular functions, and their potential as therapeutic targets. G-Quadruplex sequence motifs are prevalent in gene promoter regions and it has been hypothesized that G-quadruplex structure formation is associated with the transcriptional status of the downstream gene. Using a functional cell-based assay, we have identified two novel G-quadruplex ligands that reduce the transcription of a luciferase reporter driven from the G-quadruplex-containing c-KIT promoter. We have further shown that endogenous c-KIT expression in a human gastric carcinoma cell line is also reduced on treatment with these molecules. Biophysical analysis using surface plasmon resonance has shown that these molecules preferentially bind with high affinity to one of the two G-quadruplex sequences in the c-KIT promoter over double-stranded DNA. This work highlights the utility of cell-based reporter assays to identify new G-quadruplex binding molecules that modulate transcription and identifies benzo[a]phenoxazine derivatives as potential antitumor agents.
INTRODUCTION
Promoter regions of protein coding genes are significantly enriched in potential G-quadruplex-forming sequences relative to the rest of the genome, with >40% of human gene promoters containing one or more putative G-quadruplex forming sequences.1 G-Quadruplex-forming sequences have been identified in the gene promoters of a range of well-documented oncogenes: for example, c-MYC,2a KRAS,2b and c-KIT.2c,2d Siddiqui-Jain et al. have shown that the G-quadruplex found within the P1 promoter of c-MYC regulates transcription.2a Small molecules that bind and stabilize the c-MYC G-quadruplex also suppress c-MYC transcription in cancer cells.3 Furthermore, our own studies have shown the potential of G-quadruplex-recognizing small molecules to attenuate the transcriptional activity of the proto-oncogene c-KIT.4 c-KIT encodes a receptor tyrosine kinase for stem cell factor (SCF) and plays a role in cell survival, proliferation, and differentiation.5 Overexpression and/or mutation of c-KIT has been implicated in a wide range of cancers, including gastrointestinal stromal tumors (GIST), pancreatic cancers, seminomas (a subtype of testicular germ cell tumors), leukemias, and melanoma.6 The kinase inhibitor Imatinib (Glivec) is a significant therapy for gastrointestinal stromal tumors (GIST),7 but kinase domain mutations acquired during treatment can impart drug resistance.8 As a result, c-KIT remains activated in Glivec-resistant GIST cells, thus maintaining critical oncogenic signaling pathways.9 The human c-KIT promoter contains two G-quadruplex-forming sequences, positioned between −12 and −33 bp (c-kit1),2c and −64 and −83 bp (c-kit2),2d upstream of the transcription initiation site along with potential transcription factor-binding sites for Sp-1 and AP-2.10 Small molecules developed to target c-KIT G-quadruplexes and analyzed by biophysical methods show a range of binding affinities.4,11 Recently, a naphthalene di-imide derivative and 6-substituted indenoisoquinolines targeting c-KIT G-quadruplexes have been shown to impart growth arrest in a GIST cell line.12 The development of new c-KIT inhibitors with a mode of action at the transcriptional level rather than targeting the protein could offer therapeutic advantages.
To evaluate candidate small molecules that target the c-KIT promoter, we employed a cell-based approach that uses a luciferase gene reporter assay in a human gastric carcinoma cell line, HGC-27. The activity of a minimal c-KIT promoter after addition of small molecules is normalized to the expression of a second reporter under the control of a promoter that does not contain any G-quadruplexes (pRL-TK) and is therefore insensitive to G-quadruplex-related effects of the small molecule. Using this assay we have interrogated a library of small molecules, enriched for those which show an affinity for G-quadruplex secondary structure over duplex DNA,4,11 along with novel unpublished molecules. Positives were subjected to further validation using quantitative polymerase chain reaction (PCR) to measure c-KIT mRNA levels in treated HGC-27 cells and surface plasmon resonance (SPR) to measure small molecule binding to G-quadruplex sequences. As a result of this assay cascade, we report here the discovery of two novel G-quadruplex specific small molecules that down-regulate c-KIT expression.
RESULTS
Design of a Cell-Based G-Quadruplex Ligand-Screening Assay
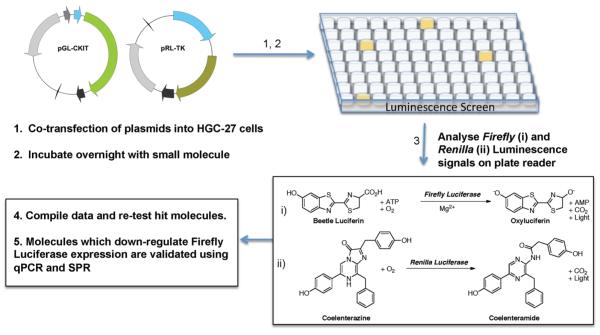
In order to develop a cell-based reporter assay for evaluating small molecules (Figure 1),13 we generated a pGL4.10 (Promega) plasmid construct containing the minimal c-KIT promoter driving expression of the synthetic firefly Photinus pyralis (luc2) gene (Figures S1 and S3, Supporting Information). Systems like this have been used to good effect, for example, in screening for small molecules which down-regulate interleukin-4 (IL4), the cytokine important in various allergic diseases.13b By using this dual luciferase system, we are able to measure the c-KIT promoter-linked expression of firefly luciferase at the same instance as a second control plasmid. The plasmid was optimally transiently transfected into human HGC-27 gastric carcinoma cells using Lipofectamine 2000 (Invitrogen). HGC-27 cells were chosen, as they have a measurable level of c-KIT expression, so factors needed for c-KIT transcription would be available to drive the plasmid-based c-KIT promoter-dependent reporter.14 A control plasmid, pRL-TK [Renilla luciferase, Rluc, linked to the herpes simplex virus thymidine kinase promoter to allow a low level constitutive expression (Figure S2, Supporting Information)] was cotransfected at a range of c-KIT:pRL-TK ratios to determine the optimal level of reporter to control expression. The final ratio of 5 parts c-KIT plasmid with 1 part pRL-TK gave the highest output level, taken as the ratio of firefly luciferase luminescence signal to Rluc luminescence signal (FF/RL), measured using a luminescence plate reader, which also gave a reproducible low error (approximately 10%). In order to reduce error between test wells, cells were transfected in 10 cm tissue culture plates and split into each 96-well test plate; this enabled each experimental plate to have equivalent transfection efficiencies whereupon changes in Renilla signal could be used to gauge the cell viability rather than to normalize for transfection efficiency.
Figure 1.
Schematic of the assay. See Figures S1–S3 for details of promoter and plasmid constructs.
In total, 173 in-house synthesized small molecules were evaluated at 0.2, 1, and 5 μM concentrations using the dual luciferase screen (as summarized in Figure 1). A reduction in the control Renilla expression level can arise from systemic effects such as cell death; therefore, any ligand that reduced the Renilla level to below 50% of control was deemed cytotoxic and was disregarded. In order to compare ligands from different experiments, all luminescence data (measured as a ratio of firefly/Renilla {FF/RL} signals) was normalized to the solvent/vehicle only control, giving a relative expression (%) for each ligand dose. Data were compiled based on the relative expression (%) at the maximum 5 μM dose, as no significant effects at 0.2 μM and 1 μM treatments were observed. Using the student t test (unpaired data), we found that five molecules gave a reduction in c-KIT promoter-linked firefly luciferase expression that was statistically different from that of control. This is summarized in Table 1, along with the number of small molecules that were cytotoxic at the different treatment concentrations and those that had no measurable response compared to control.
Table 1.
Outcomes of Dual Luciferase Assays: A Pool of 173 Small Molecules Was Assessed at 0.2, 1, and 5 μM Dosesa
| no. of molecules | |
|---|---|
| total small molecules screened | 173 |
| cytotoxic at 0.2 μM+ | 1 |
| cytotoxic at 1 μM+ | 11 |
| cytotoxic at 5 μM+ | 28 |
| down-regulation | 5 |
| no response | 128 |
Small molecules were scored as follows: giving no response, cytotoxic, or down-regulating c-KIT promoter activity. The five small molecules that down-regulated expression were further investigated by qPCR and SPR analysis.
The ideal method for statistical analysis of medium to high throughput screening data is via calculation of the Z-factor.15 This requires a positive and negative control to be tested on each plate. Since we had no notion of the outcome from our screen, this was not possible. We therefore chose to use a standardization approach using the Z-score (or Z-value). Z-Score analysis allows us to determine the magnitude of separation from the average population of the plate for each test ligand, as the number of standard deviations deflected from the mean (in this instance: −ve for down-regulation).
Data from the five ligands which were shown to reduce c-KIT promoter-linked firefly luminescence are summarized in Table 2. These small molecules reduced c-KIT promoter-linked expression in the range of 24% (±3%) to 74% (±5%) compared to control, i.e., up to 4-fold (with p-values in the range 2 × 10−4 to 2.4 × 10−2, and Z-scores were in the range of −5.2 to −1.6). While in some cases there was a slight dose-dependent reduction in expression, this was not statistically significant, and for the majority of molecules the maximum response was at the 5 μM dose. Ligands inhibiting c-KIT expression were then retested at 5 μM to confirm reproducibility of inhibition. Analysis of the absolute Renilla signal confirmed that any change in luciferase ratio was due to a reduction in firefly signal and not due to an increase in Renilla levels (Figure S4, Supporting Information). Based on these criteria, ligands A, B, and C were removed from the screen at this stage.
Table 2.
Expression of the c-KIT Quadruplex Reporter in the Dual-Luciferase Assay after Treatment with the Top Five Small Moleculesa
| dose (μM) |
||||||
|---|---|---|---|---|---|---|
| ligand | control | 0.2 | 1 | 5 | p | Z-score |
| A | 100 ± 9 | 96 ± 5 | 132 ± 6 | 24 ± 3 | 0.0002 | −4.3 |
| B | 100 ± 9 | 8.9 ± 7 | 127 ± 7 | 26 ± 1 | 0.0001 | −4.2 |
| 1 | 100 ± 10 | 108 ± 16 | 87 ± 2 | 38 ± 12 | 0.0001 | −5.2 |
| C | 100 ± 9 | 92 ± 1 | 89 ± 6 | 71 ± 11 | 0.024 | −3.7 |
| 2 | 100 ± 7 | 104 ± 9 | 96 ± 2 | 74 ± 5 | 0.0064 | −1.6 |
Ratios of firefly:Renilla signals (FF/RL) were calculated and normalized to the average control FF/RL ratio for each plate by expression as a percentage of this control value. p is calculated using the Student’s t-test at 5 μM dose. Z-scores were calculated as described in the Experimental Section with negative values indicating down-regulation.
qPCR Analysis of Candidate Small Molecules
Only two molecules showed a robust reduction in c-KIT promoter-linked expression without changes in Renilla signal. These molecules, 1 and 2, were taken forward for further analysis on the effects of endigenous c-KIT transcript levels using quantitative PCR (qPCR). HGC-27 cells were incubated with a single dose of each test molecule (5 μM, chosen because of significant response in luminescence screen) for 4, 8, or 24 h before cellular RNA was harvested and analyzed by qPCR. All expression was normalized to the constitutively expressed control genes; UBC, GusB, and YWHAZ and the geometric means calculated for each test molecule.16 Data from this qPCR analysis showed that the two molecules with a benzo[a]phenoxazine backbone, 1 and 2 (Figure 2), were able to reproducibly down-regulate c-KIT expression at all three time points tested (Figure 3). For molecule 1, expression of c-KIT at 8 and 24 h was significantly less than that at 4 h (~2.6 and 2.9, cf. ~1.5-fold, respectively). In contrast, treatment with 2 reduced c-KIT expression to similar levels at all three time points (Figure 3, Table S1, Supporting Information).
Figure 2.
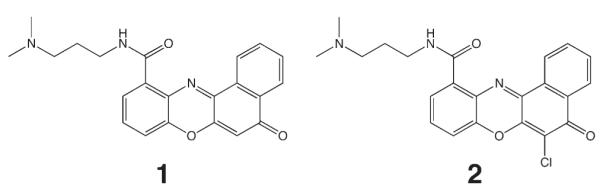
Structures of the small molecule benzo[a]phenoxazine derivatives that were positive hits from the screening cascade.
Figure 3.
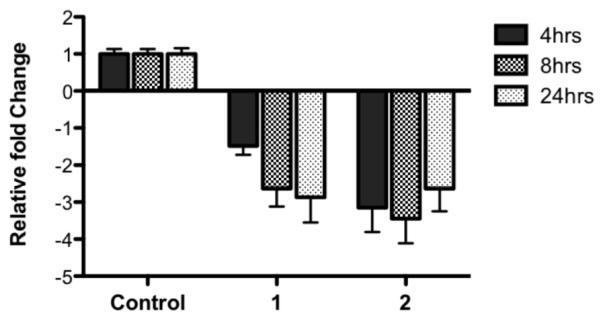
Expression of c-KIT in HGC-27 cells after treatment with benzo[a]phenoxazine derivatives 1 and 2. Expression is shown normalized relative to control gene expression. (n = 3, error bars represent SD).
Biophysical Analysis of Candidate Small Molecule–G-Quadruplex Affinity
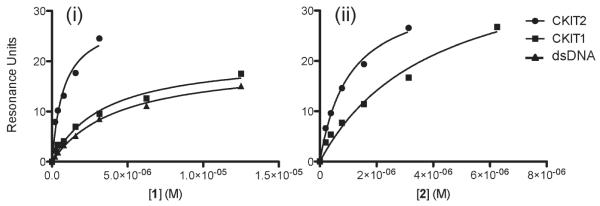
Equilibrium were performed using SPR to measure the binding affinities (Kd) of the compounds 1 and 2 with respect to the c-KIT G-quadruplex-forming DNA sequences c-kit1 and c-kit2 (Figure 4, illustrative sensorgram shown in Figure S5, Supporting Information). The association and dissociation profile indicates that neither 1 nor 2 formed covalent bonds with test DNA. It was found that within experimental error both ligands have a similar binding affinity for the c-KIT G-quadruplex-forming sequences. Both ligands had a stronger affinity for c-kit2 (Kd = 1.0 ± 0.3 μM for 1 and 1.1 ± 0.3 μM for 2) compared to c-kit1 (Kd = 9.6 ± 4 μM for 1 and 8.3 ± 0.1 μM for 2) resulting in a 9.6- and 7.5-fold preference for binding c-kit2 over c-kit1, for 1 and 2, respectively (Table S2, Supporting Information). For comparative purposes c-MYC and the human telomeric G-quadruplex sequence (hTelo) gave binding constants in the range of 2–5 μM (Table S2, Supporting Information), and thus ligands 1 and 2 show a specificity for c-kit2 in the range of 2–10-fold over other G-quadruplex-forming sequences. Binding of 1 to double-stranded DNA exhibited a 10-fold higher Kd than with c-kit2 (Kd = 9.8 ± 1.7{dsDNA}); however, no binding of 2 to dsDNA was detectable using this technique (no specific interactions up to 50 μM). Thus, in biophysical assays both ligands show a selectivity for the c-kit2 G-quadruplex over dsDNA, particularly so for 2. Both ligands were found to bind with approximately the same binding affinity for each G-quadruplex structure; thus, removal of the chlorine from 2 to give 1 did not alter the affinity for any of the G-quadruplexes within the error of the experiment.
Figure 4.
Representative surface plasmon resonance data plots for 1 (i) and 2 (ii) molecules binding with c-kit2, c-kit1, and dsDNA oligonucleotides (illustrative sensorgram in Figure S5). For 2, specific binding to dsDNA was not detectable up to 50 μM ligand concentration.
DISCUSSION
Using a cell-based assay approach, we have identified two benzo-[a]phenoxazine (BPO) molecules (1 and 2) which regulate c-KIT expression through binding with a G-quadruplex in the minimal core promoter. As judged by SPR analysis, these ligands showed binding to the two G-quadruplexes in the core c-KIT promoter, with a preference for c-kit2 over c-kit1. A preference for G-quadruplex over duplex binding was also demonstrated for 1 and 2, which was consistent with the observations from cellular studies that there was no change in expression of the Renilla control or of the indigenous control genes (GusB, UBC, and YWHAZ) on treatment of HGC-27 cells. Our data are consistent with a model in which the ligands bind to the c-KIT G-quadruplexes and modulate gene transcription. Both small molecules share an N-(3-dimethylaminopropyl)aminocarbonyl side chain at the 11 position of the benzophenoxazine core. Other molecules with similar basic side chains (including for example, morpholine, N-methylpiperazine, imidazole, or pyrrolidine in place of the dimethylamine) at the 11 position were negative in our screen (data not shown). A BPO derivative with a carboxylic acid group at the 11 position (compound 4, Scheme 1, Supporting Information) interestingly did not show any change in expression in the luciferase screen, nor was it cytotoxic. It is noteworthy that the nonchlorinated molecule 1 has a moderately improved potency in the biological screening assay, for example at 5 μM, firefly gene expression was reduced by 2.6-fold compared to the untreated control, compared to 1.4-fold for 2 at the same concentration. A slight difference in activity between the molecules was also observed in the gene expression analysis, where 2 has a slightly improved potency over 1. As discussed, removal of the chlorine did not alter the affinity for any of the G-quadruplexes within the error of the biophysical experiments. Therefore, the variation in results observed in the cell-based screen is unlikely to be because of a difference in affinity of the BPOs for the respective targets; rather the variation may occur because of other reasons such as differences in cell uptake, alternative binding positions, cellular modification, or breakdown pathways.
1 and 2 have a high affinity for the c-kit2 G-quadruplex secondary structure as measured by SPR (1.0 μM and 1.1 μM, respectively). This compares well with previously published c-KIT targeting ligand families: trisubstituted isoalloxazines,4a triarylpyridines,11a and oxazole-based peptide macrocycles,11c where equivalent c-KIT binding Kds are in the range of 2.8–9.7 μM,4a 0.2–25 μM,11a and 4–31 μM,11c respectively. These studies all used the c-kit2 sequence for binding experiments. Similarly where c-kit1 has been used for biophysical analysis, 1 and 2 again show similar affinities: 9.6 μM and 8.3 μM, respectively, compared with a range of 4.9–31 μM.4a The biological activity of trisubstituted isoalloxazines was previously investigated using qPCR in HGC-27 cells, where c-KIT mRNA levels were reduced to a maximum 2.5-fold compared to control after 24 h treatment, using the same concentration as in this study.4a 2 reduces c-KIT mRNA in the range 2.6–3.4-fold in our experiments, and 1 shows a time dependence with a maximum reduction in mRNA of 2.9-fold at 24 h. These data suggest that using both biophysical and qPCR methodology, 1 and 2 are at least as potent as previously developed ligands targeting c-KIT and have an improved biological activity and affinity for G-quadruplex structures compared with other compounds developed to date. These molecules have good aqueous solubility and minimal mechanism-unrelated cytotoxic effects and are easy to synthesize from commercially available building blocks.
In summary, we have used a cell-based assay to discover two new G-quadruplex ligands that can inhibit c-KIT transcriptional activity. This study highlights the potential of using a functional, phenotype-based assay to identify functional G-quadruplex ligands. The activity on c-KIT expression in human gastric carcinoma (HGC-27) cells demonstrates that these molecules warrant further consideration as potential therapeutics and will be vital as chemical biological tools for understanding the role of G-quadruplexes in gene expression.
EXPERIMENTAL SECTION
Chemicals and Reagents
All chemicals were from Sigma Aldrich, Poole, Dorset, UK, unless otherwise stated.
Cloning of Minimal c-KIT Promoter
The c-KIT core promoter region was taken as the region −124 to transcription start site (−1/+1).2d,10a,12a This c-KIT insert was constructed in a four-part ligation (see Figure S3, Supporting Information), to minimize any secondary structure formation and inserted into the commercially available pGL4.10 vector (Promega, Southampton, UK) at Acc65I and EcoRV restriction sites (New England Biolabs, Hitchin, UK). Three coding-strand oligonucleotides (AccEcoTop1–3) were annealed with a single template strand oligonucleotide (AccEcoBOT2) to give the Acc65I 5′ sticky end and EcoRV 3′ blunt end. This was ligated into the pGL4.10 vector digested with Acc65I and EcoRV using Quick-Stick Ligase (Bioline, London, UK). See Supporting Information for details.
Cells and Cell Culture
Human HGC-27 gastric carcinoma cells (ECACC # 94042256) were grown in Minimum Essential Media (supplemented with nonessential amino acids, 10% fetal calf serum, and 2 mM GlutaMAX (Invitrogen, Paisley, UK)) at 37 °C in 5% CO2 in air.
Small Molecule G-Quadruplex Ligand Screening Assay
Transient transfection of HGC-27 cells was carried out as follows. Plasmid (20 μg in total; ratio of 5:1 pGL-CKIT:pRL-TK) and Lipofectamine 2000 reagent (40 μL, Invitrogen) were each diluted with 1.5 mL of Optimem media (Invitrogen). DNA and Lipofectamine solutions were combined and incubated at room temperature for 20 min, as per manufacturer’s supplied protocol. DNA:Lipofectamine complexes were added dropwise to 90–95% confluent cells grown in 10 cm diameter tissue culture plates and incubated at 37 °C in 5% CO2 in air overnight (16 h). Cells were washed, counted, and seeded in standard media on white 96-well plates at 1.5 × 104 per well (Nunc, Thermo Fisher Scientific, Roskilde, Denmark). Cells were allowed to readhere for 6–7 h prior to treatment.
For the primary screen, cells (in 96-well plates) were treated with three doses of each test ligand (final concentrations of 0.2 nM, 1 μM, or 5 μM, diluted from 5 mM or 10 mM stocks) and vehicle only control, in triplicate, and incubated at 37 °C in 5% CO2 in air overnight (16 h). For further validation studies, single 5 μM doses of each test ligand plus vehicle-only control in triplicate were used. To avoid possible solvent effects, the maximum cellular exposure to DMSO was set at 0.1% in all cases. After incubation, cells were washed and lysed and levels of firefly and Renilla luciferase luminescence measured using the Promega Dual-Luciferase assay system according to the manufacturer’s protocol using an Orion II microplate luminometer (Berthold, Harpenden, UK) with reagent injection capabilities. Ratios of firefly signal:Renilla signal (FF/RL) were calculated and normalized to the average control ratio for each plate. They are expressed as a percentage of this control value.
qPCR Analysis of c-KIT Expression
For c-KIT transcript level analysis, cells were seeded in six-well plates at 2.5 × 105 per well (Nunc) and allowed to readhere (24 h) before they were treated at a single 5 μM dose of each test ligand, in triplicate, and incubated at 37 °C in 5% CO2 in air for the required time period (4, 8, and 24 h). Wells were washed with ice-cold PBS, and total RNA was extracted using an RNeasy kit (Qiagen, Crawley, UK) with supplied protocol. RNA levels were quantified using a Nanodrop spectrophotometer (Thermo Fisher Scientific, Wilmington, DE) and aliquots stored at −80 °C until required. cDNA was prepared from 1 μg of total RNA using QuantiTect Reverse Transcription Kit (Qiagen), using the supplied protocol. cDNA samples were analyzed quantitatively using the QuantiTect SYBR Green PCR Kit (20 μL total sample volume, Qiagen), in triplicate on a Roche Light Cycler 480 system (Roche Diagnostics Ltd., Burgess Hill, UK) using SG QuantiTect primers (Qiagen). Samples were analyzed in triplicate per experiment and normalized to the housekeeping genes UBC, GusB, and YWHAZ. Template-free controls were analyzed for each gene, and each experiment was carried out three times. Qiagen SG QuantiTect primers were used for all four genes. Analysis of the results was performed using qbase (Biogazelle), allowing normalization to multiple reference genes.16
Statistics
Graphpad Prism (version 5; Graphpad Software, San Fransisco, CA) was used to perform statistical analysis. Unpaired, two-tailed t-tests were used to determine statistical differences in relative luciferase activities of the control and small molecule ligand-treated c-KIT reporter constructs, and a value of p < 0.05 was considered significant. Z-score analysis was carried out for all screening data.
where z is the Z-score value, x is a raw value to be standardized (i.e., FF/RL ratio), and μ and σ are the mean and standard deviation, respectively, of the population (i.e., 96-well plate). Z-Scores show how far from the mean is the given test value. Negative Z-scores denote a down-regulation in expression, and positive would denote an up-regulation of expression.
Surface Plasmon Resonance
Surface plasmon resonance measurements were performed on a four-channel BIAcore 3000 optical biosensor system (GE Healthcare UK Ltd., Little Chalfont, UK) as detailed previously.11b SPR experiments were performed using five different immobilized DNA targets:c-kit1, c-kit2, c-MYC, hTelo, and double-stranded DNA (dsDNA). Each sample was repeated in duplicate, and the average response at equilibrium (Req) was plotted against concentration of analyte to generate a hyperbolic binding curve. The final graphs were obtained by subtracting blank sensorgrams from the appropriate duplex or G-quadruplex sensorgrams. Dissociation constants were determined by fitting the binding curve (from at least five concentrations) using the steady-state affinity algorithm (Biaevaluation 3.0.2). Each experiment was repeated in duplicate and the average taken; the errors represent the variance between the values.
General Procedures for Preparation of Substituted Benzo-[a]phenoxazines
Benzo[a]phenoxazines 1 and 2 were synthesized in two to three steps (Scheme S1, Supporting Information) using a procedure derived from Agarwal et al.17 The synthesis is described in detail in Supporting Information.
Supplementary Material
ACKNOWLEDGMENT
We acknowledge Dr. Mallesham Bejugam and Sebastian Müller for their kind donation of compounds to our collection. We thank Drs. Chris Lowe and David Tannahill for their critical reading of the manuscript and Dr. Sina Berndl for her assistance with NMR spectra. We thank Cancer Research UK for program funding and the BBSRC for project funding. We thank the EPSRC Mass Spectrometry Service for MS analysis.
Footnotes
Supporting Information. Details of all methods used, including full chemical syntheses,17 and all supplementary tables and figures cited in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Huppert JL, Balasubramanian S. Nucleic Acids Res. 2007;35:406. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2) (a).Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Proc. Natl. Acad. Sci. U.S.A. 2002;99:11593. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cogoi S, Xodo LE. Nucleic Acids Res. 2006;34:2536. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. J. Am. Chem. Soc. 2005;127:10584. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. Biochemistry. 2006;45:7854. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Liu JN, Deng R, Guo JF, Zhou JM, Feng GK, Huang ZS, Gu LQ, Zeng YX, Zhu XF. Leukemia. 2007;21:1300. doi: 10.1038/sj.leu.2404652. [DOI] [PubMed] [Google Scholar]
- (4) (a).Bejugam M, Sewitz S, Shirude PS, Rodriguez R, Shahid R, Balasubramanian S. J. Am. Chem. Soc. 2007;129:12926. doi: 10.1021/ja075881p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Waller ZAE, Sewitz SA, Hsu SD, Balasubramanian S. J. Am. Chem. Soc. 2009;131:12628. doi: 10.1021/ja901892u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Edling CE, Hallberg B. Int. J. Biochem. Cell Biol. 2007;39:1995. doi: 10.1016/j.biocel.2006.12.005. [DOI] [PubMed] [Google Scholar]
- (6) (a).Sakurai S, Fukasawa T, Chong JM, Tanaka A, Fukayama M. Jpn. J. Cancer Res. 1999;90:1321. doi: 10.1111/j.1349-7006.1999.tb00715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yasuda A, Sawai H, Takahashi H, Ochi N, Matsuo Y, Funahashi H, Sato M, Okada Y, Takeyama H, Manabe T. Mol. Cancer. 2006;5:46. doi: 10.1186/1476-4598-5-46. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) McIntyre A, Summersgill B, Grygalewicz B, Gillis AJ, Stoop J, van Gurp RJ, Dennis N, Fisher C, Huddart R, Cooper C, Clark J, Oosterhuis JW, Looijenga LH, Shipley J. Cancer Res. 2005;65:8085. doi: 10.1158/0008-5472.CAN-05-0471. [DOI] [PubMed] [Google Scholar]; (d) Miettinen M, Lasota J. Appl. Immunohistochem. 2005;13:205. doi: 10.1097/01.pai.0000173054.83414.22. [DOI] [PubMed] [Google Scholar]; (e) Smalley KSM, Nathanson KL, Flaherty KT. Cancer Res. 2009;69:3241. doi: 10.1158/0008-5472.CAN-08-4305. [DOI] [PubMed] [Google Scholar]
- (7).Dematteo RP, Heinrich MC, El-Rifai WM, Demetri G. Hum. Pathol. 2002;33:466. doi: 10.1053/hupa.2002.124122. [DOI] [PubMed] [Google Scholar]
- (8).Antonescu CR, Besmer P, Guo TH, Arkun K, Hom G, Koryotowski B, Leversha MA, Jeffrey PD, Desantis D, Singer S, Brennan MF, Maki RG, DeMatteo RP. Clin. Cancer Res. 2005;11:4182. doi: 10.1158/1078-0432.CCR-04-2245. [DOI] [PubMed] [Google Scholar]
- (9).Heinrich MC, Corless CL, Blanke CD, Demetri GD, Joensuu H, Roberts PJ, Eisenberg BL, von Mehren M, Fletcher CDM, Sandau K, McDougall K, Ou WB, Chen CJ, Fletcher JA. J. Clin. Oncol. 2006;24:4764. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- (10) (a).Park GH, Plummer HK, III, Krystal GW. Blood. 1998;92:4138. [PubMed] [Google Scholar]; (b) Chu TY, Besmer P. Proc. Natl. Sci. Counc., Repub. China, Part B: Life Sci. 1995;19:8. [PubMed] [Google Scholar]; (c) Huang S, Jean D, Luca M, Tainsky MA, Bar-Eli M. EMBO J. 1998;17:4358. doi: 10.1093/emboj/17.15.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11) (a).Waller ZAE, Shirude PS, Rodriguez R, Balasubramanian S. Chem. Commun. 2008:1467. doi: 10.1039/b718854d. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Dash J, Shirude PS, Hsu S-TD, Balasubramanian S. J. Am. Chem. Soc. 2008;130:15950. doi: 10.1021/ja8046552. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Jantos K, Rodriguez R, Ladame S, Shirude PS, Balasubramanian S. J. Am. Chem. Soc. 2006;128:13662. doi: 10.1021/ja064713e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12) (a).Gunaratnam M, Swank S, Haider SM, Galesa K, Reszka AP, Beltran M, Cuenca F, Fletcher JA, Neidle S. J. Med. Chem. 2009;52:3774. doi: 10.1021/jm900424a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bejugam M, Gunaratnam M, Müuller S, Sanders DA, Sewitz S, Fletcher JA, Neidle S, Balasubramanian S. ACS Med. Chem. Lett. 2010;1:306. doi: 10.1021/ml100062z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13) (a).Fan F, Wood KV. Assay Drug Dev. Technol. 2007;5:127. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]; (b) Choi JJ, Park BK, Song GY, Kim JS, Kim JH, Kim DH, Jin M. Arch. Pharm. Res. 2007;30:1102. doi: 10.1007/BF02980244. [DOI] [PubMed] [Google Scholar]
- (14).Hassan S, Kinoshita Y, Kawabami C, Kishi K, Matsushima Y, Ohashi A, Funasaka Y, Okada A, Maekawa T, He-Yao W, Chiba T. Dig. Dis. Sci. 1998;43:8. doi: 10.1023/a:1018851415704. [DOI] [PubMed] [Google Scholar]
- (15).Zhang JH, Chung TD, Oldenburg KR. J. Biomol. Screen. 1999;4:67. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- (16).Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. Gen. Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Agarwal NL, Schafer W. J. Org. Chem. 1980;45:2155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.