Summary
SIRPα is an inhibitory receptor present on myeloid cells that interacts with a widely distributed membrane protein CD47. The activating member SIRPβ, despite extensive sequence similarity to SIRPα in the extracellular region, shows negligible binding to CD47. The SIRPα / CD47 interaction is unusual in that it can lead to bidirectional signalling through both SIRPα and CD47. This review concentrates on the interactions of SIRPα with CD47 where recent data have shed light on the structure of the proteins including determining why the activating SIRPβ does not bind CD47, evidence of extensive polymorphisms and implication for the evolution and function of this protein and paired receptors in general. The interaction may be modified by endocytosis of the receptors, cleavage by proteolysis and through interactions of surfactant proteins.
Introduction
The regulation of the immune system is remarkable in being able to get the right sort of response to the required site and then to switch it off once the hazard is removed. Not surprisingly this is reflected in the large number of proteins at the surface of leukocytes that can affect their activity. These can be split into two broad categories, receptors for cell surface proteins and receptors for soluble factors such as chemokines and cytokines. The former have the advantage in that activity can be controlled by the requirement for cell cell contact and hence are ideal for sensing the environment. This review discusses recent advances in the inhibitory receptor signal regulatory protein alpha (SIRPα but also known by a variety of names such as CD172a, SHPS-1, BIT [1]) and its ligand CD47 (See BOX 1). Membrane receptors that can transmit signals are often highly restricted to a subset of cells whereas their ligands can be widespread. This is the case for SIRPα that is present mainly on myeloid cells although is also present on neuronal cells. SIRPα interacts with the widely distributed membrane protein CD47 but the outcome is complicated by the ability of CD47 to interact with other ligands and signal itself (Figure 1). This review will concentrate on publications in the last two years on the SIRPα recognition and constraints on the functional outcome. Detailed analysis of signalling mechanisms is outside the scope of the review although there are recent data [2-8]; however molecular interactions that influence whether signalling will occur will be discussed.
BOX 1. The SIRP family and possible ligands that may affect its function.
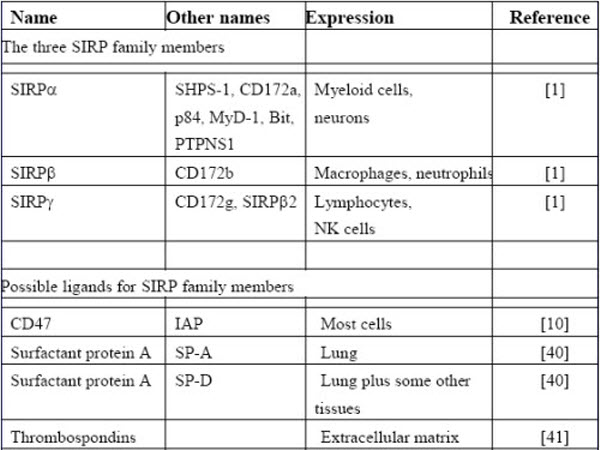
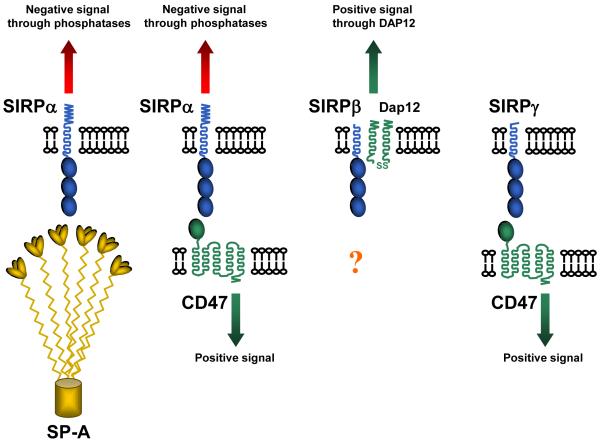
Figure 1.
Cartoon to show interactions of the extracellular region of the SIRP family. The three IgSF domains of the three SIRPs are shown as ovals (blue) and the single domain of CD47 as an oval (green). SIRPα and SIRPγ can both bind CD47 but SIRPβ does not. SP-A (yellow) denotes lung surfactant protein A that belongs to the collectin family that binds SIRPα giving signals through this protein and potentially blocking its interaction with CD47. A related protein SP-D (not shown) also binds SIRPα [32,33].
In addition to SIRPα, there are two closely related proteins in the SIRP family namely SIRPβ and SIRPγ. All three have three immunoglobulin superfamily (IgSF) domains in their extracellular region, SIRPβ has the potential to give activating signals through its association with the transmembrane adaptor protein DAP12 but does not react with CD47 – this makes the SIRP family a member of the class of proteins called paired receptors [1]. The third member SIRPγ binds CD47 albeit weaker than SIRPα but it lacks an extensive cytoplasmic region and is unlikely to signal. Recent data suggest it has a role in T cell migration in endothelium [9]. In considering the CD47 / SIRPα interaction it is necessary to consider signals through SIRPα, why SIRPβ does not bind along with how CD47 signals itself and the role of additional ligands for the SIRPs and CD47.
CD47 is involved in interactions other than with SIRPs such as in cis with integrins and trans with thrombospondins [10]. One puzzling aspect concerning CD47 is that much of the data on thrombospondin binding is from peptide binding analysis but the structure of the relevant thrombospondin domain shows that the peptides are in the core of the domain and not readily accessible [11]. This domain shows no affinity for the extracellular IgSF domain of CD47 [12] but direct binding of comparable proteins to CD47 at the cell surface has recently been shown [13] which might indicate that other parts of CD47 are involved. CD47 interactions and signalling are complex and beyond detailed analysis in this review.
The structure of SIRPα and its ligand CD47
X-ray crystallography structures have been determined for the ligand binding domain of mouse, rat and human SIRPα [14-16], the single IgSF domain of CD47, a complex of SIRPα domain 1 and CD47, the N terminal domains of two alleles of SIRPβ (the activating receptor) and SIRPγ [16] and a NMR structure for SIRPβ (Protein Data Bank Code; 2D9C). The interacting domains are typical IgSF V-set domains but the binding site of SIRPα is highly convoluted and made up from loops at the end of the domain in a manner analogous to binding of antigens by immunoglobulins and the T cell receptor rather than involving the faces of the domain as in most cell cell interaction proteins (see [17]). As expected from the sequence similarity of the SIRPα and SIRPβ there is little difference in the overall structures of the domains but subtle differences in the loops were found with evidence for considerable mobility in these [17]. Sequence analysis and mutagenesis have failed to provide a simple explanation for the failure of SIRPβ to bind to CD47 [14,15,17-19] but the structures suggest that this failure is due to subtle differences in the loops and involving indirect changes and not solely contact residues [17].
Polymorphisms in SIRPα and ligand recognition
Attention was drawn to the high level of polymorphisms in SIRPα N-terminal domain by Takenaka et al. [20] who showed that it was the Sirpa locus that was responsible for the observation that human bone marrow stem cells could be engrafted more efficiently into the SCID (severe combined immunodeficiency) NOD (non-obese) mouse strain than other strains. In general there is little cross reaction between CD47 from one species and SIRPα of another [21,22]. However the allele of SIRPα in the NOD mouse is sufficiently different from other mouse strains that it can bind human CD47 and give protection of the human cells in this mouse strain from host macrophages and xenogeneic engraftment occurs.
The SIRPα / CD47 interaction may have implications for xenotransplantation as pig tissues are an attractive source of organs for human tissue transplantation given the shortage of cadavers. Pig CD47 does not interact with human SIRPα and hence is not able to give inhibitory signals [21]. This has been shown to be relevant as manipulation of porcine cells to express human CD47 or addition of soluble human CD47-Fc fusion proteins reduces susceptibility of the porcine cells to phagocytosis by human macrophages [23]. Thus transgenic pigs expressing human CD47 may be advantageous as donors for human organ transplantation.
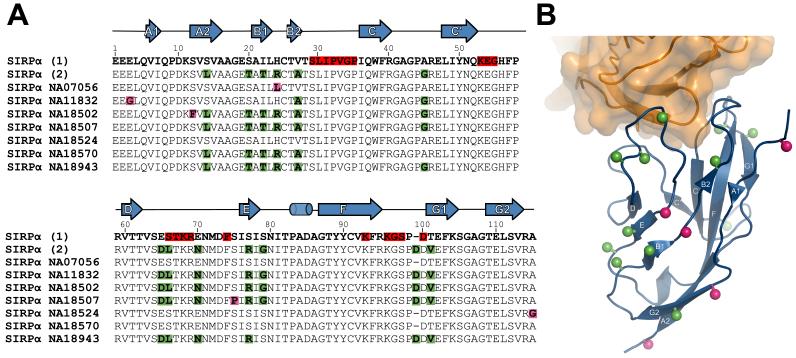
Human SIRPα domain 1, but not the other domains, is highly polymorphic and it was suggested that these polymorphisms in the SIRPα might affect its interaction with CD47 and be important in obtaining optimal engraftment of transplanted tissue [20,24]. However the recent data on the structure of the SIRPs and CD47 suggest that the polymorphisms in SIRPα are unlikely to affect CD47 binding in humans as they are distant from the binding site as illustrated in Figure 2. In addition two quite divergent alleles of SIRPα have same affinity of binding to CD47 [16].
Figure 2.
Polymorphisms and structure of SIRPα. (A) Alignments of the amino acid sequences of domain 1 of the two commonly studied SIRPα sequences (accession numbers CAA71403 and NP_542970 for sequence 1 and 2 respectively) together with polymorphisms identified in [20]. The positions of polymorphic residues are indicated by giving the residue for each sequence and indicating those that differ from SIRPα (1) by green or for those different from either the two common alleles SIRPα (1) or SIRPα (2) by magenta. Both these proteins bind CD47 with the same affinity [16]. The residues that form contacts with CD47 are highlighted in red and the positions of the beta strands and one helical region are shown above the alignment. (B) The positions of the polymorphic residues are mapped onto the SIRPα domain 1 - CD47 extracellular domain co-crystal structure [17]. The sidechains of the polymorphic residues are indicated with spheres using green and magenta to distinguish the polymorphic residues as in Figure 1A. This figure is from [16] and is reproduced with permission from Cell Press.
Why is the ligand binding domain of SIRPα so polymorphic if it does not affect ligand binding? The polymorphisms involve outpointing residues and it is likely the selective pressure involves binding. Inhibitory receptors may be good targets for pathogens and one idea is that the polymorphisms are important in helping the receptor avoid binding of pathogens and giving down regulation through the inhibitory receptor [16]. This concept is extended into suggesting that the role of the activating receptor is to provide a counterbalance in that pathogens that target the inhibitory receptor have a good chance of reacting with the activating receptor (because of its sequence similarity) and hence nullifying the effect. This counterbalance theory may be applicable to the evolution and function of other paired receptors [16].
When are interactions between SIRPα and CD47 functional? Areas of close contact
It is apparent that the leukocyte cell surface is highly complex with many different proteins that can interact with other cell surface proteins. There are constraints as to when these interactions can occur and be functional as illustrated by the extensive work on T cell and NK signalling involving immunological synapses [25] and other regions of close contact that precede the synapse itself [26]. In addition the Tyr residues in the cytoplasmic region of SIRPα need to be phosphorylated before they can bind phosphatases and mediate inhibition.
One restriction may be that cells have to be in close contact for sufficient time together with appropriate signals to lead to SIRPα phosphorylation. The dimensions of the extracellular regions of SIRPα and CD47 are compatible with the interaction occurring at sites of close contact between cells as the single domain of CD47 interacts with the N-terminal domain of the 3 domains of SIRPα (Fig. 1). Microscopy data show that SIRPα accumulates at a contact region with red blood cells that express CD47 [27]. A decrease in phosphorylation at areas of close contact between macrophages phagocytosing erythrocytes expressing CD47 and SIRPα is compatible with the attraction of phosphatases to SIRPα [27].
What regulates SIRPα phosphorylation? The roles of CD47 and integrin mediated effects has been clarified using CD47 knockout cells [28]. SIRPα was not constitutively phosphorylated in leukocytes but phosphorylation occurred on adherence. This effect occurred in CD47 deficient leukocytes so was not dependent on CD47 engagement although the presence of CD47 enhanced the effect. However in SIRPα expressing endothelial cells SIRPα phosphorylation was dependent on CD47 pointing to subtle differences in SIRPα activation according to cell type [28]. In neuronal cells Csk homologous kinase (CHK) can bind and phosphorylate SIRPα [29].
When are interactions between SIRPα and CD47 functional? Effects of other cells and proteins
Red blood cells from CD47 knockout mice are cleared rapidly when transferred to wild type mice which is compatible with CD47 giving an inhibitory signal to phagocytic SIRPα positive cells [30]. However this result raises questions as to why this CD47−/− mouse does not eat its own blood cells and become non-viable? The use of bone marrow chimeras showed that the absence of CD47 on the non-hematopoietic cells is vital in inducing the macrophages to be tolerant of the CD47−/− red cells [31]. Thus this signalling system in macrophages can be controlled by nonhematopoietic cells providing evidence for complex interrelationships in regulation of macrophages in tissues [31].
The lung surfactant proteins A and D (SP-A and SP-D) had been shown to interact with SIRPα (Fig. 1) and block its interaction with CD47 [32]. Further studies showed that SIRPα engagement through SP-A or SP-D lead to decreased uptake of apoptotic cells by alveolar cells. However during inflammation newly recruited macrophages escape this blocking and can then give efficient clearance of apoptotic cells [33]. This is an important concept as one is not be able to assume a receptor is available for ligand engagement just because of its presence.
A factor in the ability of receptor ligand interactions to occur is the close contact necessary as discussed above, but for continued signalling the proteins must continue to be present at the surface. In neural cells, confocal microscopy showed that SIRPα / CD47 can be removed from the cell surface by endocytosis providing a mechanism for switching off this interaction once it has occurred [34] .Whether this occurs in myeloid cells and indeed how general a mechanism this is remains to be resolved. The level of the receptor is important in determining the level of signalling as shown by the reduction of SIRPα expression with LPS [35]. This results in releasing the down regulation of macrophage activity that can also be achieved using SIRPα knockdowns [35].
The level of the ligand also affects the outcome. The use of heterozygous CD47−/+ mice showed that the level of CD47 on red blood cells was also critical in determining their phagocytosis if they were opsonised but not with unopsonised cells [36]. Another interesting result from neural cells on levels of expression is the finding that CD47 can be released from the cell surface by proteolysis [37]. One point that is not resolved in this study is where the cleavage occurs? Presumably it is between the extracellular domain and first transmembrane region but one would still expect the domain to be attached through the disulphide that links this domain to a short extracellular loop between transmembrane regions [38]. Whether cleavage of CD47 occurs on myeloid cells is an open question.
Again in the nervous system, a soluble version of SIRPα was identified biochemically as a component with presynaptic organizer activity [39]. Only part of this activity could be inhibited with CD47 suggesting a novel interaction. Recombinant proteins corresponding to SIRPβ and SIRPγ also worked in this assay pointing to a function dependent on part of the SIRP proteins distinct for that determining the specificity of SIRPα for CD47.
Conclusions
The SIRPα /CD47 interaction has been extensively characterised at molecular and cellular levels giving a greater appreciation of the roles of the proteins involved. The results have more general implications for the function and evolution of paired receptors and in understanding the factors that may restrict the availability of active receptors.
Acknowledgements
I am grateful to Marion H Brown and Deborah Hatherley for helpful comments and support from the MRC and the Wellcome Trust.
References
- 1.Barclay AN, Brown MH. The SIRP family of receptors and immune regulation. Nat Rev Immunol. 2006;6:457–464. doi: 10.1038/nri1859. [DOI] [PubMed] [Google Scholar]
- 2.Maile LA, Capps BE, Miller EC, Aday AW, Clemmons DR. IAP association with SHPS-1 regulates IGF-I signaling in vivo. Diabetes. 2008 doi: 10.2337/db08-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radhakrishnan Y, Maile LA, Ling Y, Graves LM, Clemmons DR. Insulin-like growth factor-I stimulates Shc-dependent phosphatidylinositol 3-kinase activation via Grb2-associated p85 in vascular smooth muscle cells. J Biol Chem. 2008;283:16320–16331. doi: 10.1074/jbc.M801687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sobota RM, Muller PJ, Khouri C, Ullrich A, Poli V, Noguchi T, Heinrich PC, Schaper F. SHPS-1/SIRP1alpha contributes to interleukin-6 signalling. Cell Signal. 2008;20:1385–1391. doi: 10.1016/j.cellsig.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Xi G, Shen X, Clemmons DR. p66shc Negatively regulates IGF-I signal transduction via inhibition of p52shc binding to SHPS-1 leading to impaired Grb2 membrane recruitment. Mol Endocrinol. 2008 doi: 10.1210/me.2008-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong LW, Kong XN, Yan HX, Yu LX, Chen L, Yang W, Liu Q, Huang DD, Wu MC, Wang HY. Signal regulatory protein alpha negatively regulates both TLR3 and cytoplasmic pathways in type I interferon induction. Mol Immunol. 2008;45:3025–3035. doi: 10.1016/j.molimm.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Dance M, Montagner A, Salles JP, Yart A, Raynal P. The molecular functions of Shp2 in the Ras/Mitogen-activated protein kinase (ERK1/2) pathway. Cell Signal. 2008;20:453–459. doi: 10.1016/j.cellsig.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Maile LA, Capps BE, Miller EC, Aday AW, Clemmons DR. Integrin-Associated Protein Association With Src Homology 2 Domain Containing Tyrosine Phosphatase Substrate 1 Regulates IGF-I Signaling In Vivo. Diabetes. 2008;57:2637–2643. doi: 10.2337/db08-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stefanidakis M, Newton G, Lee WY, Parkos CA, Luscinskas FW. Endothelial CD47 interaction with SIRPgamma is required for human T-cell transendothelial migration under shear flow conditions in vitro. Blood. 2008;112:1280–1289. doi: 10.1182/blood-2008-01-134429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown EJ, Frazier WA. Integrin-associated protein (CD47) and its ligands. Trends in Cell Biology. 2001;11:130–135. doi: 10.1016/s0962-8924(00)01906-1. [DOI] [PubMed] [Google Scholar]
- 11.Kvansakul M, Adams JC, Hohenester E. Structure of a thrombospondin C-terminal fragment reveals a novel calcium core in the type 3 repeats. EMBO J. 2004;23:1223–1233. doi: 10.1038/sj.emboj.7600166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams JC, Bentley AA, Kvansakul M, Hatherley D, Hohenester E. Extracellular matrix retention of thrombospondin 1 is controlled by its conserved C-terminal region. J Cell Sci. 2008;121:784–795. doi: 10.1242/jcs.021006. [DOI] [PubMed] [Google Scholar]
- 13.Isenberg JS, Annis DS, Pendrak ML, Ptaszynska M, Frazier WA, Mosher DF, Roberts DD. Differential interactions of thrombospondins-1, -2 and -4 with CD47 and effects on cGMP signaling and Ischemic injury responses. J. Biol. Chem. 2008 doi: 10.1074/jbc.M804860200. M804860200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatherley D, Harlos K, Dunlop DC, Stuart DI, Barclay AN. The structure of the macrophage signal regulatory protein alpha (SIRPalpha) inhibitory receptor reveals a binding face reminiscent of that used by T cell receptors. J Biol Chem. 2007;282:14567–14575. doi: 10.1074/jbc.M611511200. [DOI] [PubMed] [Google Scholar]
- 15.Nakaishi A, Hirose M, Yoshimura M, Oneyama C, Saito K, Kuki N, Matsuda M, Honma N, Ohnishi H, Matozaki T, et al. Structural insight into the specific interaction between murine SHPS-1/SIRP alpha and its ligand CD47. J Mol Biol. 2008;375:650–660. doi: 10.1016/j.jmb.2007.10.085. [DOI] [PubMed] [Google Scholar]
- 16.* Barclay AN, Hatherley D. The Counterbalance theory for Evolution and Function of Paired Receptors. Immunity. 2008;29:675–678. doi: 10.1016/j.immuni.2008.10.004. A perspective analysing new structural data on paired receptors and suggesting a theory for the evolution of paired receptors.
- 17.** Hatherley D, Graham SC, Turner J, Harlos K, Stuart DI, Barclay AN. Paired receptor specificity explained by structures of signal regulatory proteins alone and complexed with CD47. Mol Cell. 2008;31:266–277. doi: 10.1016/j.molcel.2008.05.026. Comprehensive X-ray crystallography analysis of SIRPα binding CD47 and also equivalent domains in SIRPβ and SIRPγ that characterise the specificity for CD47 binding.
- 18.Lee WY, Weber DA, Laur O, Severson EA, McCall I, Jen RP, Chin AC, Wu T, Gernert KM, Parkos CA. Novel structural determinants on SIRP alpha that mediate binding to CD47. J Immunol. 2007;179:7741–7750. doi: 10.4049/jimmunol.179.11.7741. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Tong Q, Zhou Y, Lee HW, Yang JJ, Buhring HJ, Chen YT, Ha B, Chen CX, Yang Y, et al. Functional elements on SIRPalpha IgV domain mediate cell surface binding to CD47. J Mol Biol. 2007;365:680–693. doi: 10.1016/j.jmb.2006.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.* Takenaka K, Prasolava TK, Wang JC, Mortin-Toth SM, Khalouei S, Gan OI, Dick JE, Danska JS. Polymorphism in Sirpa modulates engraftment of human hematopoietic stem cells. Nat Immunol. 2007;8:1313–1323. doi: 10.1038/ni1527. Showed that xenotransplantation of human bone marrow into mice was dependent on SIRPα and that the N-terminal domain of SIRPα was particularly polymorphic. .
- 21.Subramanian S, Parthasarathy R, Sen S, Boder ET, Discher DE. Species- and cell type-specific interactions between CD47 and human SIRPalpha. Blood. 2006;107:2548–2556. doi: 10.1182/blood-2005-04-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian S, Boder ET, Discher DE. Phylogenetic divergence of CD47 interactions with human signal regulatory protein alpha reveals locus of species specificity. Implications for the binding site. J Biol Chem. 2007;282:1805–1818. doi: 10.1074/jbc.M603923200. [DOI] [PubMed] [Google Scholar]
- 23.* Ide K, Wang H, Tahara H, Liu J, Wang X, Asahara T, Sykes M, Yang YG, Ohdan H. Role for CD47-SIRPalpha signaling in xenograft rejection by macrophages. Proc Natl Acad Sci U S A. 2007;104:5062–5066. doi: 10.1073/pnas.0609661104. Shows that failure of SIRPα to recognise CD47 across species has implications for successful xenotransplantation.
- 24.van den Berg TK, van der Schoot CE. Innate immune ‘self’ recognition: a role for CD47-SIRPalpha interactions in hematopoietic stem cell transplantation. Trends Immunol. 2008;29:203–206. doi: 10.1016/j.it.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 25.Bromley SK, Burack WR, Johnson KG, Somersalo K, Sims TN, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML. The immunological synapse. Annu Rev Immunol. 2001;19:375–396. doi: 10.1146/annurev.immunol.19.1.375. [DOI] [PubMed] [Google Scholar]
- 26.Douglass AD, Vale RD. Single-molecule microscopy reveals plasma membrane microdomains created by protein-protein networks that exclude or trap signaling molecules in T cells. Cell. 2005;121:937–950. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.** Tsai RK, Discher DE. Inhibition of “self” engulfment through deactivation of myosin-II at the phagocytic synapse between human cells. J Cell Biol. 2008;180:989–1003. doi: 10.1083/jcb.200708043. Evidence for the accumulation of SIRPα at synapses when interacting with CD47 and that this is associated with decreased tyrosine phosphorylation in the close proximity.
- 28.Johansen ML, Brown EJ. Dual regulation of SIRPalpha phosphorylation by integrins and CD47. J Biol Chem. 2007;282:24219–24230. doi: 10.1074/jbc.M701565200. [DOI] [PubMed] [Google Scholar]
- 29.Mitsuhashi H, Futai E, Sasagawa N, Hayashi Y, Nishino I, Ishiura S. Csk-homologous kinase interacts with SHPS-1 and enhances neurite outgrowth of PC12 cells. J Neurochem. 2008;105:101–112. doi: 10.1111/j.1471-4159.2007.05121.x. [DOI] [PubMed] [Google Scholar]
- 30.Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP. Role of CD47 as a marker of self on red blood cells. Science. 2000;288:2051–2054. doi: 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- 31.** Wang H, Madariaga ML, Wang S, Van Rooijen N, Oldenborg PA, Yang YG. Lack of CD47 on nonhematopoietic cells induces split macrophage tolerance to CD47null cells. Proc Natl Acad Sci U S A. 2007;104:13744–13749. doi: 10.1073/pnas.0702881104. Intriguing result showing that macrophage activity can be regulated by nonhematopoietic cells
- 32.Gardai SJ, Xiao YQ, Dickinson M, Nick JA, Voelker DR, Greene KE, Henson PM. By binding SIRPalpha or calreticulin/CD91, lung collectins act as dual function surveillance molecules to suppress or enhance inflammation. Cell. 2003;115:13–23. doi: 10.1016/s0092-8674(03)00758-x. [DOI] [PubMed] [Google Scholar]
- 33.* Janssen WJ, McPhillips KA, Dickinson MG, Linderman DJ, Morimoto K, Xiao YQ, Oldham KM, Vandivier RW, Henson PM, Gardai SJ. Surfactant proteins A and D suppress alveolar macrophage phagocytosis via interaction with SIRP alpha. Am J Respir Crit Care Med. 2008;178:158–167. doi: 10.1164/rccm.200711-1661OC. Surfactants in the lung may give constitutive signalling through SIRPα in the resting state and hence may restrict the availibility of SIRPα to interact with CD47.
- 34.* Kusakari S, Ohnishi H, Jin FJ, Kaneko Y, Murata T, Murata Y, Okazawa H, Matozaki T. Trans-endocytosis of CD47 and SHPS-1 and its role in regulation of the CD47-SHPS-1 system. J Cell Sci. 2008;121:1213–1223. doi: 10.1242/jcs.025015. Shows that SIRPα CD47 engagement leads to endocytosis
- 35.Kong XN, Yan HX, Chen L, Dong LW, Yang W, Liu Q, Yu LX, Huang DD, Liu SQ, Liu H, et al. LPS-induced down-regulation of signal regulatory protein {alpha} contributes to innate immune activation in macrophages. J Exp Med. 2007;204:2719–2731. doi: 10.1084/jem.20062611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsson M, Nilsson A, Oldenborg PA. Dose-dependent inhibitory effect of CD47 in macrophage uptake of IgG-opsonized murine erythrocytes. Biochem Biophys Res Commun. 2007;352:193–197. doi: 10.1016/j.bbrc.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Maile LA, Capps BE, Miller EC, Allen LB, Veluvolu U, Aday AW, Clemmons DR. Glucose regulation of integrin-associated protein cleavage controls the response of vascular smooth muscle cells to insulin-like growth factor-I. Mol Endocrinol. 2008;22:1226–1237. doi: 10.1210/me.2007-0552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rebres RA, Vaz LE, Green JM, Brown EJ. Normal ligand binding and signaling by CD47 (integrin-associated protein) requires a long range disulfide bond between the extracellular and membrane-spanning domains. J Biol Chem. 2001;276:34607–34616. doi: 10.1074/jbc.M106107200. [DOI] [PubMed] [Google Scholar]
- 39.Umemori H, Sanes JR. Signal regulatory proteins (SIRPs) are secreted presynaptic organizing molecules. J Biol Chem. 2008 doi: 10.1074/jbc.M805729200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akiyama J, Hoffman A, Brown C, Allen L, Edmondson J, Poulain F, Hawgood S. Tissue Distribution of Surfactant Proteins A and D in the Mouse. J. Histochem. Cytochem. 2002;50:993–996. doi: 10.1177/002215540205000713. [DOI] [PubMed] [Google Scholar]
- 41.Adams JC. Functions of the conserved thrombospondin carboxy-terminal cassette in cell-extracellular matrix interactions and signaling. Int J Biochem Cell Biol. 2004;36:1102–1114. doi: 10.1016/j.biocel.2004.01.022. [DOI] [PubMed] [Google Scholar]