Abstract
The development of the human cerebral cortex is an orchestrated process involving the birth of neural progenitors in the peri-ventricular germinal zones, cell proliferation characterized by both symmetric and asymmetric mitoses, followed by migration of post-mitotic neurons to their final destinations in 6 highly ordered, functionally-specialized layers1,2. An understanding of the molecular mechanisms guiding these intricate processes is in its infancy, substantially driven by the discovery of rare mutations that cause malformations of cortical development (MCD)3-6. Mapping of disease loci in putative Mendelian forms of MCD has been hindered by marked locus heterogeneity, small kindred sizes and diagnostic classifications that may not reflect molecular pathogenesis. Here we demonstrate the use of whole-exome sequencing to overcome these obstacles by identifying recessive mutations in WDR62 as the cause of a wide spectrum of severe cerebral cortical malformations including microcephaly, pachygria with cortical thickening as well as hypoplasia of the corpus callosum. Some patients with WDR62 mutations had evidence of additional abnormalities including lissencephaly, schizencephaly, polymicrogyria and, in one instance, cerebellar hypoplasia, all traits traditionally regarded as distinct entities. In mouse and humans, WDR62 transcripts and protein are enriched in neural progenitors within the ventricular and subventricular zones. WDR62 expression in the neocortex is transient, spanning the period of embryonic neurogenesis. Unlike other known microcephaly genes, WDR62 does not apparently associate with centrosomes and is predominantly nuclear in localization. These findings unify previously disparate aspects of cerebral cortical development and highlight the utility of whole-exome sequencing to identify disease loci in settings in which traditional methods have proved challenging.
Malformations of cortical development (MCD) are a diverse group of often-devastating structural brain disorders reflecting deranged neuronal proliferation, migration or organization. Application of traditional mapping approaches have proven to be particularly challenging for gene discovery in MCD syndromes, where kindreds with a single affected member are most common, linkage studies support high locus heterogeneity, and recent genetic findings have fundamentally challenged prior diagnostic nosology3,7,8. Based on the expectation that whole exome sequencing using next generation platforms9-11 can markedly improve gene discovery efforts in these situations, we applied this technology to the index case of a small consanguineous kindred (NG 26) (Fig. 2a) from Eastern Turkey who presented to medical attention due to failure to reach developmental milestones and was found on clinical exam to have microcephaly. Neuroimaging studies identified a complex array of developmental abnormalities including pachygria and thickened cortex (Figs. 1a-d, 3c and Supplementary Videos 1 and 2).
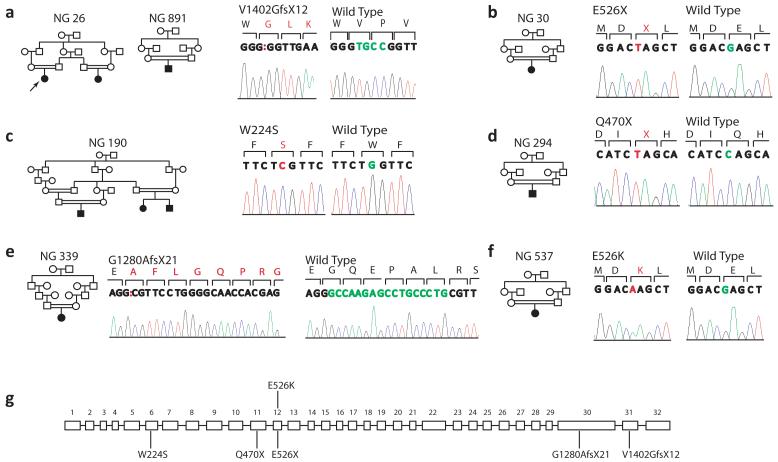
Fig. 2. Additional WDR62 mutations.
a-f, Pedigree structures along with mutated bases (red) and the corresponding normal alleles (green) are marked on the chromatograms (mutant–left; wild type-right). a, Families NG 26 and 891 harbor the identical 4 base pair deletion, whereas nonsense mutations leading to premature stop codons (X) are observed in NG 30 (b) and NG 294 (d). Missense mutations affecting conserved amino acids are seen in NG 190 (c) and NG 537 (f). In NG 339 (e), a 17 base pair deletion leads to a premature stop codon. (g) The locations of independent mutations are indicated on the genomic organization of WDR62.
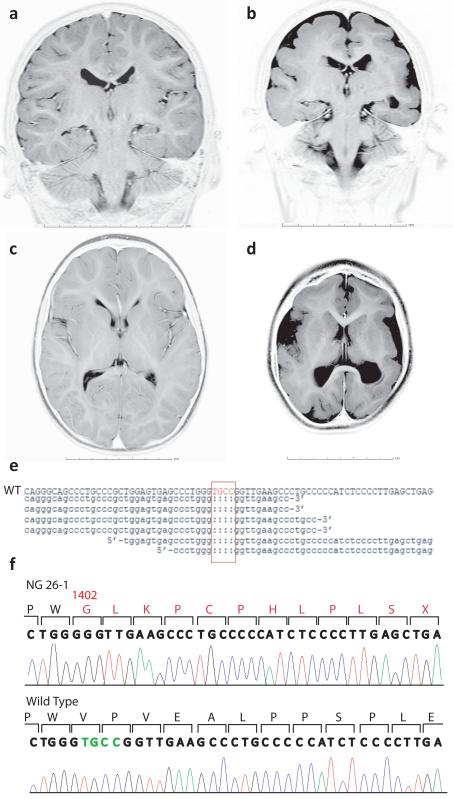
Fig. 1. Identification of a 4 base pair deletion in the WDR62 gene in a family with microcephaly and pachygria.
a-d, Coronal (a) and axial (c) MR images of a control subject as compared to NG 26-1 (b, d) confirms the clinical diagnosis of microcephaly and shows a diffusely thickened cortex, an indistinct gray-white junction, pachgyria and underoperculization. All images are T2 weighted (photographically inverted). Scale bars in centimeters are shown. e, A four base-pair deletion (red box) in the WDR62 is identified through exome sequencing (WT: wild type). f, Sanger sequencing confirms the deleted bases (in green). The altered amino acid sequence (starting at position 1402) leading to a premature stop-codon (X) is shown in red.
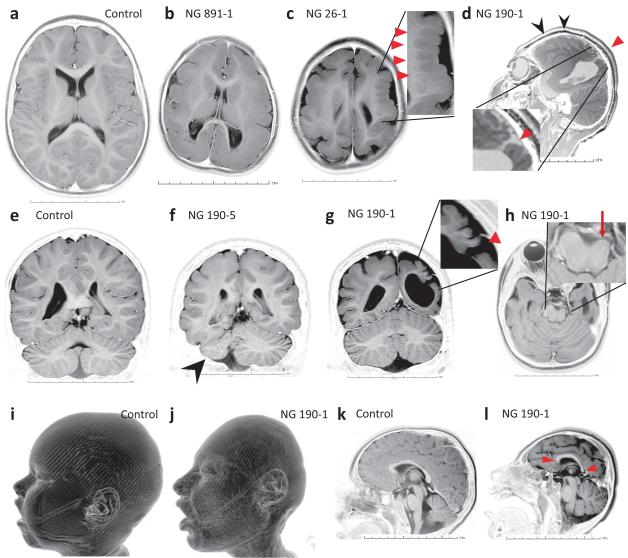
Fig. 3. Representative MRI images from patients demonstrating the wide spectrum of findings associated with mutations in WDR62.
a, e, i, k, Axial (a), coronal (e), sagittal (k) MRI images and three-dimensional (3D) surface rendering (i) of a control subject are shown. b, Microlissencephalic features with microcephaly, diffusely thickened cortex, loss of gray-white junction and pachygyria. c, Asymmetric microcephalic hemispheres with marked polymicrogyria (arrowheads). d, Significant polymicrogyria (black arrowheads) and open-lip schizencephaly (red arrowhead). f, Unilateral cerebellar hypoplasia (arrowhead). g, Open-lip schizencephaly (red arrowhead) and the polymicrogyric cortex. h, Unilateral brain stem atrophy (arrow). j, 3D surface rendering demonstrating craniofacial dysmorphology. l, Microcephaly, pachygyria and abnormally shaped corpus callosum (arrowheads).
We initially performed whole genome genotyping of the two affected members to identify shared homozygous segments (each > 2.5 centimorgans (cM)) that together comprised 80.11 cM (Supplementary Table 1). Given the substantial length of these shared segments, we next performed whole-exome sequencing of the index case using Nimblegen solid phase arrays and the Illumina Genome Analyzer IIx instrument9. We achieved a mean coverage of 44X and 94% of all targeted bases were read more than 4 times, sufficient to identify novel homozygous variants with high specificity (Supplementary Table 2). We identified two novel homozygous missense variants and one novel homozygous frameshift mutation within the shared homozygozity intervals (Supplementary Fig. 1 and Supplementary Table 3). The frameshift mutation occurred in WD repeat domain 62 (WDR62), deleting 4 base pairs in exon 31 (Fig. 1e). The full length WDR62 (NM_001083961) maps to chromosome 19q13.12 and encodes 1,523 amino acids. The identified mutation causes a frameshift in codon 1,402, resulting in a premature stop codon at position 1,414 (Fig. 1f). The mutation was confirmed to be homozygous in both affected subjects and to beheterozygous in both parents using Sanger sequencing (Fig. 2a and Supplementary Fig. 2). It was not observed in 1,290 Turkish control chromosomes.
Since this homozygous mutation in WDR62 was particularly compelling, we sought to determine whether mutations in this gene might account for additional cases of MCD. As the index case was ascertained with an initial diagnosis of pachygyria, we focused on a group of 30 probands who carried diagnoses of agyria or pachygyria and were products of consanguineous unions (inbreeding coefficient > 1.5%12). Among these patients, whole genome genotyping identified 8 with homozygosity of at least 2 cM spanning the WDR62 locus (Supplementary Notes). One of these affected subjects, NG 891-1, was found to have the identical homozygous haplotype spanning the WDR62 locus and had the same 4 base pair deletion (Fig. 2a and Supplementary Fig. 2). Although there was no known relatedness between the two pedigrees, the kinship coefficient of NG 891-1 with NG 26-1 and NG 26-4 was 5.47% and 3.72%, respectively, consistent with 4th degree relatedness (e.g. first cousins once removed).
Further Sanger sequencing of the complete coding region of WDR62 in the 7 remaining kindreds revealed five additional novel homozygous mutations (Fig. 2b-f). The affected member of kindreds NG 30 and NG 294 had homozygous nonsense mutations at codons 526 (E526X) and 470 (Q470X), respectively (Fig. 2b and d); subject NG 339-1 had a homozygous 17 base pair deletion leading to a frameshift at codon 1,280 that resulted in a premature termination codon following a novel peptide of 20 amino acids (Fig. 2e); subjects NG 190-1 and NG 537-1 respectively had novel homozygous missense variants W224S and E526K (Fig. 2c and f), which occurred at positions highly conserved among vertebrates, and were predicted to be deleterious by the Polyphen algorithm (Supplementary Fig.3). Moreover, following identification of the W224S mutation in NG 190, we ascertained two additional relatives affected with microcephaly and mental retardation (kinship coefficients of 7.47% and 5.81%) both of whom also proved to be homozygous for the same mutation. The resulting lod score for linkage to the trait within the expanded kindred was 3.64; the chromosome segment containing WDR62 was the sole homozygous region shared among all 3 affected subjects (Supplementary Fig. 4).
All of the newly identified mutations, except E526K, were absent from 1,290 Turkish and 1,500 Caucasian control chromosomes. The heterozygous E526K variant was detected in 3 apparently unrelated Turkish individuals who were neurologically normal (allele frequency 0.2%). As an additional control measure in the evaluation of these homozygous mutations, we sequenced the coding region of the gene in 12 consanguineous patients with non-neurological conditions who were found to have segments of homozygosity of at least 1 million base pairs spanning WDR62. None of these 12 individuals had protein coding changes in WDR62 (data not shown). Similarly, we identified only four heterozygous novel missense variants in WDR62 in the sequence of 100 whole exomes of subjects with non-neurological diseases (Supplementary Table 4). Public databases (dbSNP) showed no validated nonsense or frameshift alleles at this locus. Finally, we have not observed any copy number variants (CNVs) overlapping the coding regions of WDR62 in our own set of 11,320 whole genome genotypes (data not shown) and only one deletion identified by BAC array is reported in the Database of Genomic Variants (http://projects.tcag.ca/variation/).
All of the index cases with WDR62 mutations presented to medical attention with mental retardation and were found to have prominent microcephaly on physical exam; some also suffered from seizures (Supplementary Notes). Re-examination of the high field strength (3 Tesla) magnetic resonance imaging (MRI) scans of the affected subjects by independent neuroradiologists blinded to prior diagnoses identified hallmarks of a wide range of severe cortical malformations (summarized in Supplementary Table 5 and shown in Supplementary Videos). All 9 patients had extreme microcephaly, pachygyria and hypoplasia of the corpus callosum (Fig. 3). In addition, they demonstrated radiographic features consistent with lissencephaly, including varying degrees of cortical thickening and loss of gray-white junction (Fig. 3). Underopercularization (shallow Sylvian fissures) (Fig. 1b) was observed in 6 affected subjects. Two of the subjects had striking polymicrogyria that predominantly affected one hemisphere (Fig. 3c, d and g); in one this was associated with a unilateral open-lip schizencephaly characterized by a cleft surrounded by gray matter that extended into the ventricle (Fig. 3d and g). Other malformations observed included hippocampal dysmorphology with vertical orientation in 6 cases and a single case of unilateral dysgenesis of the cerebellum (Fig. 3f). There were no abnormalities of the brainstem with the exception of unilateral atrophy observed in one patient, most likely secondary to Wallerian degeneration from the severe cerebral abnormalities observed (Fig. 3h).
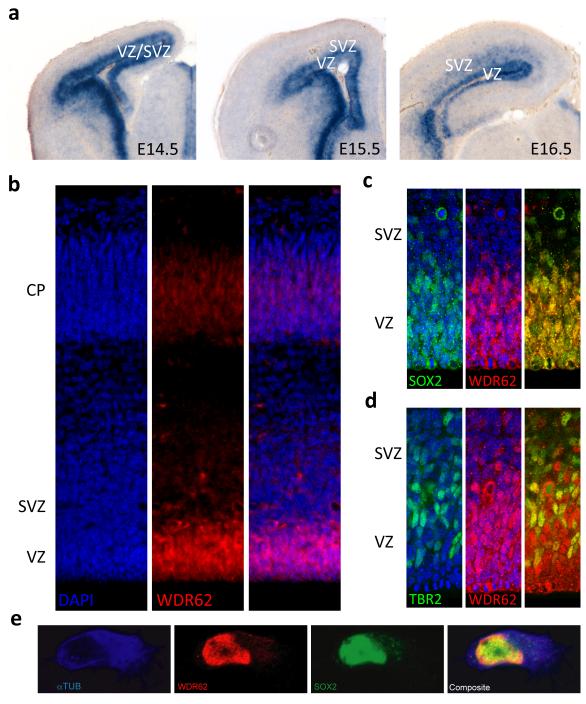
Given the wide range of cortical malformations associated with WDR62 mutations, we next investigated its expression in the developing mouse brain. Notably, during early development, in whole-mount embryos from E9.5 to E11.5, Wdr62 expression is prominent in neural crest lineages (Supplementary Fig. 5a-c). Wdr62 also shows striking expression in the ventricular and subventricular zones (VZ and SVZ, respectively) during the period of cerebral cortical neurogenesis (embryonic days 11.5 to 16.5), with expression decreasing in intensity by E17.5 (Fig. 4a and data not shown). In the cerebellum, Wdr62 is strongly expressed in precursors of granule neurons (GNPs) at late embryonic and early postnatal stages and by postnatal day 9 (P9), Wdr62 expression is dramatically reduced (Supplementary Fig. 5g and h). By postnatal day 21 (P21), low levels of Wdr62 expression are detected only in the hippocampus and the piriform cortex and transcription is absent among differentiated cortical neurons (Supplementary Fig. 5i).
Fig 4. Wdr62 expression in the developing mouse brain.
a, Wdr62 expression is enriched in the VZ and SVZ as seen with in-situ hybridization. b, WDR62 protein(red) distribution reveals a similar pattern. c, d, WDR62 (red) localizes to the nuclei and is expressed by neural stem cells and intermediate progenitors, as marked by SOX2 and TBR2 expression (green), respectively. e, Immunofluorescent staining for α-tubulin (cytoplasmic, blue), S0X2 (nuclear, green) and WDR62 (red) in E12.5 cortical neural progenitor cells reveals that the distribution of the WDR62 overlaps with that of SOX2 and is predominantly nuclear. (Nuclear staining by DAPI (blue) in b-d; Right most panels are composite images in b-e).
We next examined WDR62 protein expression using a previously characterized antibody13 (Fig. 4b-d). Both in the mouse and human fetal brain, WDR62 was enriched within the VZ and SVZ, consistent with our in-situ hybridization findings (Fig. 4 and Supplementary Figure 6). These stainings suggested that WDR62 localizes predominantly to the nucleus in neuronal cells, which we confirmed by immunofluorescence microscopy using cell cultures and western blotting with sub-cellular fractionation of cortical embryonic mouse cells with a second antibody (Fig. 4e and Supplementary Fig. 7). Genes previously implicated in microcephaly encode centrosomal proteins14-16, thus it is noteworthy that WDR62 is apparently not associated with the centrosome during mitosis (Supplementary Fig. 8).
Our findings implicate WDR62 in the pathogenesis of a spectrum of cortical abnormalities that heretofore have largely been conceptualized to be distinct3,7,8, suggesting that these diverse features can have unified underlying causation. It is noteworthy that WDR62 lies in a 10 million base pair interval that had previously been identified as a microcephaly locus, MCPH217. Although there was no imaging studies presented in the prior mapping of this locus, our findings suggest that WDR62 is the MCPH2 gene and extend the phenotype beyond microcephaly.
To seek further insight into the biological function of WDR62, we examined expression data of early embryonic development of mouse brain (GSE8091)18 for genes with expression profiles significantly correlated with that of WDR62 (Bonferroni corrected P < 0.01, n=1,104). Functional annotation suggested that positively correlated genes were enriched for those encoding nuclear proteins (Benjamini adjusted P = 6.23 ×10−30), RNA processing proteins (Benjamini adjusted P = 1.90 ×10−31) and cell cycle proteins (Benjamini adjusted P = 3.25 ×10−18). Negatively correlated genes encoded neuronal differentiation proteins (Benjamini adjusted P = 1.40 ×10−7). Several genes linked to developmental brain malformations, such as DCX, DCC, and BURB1B, are found in these enrichment sets (Supplementary Table 6). Further work will be required to extend these expression findings and clarify the normal role of WDR62.
To date, whole exome sequencing has led to the identification of a single new Mendelian locus for a genetically homogeneous condition10. Our results demonstrate that this technology will be particularly valuable for gene discovery in those conditions in which mapping has been confounded by locus heterogeneity and uncertainty regarding the boundaries of diagnostic classification, pointing to a very bright future for its broad application to medicine.
Methods Summary
Human Subjects
The study protocol was approved by the Yale Human Investigation Committee. Institutional review board approvals for genetic studies, along with written consent from all study subjects, were obtained at the participating institutions (see Supplementary Notes).
Targeted Exome Sequencing
Genomic DNA of sample NG 26-1 was captured on a NimbleGen 2.1M human exome array with modifications to the manufacturer’s protocol9, followed by single-read cluster generation on the Cluster Station (Illumina, San Diego, CA, USA). The captured, purified, and clonally amplified library targeting the exome from patient NG 26-1 was then sequenced on Genome Analyzer IIx. Two lanes of single-read sequencing at a read length of 74 base pairs was performed following the manufacturer’s protocol.
Exome Sequence Analysis
The sequence reads obtained were aligned to the human genome (hg18) using Maq 19and BWA 20 software. The percent alignment of the reads to both the reference genome as well as the targeted region, exome, was calculated using perl scripts9. Similarly, perl scripts were used for the detection of mismatch frequencies and error positions. SAMtools 21 was used for the detection of single nucleotide variations on the reads aligned with Maq. The indels were detected on the reads aligned with BWA for its ability to allow for gaps during the alignment. Shared homozygous segments of the affected individuals were detected using Plink software version 1.0612, and the variants were filtered for shared homozygosity. The variants were annotated for novelty with comparison to both dbSNP (build 130) and nine personal genome databases and previous exome sequencing experiments performed by our human genomics group. Novel variants were further evaluated for their impact on the encoded protein, conservation across 44 vertebrate species and C. elegans and D. melanogaster, expression patterns and potential overlap with known miRNAs.
Supplementary Material
Acknowledgments
We are deeply indebted to the patients and families who have contributed to this study with the hope that one day we can prevent childhood diseases, especially those due to consanguineous unions. We would like to thank Dr. James Noonan for expert advice and Cathy Camputaro, BSRT for her help with 3D reconstruction of the MR images. This study was supported by the Yale Program on Neurogenetics, Yale Center for Human Genetics and Genomics and NIH grants RC2 NS070477 (to MG), UL1 RR024139NIH (Yale CTSA) and UO1MH081896 (to NS). SNP genotyping was supported in part by an NIH Neuroscience Microarray Consortium award U24 NS051869-02S1 (to SM). RPL is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Competing financial interests The authors have a provisional patent application under consideration based on the findings of this work.
References
- 1.Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–22. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- 2.Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–35. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerrini R, Dobyns WB, Barkovich AJ. Abnormal development of the human cerebral cortex: genetics, functional consequences and treatment options. Trends Neurosci. 2008;31:154–62. doi: 10.1016/j.tins.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Guerrini R. Genetic malformations of the cerebral cortex and epilepsy. Epilepsia. 2005;46(Suppl 1):32–7. doi: 10.1111/j.0013-9580.2005.461010.x. [DOI] [PubMed] [Google Scholar]
- 5.Guerrini R, Carrozzo R. Epilepsy and genetic malformations of the cerebral cortex. Am J Med Genet. 2001;106:160–73. doi: 10.1002/ajmg.1569. [DOI] [PubMed] [Google Scholar]
- 6.Mochida GH, Walsh CA. Molecular genetics of human microcephaly. Curr Opin Neurol. 2001;14:151–6. doi: 10.1097/00019052-200104000-00003. [DOI] [PubMed] [Google Scholar]
- 7.Barkovich AJ, Kuzniecky RI, Jackson GD, Guerrini R, Dobyns WB. Classification system for malformations of cortical development: update 2001. Neurology. 2001;57:2168–78. doi: 10.1212/wnl.57.12.2168. [DOI] [PubMed] [Google Scholar]
- 8.Barkovich AJ, Kuzniecky RI, Jackson GD, Guerrini R, Dobyns WB. A developmental and genetic classification for malformations of cortical development. Neurology. 2005;65:1873–87. doi: 10.1212/01.wnl.0000183747.05269.2d. [DOI] [PubMed] [Google Scholar]
- 9.Choi M, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng SB, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–5. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng SB, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wasserman T, et al. A novel c-Jun N-terminal kinase (JNK)-binding protein WDR62 is recruited to stress granules and mediates a nonclassical JNK activation. Mol Biol Cell. 21:117–30. doi: 10.1091/mbc.E09-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bond J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–5. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Girimaji SC, Duvvari MR, Blanton SH. Mutations in STIL, encoding a pericentriolar and centrosomal protein, cause primary microcephaly. Am J Hum Genet. 2009;84:286–90. doi: 10.1016/j.ajhg.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thornton GK, Woods CG. Primary microcephaly: do all roads lead to Rome? Trends Genet. 2009;25:501–10. doi: 10.1016/j.tig.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts E, et al. The second locus for autosomal recessive primary microcephaly (MCPH2) maps to chromosome 19q13.1-13.2. Eur J Hum Genet. 1999;7:815–20. doi: 10.1038/sj.ejhg.5200385. [DOI] [PubMed] [Google Scholar]
- 18.Hartl D, et al. Transcriptome and proteome analysis of early embryonic mouse brain development. Proteomics. 2008;8:1257–65. doi: 10.1002/pmic.200700724. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.