Abstract
Variable interaction between the Bw4 epitope of HLA-B and the polymorphic KIR3DL1/S1 system of inhibitory and activating NK cell receptors diversifies the development, repertoire formation and response of human NK cells. KIR3DL1*004, a common KIR3DL1 allotype, in combination with Bw4+, HLA-B slows progression of HIV infection to AIDS. Analysis here of KIR3DL1*004 membrane traffic in NK cells shows this allotype is largely misfolded but stably retained in the endoplasmic reticulum, where it binds to the chaperone calreticulin and does not induce the unfolded protein response. A small fraction of KIR3DL1*004 folds correctly and leaves the endoplasmic reticulum to be expressed on the surface of primary NK and transfected NKL cells, in a form that can be triggered to inhibit NK cell activation and secretion of interferon-γ. Consistent with this small proportion of correctly-folded molecules, trace amounts of MHC Class I co-immunoprecipitated with KIR3DL1*004. There was no indication of any extensive intracellular interaction between unfolded KIR3DL1*004 and cognate Bw4+ HLA-B. A similarly limited interaction of Bw4 with KIR3DL1*002, when both were expressed by the same cell, was observed despite the efficient folding of KIR3DL1*002 and its abundance on the NK cell surface. Several positions of polymorphism modulate KIR3DL1 abundance at the cell surface, differences that do not necessarily correlate with the potency of allotype function. In this context our results suggest the possibility that the effect of Bw4+ HLA-B and KIR3DL1*004 in slowing progression to AIDS is mediated by interaction of Bw4+ HLA-B with the small fraction of cell surface KIR3DL1*004.
Introduction
Natural killer (NK) cells are important mediators of cytotoxicity and cytokine secretion in innate immunity. They also influence the adaptive immune response by secreting immunoregulatory cytokines that stimulate T cells and dendritic cells (1, 2). NK cells can recognize and lyse pathogen-infected, tumorigenic or allogeneic cells. Regulation of these effector functions stems from a balance of signaling through activating and inhibitory receptors. On human NK cells, the killer cell Ig-like receptor (KIR) family of receptors can both inhibit and activate the NK cell response through recognition of HLA class I proteins of the major histocompatibility complex (MHC) (3, 4). MHC class I interaction with inhibitory KIR is also critical for NK cell education and development of the NK cell repertoire (5, 6). KIR evolve rapidly and can exhibit a high degree of allelic polymorphism, approaching that of classical MHC class I genes (7). Genetic studies link specific KIR-HLA allele combinations with the clinical outcome of a diverse range of diseases, including viral and protozoan infection, autoimmune and inflammatory disease, tumor development, pregnancy-related disorders and the success of bone marrow transplantation (8–11). This broad influence on disease is consistent with a role for pathogen-driven, rapid KIR gene evolution and diversity balanced by the distinctive demands of reproductive selection (12, 13).
Several studies link disease progression from HIV to AIDS to the KIR3DL1 gene (reviewed in (8, 14)). The KIR3DL1/KIR3DS1 locus is highly polymorphic with more than 60 different alleles currently described (http://www.ebi.ac.uk/ipd/kir (15, 16). Of these, approximately 80% encode inhibitory (KIR3DL1) receptors, which recognize HLA-A and HLA-B containing the Bw4 serological epitope (17, 18) and the remaining alleles encode activating (KIR3DS1) receptors. Inhibitory allotypes differ in their level of expression at the cell surface (19, 20) and in their functional capacity to inhibit NK cell activation (21–24). Genetic studies indicate that the combination of KIR3DS1 and HLA-Bw4 is protective against progression to AIDS in HIV-infected individuals, but a ligand-receptor relationship between HLA-Bw4 and KIR3DS1 has not been demonstrated (discussed in (14, 25)). Similarly, the combination of HLA-Bw4 and inhibitory KIR3DL1*004 has been associated with slowed progression to AIDS (26), but neither a physical nor functional interaction between KIR3DL1*004 and HLA-Bw4 has been defined. That KIR3DL1*004 is largely retained inside the cell and is barely detectable at the cell surface (27), raises questions about the mechanism of action of this receptor that are investigated here.
The intracellular retention of KIR3DL1*004 depends on two amino-acid substitutions, leucine 86 and serine 182, respectively in the D0 and D1 Ig-like extracellular domains (27). These two substitutions are predicted to influence protein folding, which could affect both receptor transport and ligand binding (27, 28). The selective advantage of KIR3DL1*004 and HLA-Bw4 in responding to HIV infection (26) suggested the possibility that KIR3DL1*004 functions by binding HLA-Bw4 ligand inside the cell, as has been observed for endosome-localized KIR2DL4 (29). While, membrane traffic pathways for some human NK cell receptors have been characterized (30–36), little is known about inhibitory KIR export pathways that might influence their potential intracellular function. Whether KIR molecules can have intracellular interactions with their ligands, as reported for the Ly49 family of NK cell receptors in mice (37), is also not known. To address these issues and determine how KIR3DL1*004 might interact functionally with HLA-Bw4, we set out to define the molecular basis for intracellular retention of KIR3DL1*004 and determine whether it is capable of intracellular binding to HLA-Bw4. We confirm that KIR3DL1*004 is largely misfolded in the cell and establish that this misfolded protein does not induce a cellular stress response. We identify calreticulin as a key chaperone protein involved in the folding of KIR3DL1*004 and KIR3DL1*002, and presumably in intracellular retention of the misfolded KIR3DL1*004. We find no evidence of extensive intracellular receptor-ligand interactions with HLA-Bw4 for either KIR3DL1*004 or KIR3DL1*002. However, we do observe that small amounts of properly folded KIR3DL1*004 are present at the surface of both transfected NKL cells and primary NK cells. These KIR3DL1*004 molecules are functionally active and can mediate inhibition of IFN-γ production by NKL cells. We therefore conclude that the low abundance of KIR3DL1*004 on NK cell surfaces is more likely the cause of its favorable influence on HIV infection, rather than the intracellular activity of this enigmatic receptor.
Materials and Methods
Cell Lines and Transfectants
Human Natural Killer cell Line NKL (38) was maintained in RPMI 1640 supplemented with 10% Bovine Growth Serum (Hyclone), 100 U/ml Penicillin, 100 μg/ml Streptomycin, and 200 U/ml IL-2 (NCI BRB Preclinical Repository) at 37 °C/5% CO2. NKL transfectants expressing KIR3DL1*002 were a gift from L. L. Lanier (University of California San Francisco, CA, USA). Constructs encoding EGFP-tagged KIR3DL1*002 and KIR3DL1*004 (27) were transfected into NKL by electroporation (Amaxa). Stable transfectants were selected by G418 resistance and sorted by flow cytometry (FACSAria™; BD Biosciences). Using the sequence based AlleleSEQR kit (Atria Genetics), the NKL cell line was shown to have the following HLA type: HLA-A*02, A*11, B*35, B*51, C*04, C*15. HLA-B*51 has the Bw4 sequence motif at position 79–83 (RIALR), with isoleucine at position 80, that is associated with slower progression to AIDS (26, 39).
Human PBMC and KIR Genotyping
Human PBMC were collected from healthy donors in accordance with institutional review board-approved protocols. PBMC were isolated by density gradient centrifugation (Ficoll-Hypaque; GE Healthcare) and cryopreserved in Fetal Bovine Serum supplemented with 10% DMSO. Presence or absence of the 16 individual KIR genes was analyzed using a sequence-specific priming-PCR approach, described previously (40). No distinction could be made between KIR2DL5A or KIR2DL5B. The complete KIR3DL1/KIR3DS1 gene was amplified and sequenced using a high-resolution sequence-based typing approach described previously (41), with additional changes to improve sequence coverage and quality (J. Schellekens, unpublished data). Sequences were analyzed (SeqScape version 2.5; Applied Biosystems) and KIR3DL1/KIR3DS1 alleles were identified after comparison to the KIR Immuno Polymorphism Database (http://www.ebi.ac.uk/ipd/kir (42)).
Flow Cytometry
For NKL cell transfectants, 5×105 cells/ml were incubated in culture medium at various temperatures between 16 and 37 °C as indicated for 16 h. Cells were stained with PE-conjugated anti-KIR3DL1 mAb (177407; R&D Systems) or isotype-matched control (20102; R&D Systems) in PBS/1% BSA/0.01% sodium azide for 1 h at 4 °C. The 177407 mAb was made against KIR3DL1 and its specificity has been characterized as binding both KIR3DL1 and KIR3DL2, but only in the conformation of properly folded protein (28). Total (GFP) versus cell surface (mAb stained) KIR3DL1 protein expression was analyzed by flow cytometry (FACScan™; BD Biosciences) using FlowJo software (Tree Star).
Cell surface MHC class I protein was stained with pan MHC class I (W6/32; previously described (43)) or biotin-conjugated HLA-Bw4-specific (BIH0007; One Lambda) mAb. Isotyped-matched control (unlabeled or biotin-labeled G155-178; BD Biosciences), Alexa Fluor 647-PE-conjugated goat anti-mouse IgG (Invitrogen), and PE-Cy5-conjugated Streptavidin (BD Biosciences) were also used. To confirm Ab specificity, anti-HLA-Bw4 mAb was tested for binding to 721.221 cells that express either HLA-B*58:01 or HLA-B*15:01 and the mAb bound to cells expressing HLA-B*58:01, but not HLA-B*15:01.
For intracellular staining, cells were cultured at 25 or 37 °C for 12 h in the presence of 1 μl/106 cells/ml DMSO or GolgiPlug™ (BD Biosciences). Cells were fixed, permeabilized and stained with PE-conjugated anti-KIR3DL1 mAb (177407; R&D Systems) or the isotype-matched control (20102; R&D Systems) according to the manufacturer’s instructions (Cytofix/Cytoperm™ Plus Fixation/Permeabilization Kit; BD Biosciences). Surface epitopes were blocked by staining cells with 10μg/ml unlabeled anti-KIR3DL1 mAb or isotype-matched control for 30 min at 4 °C prior to fixation.
To detect transiently expressed protein, 5×105 NKL cell transfectants/ml were incubated in culture medium at 37 °C/5% CO2 for various times up to 8 h (as indicated) in the presence of PE-conjugated anti-KIR3DL1 mAb (177407, R&D Systems) or isotype-matched control (20102; R&D Systems). Cells were then placed on ice, washed in cold PBS, and stained with the same mAb to label surface protein (as above). The 0 h time point represents steady-state surface protein expression.
For PBMC analysis, cells were incubated at 25 and 37 °C for 16 h in DMEM supplemented with 10% Fetal Bovine Serum (Hyclone), 100 U/ml Penicillin, 100 μg/ml Streptomycin, 2 mM L-glutamine, 110μg/ml sodium pyruvate, 1X non-essential amino acids, and 55 μM β-mercaptoethanol. Cells were stained with FITC-conjugated anti-CD56 (NCAM16.2), PE-conjugated anti-KIR3DL1 (DX9) and PerCP-Cy5.5-conjugated CD3 (UCHT1) mAb (all BD Biosciences) or isotype-matched controls.
Immunoprecipitation and Immunoblotting
Chaperone proteins were immunoprecipitated using a protocol modified from one previously described (44). Cells were lysed on ice for 30 min in 2% CHAPS/50 mM Hepes, pH 6.8 or 7.4/200 mM NaCl/1 mM CaCl2 containing the following protease inhibitors: 1 mM PMSF and 10 μg/ml each of Antipain, Aprotinin, Chymostatin, Leupeptin and Pepstatin A (Sigma-Aldrich). Lysates (100 μg/lysate adjusted to equal volumes with lysis buffer) were pre-cleared with Protein G Sepharose™ (GE Healthcare), incubated with antibody for 1 h at 4 °C, and precipitated with protein G for 1 h at 4 °C. Beads were washed three times in 0.5% CHAPS/50 mM Hepes, pH 7.4/200 mM NaCl/1 mM CaCl2 and proteins eluted by boiling in 1X Laemmli sample buffer (45). Proteins were resolved by reducing SDS-PAGE (8% gel), transferred to nitrocellulose (Protran® BA83; Fisher Scientific), and probed by immunoblotting for the proteins indicated, using a standard protocol. Bands were visualized with enhanced chemiluminescence (GE Healthcare) and quantified using Quantity One® software (Bio-Rad). Blots were stripped and re-probed where indicated. For Endo H treatment, immunoprecipitated protein was digested with 2000–5000 U/reaction for 1 h at 37 °C according to the manufacturer’s instructions (NEB). For immunoprecipitation, 1 μg of mAb specific for KIR3DL1 (177407), MHC class I (W6/32), or isotype-matched control (G155-178; BD Biosciences) was used. 0.5 μl anti-GFP antibody (rabbit polyclonal; a gift from R. Parton, University of Queensland), and 4 μl anti-Calnexin (rabbit polyclonal, SPA-865; Assay Designs) and anti-Calreticulin (rabbit polyclonal, SPA-600; Assay Designs) antibodies were also used. For Western blotting, mAb specific for the following proteins were used: GFP (B34; Covance), β-actin (AC-15; Sigma), Calnexin (37/Calnexin; BD Biosciences), Calreticulin (FMC 75; Assay Designs), Bip/Grp78/KDEL (10C3; Assay Designs), Ubiquitin (horseradish peroxidase-conjugated, P4D1; Santa Cruz), and MHC class I (HC10; previously described (46)). Horseradish peroxidase-conjugated anti-mouse IgG (Thermo Scientific) and mouse IgG TrueBlot™ (eBioscience) were used where appropriate.
RT-PCR
To analyze NKL cell transfectants for an unfolded protein response, 106 cells/ml were cultured for 14 h in the presence of 2 μg/ml tunicamycin or DMSO (Sigma-Aldrich). Total RNA was isolated from 3×106 cells (RNeasy Mini Kit; Qiagen) and cDNA synthesized from 1 μg RNA by reverse-transcription (First Strand cDNA Synthesis Kit using SuperScript™ II; Invitrogen). Human XBP-1 and β-actin DNA was amplified by PCR using the following primers: XBP-1-forward (5′-AAACAGAGTAGCAGCTCAGACTGC-3′), XBP-1-reverse (5′-TCCTTCTGGGTAGACCTCTGGGAG-3′), β-actin-forward (5′-CAAGAGATGGCCACGGCTGCT-3′), and β-actin-reverse (5′-TCCTTCTGCATCCTGTCGGCA-3′). PCR products were purified (QIAquick PCR Purification Kit; Qiagen), digested with PstI (5 U/μg DNA), and resolved by 2–3% agarose gel electrophoresis.
Laser Scanning Confocal Microscopy
NKL cell transfectants were stained using a protocol modified from one previously described (47). 106 cells were fixed (Cytofix/Cytoperm™ Plus Fixation/Permeabilization Kit; BD Biosciences) for 12 min at 4 °C, washed twice in cold PBS/0.1% Tween® 20 (Sigma-Aldrich), and blocked in saponin-containing buffer (Perm/Wash; BD Biosciences) supplemented with 3% BSA (Sigma-Aldrich) and 5% Horse Serum (Hyclone) for 1 h at 4 °C. Cells were stained with 10 μg/ml mAb against MHC class I (W6/32) or isotyped-matched control (G155-178; BD Biosciences) in blocking buffer for 1 h at 4 °C, washed three times, stained with 2 μg/ml Alexa Fluor 555-conjugated donkey anti-mouse IgG (Invitrogen) for 1 h at 4 °C, followed by three washes in PBS/0.1% Tween® 20 and one wash in PBS. Cells were imaged by laser scanning confocal microscopy using a 63X objective lens and excitation wavelengths of 488 and 543 nm (SP5; Leica). To quantitate co-localization, Pearson’s correlation coefficients were calculated for cells in which the mean fluorescence intensity of the image for each channel was within 10% (LAS AF SP5; Leica).
Antibody mediated NK cell activation and inhibition and detection of IFN-γ release
For plate-bound mAb stimulation, 105 cells were cultured for 16 h in triplicate in 96-well, Nunc MaxiSorp plates (Fisher Scientific) coated with mAb as appropriate. To stimulate cells, 0.5 μg/ml 2B4 mAb (C1.7; Beckman Coulter) or isotype-matched control (107.3; BD Biosciences) was used. Activation resulted in an 11-fold to 68-fold increase in IFN-γ release (absolute values varied between independent experiments: 44–294 pg/ml for IgG1, 4987–13237 pg/ml for 2B4 ligation of KIR3DL1*002-GFP, and 860–3617 pg/ml for 2B4 ligation of KIR3DL1*004-GFP; Supplemental Fig. 1). To assess inhibition through KIR3DL1, plates were coated with 0.5 μg/ml 2B4 and either 0.1 – 1 μg/ml KIR3DL1 mAb (177407; R&D Systems) or isotype-matched control (G155-178; BD Biosciences). Human IFN-γ release into the supernatant was assessed by ELISA according to the manufacturer’s instructions (Ready-SET-Go! Kit; eBioscience). To allow comparison of independent experiments, absolute values of IFN-γ were normalized to isotype-matched controls.
Statistical Analysis
One-way analysis of variance (ANOVA) with randomized block followed by post test for linear trend, unpaired Student’s t tests, and two-way ANOVA with randomized block followed by Bonferroni post tests were performed as appropriate (GraphPad Prism 5.01). A threshold significance level of 0.05 was used.
Results
A defect in protein folding retains KIR3DL1*004 inside the cell
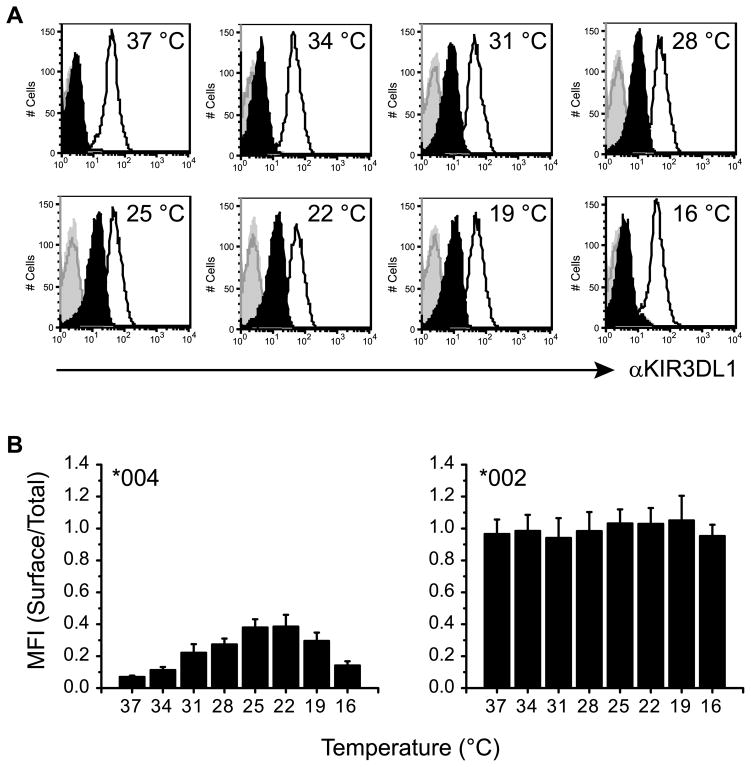
In order to characterize the mechanism for intracellular retention of KIR3DL1*004, NKL cells were transfected to express KIR3DL1*004 tagged with enhanced green fluorescent protein (GFP) and a stable cell line was produced. For comparison, a second stable cell line expressing a GFP-tagged allotype KIR3DL1*002 that is detectable in high amounts at the cell surface was produced. Although NKL cells have many properties of human NK cells, they do not express KIR, so provide a powerful vehicle for studying KIR allotypes in isolation. Each KIR variant expressed in these cells behaved as expected from previous analyses of allotype expression on peripheral blood NK cells or after transfection into the Jurkat T cell line (19, 27). At physiological temperature (37 °C), KIR3DL1*004 was barely detected at the cell surface using mAb 177407 that recognizes KIR3DL1 (28), compared to the total amount of KIR3DL1 expressed in the cell which was measured by GFP fluorescence (Fig. 1). In contrast, a robust cell surface signal was detected for KIR3DL1*002, relative to the amount of GFP signal from the transfected protein.
Figure 1. Culture at lower-than-physiological temperatures increases cell surface expression of KIR3DL1*004.
A, NKL cells transfected to express KIR3DL1*002-GFP (line histograms) or KIR3DL1*004-GFP (filled histograms) were incubated for 16 h at the temperatures indicated (top right of histogram). Cell surface expression of KIR3DL1 was detected by flow cytometry after staining with anti-KIR3DL1 mAb 177407 (black) or an isotype-matched control (grey). B, Mean fluorescence intensity (MFI) ratios for histograms shown in A, are plotted as surface (mAb)/total (GFP) KIR3DL1. Mean and SD across 3 independent experiments are shown. Temperature induced changes in KIR3DL1*004 but not KIR3DL1*002 expression are statistically significant as assessed by one-way ANOVA (P < 0.0001 and P = 0.2054 respectively). For KIR3DL1*004 expression, post test analysis for linear trend at temperatures permitting exocytosis (37–22 °C) is also statistically significant (P < 0.0001).
The previously observed membranous intracellular distribution of KIR3DL1*004 expressed in Jurkat cells was consistent with localization to the endoplasmic reticulum (ER) (27). As misfolded proteins are commonly sequestered in the ER we investigated whether defective protein folding could be responsible for the intracellular retention of KIR3DL1*004. Culturing cells at reduced temperature has been shown to promote folding of class I MHC molecules lacking bound peptide, resulting in increased expression at the cell surface (48). Incubation of NKL transfectants at reduced temperatures ranging from 16–34 °C similarly led to a marked increase in cell surface expression of KIR3DL1*004 with maximal expression at 22–25 °C (Fig. 1A). The decline in expression observed at 16–19 °C is consistent with a block in exocytosis (49–51). Comparing cell surface expression of KIR3DL1 (detected by staining with mAb) to the total amount of KIR3DL1 present in the cell (marked by GFP) confirmed that a greater proportion of total KIR3DL1*004 was expressed at the cell surface at reduced temperature (Fig. 1B). While incubation at lower temperatures significantly increased cell surface expression of KIR3DL1*004, expression levels never reached those of KIR3DL1*002. Expression of the KIR3DL1*002 allotype was not significantly increased by culturing NKL at 22–25 °C, consistent with the high cell surface expression of this receptor and indicative of efficient folding and export through the secretory pathway. The temperature dependence for cell surface expression of KIR3DL1*004 further supports a defect in protein folding and/or stability that can be overcome by allowing folding to take place at a slower rate.
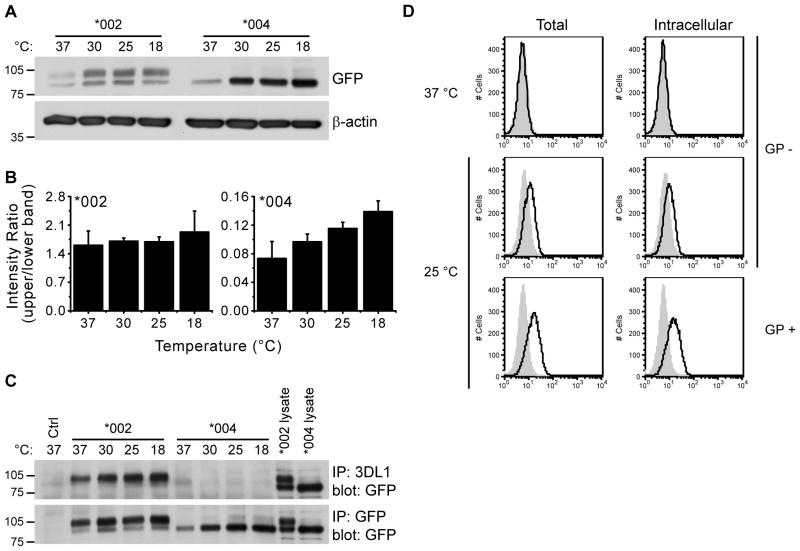
NKL cell lines expressing either KIR3DL1*004-GFP or KIR3DL1*002-GFP were incubated at different temperatures and lysates made from these cells were analyzed by immunoblotting with mAb against GFP to detect transfected KIR proteins. Following culture at physiological temperature, two protein species were detected for KIR3DL1*002, but only one for KIR3DL1*004 (Fig. 2A). The single band for KIR3DL1*004 ran at an equivalent molecular mass to the lower band present in the KIR3DL1*002 doublet. Lower temperatures induced the appearance of a faint but measurable upper band in KIR3DL1*004-GFP-expressing cells (Fig. 2A and 2B). This second KIR3DL1*004 band ran at an equivalent molecular mass to the upper band of the KIR3DL1*002 doublet. These two bands likely represent two stages of maturation during KIR3DL1 biosynthesis, with the upper band corresponding to the mature glycosylated form of KIR3DL1 that is present at the cell surface and the lower band representing unfolded immature protein. Quantification revealed a progressive increase of the faint upper band in the KIR3DL1*004-expressing cells, compared to total protein, as culture temperature was lowered. This suggested improved KIR3DL1*004 folding and progression along the secretory pathway under low temperature conditions. Reduced temperature also increased the total amount of KIR3DL1 protein present in the lower band (Fig. 2A), likely reflecting increased protein stability at lower temperatures due to reduced protease activity. In similar analysis of KIR3DL1*002-expressing cells, there were no statistically significant changes in the relative amounts of the two forms at temperatures that are favorable for Golgi export, and total protein amounts were increased, also consistent with reduced protease activity at lower temperatures. In conjunction with the FACS analysis (Fig. 1), this confirms the efficient folding and progression of KIR3DL1*002 along the secretory pathway.
Figure 2. KIR3DL1*004 is largely mis-folded.
A–C, NKL cells expressing KIR3DL1*002-GFP (*002) or KIR3DL1*004-GFP (*004) were incubated for 16 h at the temperatures indicated (°C). A, Equivalent amounts of whole cell lysates were resolved by reducing SDS PAGE and probed for the proteins indicated. B, Quantification of the Western blots shown in A. Band intensities are plotted as upper band/lower band. Mean and SD across 3 independent experiments are shown. Temperature induced changes in KIR3DL1*004 but not KIR3DL1*002 expression are statistically significant as assessed by one-way ANOVA (P = 0.0025 and P = 0.7252 respectively). For KIR3DL1*004 expression, post test analysis for linear trend is also statistically significant (P = 0.0004). C, KIR3DL1 was immunoprecipitated with anti-KIR3DL1 (upper) or -GFP (lower) antibodies, and detected by Western blotting for GFP. Ctrl = untransfected NKL (upper) or NKL transfected to express KIR3DL1*002 lacking GFP (lower). The migration position of molecular weight markers (kD) is shown at the left of panels A and C. D, NKL expressing KIR3DL1*004-GFP were incubated at 37 (upper) or 25 °C (middle and lower) for 12 h in the presence of DMSO (GP -, upper and middle) or GolgiPlug™ (GP +, lower) to inhibit protein transport through the ER and Golgi complex. Surface epitopes were blocked by staining with unlabeled anti-KIR3DL1 mAb (Intracellular, right) or an isotype-matched control (Total, left) prior to fixation and permeabilization. Cells were then stained for KIR3DL1 with PE-conjugated mAb (black line histogram) or an isotype-matched control (grey filled histogram).
To investigate KIR3DL1*004 folding further, we used mAb 177407 to immunoprecipitate KIR3DL1*002 and KIR3DL1*004 from NKL transfectants incubated at 18–37 °C (Fig. 2C). The binding specificity of mAb 177407 was recently shown to be sensitive to the folding conformation of KIR3DL1 (28). Immunoprecipitation of KIR3DL1*002 resulted in detection of the upper band only, while neither doublet species could be readily immunoprecipitated from KIR3DL1*004-GFP-expressing cells. This result from immunoprecipitation further establishes that mAb 177407 preferentially recognizes correctly folded, mature KIR3DL1. In contrast, both KIR3DL1*002 and KIR3DL1*004 were readily immunoprecipitated with anti-GFP antibody from cells incubated at all temperatures (Fig. 2C). In agreement with these findings is the observation that intracellular KIR3DL1*004 is not detected in whole cells by flow cytometry using anti-KIR3DL1 mAb 177407 (Fig. 2D) or mAb DX9 (27). However, incubating KIR3DL1*004-GFP-expressing cells at 25 °C resulted in the detection of some intracellular KIR3DL1*004 using mAb 177407 after fixation and permeabilization (Fig. 2D). The total increase in staining for folded KIR3DL1*004 was not as dramatic as observed for cell surface staining of KIR3DL1*004 on live cells at 25 °C (Fig. 1), suggesting reduced mAb recognition of cell surface receptors after cell fixation and permeabilization. While some intracellular staining for folded KIR3DL1*004 was detected at 25°C, it was further enhanced when cells were incubated in the presence of GolgiPlug™, at 25 °C. This treatment inhibits protein trafficking through the Golgi apparatus, such that correctly folded KIR3DL1*004 can be trapped en route to the cell surface and detected intracellularly (Fig. 2D). Thus, mAb recognition of intracellular KIR3DL1*004 parallels increased protein stability and improved folding at 25 °C. Together, these results suggest that poor folding is responsible for reduced delivery of KIR3DL1*004 to the cell surface.
KIR3DL1*004 is retained in the ER by calreticulin
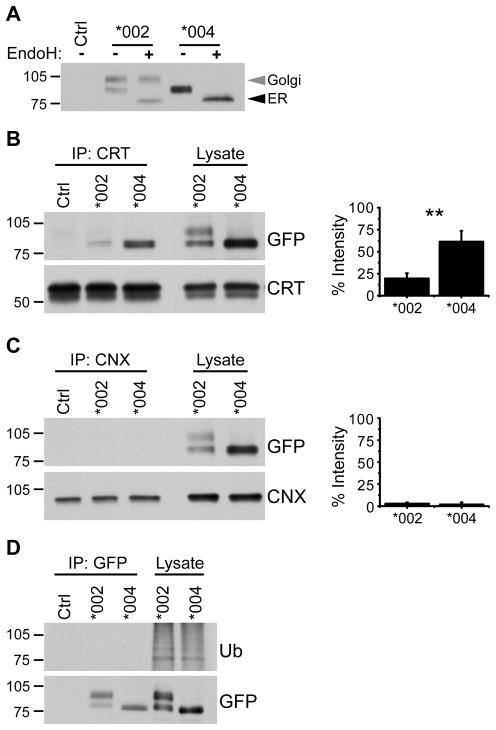
Misfolded proteins are usually retained in the ER where they are bound to chaperone proteins that assist with protein folding and assembly. To determine if any forms of KIR3DL1 are sequestered in the ER, GFP-tagged KIR3DL1*002 and KIR3DL1*004 proteins were immunoprecipitated from NKL transfectants and digested with Endoglycosidase H (Endo H) (Fig. 3A). Endo H specifically cleaves asparagine-linked high mannose oligosaccharides and can therefore be used to monitor post-translational modification of these oligosaccharides in the Golgi apparatus. Proteins retained in the ER are sensitive to Endo H digestion, while mature receptors that have trafficked through the Golgi en route to the cell surface are Endo H-resistant. Treatment with Endo H revealed that the high molecular mass species of the KIR3DL1*002 doublet is resistant to digestion and has undergone post-translational modification in the Golgi (Fig. 3A), representing mature glycosylated receptor that traffics to the cell surface. In contrast, the low molecular mass species of KIR3DL1*002 was sensitive to Endo H and therefore represents the immature ER-resident form. For KIR3DL1*004, all detectable protein was sensitive to Endo H digestion establishing that the receptor is predominantly retained in the ER.
Figure 3. KIR3DL1*004 is retained in the ER.
A, KIR3DL1 was immunoprecipitated from lysate of NKL cells expressing KIR3DL1*002-GFP (*002) or KIR3DL1*004-GFP (*004) using anti-GFP antibody. Immunoprecipitated protein was treated with Endoglycosidase H (Endo H) and KIR3DL1 was detected by Western blotting for GFP. Ctrl = NKL transfected to express KIR3DL1*002 lacking GFP. Endo H-resistant (Golgi) and -sensitive (ER) species are marked (grey and black arrowheads respectively). B, Calreticulin (CRT) or C, Calnexin (CNX) was immunoprecipitated from NKL transfectants. Ctrl = NKL transfected to express KIR3DL1*002 lacking GFP. Co-immunoprecipitated KIR3DL1 was detected by Western blotting for GFP (upper). Membranes were stripped and re-probed for the immunoprecipitated chaperone protein (lower). Graphs show quantification of Western blots (right). Band intensities are plotted with co-immunoprecipitated KIR3DL1 expressed as a % of signal from lysate. Mean and SD across 3 independent experiments are shown. Statistical significance was assessed by unpaired Student’s t test. ** P = 0.0062. D, KIR3DL1 was immunoprecipitated as in A followed by Western blotting for ubiquitin (upper). Membrane was stripped and re-probed for GFP (lower). The migration position of molecular weight markers (kD) is shown at the left of each panel.
To identify the ER retention mechanism for the immature forms of KIR3DL1, cell lysates were prepared from NKL transfectants expressing KIR3DL1*004-GFP or KIR3DL1*002-GFP. Candidate chaperone proteins were then immunoprecipitated and probed for associated KIR3DL1 protein by Western blotting for their GFP tag (Fig. 3). Calreticulin, a soluble ER chaperone protein, bound the low molecular mass form of KIR3DL1*002 and the only detectable species of KIR3DL1*004, consistent with these forms of KIR3DL1 being ER resident (Fig. 3B). Significantly, a higher proportion of the total amount of KIR3DL1*004 expressed in the cell bound to calreticulin, compared to the proportion of KIR3DL1*002 associated with calreticulin. Binding of KIR3DL1 to the related integral membrane homologue calnexin was not detected, at least for the conditions used here (Fig. 3C). This is not surprising as calreticulin and calnexin have been shown to recognize distinct subsets of glycoproteins, which is attributed to differences in chaperone and/or substrate localization within the ER (52–54). Thus, calreticulin appears to be the major chaperone mediating KIR3DL1 folding and retention. More KIR3DL1*004 is retained in the ER than KIR3DL1*002, confirming that a higher proportion of KIR3DL1*004 is in an unfolded state.
Mono/multiubiquitination has been shown to cause ER retention of unmodified proteins that are normally modified by palmitoylation (55). Whilst it has not been shown whether KIR3DL1 is palmitoylated, comparison of the amino-acid sequences of KIR3DL1*002 and KIR3DL1*004 revealed a putative palmitoylation site at cysteine 343, directly following the transmembrane domain of KIR3DL1*002 but not KIR3DL1*004. To test whether monoubiquitination might play a role in KIR3DL1*004 retention, we probed immunoprecipitates of KIR3DL1*002-GFP and KIR3DL1*004-GFP for the presence of ubiquitin (Fig. 3D). Both KIR3DL1*002 and KIR3DL1*004 lacked detectable ubiquitination suggesting that the primary mechanism for sequestering KIR3DL1*004 in the ER is protein misfolding and persistence of calreticulin binding.
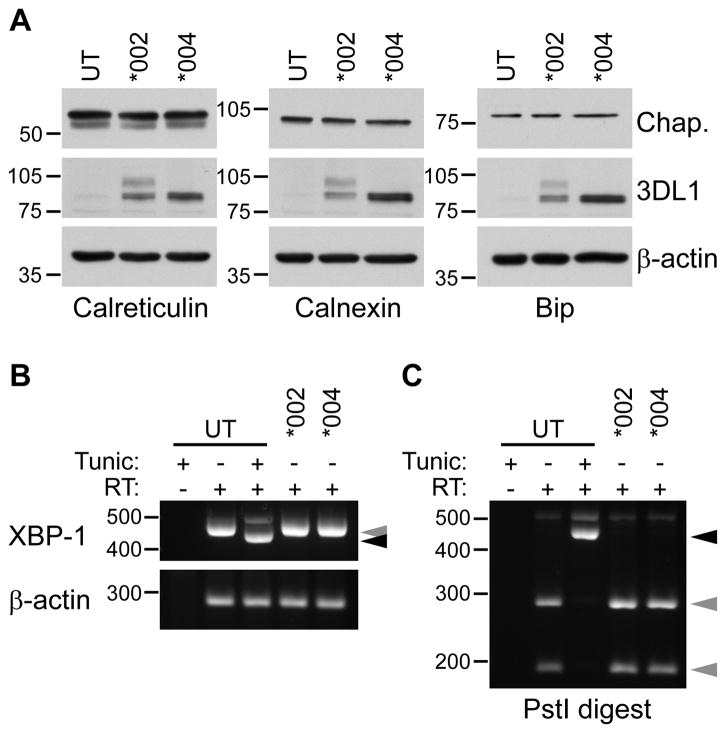
Accumulation of misfolded protein and increased synthesis of chaperone proteins can cause ER stress and induce the unfolded protein response (UPR) (56). This results in profound intracellular changes to accommodate expansion of ER activity and to degrade intracellular proteins, which could potentially affect cellular function. We used two complementary approaches to determine whether expression of KIR3DL1*004 resulted in a change in ER homeostasis, which might in turn influence NK cell function. The first was to assess ER chaperone protein levels by Western blotting for calreticulin, calnexin and Bip/Grp78 (Fig. 4A). Protein levels for these three molecular chaperones did not change upon expression of KIR3DL1*004, and were comparable to levels in untransfected and KIR3DL1*002-GFP-expressing NKL cells. The second approach used RT-PCR to detect an mRNA splice variant of X-box binding protein-1 (XBP-1), which is produced upon induction of the UPR (57, 58) (Fig. 4B). As a control, parental NKL were treated with tunicamycin to inhibit N-linked protein glycosylation and induce ER stress. In these cells, spliced XBP-1 mRNA was clearly detected. When either KIR3DL1*002-GFP or KIR3DL1*004-GFP were expressed in NKL, only unspliced XBP-1 mRNA was detected. Unspliced XBP-1 mRNA contains a PstI recognition site in the 26 nucleotide intron excised during splicing. PstI digestion of RT-PCR products confirmed that XBP-1 splicing did not occur upon expression of either KIR3DL1 allele (Fig. 4C). In summary, these data establish that the largely unfolded KIR3DL1*004 is retained in the ER where it binds luminal ER resident chaperone protein calreticulin. However, expression of misfolded KIR3DL1*004 appears to be tolerated by the cell, indicating that the function of a KIR3DL1*004-expressing NK cell is not compromised by the unfolded protein stress response.
Figure 4. Misfolding of KIR3DL1*004 does not induce the unfolded protein response.
A, Whole cell lysates made from untransfected parental NKL (UT) or NKL transfected to express KIR3DL1*002-GFP (*002) or 3DL1*004-GFP (*004) were probed for the chaperone proteins (Chap.) indicated (upper; Calreticulin left, Calnexin middle, Bip/Grp78 right). Membranes were stripped and re-probed for GFP to detect KIR3DL1 (middle) and the loading control β-actin (lower). The migration position of molecular weight markers (kD) is shown at the left of each panel. B, RT-PCR for XBP-1 (upper) or β-actin (lower) was performed on total RNA isolated from NKL transfectants, or parental NKL treated with Tunicamycin (Tunic) or DMSO with and without reverse transcriptase (RT). Spliced (black arrowhead) and unspliced (grey arrowhead) XBP-1 is indicated. C, PstI digestion of RT-PCR products in B. Spliced XBP-1 mRNA lacks the PstI recognition site (black arrowhead) and is not detected upon expression of KIR3DL1*004. Unspliced XBP-1 is cleaved by PstI (grey arrowheads). B and C, Molecular weight markers are in base pairs.
Class I MHC is not retained in the cell by KIR3DL1*004
Our observation that most KIR3DL1*004 molecules are retained within the cell raised the possibility that this KIR has functional intracellular interactions with Bw4-positive HLA molecules. To test this hypothesis we made use of the fact that NKL cells express endogenous HLA-B*51, an allotype having the Bw4 epitope and well established as a functional ligand for KIR3DL1 (21, 59). We used flow cytometry, immunofluorescence microscopy and an examination of the maturation kinetics of HLA-B*51 to look for evidence of intracellular association of HLA-B*51 with KIR3DL1*004 or KIR3DL1*002. These two allotypes represent the range of KIR3DL1 behavior, KIR3DL1*004 being mainly intracellular and KIR3DL1*002 being well-expressed on the cell surface (Fig. 5).
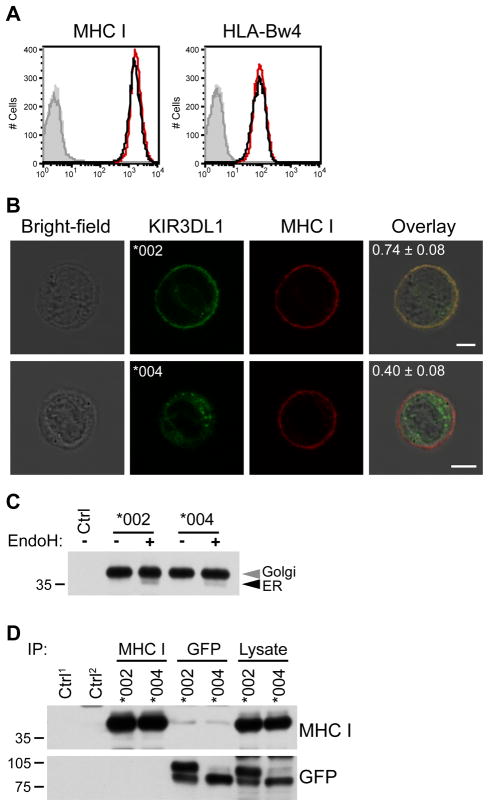
Figure 5. KIR3DL1*004 does not retain MHC class I inside the cell.
A, Cell surface expression of MHC class I protein was analyzed by flow cytometry. NKL cells transfected to express KIR3DL1*004-GFP (black) or KIR3DL1*002-GFP (red) were stained with mAb against all MHC class I (left) or HLA-Bw4 (right). Isotype-matched controls are shown in grey (KIR3DL1*004 transfectants = line, KIR3DL1*002 transfectants = fill). B, NKL expressing GFP-tagged KIR3DL1 (green, second column) were fixed and stained for MHC class I (red, third column). Fluorescence/bright-field overlays are also shown (fourth column) and overlap of red and green signals (yellow) was quantified with Pearson’s correlation coefficient (mean ± SD shown in panel). 62 and 78 cells were imaged for KIR3DL1*002 (*002) and KIR3DL1*004 (*004) respectively. Scale bars = 5 μm. C, MHC class I was immunoprecipitated from NKL transfectants. Ctrl = immunoprecipitation with isotype-matched control in KIR3DL1*002-GFP transfectants. Immunoprecipitated protein was treated with Endoglycosidase H (Endo H) and detected by Western blotting for MHC class I. D, Co-immunoprecipitation of GFP-tagged KIR3DL1 and MHC class I was assessed in NKL transfectants. Immunoprecipitation using GFP and MHC class I-specific antibodies (top) was followed by Western blotting for the reciprocal protein (right). Endo H-resistant (Golgi) and -sensitive (ER) species are marked (grey and black arrowheads respectively). Ctrl1 = isotype-matched control immunoprecipitation from NKL transfected to express KIR3DL1*002-GFP. Ctrl2 = anti-GFP immunoprecipitation from NKL transfectants expressing KIR3DL1*002 which lack GFP. The migration position of molecular weight markers (kD) is shown at the left of panels in C and D.
By flow cytometry, NKL transfected to stably express equivalent levels of KIR3DL1*002-GFP or KIR3DL1*004-GFP, were observed to express equivalent amounts of MHC class I protein at the cell surface (Fig. 5A). Similar results were obtained using either a pan MHC class I mAb (W6/32) or an HLA-Bw4-specific mAb (BIH0007) that detects HLA-B*51. This result was further analyzed by laser scanning confocal microscopy following mAb staining of MHC class I in fixed and permeabilized cells. Class I MHC and KIR3DL1*002-GFP were clearly both present at the plasma membrane (Fig. 5B), as indicated by yellow signal revealing overlap of the red anti-class I antibody and the green GFP tag on KIR3DL1*002. On the other hand, in cells expressing KIR3DL1*004, green GFP tagging the receptor was visible inside the cell, while red labeling of MHC class I was localized to the plasma membrane and not retained with the intracellular KIR3DL1*004. Using Pearson’s correlation coefficients to analyze >60 cells for each transfectant, fluorescence co-localization of MHC class I with KIR3DL1*002 (0.74) but not KIR3DL1*004 (0.40) was confirmed. HLA glycoproteins undergo N-linked glycosylation at asparagine 86 (60) allowing their transport through the secretory pathway to be monitored by Endo H digestion. If KIR3DL1*004 retains class I MHC molecules in the ER, then protein sensitivity to Endo H should be greater in KIR3DL1*004-expressing NKL compared to KIR3DL1*002-expressing cells. This was not observed, as virtually all class I MHC in NKL transfectants expressing either allele was resistant to Endo H digestion (Fig. 5C).
These results so far suggest a lack of intracellular interaction between KIR3DL1 and MHC class I molecules. To further investigate potential intracellular receptor-ligand interaction, association between both KIR3DL1 allotypes and MHC class I was assessed by immunoprecipitation (Fig. 5D). Using anti-GFP antibody to isolate either transfected KIR3DL1 allotype co-precipitated a small amount of MHC class I. In contrast, the reciprocal isolation of MHC class I using the W6/32 antibody did not yield any associated KIR3DL1 of either allotype. Given that W6/32 blocks class I-KIR interaction between cells (61), this antibody can be considered as a negative control for detecting any co-association. Thus the co-precipitation by anti-GFP could reflect a real, but minimal interaction of KIR3DL1 with MHC class I from the same cell. Arguing against this interpretation, the degree of co-association detected for the well-folded KIR3DL1*002 allotype was no better than for KIR3DL1*004. In either case, the detected association with KIR3DL1*004 does not affect MHC class I processing or cell surface expression (Fig. 5A–C).
Cell surface KIR3DL1*004 can deliver an inhibitory signal
In light of the above results, it was of interest to determine whether trace amounts of folded KIR3DL1*004 might be expressed transiently at the cell surface. To this end, cells were incubated with PE-labeled mAb 177407 at 37 °C for various times up to 8 h, followed by exposure to the same antibody for surface protein detection (Fig. 6A). Any KIR3DL1 that is transiently expressed at the cell surface will bind and internalize antibody during the incubation period, generating a fluorescent signal to supplement cell surface staining, both of which are distinguishable from the green fluorescence of the GFP tag on the transfected proteins. A similar method is used to detect transient surface expression of LAMP1/CD107a during NK cell degranulation (62). At steady state, levels of KIR3DL1*004 are barely detectable compared to KIR3DL1*002. Thus at the start of the experiment (time 0 h) the mean fluorescence intensity (MFI) ratios between mAb 177407 recognizing KIR3DL1 and isotype control labeling were 1.11 and 6.81, respectively for cells expressing KIR3DL1*004 or KIR3DL1*002 (Fig. 6A). After timed incubation with mAb 177407, the MFI of cells expressing KIR3DL1*002 increased about ten-fold, with a six-fold increase within 2 h. For cells expressing KIR3DL1*004, a 35% increase in MFI was observed within 2 h, with only a slight further increase upon continued incubation. The small, but significant, increase in MFI for KIR3DL1*004 over time suggests that even at 37°C, a low amount of correctly folded receptor can transiently reach the cell surface and is accessible to external antibody binding.
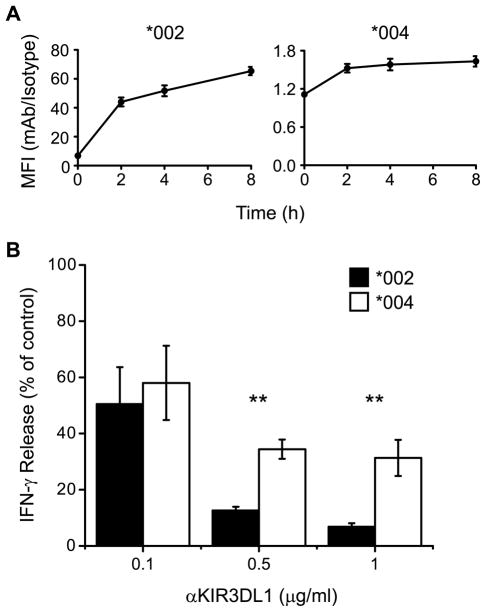
Figure 6. Transient surface expression of KIR3DL1*004 is functional for inhibition of NK cell activation.
A, Transient expression of KIR3DL1 and B, IFN-γ production were assessed in NKL cells transfected to express GFP-tagged KIR3DL1*002 (*002) or KIR3DL1*004 (*004). A, Cells were incubated at 37 °C for the times indicated in the presence of PE-labeled anti-KIR3DL1 mAb 177407 or isotype-matched control. KIR3DL1 expression (surface + internalized) was analyzed by flow cytometry and mean fluorescence intensity (MFI) ratios for mAb/isotype control are shown. Increase in expression of KIR3DL1*002 and KIR3DL1*004 over time are both statistically significant as assessed by one-way ANOVA (P < 0.0001 for both). Mean and SD across 4 independent experiments are shown. B, IFN-γ production after simultaneous receptor cross-linking with plate-bound mAb against 2B4 (0.5 μg/ml) and KIR3DL1 (0.1, 0.5, or 1 μg/ml). IFN-γ release into the supernatant was assayed by ELISA and is expressed as a percentage of the amount released when equivalent concentrations of isotype-matched control mAb was used instead of anti- KIR3DL1 mAb. Mean and SD across 4 independent experiments are shown. Statistical significance was assessed by two-way ANOVA. Antibody concentration (P < 0.0001) and KIR3DL1 allotype (P = 0.0037) both affect IFN-γ release. Bonferroni post test analysis comparing inhibition through KIR3DL1*002 and KIR3DL1*004 at 0.5 and 1.0 μg/ml anti-KIR3DL1 mAb are statistically significant (** P < 0.01).
To determine whether the low amount of folded KIR3DL1*004 that is transiently expressed at the cell surface is functional, an assay for inhibition of NK cell function was performed (Fig. 6B). NKL transfectants were stimulated by ligation of the activating receptor 2B4 using plate-bound antibody. Then KIR3DL1-mediated inhibition of activation was measured for cells expressing either KIR3DL1*004 or KIR3DL1*002 by addition of increasing amounts of mAb against KIR3DL1 to the stimulation assay. IFN-γ secretion was measured as a marker of NK cell activation. Stimulation of NKL transfectants with 0.5 μg/ml anti-2B4 mAb resulted in an 11- to 68-fold increase in IFN-γ release compared to exposure to control IgG1 (supplemental figure 1). When KIR3DL1*002 or KIR3DL1*004 were cross-linked with 0.1μg/ml mAb 177407 (in the presence of 0.5 μg/ml anti-2B4 mAb), IFN-γ release was reduced by approximately 50% compared to cells incubated with anti-2B4 mAb plus isotype control IgG2A (Fig. 6B). Cross-linking KIR3DL1*002 with increasing concentrations of mAb led to dose-dependent inhibition of IFN-γ release. In contrast, ligation of KIR3DL1*004 with higher concentrations of mAb only slightly decreased IFN-γ release further. Inhibition through KIR3DL1*004 was comparable when either 0.5 or 1 μg/ml mAb were used. At mid to high concentrations of mAb-mediated receptor cross-linking, KIR3DL1*004 did not inhibit IFN-γ production as effectively as KIR3DL1*002. No inhibition of IFN-γ release was detected upon exposure of anti-2B4-stimulated parental NKL cells to any concentration of mAb 177407, demonstrating that its inhibitory effects on the transfectants are a function of their KIR3DL1 expression (data not shown). These data indicate that the low amounts of KIR3DL1*004 expressed at the cell surface can function to transduce inhibitory signals that affect NK activation as measured by IFN-γ release. Inhibition through KIR3DL1*004 is less effective than through KIR3DL1*002, most likely due to the comparatively low level of functional KIR3DL1*004 expressed on the cell surface.
Primary NK cells can be induced to express cell surface KIR3DL1*004
The analysis of NKL transfectants to characterize KIR3DL1*004 membrane traffic has indicated that most KIR3DL1*004 is retained in an unfolded state inside cells but that a small amount can fold correctly for transient export to the cell surface, where it is functional. To establish whether KIR3DL1*004 expressed in primary NK cells is potentially functional, we investigated whether endogenous, KIR3DL1*004 protein is expressed at the surface of human NK cells in a temperature sensitive manner.
PBMC were isolated from the peripheral blood of genotyped individuals and were incubated at 25 and 37 °C for 16 h, then stained with mAb to detect surface KIR3DL1*004 by flow cytometry (Table I; Supplemental Fig. 2). We used anti-KIR3DL1 mAb DX9, even though 177407 more readily detects correctly folded KIR3DL1*004 (data not shown), because 177407 cross-reacts with KIR3DL2 while DX9 is specific for KIR3DL1 (28). Of the four individuals analyzed, two were homozygous for KIR3DL1*004 and two were heterozygous for KIR3DL1*004 in combination with either KIR3DS1*0013 or KIR3DL1*019. KIR3DS1*013 does not react with DX9 (28), while KIR3DL1*019 is essentially a variant of 3DL1*004 (16, 63). It only differs from KIR3DL1*004 at position 30 (substitution of cysteine for tyrosine) and thus has leucine 86 and serine 182 that are responsible for intracellular retention (27). Furthermore, the substitution of cysteine for tyrosine at position 30 in KIR3DL1*015, had no effect on the high cell surface expression of this allotype as detected with DX9 (28). For PBMC from all four subjects, KIR3DL1 was not detected by DX9 on cells incubated at physiological temperature (Table I). In contrast, lowering the temperature to 25 °C resulted in the appearance of DX9-positive NK cells at frequencies ranging from 4–8% for the KIR3DL1*004 homozygotes and the KIR3DL1*004/KIR3DL1*019 heterozygote. The KIR3DL1*004/KIR3DS1*013 subject had <1% DX9-positive cells, consistent with our independent observation that expression of KIR3DS1 dominates that of KIR3DL1 in heterozygotes (unpublished observations). Overall, the behavior of KIR3DL1*004 and the related KIR3DL1*019, when expressed on primary NK cells, is similar to that observed for KIR3DL1*004 expressed by transfection on NKL cells. These data are characteristic of defective protein folding, with low-level expression of a properly-folded form having functional potential, for these KIR3DL1 allotypes.
Table I. Surface KIR3DL1*004 protein expression is induced in peripheral human NK cells upon incubation at low temperature.
Human PBMC isolated from KIR-genotyped donors were incubated for 16 h at 37 and 25 °C, and then stained with anti-KIR3DL1 mAb DX9 or an isotype-matched control. Cell surface expression of KIR3DL1 on CD3-negative, CD56-positive NK cells was detected by flow cytometry. The percentage of KIR3DL1-positive cells reported has the percentage of cells reacting with the isotype control subtracted.
| Donor | KIR3DL1/S1 | Genotype | KIR3DL1-positive NK cells (%) | |
|---|---|---|---|---|
| 37 °C | 25 °C | |||
| 102 | 3DL1/3DL1 | *004/*004 | 0 | 8.1 |
| 306 | 3DL1/3DL1 | *004/*004 | 0 | 4.0 |
| 251 | 3DL1/3DS1 | *004/*013 | 0 | 0.7 |
| 355 | 3DL1/3DL1 | *004/*019 | 0 | 4.8 |
Discussion
KIR3DL1*004 is a common KIR3DL1 allele. It is expressed at a frequency of ~ 20% in the Caucasian population and has been shown to provide a selective advantage against AIDS progression (26). KIR3DL1*004 appears to be maintained by natural selection, yet its mechanism of action has been unclear, as the translated protein is predominantly retained inside the cell (27). Here, we report that KIR3DL1*004 is largely misfolded and sequestered in the ER where it binds the molecular chaperone protein calreticulin, suggesting this is a key chaperone involved in KIR3DL1 folding and expression. Intracellular KIR3DL1*004 does not affect HLA-Bw4 ligand expression, nor does it exert an intracellular function by inducing a cellular stress response that might alter NK cell activity. Furthermore, we observed only minimal intracellular interactions between the HLA-Bw4 ligand and the KIR3DL1*002 allotype, which is highly expressed on the cell surface. Together this characterization of intracellular membrane traffic and molecular interactions for KIR3DL1 molecules does not support the hypothesized intracellular function for the KIR3DL1*004 allotype. Rather, the small amount of correctly folded KIR3DL1*004 that makes it to the cell surface is a functionally competent receptor that can transduce an inhibitory signal for NK cell activation. We therefore suggest that the selective advantage for this allele and its ligand is conferred by its capacity to function at the cell surface.
There are two ways in which expression of a low amount of KIR3DL1*004 might influence the outcome of an immune response and it is probable that both of these mechanisms play a role in modulating NK cell function. First, in the effector phase of NK cell activity, a low amount of inhibitory receptor could make the NK cell more readily activated, as observed for KIR3DL1*005 (22). Second, cell surface KIR3DL1*004 and its putative interaction with HLA-Bw4 could also exert an effect in the process of education during NK cell development (5, 6). Although the mechanism of NK cell education is incompletely understood, evidence suggests two features. First, the frequency of KIR3DL1/S1 NK cells appears to be positively influenced by ligand expression (64). Second, it seems that potent receptor activity for education is not simply correlated with abundance on the cell surface. Notably, HLA-C molecules, which are expressed at considerably lower levels than HLA-A or HLA-B (65), are potent educating ligands upon engagement of the inhibitory KIR2DL receptors (5, 6). Such observations are consistent with NK cell education being the result of a high avidity interaction that induces a particular signaling pathway favorable for future activation, rather than education relying on a particular signaling threshold determined by abundance of receptor-ligand interactions. The low amount of KIR3DL1*004 could thereby have an effect on NK cell education, depending on its avidity for ligand. Thus, the folding polymorphism that causes retention of the majority of KIR3DL1*004 and cell surface expression of a small amount, might be favorable at the NK cell education stage and reduce the inhibitory signal at the effector stage.
It is also conceivable that low level engagement of KIR3DL1*004 during maturation may influence later NK cell activity by a more subtle mechanism. For example, a heterozygous cell expressing KIR3DL1*004, as well as a more abundant or very avid KIR3DL1 allotype, might receive signals equivalent to a cell homozygous for the co-expressed allele. Selection for KIR3DL1*004 and its ligand would therefore allow the co-expressed KIR3DL1 allotype to dominate during selection. This effect would also be true for other KIR loci encoding variants with reduced or null surface expression. Ultimately, this should be detectable by evidence for selection of heterozygosity for alleles with impaired expression at these loci.
Characterization of membrane traffic pathways is highly relevant to NK cell function because secretory or endocytic modulation can fine-tune the balance of cell activation and inhibition (30) and presumably also influence NK cell education. Intracellular trafficking pathways that contribute to expression of the less polymorphic NK cell receptors have been described. For the non-polymorphic activating receptor CD94/NKG2C and its inhibitory homologue CD94/NKG2A, separate pathways of endocytosis and recycling serve to maintain constant levels of unligated receptor at the cell surface (66, 67). Thus regulation of the secretory pathway for these receptors may have considerable influence on receptor expression levels. This is certainly the case for the activating receptors NKG2D, CD16, CD94/NKG2C, KIR3DS1 and KIR2DS in humans, as well as Ly49H and Ly49D in mice, whose cell surface expression is influenced by co-expression with a signaling adaptor protein (for example DAP12, DAP10, CD3ζ, or FcεR1γ) (31–36, 68–70).
The present study is the first to characterize the secretory pathway for inhibitory KIRs. KIR3DL1*004 represents an interesting example of how receptor polymorphism can influence NK cell function by affecting membrane traffic of the receptor. Our recent mutagenesis studies (28) and the mapping of KIR3DL1*004 intracellular retention to the influence of only two residues, leucine 86 in the D0 domain and serine 182 in the D1 domain (27), as well as earlier studies (71, 72), suggest that the conformation of KIR3DL1 receptors is very sensitive to sequence variation. We demonstrate here that the unfolded form of KIR3DL1*004 is more strongly associated with calreticulin than the readily expressed KIR3DL1*002 receptor. This raises the possibility that other KIR3DL1 allotypes with variable levels of cell surface expression (19, 20) reflect variable degrees of retention by calreticulin due to polymorphisms that affect their rate of folding. Our data suggest that expression of KIR3DL1*019, which only differs from KIR3DL1*004 at position 30, is regulated similarly. Several unfolded variants of KIR2DL2 and KIR2DS receptors have also been described (70, 73, 74). It would now be of interest to determine whether these are truly null alleles for receptor expression or whether, as we have shown for KIR3DL*004, there are small amounts of these receptors that can fold properly for transport to the cell surface where they might function.
Our results demonstrate that intracellular KIR3DL1*004 is not folded properly and accumulates in the ER. This unfolded form of KIR3DL1*004 does not appear to function inside the cell, either by causing a cellular stress response as measured by chaperone synthesis or by induction of the UPR, or by influencing ligand expression and maturation. Very little co-immunoprecipitation of either KIR3DL1*004 or KIR3DL1*002 with HLA Class I ligand expressed in the same cell was detected. This trace interaction could arise from binding of solubilized ligands and receptors after cell lysis or from a very low level of intracellular co-association, which could occur between opposing intracellular membranes in a compartment or in a cis-arrangement in the same membrane. In any case, the intracellular association of either KIR allotype with MHC class I is nowhere near as extensive as reported for Ly49 and its cognate ligand, which is readily detected (37, 75, 76). Thus, our data suggest that KIR3DL1 function is confined to inhibitory signals transmitted by properly folded molecules at the cell surface upon interaction with their putative HLA-Bw4 ligand in trans, on the surface of other cells. The unexpected finding that the low amount of KIR3DL1*004 expressed on the cell surface is functional, leads to the hypothesis that folding variation is a way to modulate receptor function. Consequently, allotypes with polymorphisms that affect folding are subject to the same selection for evolutionary advantage as other KIR allotypes.
Supplementary Material
Acknowledgments
We thank J. Jarjoura for assistance with cell sorting. We are grateful to L. L. Lanier, W. H. Carr, S. Lopez-Vergès, and R. Parton for cell lines and antibodies and members of our laboratories for critical discussions.
This work was supported by the National Institutes of Health (P01 AI064520).
Abbreviations used in this paper
- GFP
(enhanced) Green fluorescent protein
- Endo H
Endoglycosidase H
- Grp78
Glucose regulated protein 78
- KIR
Killer cell Ig-like receptor
- UPR
Unfolded protein response
- XBP-1
X-box binding protein-1
Footnotes
The authors have no conflicting financial interests.
References
- 1.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cooper MA, Colonna M, Yokoyama WM. Hidden talents of natural killers: NK cells in innate and adaptive immunity. EMBO Rep. 2009;10:1103–1110. doi: 10.1038/embor.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long EO. Negative signaling by inhibitory receptors: the NK cell paradigm. Immunol Rev. 2008;224:70–84. doi: 10.1111/j.1600-065X.2008.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, Breso V, Frassati C, Reviron D, Middleton D, Romagne F, Ugolini S, Vivier E. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, Parham P. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168:2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 8.Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 9.Rajagopalan S, Long EO. Understanding how combinations of HLA and KIR genes influence disease. J Exp Med. 2005;201:1025–1029. doi: 10.1084/jem.20050499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moffett A, Loke C. Immunology of placentation in eutherian mammals. Nat Rev Immunol. 2006;6:584–594. doi: 10.1038/nri1897. [DOI] [PubMed] [Google Scholar]
- 11.Velardi A. Role of KIRs and KIR ligands in hematopoietic transplantation. Curr Opin Immunol. 2008;20:581–587. doi: 10.1016/j.coi.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 12.Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, Layrisse Z, Parham P. Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci U S A. 2009;106:18692–18697. doi: 10.1073/pnas.0906051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 14.Carrington M, Martin MP, van Bergen J. KIR-HLA intercourse in HIV disease. Trends Microbiol. 2008;16:620–627. doi: 10.1016/j.tim.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinson J, Waller MJ, Stoehr P, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 2005;33:D523–526. doi: 10.1093/nar/gki032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, Bruno D, Carrington CV, Chandanayingyong D, Chang YH, Crespi C, Saruhan-Direskeneli G, Fraser PA, Hameed K, Kamkamidze G, Koram KA, Layrisse Z, Matamoros N, Mila J, Park MH, Pitchappan RM, Ramdath DD, Shiau MY, Stephens HA, Struik S, Verity DH, Vaughan RW, Tyan D, Davis RW, Riley EM, Ronaghi M, Parham P. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 17.Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med. 1995;181:1133–1144. doi: 10.1084/jem.181.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med. 1994;180:1235–1242. doi: 10.1084/jem.180.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, Vilches C, Parham P. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166:2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 20.Thomas R, Yamada E, Alter G, Martin MP, Bashirova AA, Norman PJ, Altfeld M, Parham P, Anderson SK, McVicar DW, Carrington M. Novel KIR3DL1 alleles and their expression levels on NK cells: convergent evolution of KIR3DL1 phenotype variation? J Immunol. 2008;180:6743–6750. doi: 10.4049/jimmunol.180.10.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222–5229. doi: 10.4049/jimmunol.175.8.5222. [DOI] [PubMed] [Google Scholar]
- 22.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178:235–241. doi: 10.4049/jimmunol.178.1.235. [DOI] [PubMed] [Google Scholar]
- 24.Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P, Carrington M, Dong T, Rowland-Jones S. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33–37. doi: 10.4049/jimmunol.178.1.33. [DOI] [PubMed] [Google Scholar]
- 25.Alter G, Altfeld M. NK cells in HIV-1 infection: evidence for their role in the control of HIV-1 infection. J Intern Med. 2009;265:29–42. doi: 10.1111/j.1365-2796.2008.02045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O’Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171:6640–6649. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 28.Sharma D, Bastard K, Guethlein LA, Norman PJ, Yawata N, Yawata M, Pando M, Thananchai H, Dong T, Rowland-Jones S, Brodsky FM, Parham P. Dimorphic motifs in D0 and D1+D2 domains of killer cell Ig-like receptor 3DL1 combine to form receptors with high, moderate, and no avidity for the complex of a peptide derived from HIV and HLA-A*2402. J Immunol. 2009;183:4569–4582. doi: 10.4049/jimmunol.0901734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajagopalan S, Bryceson YT, Kuppusamy SP, Geraghty DE, van der Meer A, Joosten I, Long EO. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006;4:e9. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masilamani M, Peruzzi G, Borrego F, Coligan JE. Endocytosis and intracellular trafficking of human natural killer cell receptors. Traffic. 2009;10:1735–1744. doi: 10.1111/j.1600-0854.2009.00973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu J, Cherwinski H, Spies T, Phillips JH, Lanier LL. DAP10 and DAP12 form distinct, but functionally cooperative, receptor complexes in natural killer cells. J Exp Med. 2000;192:1059–1068. doi: 10.1084/jem.192.7.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8:693–701. doi: 10.1016/s1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 33.Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting Edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12-associated receptor expressed on NK cells that triggers NK cell activation. J Immunol. 2007;178:647–651. doi: 10.4049/jimmunol.178.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trundley A, Frebel H, Jones D, Chang C, Trowsdale J. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur J Immunol. 2007;37:780–787. doi: 10.1002/eji.200636773. [DOI] [PubMed] [Google Scholar]
- 35.Orr MT, Sun JC, Hesslein DG, Arase H, Phillips JH, Takai T, Lanier LL. Ly49H signaling through DAP10 is essential for optimal natural killer cell responses to mouse cytomegalovirus infection. J Exp Med. 2009;206:807–817. doi: 10.1084/jem.20090168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tassi I, Le Friec G, Gilfillan S, Takai T, Yokoyama WM, Colonna M. DAP10 associates with Ly49 receptors but contributes minimally to their expression and function in vivo. Eur J Immunol. 2009;39:1129–1135. doi: 10.1002/eji.200838972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Held W, Mariuzza RA. Cis interactions of immunoreceptors with MHC and non-MHC ligands. Nat Rev Immunol. 2008;8:269–278. doi: 10.1038/nri2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson MJ, Cochran KJ, Cameron C, Le JM, Tantravahi R, Ritz J. Characterization of a cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp Hematol. 1996;24:406–415. [PubMed] [Google Scholar]
- 39.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O’Brien SJ, Carrington M. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 40.Vilches C, Castano J, Gomez-Lozano N, Estefania E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70:415–422. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 41.Belle I, Hou L, Chen M, Steiner NK, Ng J, Hurley CK. Investigation of killer cell immunoglobulin-like receptor gene diversity in KIR3DL1 and KIR3DS1 in a transplant population. Tissue Antigens. 2008;71:434–439. doi: 10.1111/j.1399-0039.2008.01017.x. [DOI] [PubMed] [Google Scholar]
- 42.Robinson J, Mistry K, McWilliam H, Lopez R, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 2010;38:D863–869. doi: 10.1093/nar/gkp879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnstable CJ, Bodmer WF, Brown G, Galfre G, Milstein C, Williams AF, Ziegler A. Production of monoclonal antibodies to group A erythrocytes, HLA and other human cell surface antigens-new tools for genetic analysis. Cell. 1978;14:9–20. doi: 10.1016/0092-8674(78)90296-9. [DOI] [PubMed] [Google Scholar]
- 44.Solda T, Olivari S, Molinari M. Analyzing folding and degradation of metabolically labelled polypeptides by conventional and diagonal sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biol Proced Online. 2005;7:136–143. doi: 10.1251/bpo111. [DOI] [PubMed] [Google Scholar]
- 45.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 46.Stam NJ, Spits H, Ploegh HL. Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J Immunol. 1986;137:2299–2306. [PubMed] [Google Scholar]
- 47.McCann FE, Vanherberghen B, Eleme K, Carlin LM, Newsam RJ, Goulding D, Davis DM. The size of the synaptic cleft and distinct distributions of filamentous actin, ezrin, CD43, and CD45 at activating and inhibitory human NK cell immune synapses. J Immunol. 2003;170:2862–2870. doi: 10.4049/jimmunol.170.6.2862. [DOI] [PubMed] [Google Scholar]
- 48.Ljunggren HG, Stam NJ, Ohlen C, Neefjes JJ, Hoglund P, Heemels MT, Bastin J, Schumacher TN, Townsend A, Karre K, et al. Empty MHC class I molecules come out in the cold. Nature. 1990;346:476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- 49.Griffiths G, Pfeiffer S, Simons K, Matlin K. Exit of newly synthesized membrane proteins from the trans cisterna of the Golgi complex to the plasma membrane. J Cell Biol. 1985;101:949–964. doi: 10.1083/jcb.101.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saraste J, Kuismanen E. Pre- and post-Golgi vacuoles operate in the transport of Semliki Forest virus membrane glycoproteins to the cell surface. Cell. 1984;38:535–549. doi: 10.1016/0092-8674(84)90508-7. [DOI] [PubMed] [Google Scholar]
- 51.Matlin KS, Simons K. Reduced temperature prevents transfer of a membrane glycoprotein to the cell surface but does not prevent terminal glycosylation. Cell. 1983;34:233–243. doi: 10.1016/0092-8674(83)90154-x. [DOI] [PubMed] [Google Scholar]
- 52.Hebert DN, Zhang JX, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danilczyk UG, Cohen-Doyle MF, Williams DB. Functional relationship between calreticulin, calnexin, and the endoplasmic reticulum luminal domain of calnexin. J Biol Chem. 2000;275:13089–13097. doi: 10.1074/jbc.275.17.13089. [DOI] [PubMed] [Google Scholar]
- 54.Pieren M, Galli C, Denzel A, Molinari M. The use of calnexin and calreticulin by cellular and viral glycoproteins. J Biol Chem. 2005;280:28265–28271. doi: 10.1074/jbc.M501020200. [DOI] [PubMed] [Google Scholar]
- 55.Abrami L, Kunz B, Iacovache I, van der Goot FG. Palmitoylation and ubiquitination regulate exit of the Wnt signaling protein LRP6 from the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2008;105:5384–5389. doi: 10.1073/pnas.0710389105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 58.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 59.Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J Immunol. 2008;181:6293–6300. doi: 10.4049/jimmunol.181.9.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parham P, Alpert BN, Orr HT, Strominger JL. Carbohydrate moiety of HLA antigens. Antigenic properties and amino acid sequences around the site of glycosylation. J Biol Chem. 1977;252:7555–7567. [PubMed] [Google Scholar]
- 61.Komatsu F, Kajiwara M. Enhancement of tumor cell lysis by natural killer cells after blocking of HLA-monomorphic determinant using F(ab′)2 fragment of W6/32. Cancer Detect Prev. 1998;22:168–175. doi: 10.1046/j.1525-1500.1998.cdoa19.x. [DOI] [PubMed] [Google Scholar]
- 62.Alter G, Malenfant JM, Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J Immunol Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 63.Halfpenny IA, Middleton D, Barnett YA, Williams F. Investigation of killer cell immunoglobulin-like receptor gene diversity: IV. KIR3DL1/S1. Hum Immunol. 2004;65:602–612. doi: 10.1016/j.humimm.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 64.Morvan M, Willem C, Gagne K, Kerdudou N, David G, Sebille V, Follea G, Bignon JD, Retiere C. Phenotypic and functional analyses of KIR3DL1+ and KIR3DS1+ NK cell subsets demonstrate differential regulation by Bw4 molecules and induced KIR3DS1 expression on stimulated NK cells. J Immunol. 2009;182:6727–6735. doi: 10.4049/jimmunol.0900212. [DOI] [PubMed] [Google Scholar]
- 65.Schaefer MR, Williams M, Kulpa DA, Blakely PK, Yaffee AQ, Collins KL. A novel trafficking signal within the HLA-C cytoplasmic tail allows regulated expression upon differentiation of macrophages. J Immunol. 2008;180:7804–7817. doi: 10.4049/jimmunol.180.12.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Borrego F, Kabat J, Sanni TB, Coligan JE. NK cell CD94/NKG2A inhibitory receptors are internalized and recycle independently of inhibitory signaling processes. J Immunol. 2002;169:6102–6111. doi: 10.4049/jimmunol.169.11.6102. [DOI] [PubMed] [Google Scholar]
- 67.Masilamani M, Narayanan S, Prieto M, Borrego F, Coligan JE. Uncommon endocytic and trafficking pathway of the natural killer cell CD94/NKG2A inhibitory receptor. Traffic. 2008;9:1019–1034. doi: 10.1111/j.1600-0854.2008.00738.x. [DOI] [PubMed] [Google Scholar]
- 68.Snyder MR, Nakajima T, Leibson PJ, Weyand CM, Goronzy JJ. Stimulatory killer Ig-like receptors modulate T cell activation through DAP12-dependent and DAP12-independent mechanisms. J Immunol. 2004;173:3725–3731. doi: 10.4049/jimmunol.173.6.3725. [DOI] [PubMed] [Google Scholar]
- 69.Blery M, Delon J, Trautmann A, Cambiaggi A, Olcese L, Biassoni R, Moretta L, Chavrier P, Moretta A, Daeron M, Vivier E. Reconstituted killer cell inhibitory receptors for major histocompatibility complex class I molecules control mast cell activation induced via immunoreceptor tyrosine-based activation motifs. J Biol Chem. 1997;272:8989–8996. doi: 10.1074/jbc.272.14.8989. [DOI] [PubMed] [Google Scholar]
- 70.Steiner NK, Dakshanamurthy S, VandenBussche CJ, Hurley CK. Extracellular domain alterations impact surface expression of stimulatory natural killer cell receptor KIR2DS5. Immunogenetics. 2008;60:655–667. doi: 10.1007/s00251-008-0322-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rojo S, Wagtmann N, Long EO. Binding of a soluble p70 killer cell inhibitory receptor to HLA-B*5101: requirement for all three p70 immunoglobulin domains. Eur J Immunol. 1997;27:568–571. doi: 10.1002/eji.1830270231. [DOI] [PubMed] [Google Scholar]
- 72.Khakoo SI, Geller R, Shin S, Jenkins JA, Parham P. The D0 domain of KIR3D acts as a major histocompatibility complex class I binding enhancer. J Exp Med. 2002;196:911–921. doi: 10.1084/jem.20020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.VandenBussche CJ, Dakshanamurthy S, Posch PE, Hurley CK. A single polymorphism disrupts the killer Ig-like receptor 2DL2/2DL3 D1 domain. J Immunol. 2006;177:5347–5357. doi: 10.4049/jimmunol.177.8.5347. [DOI] [PubMed] [Google Scholar]
- 74.VandenBussche CJ, Mulrooney TJ, Frazier WR, Dakshanamurthy S, Hurley CK. Dramatically reduced surface expression of NK cell receptor KIR2DS3 is attributed to multiple residues throughout the molecule. Genes Immun. 2009;10:162–173. doi: 10.1038/gene.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doucey MA, Scarpellino L, Zimmer J, Guillaume P, Luescher IF, Bron C, Held W. Cis association of Ly49A with MHC class I restricts natural killer cell inhibition. Nat Immunol. 2004;5:328–336. doi: 10.1038/ni1043. [DOI] [PubMed] [Google Scholar]
- 76.Scarpellino L, Oeschger F, Guillaume P, Coudert JD, Levy F, Leclercq G, Held W. Interactions of Ly49 family receptors with MHC class I ligands in trans and cis. J Immunol. 2007;178:1277–1284. doi: 10.4049/jimmunol.178.3.1277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.