Abstract
Hepatocellular carcinoma (HCC) is the third most common cause of cancer deaths worldwide, and the incidence of this fatal disease is still on rise. The majority of HCCs emerge in the background of a chronic liver disease, such as chronic hepatitis and liver cirrhosis. The current understanding is that majority of HCCs evolve as a consequence of chronic inflammation and due to the presence of infection with hepatitis viruses. These underlying pathogenic stimuli subsequently induce a spectrum of genetic and epigenetic alterations in several cancer-related genes, which are involved in cell-cycle regulation, cell growth and adhesion. Such widespread genomic alterations cause disruption of normal cellular signaling and finally lead to the acquisition of a malignant phenotype in HCC. In general, the type of gene alterations, such as point mutations, deletion of chromosomal regions and abnormal methylation of gene promoters differ according to the individual targeted gene. In HCC, incidence of genetic alterations is relatively rare and is limited to a subset of few cancer-specific genes, such as the tumor suppressor p53, RB genes and oncogenes such as the CTNNB1. In contrast, epigenetic changes that involve aberrant methylation of genes and other post-transcriptional histone modifications occur far more frequently, and some of these epigenetic alterations are now being exploited for the development of molecular diagnostic signatures for HCC. In addition, recent findings of unique microRNA expression profiles also provide an evidence for the existence of novel mechanisms for gene expression regulation in HCC. In this review article, we will review the current state of knowledge on the activation of various oncogenic pathways and the inactivation of tumor suppressor pathways in HCC that result in the disruption of cancer-related gene function. In addition, we will specifically emphasize the clinical implication of some of these genetic and epigenetic alterations in the management of hepatocarcinogenesis.
Keywords: Oncogenic pathway, hepatocellular carcinoma, DNA methylation, mutation, oncogene, tumor suppressor gene.
INTRODUCTION
It is now widely accepted that stepwise accumulation of mutations in cancer-related genes and chromosomal alterations are involved in human carcinogenesis [1]. However, in the last decade we have recognized that in addition to genetic artifacts, epigenetic alterations that involve aberrant methylation of gene promoters and dysregulated expression of microRNA (miRNA), constitute equally important mechanisms of genomic instability in human cancers. Studies of these molecular alterations in hepatocellular carcinoma (HCC) have also revealed that this malignancy involves a multipathway process, and accumulation of genetic and epigenetic events leads to an abnormal activation or inactivation of multiple cellular signaling pathways including cellular proliferation, cellular survival, differentiation, and angiogenesis. Additionally, the emerging consensus is that the core biological processes including regulation of p53/ARF, RB/INK4A and Wnt/β-catenin pathways are commonly affected in a majority of HCCs regardless of the etiologies, suggesting the presence of a common oncogenic process of HCC development.
This review provides an overview of genetic and epigenetic alterations observed in HCC, and discusses how these abnormalities relate to the disruption of specific biological signaling for the maintenance of homeostasis in normal hepatocytes. It is anticipated that such precise analyses of genomic profiling will reveal a global scheme of molecular classification of HCC, aid in the development of novel molecular-targeted therapy for specific subclasses of HCC, as well as assist once in the discovery and development of diagnostic and predictive biomarkers of HCC.
P53/ARF PATHWAY
The p53/ARF pathway plays a key role in a variety of cellular functions, such as regulation of cell cycle, apoptosis and DNA repair. Cellular stress activates this pathway by activating the p53 protein as a transcription factor, and leads to the induction of the transcriptional network of p53-responsive genes [2]. The p53 circuit communicates with several fundamental cellular signals, such as the Wnt/β-catenin, RB/INK4a and p38 MAP pathways. Because the p53 protein has a variety of important functions for the maintenance of cellular responses, disruption of the p53/ARF pathway has been reported in almost every type of cancer including hepatocellular carcinoma (HCC) [3]. Notwithstanding the existence of multiple auto-regulatory loops that control p53 activity, MDM2 protein plays a central role in this process. The ARF antagonizes functions of MDM2 to induce regulatory responses that depend on the activation of p53 and its target genes. Although disruption of the p53 occurs in a subset of HCC, more than half of tumors retain a wild-type p53, suggesting that alteration of other molecules involved in this pathway might also contribute to hepatocarcinogenesis [4].
The p53 Gene
The p53 gene is located on 17p13.1 and encodes this transcription factor with a tumor suppressor function. In human HCC, common alterations of the p53 gene are point mutations within the conserved region of exons accompanied by loss of the short arm of chromosome 17. Generally, 30 - 60% of HCC carry mutations of the p53 gene [3]. The spectrum of point mutations reported in HCC varies according to the etiology of the underlying molecular pathogenesis. For example, aflatoxin B1 (AFB1) reportedly induces G:C to T:A transversion mutation in codon 249 of the p53 [5, 6]. As this type of mutation is detected in serum of HCC patients with an exposure to AFB1, this can be applied for risk assessment of HCC, and as a potential biomarker of HCC emergence and exposure to AFB1 [7]. However, it has been shown that AFB1-related liver tumor in rats didn’t carry any specific mutations that corresponded to codon 249 of human HCC [8]. On the other hand, transgenic mice with the hepatitis B virus-X (HBx) gene, in concert with AFB1 intake, develop HCC harboring G:C to T:A transversion mutation at the site corresponding to the codon 249 of the human p53 [9]. This evidence highlights the potential scenario where both AFB1 insult and infection with HBV are required for the evolution of specific mutations within the p53 gene in HCC.
On the other hand, even though the mutational spectrum of the p53 gene in AFB1-negative HCC is heterogeneous, the majority of mutations are still confined to a conserved region of DNA-binding domain encoded on exons 5 – 9 [4].
Other reports suggest that viral proteins affect the function of the p53 protein and contribute to HCC formation. For example, HBx protein itself reportedly binds to p53 and disturbs its capacity for DNA binding, transcription and induction of apoptosis [10]. None of the previous reports have provided any evidence for the direct action of HCV-related protein to p53. However, it has been shown that, nitric oxide (NO), which is induced by inflammatory cytokines such as TNF-α a and IFN-γ in chronic hepatitis C virus (HCV) infection, can induce mutation of cancer-related genes including the p53. In cases of colon cancer and lung cancer, nitric oxide synthetase 2 activity in the tumor closely related to the G:C to A:T mutations in the p53 gene within the CpG sites [11], which is the major type of mutation in the p53 gene found in HCC without AFB1 contamination.
The CDKN2A (p14ARF) Gene
The CDKN2A gene encodes two tumor suppressors, p14ARF and p16INK4a, which share the same second exons. The p14ARF transcript functions as a stabilizer of the p53 protein. The p14ARF interacts with the MDM2 protein, which is responsible for the degradation of p53 and in regulating the control of cell cycle during G1-S phase [12]. This gene is frequently mutated or deleted in a wide variety of human tumors including HCC. Generally, 7% of HCC carry homozygous deletion of the CDKN2A gene, and inactivation of the p14ARF gene via aberrant methylation its gene promoter is a frequent event in HCC [13]. However, according to our analysis, methylation of the promoter of p14ARF was also evident in small proportion of non-cancerous liver tissues as well, but it was far more frequent in HCC tissues [14]. In addition, a decrease of mRNA level was detected exclusively in HCC with homozygous deletion of this gene. From this point of view, the role of methylation at the promoter of p14ARF may still be viewed somewhat controversial in pathogenesis of HCC [14].
The CDKN1A and CDKA1B Genes
Although alterations in the genomic sequences of the tumor suppressor CDKN1A and CDKN1B genes are rare in HCC, the down-regulation of these cell-cycle regulatory proteins has been frequently reported [15-17]. Expression of p21WAF1, the product of the CDKN1A gene, is regulated by the tumor suppressor p53, and p21WAF1 in turn regulates the activities of cyclin-dependent kinases (CDK) 2 and 4, which control the G1-S and G2-M phase of cell cycle, respectively. In human HCC, down-regulation of p21WAF1 has been observed and it associates with tumor progression and bad prognosis of the disease [15]. The virus-related protein of HBx and core protein of HCV are also known to suppress transcription of p21WAF1 [16].
P27kip1, the product of the CDKN1B gene, is also a member of cyclin dependent kinase inhibitors (CDKI) and inhibits kinase activity of cyclin E-CDK2 and cyclin D-CDK4. The expression level of this CDKI was also correlated to the poor outcomes for disease-free survival of HCC cases [17]. The protein level of p27kip1 was reportedly lower in cirrhotic liver of non-cancerous tissues of HCC cases than in those without HCC, and the down-regulation of its expression was associated with promoter methylation of the CDKN1B gene [17, 18].
RB/INK4A PATHWAY
The RB/INK4A pathway plays a central role in the regulation of the G1-S phase of cell cycle progression. It is known that RB activities associate with its phosphorylation status, and phosphorylation of RB depends on cell cycle progression coupled with the activation of CDK. The unphosphlyrated RB binds to the E2F-1 transcription factor, preventing it from interacting with the transcriptional machinery within the cell. The phosphorylated RB sequesters E2Fs, which are responsible for the transcription of many genes [19]. Among these genes, cyclin E, which binds to the CDK2, induces DNA replication during cell cycle. The majority of tumors, including HCC, frequently harbor aberrations of the members of this pathway. Our analyses indicate that 81% of HCC showed alteration of at least one component of the RB/INK4A pathway [20-22].
The RB Gene
Loss of RB activity has been identified in many types of cancers including retinoblastoma, osteosarcomas, small cell and non-small cell lung cancers, breast cancer and HCC. In human HCC, 18–48% of tumors represent chromosomal loss of 13q14 region, where the RB gene resides. Our previous reports demonstrate that some HCCs with loss of heterozygosity on 13q14 carry additional structural alterations of the interstitial deletion in the RB gene, suggesting that a double hit on this locus leads to complete functional inactivation of RB [23]. On the other hand, down-regulation of RB protein is observed in 30–50% of HCC tissues compared to their corresponding non-neoplastic liver tissues [24]. Although the precise mechanism of the down-regulation of RB has not been clarified completely, some reports have suggested that abnormal methylation of the RB promoter may cause transcriptional inactivation of the gene in cancer cells [25].
The CDKN2A (p16INK4a ) Gene
As described previously, p16INK4a is a tumor suppressor protein, which is encoded by the CDKN2A gene. This molecule plays an important role in regulating the cell cycle, by inhibiting CDK4 and controlling cell cycle progression at the G1 phase. Mutation of p16INK4a is also involved in the development of a variety of cancers including HCC.
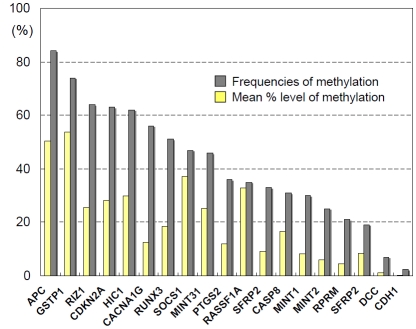
Some HCCs carry homozygous deletions or point mutations of p16INK4a [26]. However, recent data suggests that the major mechanism of inactivation of this gene is abnormal methylation of its gene promoter. As much as 40-70% of HCCs demonstrate p16INK4a methylation, and this aberrant methylation associates with the down-regulation of the protein expression [27]. Since low-levels of methylation were also detected in the background non-cancerous liver of the HCC patients, we normalized methylation levels in HCC tissues to the background aberrant methylation and determined that p16INK4a methylation inactivates the gene in 63% of human HCC cases (Fig. 1) [28]. Previous reports suggest that p16INK4a methylation also associates with the presence of HBV or HCV infection [29].
Fig. (1).
Mean methylation levels and frequencies of hypermethylation of several tumor suppressor genes (TSGs) in HCC. In contrast to genetic alterations such as point mutation, a variety of TSGs are frequently inactivated via epigenetic mechanism, suggesting that DNA methylation is a major alteration as a driver for human hepatocarcinogenesis.
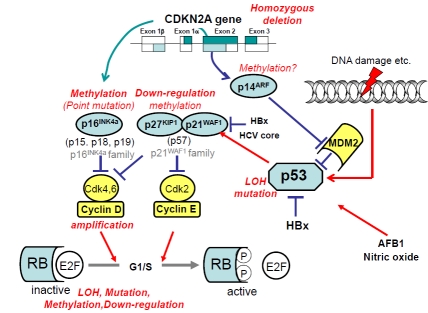
As described above, there is a close association between RB/INK4A pathway and p53/ARF pathway, especially in the regulation of cell cycle progression and apoptosis. We summarized the association of the responsible molecules in these pathways and the type of their alterations found in HCC in Fig. (2).
Fig. (2).
Schematic representation of alterations in p53/ARF and RB/INK4A pathways in HCC. Molecules with oncogenic function are shown in yellow and those with tumor suppressive function are shown in blue. Specific type of genetic and epigenetic alterations found in HCC are shown in red letter.
WNT/β-CATENIN PATHWAY
Activation of Wnt/β-catenin pathway facilitates the expression of several genes indispensable for cell growth, such as c-myc and cyclin D1, through translocation of β-catenin to nucleus and its interaction with various transcription factors.
In general, β-catenin within cells is present in the form of complexes with several proteins, and the ability of β-catenin to bind to other proteins is regulated by tyrosine kinases and serine kinases, such as glycogen synthase kinase (GSK)-3β. The binding proteins include cell adhesion molecules, several transcription factors, and axin, which is a component of the Wnt signalling pathway. β-catenin is associated with the axin complex together with GSK-3β and APC (adenomatosis polyposis coli) [30, 31]. This complex substantially induces the increase in phosphorylation of β-catenin by facilitating the action of GSK-3β, which leads to ubiquitin-dependent degradation of β-catenin by proteosomes. The action of GSK-3β is inhibited by the binding of Wnt to its receptors, which leads to the translocation of β-catenin to the nucleus and induction of a variety of functions within this pathway [32]. β-catenin also acts in conjunction with the T-cell factor (TCF)/lymphoid enhancing factor (LEF) family transcription factors to activate several target genes.
Among the components of the Wnt/β-catenin pathway, the APC gene is known to be the primary target in patients with familial adenomatous polyposis (FAP) colon cancers [33]. Similarly, mutation of the AXIN2 is a cause of attenuated polyposis [34], and mutation of the CDH1 gene is known to induce familial gastric cancer [35]. Alterations of several components of this pathway are also common events in human HCC (Fig. 3).
Fig. (3).
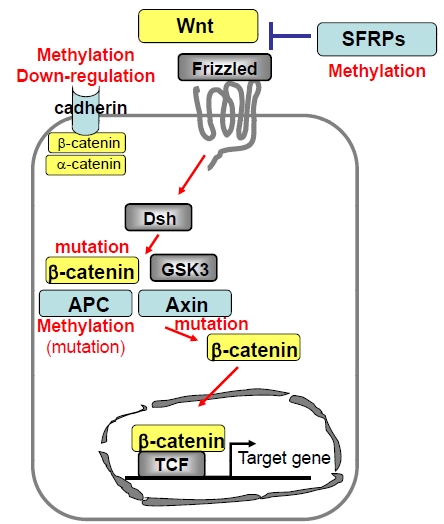
Schematic representation of Wnt/β-catenin pathway and the alterations reported in HCC. Functions of molecules shown in yellow and blue represent oncogenic and tumor suppressive respectively. Specific types of genetic and epigenetic alterations found in HCC are shown in red letter.
The CTNNB1 Gene
β-catenin is encoded by the CTNNB1 gene and plays an important role in various aspects of liver biology including pathogenesis of liver cancer. Mutations in this gene are the cause of several cancers, such as colorectal cancer and ovarian cancer [36, 37]. In human HCC, point mutations or deletions at the phosphorylation site on neighboring codons in the exon 3 of the CTNNB1 have been proposed to cause deregulation of the signaling function and contribute towards HCC pathogenesis. Generally speaking, 10–30 % of HCC carries mutations of the CTNNB1 gene, which induces the accumulation of β-catenin in the nucleus [28].
The AXIN Genes
As described above, the axin protein interacts with APC, β-catenin, GSK-3β, and protein phosphate 2. Mutations in this gene are also associated with hepatocellular carcinoma, hepatoblastomas, and several other cancers. Mutations of the AXIN1 gene can be detected in 5–9 %, and mutations of the AXIN2 are reported in ~3% of human HCC or HCC cell lines that carry the wild type CTNNB1 gene [38]. Gene transfer of wild-type AXIN1 resulted in enhanced apoptosis in HCC cell lines as a consequence of increased accumulation of β-catenin in the nucleus. On the other hand, some target genes of β-catenin, such as glutamine synthetase, did not show an enhanced expression even in cell lines with the mutated AXIN genes, suggesting that the tumor suppressor function of this protein might act via additional pathways other than Wnt/β-catenin [39].
The APC Gene
APC also forms complexes with axin and GSK-3β , and contributes in controlling cellular β-catenin levels through ubiquitin proteasome-dependent degradation of β-catenin. In human HCC, biallelic inactivation of the APC gene (mutation of the gene accompanied by loss of chromosomal region of the APC locus) was reported in a HCC case [40]. In our analysis, 84% of HCC carried dense methylation on the promoter of this gene in HCC (Fig. 1), indicating that promoter methylation was the major mechanism of inactivation of the APC gene in human HCC [28].
The CDH1 (E-Cadherin) Gene
E-cadherin is a calcium-dependent cell-cell adhesion glycoprotein that plays an important role in the formation of the epithelial cell layer. Cadherin can be found in complexes with catenin, which is important for the biological function of this gene. The oncogenic signals from EGF and Src also target β-catenin. It has been shown that phosphorylation of β-catenin through the EGF pathway associated with invasion and metastasis of cancer because of suppression of the adhesion function of Cadherin [41]. Mutations of this gene are reported in gastric, colorectal and breast cancers. Loss of function of this gene contributes to the progression of HCC by permitting increased proliferation, invasion, and metastasis of the neoplastic cells. Expression analysis of E-cadherin in hepatocellular carcinomas demonstrated that 56% of HCC showed the down-regulation of E-cadherin expression, and this phenomenon directly correlated with the size of tumors, as well as the mitotic index and survival [42].
Several reports suggested that the CDH1 gene can also be inactivated through the promoter hypermethylation of its promoter region. Interestingly, it has also been reported that reactive oxygen species (ROS) induce hypermethylation of the CDH1promoter. In this context, activation of PI3K/Akt/GSK-3β by ROS has been shown to recruitment of histone deacethylase and DNA methyltransferase on its promoter, which causes the downregulation of CDH1expression [43]. Because longstanding chronic inflammation enhances the production of ROS in the background liver of HCC, it is reasonable to speculate that these epimutations play an important role in the initial steps of inflammation-related hepatocarcinogenesis.
Secreted Frizzled-Related Protein (SFRP) 2
Frizzled are membrane proteins which are known as receptors of Wnt. SFRPs act as soluble modulators of Wnt signaling as SFRPs containing the putative Wnt-binding site of frizzled proteins. Because SFRPs are able to down-regulate Wnt signaling by forming an inhibitory complex with the frizzled receptors, SFRPs are considered as tumor suppressors. We have reported that one of the SFRP family genes, SFRP2, showed hypermethylation of its promoter and was frequently inactivated in human HCC (Fig. 1) [28].
ABNORMAL METHYLATION OF TUMOR SUPPRESSOR GENES
It is now increasingly being recognized that the patterns of DNA methylation, which are ordinarily maintained during normal cell division, are often disturbed in many human tumors, including HCC. Since low levels of DNA methylation can also be detected in the non-cancerous liver of patients with chronic liver damage or HCC, it is now believed that this epigenetic defect emerges at an early stage during hepatocarcinogenesis [44]. In human HCC, both regional hypermethylation of tumor suppressor gene (TSG) promoters and hypomethylation of repetitive DNA sequences are commonly observed [45]. The former is an important mechanism for the inactivation of corresponding gene, while the latter plays a role in maintaining chromosomal fragility and activation of retrotransposons and microRNA (miRNA), both of which could be critical for HCC development and progression. Although the precise mechanisms for the alteration of DNA methylation during hepatocarcinogenesis are still unclear, both HBx and HCV core proteins have been shown to induce the expression of DNA methyltransferases, which then leads to abnormal methylation of the CDH1 gene [46, 47].
We have previously reported that DNA methylation of TSGs was closely associated with the presence of HCV not only in HCCs but also in chronic hepatitis tissues, suggesting that the chronic inflammation or presence of HCV may contribute to the emergence of abnormal methylation [48]. The numbers and frequencies of genetic alterations of cancer-related genes in HCC are limited, however, epigenetic alterations are frequently observed in several TSGs (Fig. 1) [49].
CLINICAL IMPLICATIONS OF GENETIC AND EPIGENETIC ALTERATIONS
Cancer specific alterations of several genes are promising candidates for developing molecular markers of HCC, such as diagnosis and prediction of prognosis. Among them, alteration of DNA methylation is widely and frequently observed in almost every HCC, and DNA methylation can be easily and quantitatively measured in a variety of clinical specimens. Detection of methylation of the CDKN2A and GSTP1 genes in serum has been used for the early diagnosis of HCC [50, 51]. Furthermore, detection of the methylated CDKN2A and RASSF1A genes in serum of cases at a high risk of HCC could predict early occurrence of HCC [52]. Similarly, alteration of certain cancer-related genes or oncogenic pathways could be a target of developing a novel therapy of HCC. For example, a high level of copy-number gain and overexpression of vascular endothelial growth factor A was reported in a subset of HCC, suggesting that antiangiogenic therapies could be exclusively effective for this type of tumor [53].
ALTERED EXPRESSION OF miRNAs
Expression of miRNAs, which has been identified as a new class of small non-coding family of genes that are involved in post-transcriptional gene regulation, is also shown to be frequently altered in HCC. Several comprehensive analyses of miRNA have identified differentially expressed miRNAs in various subsets of HCCs. This type of alteration can also modulate several important signaling pathways, which is critical for hepatocarcinogenesis.
In human HCCs, miRNA (miR)-122 expression is significantly reduced in a subset of HCC compared to the non-cancerous tissues [54]. miR-122 can modulate cyclin G1 expression in HCC-derived cell lines, and an inverse correlation between miR-122 and cyclin G1 expression has been shown in human HCC, suggesting that cyclin G1 is a target of miR-122 [55]. On the other hand, miR-221 also targets cell-cycle inhibitors CDKN1B (p27kip1) and CDKN1C (p57kip2) and its altered expression also results in a disturbed normal cell-cycle regulation in hepatocytes [56].
MiR-21 is shown to be overexpressed in HCC as well as HCC cell lines, and inhibition of miR-21 increases expression of the tumor suppressor PTEN which helps in decreased tumor proliferation and invasion [57]. On the other hand, Ji, et al. showed that tumors with reduced miR-26 expression had a distinct transcriptomic pattern, with the activation of NF-κB and IL-6 signaling pathways [58]. Interestingly, induction of IL-6 is reported as an important step for inflammation-related HCC pathogenesis [59], and high levels of serum IL-6 can be a risk factor for HCC in HCV-positive patients [60]. Activation of this pathway also accounts for the gender differences and risk of HCC, as estrogen is known to suppress MyD88-dependent IL-6 production, and miR-26 is expressed at a higher level in women than men in the liver [58, 59]. Reportedly, this miRNA is also a good candidate for therapeutic target of HCC. Patients whose tumors had low miR-26 expression had shorter survival but a better response to interferon therapy than did patients whose tumors had high expression of this miRNA [58]. In addition, Kota, et al. reported the significance of this miRNA on treatment response using a mouse model [61]. Expression of miR-26 in liver cancer cells in vitro induces cell cycle arrest via suppression of cyclin D2 and cyclin E2, and systemic administration of miR-26 in mouse HCC model induces inhibition of cancer cell proliferation and induction of tumor-specific apoptosis without toxicity [61]. However, as individual miRNAs regulate hundreds of transcripts, antiproliferative effects of miR-26 in HCC, might not attribute to a single oncogeneic pathway but a regulation of multiple pathways, such as c-Myc and p53-dependent pathway.
On the other hand, several other studies revealed the association between expression profile of miRNA and etiology as well as specific genetic alterations. The miR-96 is overexpressed in HBV-related tumors, and miR-126 is down-regulated in alcohol-related hepatocellular carcinoma. Similarly, several reports suggest that the expression profile of miRNA can be applied to evaluate the etiology of tumor and predict tumor behavior, such as metastasis and prognosis [58, 62-66]. These evidences support the idea that in addition to genetic and epigenetic alteration of oncogenes and TSGs, the altered expression of miRNAs is an additional mechanism critical for human hepatocarcinogenesis. This concept is bound to receive a lot of attention in the coming years, as it not only helps in a better understanding of the molecular mechanisms of HCC, but also has a potential for clinical use including the development of biomarkers and therapeutic targets of this malignancy.
CONCLUSION
We have summarized in this review article the spectrum of various genetic and epigenetic alterations in a variety of genes and their corresponding oncogeneic pathways that are operational in human hepatocarcinogenesis (Figs. 2 and 3). What we now understand is that similar to many other cancers, development of HCC associates with a step-wise accumulation of genetic and epigenetic defects in cancer-related genes that includes point mutations, chromosomal alterations, aberrant DNA methylation and alterations of miRNA expression. Although the precise underpinnings of these various processes are still hazy, nonetheless, it is very likely that the defects in multiple signaling pathways might overlap and crosstalk exists among these pathways, as the premalignant clones progress towards a more advanced stage of HCC. Given the high prevalence of chronic liver disease and HCC patients, knowledge of these molecular events is very exciting and timely, because an understanding of these events will help tailor approaches that can be best exploited for the early detection, management and therapy of HCC.
REFERENCES
- 1.Nishida N, Nishimura T, Ito T, Komeda T, Fukuda Y, Nakao K. Chromosomal instability and human hepatocarcinogenesis. Histol. Histopathol. 2003;18:897–909. doi: 10.14670/HH-18.897. [DOI] [PubMed] [Google Scholar]
- 2.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat. Rev. Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 3.Hussain SP, Schwank J, Staib F, Wang XW, Harris CC. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene. 2007;26:2166–2176. doi: 10.1038/sj.onc.1210279. [DOI] [PubMed] [Google Scholar]
- 4.Nishida N, Fukuda Y, Kokuryu H, Toguchida J, Yandell DW, Ikenega M, Imura H, Ishizaki K. Role and mutational heterogeneity of the p53 gene in hepatocellular carcinoma. Cancer Res. 1993;53:368–372. [PubMed] [Google Scholar]
- 5.Aguilar F, Harris CC, Sun T, Hollstein M, Cerutti P. Geographic variation of p53 mutational profile in nonmalignant human liver. Science. 1994;264:1317–1319. doi: 10.1126/science.8191284. [DOI] [PubMed] [Google Scholar]
- 6.Aguilar F, Hussain SP, Cerutti P. Aflatoxin B1 induces the transversion of G-->T in codon 249 of the p53 tumor suppressor gene in human hepatocytes. Proc. Natl. Acad. Sci. USA. 1993;90:8586–8590. doi: 10.1073/pnas.90.18.8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirk GD, Camus-Randon AM, Mendy M, Goedert JJ, Merle P, Trepo C, Brechot C, Hainaut P, Montesano R. Ser-249 p53 mutations in plasma DNA of patients with hepatocellular carcinoma from The Gambia. J. Natl. Cancer Inst. 2000;92:148–153. doi: 10.1093/jnci/92.2.148. [DOI] [PubMed] [Google Scholar]
- 8.Hulla JE, Chen ZY, Eaton DL. Aflatoxin B1-induced rat hepatic hyperplastic nodules do not exhibit a site-specific mutation within the p53 gene. Cancer Res. 1993;53:9–11. [PubMed] [Google Scholar]
- 9.Madden CR, Finegold MJ, Slagle BL. Altered DNA mutation spectrum in aflatoxin b1-treated transgenic mice that express the hepatitis B virus x protein. J. Virol. 2002;76:11770–11774. doi: 10.1128/JVI.76.22.11770-11774.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang XW, Gibson MK, Vermeulen W, Yeh H, Forrester K, Sturzbecher HW, Hoeijmakers JH, Harris CC. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res. 1995;55:6012–6016. [PubMed] [Google Scholar]
- 11.Ambs S, Bennett WP, Merriam WG, Ogunfusika MO, Oser SM, Harrington AM, Shields PG, Felley-Bosco E, Hussain S P, Harris CC. Relationship between p53 mutations and inducible nitric oxide synthase expression in human colorectal cancer. J. Natl. Cancer Inst. 1999;91:86–88. doi: 10.1093/jnci/91.1.86. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 13.Tannapfel A, Busse C, Weinans L, Benicke M, Katalinic A, Geissler F, Hauss J, Wittekind C. INK4a-ARF alterations and p53 mutations in hepatocellular carcinomas. Oncogene. 2001;20:7104–7109. doi: 10.1038/sj.onc.1204902. [DOI] [PubMed] [Google Scholar]
- 14.Ito T, Nishida N, Fukuda Y, Nishimura T, Komeda T, Nakao K. Alteration of the p14(ARF) gene and p53 status in human hepatocellular carcinomas. J. Gastroenterol. 2004;39:355–361. doi: 10.1007/s00535-003-1302-9. [DOI] [PubMed] [Google Scholar]
- 15.Kao J.T Chuah SK, Huang C.C Chen CL, Wang C.C Hung CH, Chen C.H Wang JH, Lu S.N Lee CM, Changchien C.S Hu TH. P21/WAF1 is an independent survival prognostic factor for patients with hepatocellular carcinoma after resection. Liver Int. 2007;27:772–81. doi: 10.1111/j.1478-3231.2007.01499.x. [DOI] [PubMed] [Google Scholar]
- 16.Han HJ, Jung EY, Lee WJ, Jang KL. Cooperative repression of cyclin-dependent kinase inhibitor p21 gene expression by hepatitis B virus X protein and hepatitis C virus core protein. FEBS Lett. 2002;518:169–172. doi: 10.1016/s0014-5793(02)02694-7. [DOI] [PubMed] [Google Scholar]
- 17.Ito Y, Matsuura N, Sakon M, Miyoshi E, Noda K, Takeda T, Umeshita K, Nagano H, Nakamori S, Dono K, Tsujimoto M, Nakahara M, Nakao K, Taniguchi N, Monden M. Expression and prognostic roles of the G1-S modulators in hepatocellular carcinoma: p27 independently predicts the recurrence. Hepatology. 1999;30:90–99. doi: 10.1002/hep.510300114. [DOI] [PubMed] [Google Scholar]
- 18.Lei PP, Zhang ZJ, Shen LJ, Li JY, Zou Q, Zhang HX. Expression and hypermethylation of p27 kip1 in hepatocarcinogenesis. World J. Gastroenterol. 2005;11:4587–4591. doi: 10.3748/wjg.v11.i29.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukas J, Parry D, Aagaard L, Mann DJ, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent cell-cycle inhibition by the tumour suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 20.Nishida N, Fukuda Y, Komeda T, Kita R, Sando T, Furukawa M, Amenomori M, Shibagaki I, Nakao K, Ikenaga M. Amplification and overexpression of the cyclin D1 gene in aggressive human hepatocellular carcinoma. Cancer Res. 1994;54:3107–3110. [PubMed] [Google Scholar]
- 21.Nishida N, Fukuda Y, Ishizaki K, Nakao K. Alteration of cell cycle-related genes in hepatocarcinogenesis. Histol. Histopathol. 1997;12:1019–1025. [PubMed] [Google Scholar]
- 22.Azechi H, Nishida N, Fukuda Y, Nishimura T, Minata M, Katsuma H, Kuno M, Ito T, Komeda T, Kita R, Takahashi R, Nakao K. Disruption of the p16/cyclin D1/retinoblastoma protein pathway in the majority of human hepatocellular carcinomas. Oncology. 2001;60:346–354. doi: 10.1159/000058531. [DOI] [PubMed] [Google Scholar]
- 23.Nishida N, Fukuda Y, Kokuryu H, Sadamoto T, Isowa G, Honda K, Yamaoka Y, Ikenaga M, Imura H, Ishizaki K. Accumulation of allelic loss on arms of chromosomes 13q, 16q and 17p in the advanced stages of human hepatocellular carcinoma. Int. J. Cancer. 1992;51:862–868. doi: 10.1002/ijc.2910510605. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Xu HJ, Murakami Y, Sachse R, Yashima K, Hirohashi S, Hu SX, Benedict WF, Sekiya T. Deletions of chromosome 13q, mutations in Retinoblastoma 1, and retinoblastoma protein state in human hepatocellular carcinoma. Cancer Res. 1994;54:4177–4182. [PubMed] [Google Scholar]
- 25.Roncalli M, Bianchi P, Bruni B, Laghi L, Destro A, Di Gioia S, Gennari L, Tommasini M, Malesci A, Coggi G. Methylation framework of cell cycle gene inhibitors in cirrhosis and associated hepatocellular carcinoma. Hepatology. 2002;36:427–432. doi: 10.1053/jhep.2002.34852. [DOI] [PubMed] [Google Scholar]
- 26.Kita R, Nishida N, Fukuda Y, Azechi H, Matsuoka Y, Komeda T, Sando T, Nakao K, Ishizaki K. Infrequent alterations of the p16INK4A gene in liver cancer. Int. J. Cancer. 1996;67:176–180. doi: 10.1002/(SICI)1097-0215(19960717)67:2<176::AID-IJC4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 27.Matsuda Y, Ichida T, Matsuzawa J, Sugimura K, Asakura H. p16(INK4) is inactivated by extensive CpG methylation in human hepatocellular carcinoma. Gastroenterology. 1999;116:394–400. doi: 10.1016/s0016-5085(99)70137-x. [DOI] [PubMed] [Google Scholar]
- 28.Nishida N, Nishimura T, Nagasaka T, Ikai I, Goel A, Boland CR. Extensive methylation is associated with beta-catenin mutations in hepatocellular carcinoma: evidence for two distinct pathways of human hepatocarcinogenesis. Cancer Res. 2007;67:4586–4594. doi: 10.1158/0008-5472.CAN-06-3464. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Hui AM, Sun L, Hasegawa K, Torzilli G, Minagawa M, Takayama T, Makuuchi M. p16INK4A hypermethylation is associated with hepatitis virus infection, age, and gender in hepatocellular carcinoma. Clin. Cancer Res. 2004;10:7484–7489. doi: 10.1158/1078-0432.CCR-04-1715. [DOI] [PubMed] [Google Scholar]
- 30.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 31.Matsumine A, Ogai A, Senda T, Okumura N, Satoh K, Baeg GH, Kawahara T, Kobayashi S, Okada M, Toyoshima K, Akiyama T. Binding of APC to the human homolog of the Drosophila discs large tumor suppressor protein. Science. 1996;272:1020–1023. doi: 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- 32.Boutros M, Mihaly J, Bouwmeester T, Mlodzik M. Signaling specificity by Frizzled receptors in Drosophila. Science. 2000;288:1825–1828. doi: 10.1126/science.288.5472.1825. [DOI] [PubMed] [Google Scholar]
- 33.Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, Horii A, Koyama K, Utsunomiya J, Baba S, Hedge P. Mutations of chromosome 5q21 genes in FAP and colorectal cancer patients. Science. 1991;253:665–669. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- 34.Renkonen ET, Nieminen P, Abdel-Rahman WM, Moisio AL, Jarvela I, Arte S, Jarvinen HJ, Peltomaki P. Adenomatous polyposis families that screen APC mutation-negative by conventional methods are genetically heterogeneous. J. Clin. Oncol. 2005;23:5651–5659. doi: 10.1200/JCO.2005.14.712. [DOI] [PubMed] [Google Scholar]
- 35.Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, Taite H, Scoular R, Miller A, Reeve AE. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 36.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K.W. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 37.Gamallo C, Palacios J, Moreno G, Calvo de Mora J, Suarez A, Armas A. beta-catenin expression pattern in stage I and II ovarian carcinomas: relationship with beta-catenin gene mutations, clinicopathological features, and clinical outcome. Am. J. Pathol. 1999;155:527–536. doi: 10.1016/s0002-9440(10)65148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniguchi K, Roberts LR, Aderca IN, Dong X, Qian C, Murphy LM, Nagorney DM, Burgart LJ, Roche PC, Smith DI, Ross JA, Liu W. Mutational spectrum of beta-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene . 2002;21:4863–4871. doi: 10.1038/sj.onc.1205591. [DOI] [PubMed] [Google Scholar]
- 39.Zucman-Rossi J, Benhamouche S, Godard C, Boyault S, Grimber G, Balabaud C, Cunha AS, Bioulac-Sage P, Perret C. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene. 2007;26:774–780. doi: 10.1038/sj.onc.1209824. [DOI] [PubMed] [Google Scholar]
- 40.Katoh H, Shibata T, Kokubu A, Ojima H, Kosuge T, Kanai Y, Hirohashi S. Genetic inactivation of the APC gene contributes to the malignant progression of sporadic hepatocellular carcinoma: a case report. Genes Chromosomes Cancer. 2006;45:1050–1057. doi: 10.1002/gcc.20362. [DOI] [PubMed] [Google Scholar]
- 41.Fujii K, Furukawa F, Matsuyoshi N. Ligand activation of overexpressed epidermal growth factor receptor results in colony dissociation and disturbed E-cadherin function in HSC-1 human cutaneous squamous carcinoma cells. Exp. Cell Res. 1996;223:50–62. doi: 10.1006/excr.1996.0057. [DOI] [PubMed] [Google Scholar]
- 42.Garcia S, Martini F, De Micco C, Andrac L, Hardwigsen J, Sappa P, Lavaut MN, Le Treut YP, Charpin C. Immunoexpression of E-cadherin and beta-catenin correlates to survival of patients with hepatocellular carcinomas. Int. J. Oncol. 1998;12:443–447. doi: 10.3892/ijo.12.2.443. [DOI] [PubMed] [Google Scholar]
- 43.Lim SO, Gu JM, Kim MS, Kim HS, Park YN, Park CK, Cho JW, Park YM, Jung G. Epigenetic changes induced by reactive oxygen species in hepatocellular carcinoma: methylation of the E-cadherin promoter. Gastroenterology. 2008;135:2128–2140. doi: 10.1053/j.gastro.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 44.Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis--A comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32:970–979. doi: 10.1053/jhep.2000.19797. [DOI] [PubMed] [Google Scholar]
- 45.Park IY, Sohn BH, Yu E, Suh DJ, Chung YH, Lee JH, Surzycki SJ, Lee YI. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476–1494. doi: 10.1053/j.gastro.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 46.Jung JK, Arora P, Pagano JS, Jang KL. Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res. 2007;67:5771–5778. doi: 10.1158/0008-5472.CAN-07-0529. [DOI] [PubMed] [Google Scholar]
- 47.Arora P, Kim EO, Jung JK, Jang KL. Hepatitis C virus core protein downregulates E-cadherin expression via activation of DNA methyltransferase 1 and 3b. Cancer Lett. 2008;261:244–252. doi: 10.1016/j.canlet.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 48.Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47:908–918. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nishida N. Impact of hepatitis virus and aging on DNA methylation in human hepatocarcinogenesis. Histol. Histopathol. 2010;25:647–654. doi: 10.14670/HH-25.647. [DOI] [PubMed] [Google Scholar]
- 50.Wong IH, Lo YM, Zhang J, Liew CT, Ng MH, Wong N, Lai PB, Lau WY, Hjelm NM, Johnson PJ. Detection of aberrant p16 methylation in the plasma and serum of liver cancer patients. Cancer Res. 1999;59:71–73. [PubMed] [Google Scholar]
- 51.Wang J, Qin Y, Li B, Sun Z, Yang B. Detection of aberrant promoter methylation of GSTP1 in the tumor and serum of Chinese human primary hepatocellular carcinoma patients. Clin. Biochem. 2006;39:344–348. doi: 10.1016/j.clinbiochem.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 52.Zhang YJ, Wu HC, Shen J, Ahsan H, Tsai WY, Yang HI, Wang LY, Chen SY, Chen CJ, Santella RM. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clin. Cancer Res. 2007;13:2378–2384. doi: 10.1158/1078-0432.CCR-06-1900. [DOI] [PubMed] [Google Scholar]
- 53.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, LeBlanc AC, Donovan DJ, Thung SN, Sole M, Tovar V, Alsinet C, Ramos AH, Barretina J, Roayaie S, Schwartz M, Waxman S, Bruix J, Mazzaferro V, Ligon AH, Najfeld V, Friedman SL, Sellers WR, Meyerson M, Llovet JM. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer Res. 2008;68:6779–6788. doi: 10.1158/0008-5472.CAN-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, Jacob ST, Ghoshal K. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J. Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gramantieri L, Ferracin M, Fornari F, Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E, Grazi GL, Croce CM, Bolondi L, Negrini M. Cyclin G1 is a target of miR-122a a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 56.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin G A, Grazi G L, Giovannini C, Croce CM, Bolondi L, Negrini M. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 57.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, Qin LX, Man K, Lo CM, Lee J, Ng IO, Fan J, Tang ZY, Sun HC, Wang XW. MicroRNA expression, survival, and response to interferon in liver cancer. N. Engl. J. Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naugler WE, Sakurai T, Kim S, Maeda S, Kim K, Elsharkawy AM, Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 60.Nakagawa H, Maeda S, Yoshida H, Tateishi R, Masuzaki R, Ohki T, Hayakawa Y, Kinoshita H, Yamakado M, Kato N, Shiina S, Omata M. Serum IL-6 levels and the risk for hepatocarcinogenesis in chronic hepatitis C patients: an analysis based on gender differences. Int. J. Cancer. 2009;125:2264–2269. doi: 10.1002/ijc.24720. [DOI] [PubMed] [Google Scholar]
- 61.Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang H-W, Chang T-C, Vivekanandan P, Torbenson M, Clark KR, Mendell JR, Mendell JT. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, Tang ZY, Wang XW. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 63.Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 64.Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098–1112. doi: 10.1002/hep.22749. [DOI] [PubMed] [Google Scholar]
- 65.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 66.Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, Yan M, Ge C, Zhang Z, Chen T, Wan D, Yao M, Li J, Gu J, He X. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 2010;51:846–856. doi: 10.1002/hep.23443. [DOI] [PubMed] [Google Scholar]