Abstract
ABSTRACT The complete dnaK operon of Listeria monocytogenes was isolated by chromosome walking using the previously cloned dnaK gene as a probe. Molecular analysis of the locus identified 6 genes in the order hrcA, grpE, dnaK, dnaJ, orf35, and orf29. Primer extension analysis revealed 3 transcription start sites—S1, S2, and S3—upstream of the hrcA, grpE, and dnaJ, respectively. The transcription from S1 was heat inducible. Analysis of the sequences revealed the consensus promoter sequences of gram-positive bacteria, P1 and P2 upstream of the hrcA and dnaJ, respectively. The hrcA gene and a regulatory sequence, designated CIRCE (controlling inverted repeat of chaperone expression), play a role in the regulation of expression of the dnaK locus in response to heat shock in several gram-positive bacteria. Their presence upstream of the dnaK locus in L monocytogenes suggested a similar regulatory mechanism for the transcription initiated at the promoter, P1. Northern blot analysis led to the detection of 4 mRNA species of 4.9 kb, 3.6 kb, 3.6 kb, and 1.2 kb; the first 2 species were heat inducible. The current results indicate that 4 distinct transcripts directed by 3 promoters are involved in the expression of the dnaK operon of L. monocytogenes.
INTRODUCTION
Listeria monocytogenes is a foodborne gram-positive bacterium that can cause lethal infections in immunocompromised persons. L. monocytogenes belongs to the facultative intracellular bacteria, which are able to survive and replicate in professional phagocytes such as macrophages and monocytes and in unprofessional phagocytic mammalian cells (Dabri et al 1990; Gaillard et al 1987; Portnoy et al 1988). The intracellular environment, especially that of professional phagocytes, imposes hostile conditions on invading bacteria. The diverse bactericidal factors include toxic oxidative products, lysosomal and granular peptides, low pH, and low levels of essential nutrients. It is therefore expected that intracellular bacteria require specific products to cope with intracellular stress conditions. Recent evidence shows that stress proteins play an important role in intracellular survival and growth by facultative bacteria within macrophages (Johnson et al 1991; Köhler et al 1990; Phillips et al 1995; Rouquette et al 1996; Yamamoto et al 1996).
The stress proteins are induced by a variety of different stimuli, such as heat shock, oxidative products, nutrient limitation, acidic condition, and damage caused by toxic chemicals and physical agents. We previously characterized the heat shock response of L. monocytogenes (Hanawa et al 1995). Among the stress proteins identified were homologues of DnaK, GroEL, and GroES. DnaK is well known to have an important role on cellular metabolism as a molecular chaperone. We have recently demonstrated that the DnaK chaperone of L. monocytogenes is involved in stress tolerance and efficient phagocytosis with macrophages through molecular cloning of the dnaK locus, introduction of an insertion in the locus, and characterization of the mutant (Hanawa et al 1999). Early studies in Escherichia coli revealed that the dnaK operon comprises the genes dnaK and dnaJ (for review, see Georgopoulos et al 1990) and that the expression of the genes is under the positive control of an alternative factor, σ32, which is a sigma subunit of RNA polymerase (for review, see Yura et al 1993). In contrast, recent studies in gram-positive bacteria revealed the dnaK operon is more complex and more variable in its molecular organization. Genes associated with the dnaK operon usually include hrcA, grpE, dnaK, and dnaJ (Eaton et al 1993; Narberhaus et al 1992; Ohta et al 1994; Wetzstein et al 1992). A further 3 genes, orf35, orf28, and orf50, are also associated with the dnaK operon in Bacillus subtilis to form a heptacistronic operon (Homuth et al 1997). Furthermore, the regulatory mechanisms of the expression in gram-positive bacteria are quite different from those of E coli. In B. subtilis and Streptococcus mutans, the regulation of expression of dnaK and other class I heat shock genes appears to be negatively controlled by an HrcA protein, which binds at a conserved cis-acting inverted repeat (a CIRCE element: controlling inverted repeat of chaperone expression) located near vegetative promoters (Zuber and Schumann 1994; Jayaraman et al 1997; Ballard et al 1998).
Our recent cloning and sequencing of the dnaK locus of L. monocytogenes have revealed that the hrcA, grpE, and dnaJ genes are associated with this region, suggesting that the operon consists of the genes hrcA, grpE, dnaK, and dnaJ (Hanawa et al 1999). However, the disruption of the dnaK gene by insertion of a fragment did not impair the production of a DnaJ protein, suggesting the independent expression of the dnaJ gene (Hanawa et al 1999). Furthermore, no obvious promoters were detected upstream of the dnaK translational start site. To define the molecular structure and the expression of the dnaK locus in L monocytogenes, we decided to isolate the complete dnaK operon and analyze the transcriptional regulation of the genes. In this report, we show that 6 genes are in the dnaK operon of L. monocytogenes and that 4 distinct transcripts are produced from the operon through cloning, sequencing, and transcriptional analysis.
MATERIALS AND METHODS
Bacterial strains and plasmids
L. monocytogenes serotype 1 strain 10403S (Bishop and Hinrichs 1987) was used and its cultivation was previously described (Hanawa et al 1995). The E. coli JM109 (Yanisch-Perron et al 1985) was cultured in brain-heart infusion medium (Nissui, Tokyo, Japan) supplemented with 100 μg/ml ampicillin, 25 μg/ml chloramphenicol, or 25 μgml tetracycline as required. The plasmid pACYC184 (Chang and Cohen 1978) was used in the construction of a partially genomic library, while the pBluescript SK/KS (Strategene, La Jolla, CA, USA) was used for subcloning experiments.
DNA isolation, manipulation, and sequencing
The preparation of chromosomal DNA from L. monocytogenes was described previously (Hanawa et al 1999). The plasmid DNA purification, restriction analysis, ligation, and agarose gel electrophoresis were performed according to the methods described by Sambrook et al (1989). For Southern hybridization, the digested DNAs were electrophoresed through a 0.8% agarose gel and then transferred to Gene Screen Plus membrane (NEN, Boston, MA, USA). The DNA fragment as a probe was labeled by using the Gene Image random prime labeling and detection system (Amersham, Buckinghamshire, UK), and hybridization was performed according to the manufacturer's instructions. The sequencing was carried out with Sequenase (USB, Cleveland, OH, USA) and T7 or T3 DNA primers for pBluescript plasmid or synthetic primers.
Preparation of RNA and Northern hybridization
Total cellular RNA was isolated using the RNeasy kit from Qiagen (Hilden, Germany). RNA for Northern blot analysis was separated in formaldehyde gels and transferred to Gene Screen Plus membrane. Size determination was done by using an RNA ladder (281, 623, 955, 1383, 1908, 2604, 3638, 4981, and 6583 bp; Promega, Madison, WI, USA) as the standard. The following DNA fragments were used as hybridization probes specific to hrcA covering the sequence from nt 303 to 773, grpE covering nt 1446 to 1819, dnaK covering nt 2086 to 2821, dnaJ covering nt 4027 to 4702, orf35 covering nt 5154 to 6056, and orf29 covering nt 6106 to 6767. These sequences were generated through polymerase chain reaction amplification. The hybridization was performed by using the hybridization buffer from the Gene Image random prime labeling and detection system (Amersham) at 65°C for 18 to 20 hours. The washing of the membrane after hybridization and detection of signal were performed according to the manufacturer's instructions.
Determination of the transcription start site
To define the transcription start sites, primer extension analysis was used. Synthetic oligonucleotides, 0294 (5′-CGATGATGGCACGGAAAATCAAAAG-3′), 1342 (5′-TTTCTCAGACACTTTAGCCA-3′), and 3976 (5′-GCTGAGGCGCTTTTGGAAAT-3′), which are complementary to nt 294 to 270 of the hrcA coding sequence, to nt 1342 to 1323 of the grpE coding sequence, and to nt 3976 to 3956 of the dnaJ coding sequence, respectively, were end labeled with [γ-32P]-adenosine triphosphate and annealed to 25 μg of total cellular RNA. The primers were extended with 100 units of avian myeloblastosis virus reverse transcriptase (Superscript II, BRL) and deoxyribonucleotide thiophosphates (Sigma, St. Louis, MO, USA) at 42 °C for 30 minutes. Nucleic acid was precipitated with ethanol, dried, resuspended in TE buffer (pH 8.0), and extracted by Microspin column S200 (Pharmacia, Uppsala, Sweden). The samples with formamide loading dye were subjected to electrophoresis in 5% polyacrylamide sequencing gels. DNA sequencing reactions, using the same primer that was used in the primer extension analysis and a plasmid carrying the appropriate region of the hrcA locus, grpE locus, or the dnaJ locus as a temperate, were run adjacent to the primer extension reactions to identify the transcription initiation site.
Nucleotide sequence accession number
The sequence data of complete operon reported here will appear in the DDBJ/EMBL/GenBank nucleotide sequence database under accession number AB023064. Sequence data of the dnaK gene have been submitted to the DDBJ database by us and have been assigned the accession number AB007771.
RESULTS
Isolation of the complete dnaK operon ofL. monocytogenes
To determine the nucleotide sequence of the dnaK gene of L. monocytogenes, we have previously cloned the 2.9-kb EcoRV, 2.5 kb HindIII, and 6.0 kb EcoRI fragments that gave clear hybridization signals to the dnaK gene from E. coli, as previously reported (Hanawa et al 1999). Sequencing of part of the 2.9-kb insert of pTKY263 (Fig 1) revealed the presence of the hrcA homologue near one end, but the gene was incomplete, missing its 5′ end. By using the 2.9 kb EcoRV fragment containing 828 bp of the 3′ end of the hrcA gene, we cloned the complete hrcA gene in the current study. Hybridization of this fragment to chromosomal DNA digested with different restriction enzymes revealed specific signals, including a 1.9-kb HindIII fragment (results not shown). According to the restriction cleavage map and sequence of the 2.9-kb insert of pTKY263 (Fig 1), the 1.9-kb HindIII fragment probably carried the 5′ end of hrcA gene. Thus, a partially genomic library containing HindIII fragments of L. monocytogenes DNA in pACYC184 was constructed and screened by hybridization with the 2.9 kb EcoRV fragment. A positive clone, pTKY328, carrying the 1.9-kb HindIII fragment was isolated. The restriction cleavage map of the cloned fragments containing whole genes related to possible dnaK operon of L. monocytogenes is shown in Figure 1.
Fig 1.
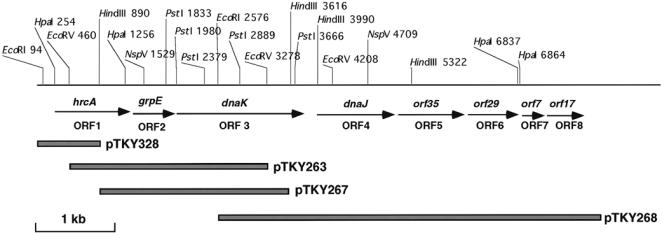
Restriction map of the Listeria monocytogenes dnaK locus and recombinant plasmids. Locations of the identified genes are indicated with arrows
Sequencing and genetic organization of the dnaK operon of L. monocytogenes
A total of 7800 bp was sequenced and analyzed. Sequencing revealed the presence of 8 Orfs (Figs 1, 2).
Fig 2.
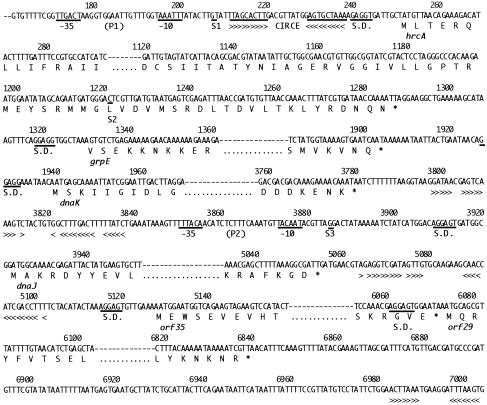
Nucleotide sequences, amino acid sequences, and potential regulatory elements of the Listeria monocytogenes dnaK locus. The nucleotide and deduced amino acid sequences of each 5′ end and 3′ end of hrcA, grpE, dnaK, dnaJ, orf35 and orf29 genes are detailed. The deduced amino acid sequences are shown below the DNA sequences with single-letter codes. The translation stop signals are marked by asterisks below the respective codons. Predicted ribosome-binding sites are underlined and labeled as SD (Shine-Dalgalno). Breaks in the sequence presented are indicated by a dashed line. The −35 and −10 regions of putative promoter sequences are underlined. The transcriptional initiation sites that were mapped by primer extension analyses are underlined and designated S1, S2, and S3, respectively. Arrowheads below the DNA sequence show regions of dyad symmetry. The CIRCE-element–like inverted repeat (see text) is indicated with arrowheads
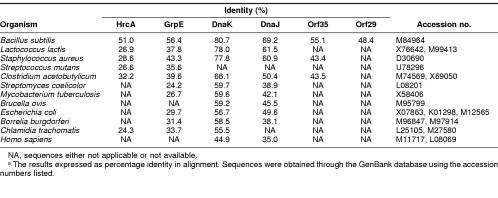
Orf1 is 1038 bp long (nt 252–1287) encoding a peptide of 345 aa with a predicted molecular mass of 39.7 kDa, with a Shine-Dalgalno–like sequence preceding the initiation codon by 8 bp. The aa sequence was similar to HrcA of B subtilis (Wetzstein et al 1992), HrcA of S mutans (Jayaraman et al 1997), OrfA of Clostridium acetobutylicum (Narberhaus et al 1992), Orf37 of Staphylococcus aureus (Ohta et al 1994), and Orf1 of Lactococcus lactis (Eaton et al 1993), as shown in Table 1. Therefore, we designated the gene hrcA.
Table 1.
Comparison of the amino acid sequences of HrcA, GrpE, DnaK, DnaJ, Orf35, and Orf29 from \i\L monocytogenes\r\ with their homologues in bacterial speciesa
Orf2 is 576 bp long (nt 1331–1904) encoding a peptide of 191 aa with a predicted molecular mass of 21.8 kDa, with a putative ribosomal binding site 8 bp 5′ to the start codon. This protein showed significant homology to GrpE of other bacteria (Table 1), and thus the gene was designated grpE.
Orf3 is 1842 bp long (nt 1940–3779) encoding a peptide of 613 aa with a predicted molecular mass of 66.2 kDa, with a putative ribosomal binding site 9 bp 5′ to the initiation codon. As already mentioned, this protein is highly homologous to DnaK of various species (Hanawa et al 1999), and thus this gene was called dnaK.
Orf4 is 1134 bp long (nt 3923–5054) encoding a peptide of 377 aa with a predicted molecular mass of 41.1 kDa, with a putative ribosomal binding site 9 bp 5′ to the initiation codon. Homology searches indicated that this protein was similar to DnaJ of other bacteria (Table 1), and thus the gene was designated dnaJ.
Orf5 is 945 bp long (nt 5129–6071) encoding a peptide of 314 aa with a predicted molecular mass of 34.8 kDa, with a putative ribosomal binding site 9 bp 5′ to the initiation codon. Homology searches indicated that this protein was similar to Orf35 of B. subtilis (Homuth et al 1997), Orf35 of S. aureus (Ohta et al 1994), and OrfB of C. acetobutylicum (Behrens et al 1993). These proteins showed significant homology to the ribosomal protein L11 metyltransferase encoded by prmA of E. coli (Vanet et al 1993).
Orf6 is 768 bp long (nt 6074–6839) encoding a peptide of 255 aa with a predicted molecular mass of 28.9 kDa, with a putative ribosomal binding site 8 bp 5′ to the initiation codon. Homology searches indicated that this protein named Orf29 was similar to Orf28 of B. subtilis (Homuth et al 1997) which exhibits significant homology to the 29.6 kDa protein YggJ of E. coli.
Orf7 is 174 bp long (nt 7039–7210) encoding a peptide of 57 aa with a predicted molecular mass of 6.9 kDa. This protein, named Orf7, exhibits significant homology to the 30S ribosomal protein S21 of B. subtilis (87.3 % identify SWISS-PROT. Rel. 34).
Orf8 is 456 bp long (nt 7223–7676) encoding a peptide of 151 aa with a predicted molecular mass of 16.8 kDa. The aa sequence was compared with sequences in the SWISS-PROT (Release 34) database. This analysis revealed that this protein, named Orf17, has a sequence homology to the transcriptional regulatory proteins TstD and PhoP of Salmonella typhimurium.
Regulatory sequences in the L monocytogenes dnaK operon
Analysis of sequences upstream of hrcA revealed sequences similar to the consensus promoter sequences of gram-positive bacteria (Graves and Rabinowitz 1986). The −35 sequence was 5′-TTGACT beginning at position 171, and the −10 sequence was 5′-TAAATT, with 17 nt in between (P1 in Fig 2 and Fig 5). Sequence analysis also revealed the presence of an inverted repeat sequence between the putative promoter element and the initiation codon of hrcA that had considerable sequence similarity with the CIRCE element found in the regulatory region of the dnaK operon and the groE operon of several bacteria (Zuber and Schumann 1994; Jayaraman et al 1997; Ballard et al 1998). The inverted repeat sequence of TTAGCACTt-nt 9-GAGTGCTAa differed from the consensus sequence of the CIRCE element at 1 position.
Fig 5.
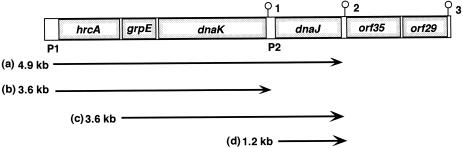
Genetic and transcriptional map of the dnaK operon of Listeria monocytogenes. This schematic representation is based on the determination of transcription start sites and Northern blot analysis. The promoters P1 and P2 were deduced from the sequences and the corresponding transcription start sites as determined by primer extension analysis. Potential stem-loop structures numbered 1 to 3 are indicated
No sequences resembling −35/−10 transcription signals were observed upstream of grpE and dnaK. The start codon of dnaJ is 141 nt downstream from the stop codon of dnaK. In this intergenic region, a second promoter (P2 in Fig 2 and Fig 5) was observed with a −35 box of 5′-TTTACA, separated 17 nt from a −10 box of 5′-TACAAT. No obvious promoter elements were observed upstream of either orf35 or orf29. However, upstream of orf7, which is 197 nt downstream from the termination codon of the orf29, a third promoter was detected with a −35 box of TTGACG separated 17 nt from a −10 box of TATAAT whose sequences are available in DDBJ/EMBL/GenBank database. No obvious promoter elements were observed upstream of Orf17. The validity of the promoters P1 and P2 was confirmed by primer extension analysis (see below).
Four palindromic structures were identified. As shown in Figures 2 and 5, the first stem and loop structure spanning nt 3798 to 3837 is located immediately downstream of the termination codon of dnaK. The second stem and loop structure was found in the region (nt 5064–5102) separating dnaJ and orf35. The third stem and loop structure was located in the region (nt 6983–7003) separating orf29 and orf7. The fourth structure spanning nt 7705–7752 was located downstream of the termination codon of orf17. The sequences of this region are available in the DDBJ/EMBL/GenBank database.
Transcriptional analysis of the L. monocytogenes dnaK operon
Sequence analysis of the dnaK region of L. monocytogenes suggested that the transcriptions are initiated at various sites. To identify the functional promoters responsible for transcription of these genes, primer extension was performed on RNA prepared from either L. monocytogenes grown at 30°C or bacteria heat shocked at 45°C for 5 minutes, 10 minutes, or 20 minutes. Synthetic oligonucleotides 0294, 1342, and 3976 corresponding to the regions immediately 5′ of hrcA, grpE, and dnaJ, respectively, were used as primers. The results are shown in Figure 3.
Fig 3.
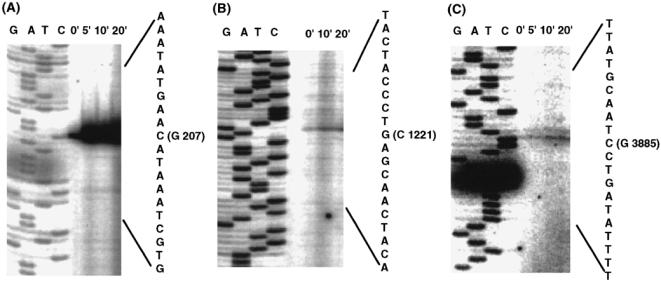
Primer extension analysis of the Listeria monocytogenes dnaK locus. Total RNAs (25 μg per lane) from L. monocytogenes 10403S grown at 30°C (0 minutes) and heat shocked at 45°C for 5 minutes, 10 minutes, or 20 minutes, were subjected to primer extension using oligonucleotides 0294 specific for hrcA (A), 1342 specific for grpE (B), and 3976 specific for dnaJ (C). The sequence of oligonucleotides used are shown in Materials and Methods. Products were compared with a sequencing ladder using the corresponding oligonucleotide primer. The sequences obtained are listed to the right of each panel and the first nucleotide of each transcript is shown with nucleotide position on the sequences
The oligo 0294 yielded a extension product identifying S1 at nt 207. S1 was located immediately downstream of the promoter element P1, which had been identified upstream of hrcA by sequence analysis. Primer extension using the oligo 1342 gave an extension product identifying S2 at nt 1221. S2 is not preceded by sequences resembling any known promoter. Furthermore, primer extension using the oligo 3976 yielded a product identifying S3 at nt 3885. S3 was located immediately downstream of the promoter element P2, identified upstream of dnaJ. These results suggest that several distinct transcripts are produced from the dnaK operon of L. monocytogenes.
The in vivo transcripts of the dnaK operon were detected using Northern blot analysis with total RNAs prepared from L. monocytogenes grown at 30°C and from heat-shocked bacteria. On Northern blots hybridized with hrcA-, grpE-, and dnaK-specific probes, 2 identical RNA species of 4.9 kb and 3.6 kb each were detected; they represent transcripts of the total operon with and without the dnaJ gene, respectively (Fig 4A–C). It is obvious that the transcription was induced by a heat shock. With the dnaJ-specific probe, as expected, the transcript of 4.9 kb was detected (Fig 4D). In addition, a 3.6-kb RNA and a 1.2-kb RNA that were not induced by heat shock appeared. We assumed that a small amount of the 3.6-kb transcript was initiated at S2 and represents a polycistronic transcript encompassing the 3 genes, grpE, dnaK, and dnaJ. To distinguish this 3.6-kb species for grpE, dnaK, and dnaJ from the 3.6-kb transcript for hrcA, grpE, and dnaK, 4 transcripts were labeled a, b, c, and d, as shown in Figure 5. Because of its low abundance, the 3.6 kb mRNA-c could be appeared as a doublet with 3.6 kb mRNA-b on hybridization blots with the probes for grpE and dnaK. The transcript of 1.2 kb could be a monocistronic dnaJ mRNA. Some blots exhibited hybridization signals at the 2 positions where the 16S and 23S rRNAs migrate.
Fig 4.
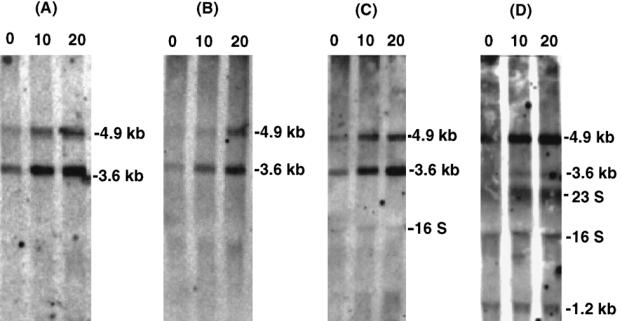
Northern blot analysis of hrcA, grpE, dnaK, and dnaJ mRNAs in response to heat shock. RNA was isolated from Listeria monocytogenes 10403S grown at 30°C (0 minutes) and at 10 or 20 minutes after a temperature shift to 45°C. The filters were hybridized with labeled DNAs corresponding to the hrcA (A), grpE (B), dnaK (C), and dnaJ (D). The size of each transcript was determined by comparison with RNA size markers
Analysis of the nucleotide sequence suggested that the orf35 and orf29 are part of the dnaK operon, because both are not separated by a large noncoding region and because no apparent promoters are observed upstream of both genes. We examined the possibility by the Northern blot analysis of total RNA with the orf35-specific and orf29-specific probes but failed to detect any band in Northern blots (data not shown; see Discussion). The transcriptional organization of the complete dnaK operon as deduced from the Northern analysis and from the primer extension is shown diagramatically in Figure 5.
DISCUSSION
This paper describes the first complete dnaK operon of L. monocytogenes. The dnaK region of L. monocytogenes contains at least 4 heat shock genes with the arrangement hrcA-grpE-dnaK-dnaJ, followed by 2 Orfs, orf35 and orf29. The chromosomal organization of these 6 genes at a single locus and the presence of the hrcA homologue along with the CIRCE element resemble the arrangement seen in other gram-positive bacteria, including C. acetobutylicum (Narberhaus et al 1992), B. subtilis (Wetzstein et al 1992), and S. aureus (Ohta et al 1994), though slight differences are observed. In C. acetobutylicum and S. aureus, orf29 has not been identified downstream of dnaJ. In B. subtilis, the dnaK operon contains a seventh large Orf, orf50, to form a heptacistronic operon (Homuth et al 1997). On the contrary, in L. monocytogenes, orf29 was followed by 2 Orfs, orf7 and orf17, which appeared to form a separate transcriptional unit from the dnaK operon (Fig 1).
Three transcriptional start sites, S1, S2, and S3, were identified upstream of hrcA, grpE, and dnaJ, respectively, by primer extension analysis (Fig 3). S1 and S3 mapped were consistent with the presence of promoter sequences P1 and P2 predicted from the sequence data. However, S2 is not preceded by sequences resembling any known promoter. It could be initiated by an alternative sigma factor of L. monocytogenes. Northern blot analysis revealed 4 mRNA species of various lengths. The 4.9-kb mRNA-a in all blots (Fig 4A–D), clearly enhanced by heat shock, could correspond by length to the transcript initiated at S1 and extending from hrcA through dnaJ. The 3.6-kb mRNA-b (Fig 4A–C), whose transcription was also induced by heat shock, could correspond by length to the genes of hrcA, grpE, and dnaK. As any additional starting sites in the region 5′ to hrcA coding sequence were not found, this transcription was initiated at S1 and could be terminated at the stem-loop structure 1 in the intergenic region between dnaK and dnaJ. Alternatively, the 3.6-kb mRNA-b could be a processing product of the 4.9 kb species as the stem-loop structure between dnaK and dnaJ could be a processing site. The following observation favors the processing hypothesis. The 3.6-kb mRNA-c (Fig 4D) that corresponds to the transcript extending to dnaJ was initiated at S2 by the length to the genes, grpE, dnaK, and dnaJ. If the stem-loop structure 1 acts as a transcriptional terminator, the 2.4 kb that was initiated at S2 and could be terminated at this structure should be observed in the blots with probes for grpE (Fig 4B) and dnaK (Fig 4C). However, the possible band was not detected in the hybridizations. Therefore, we assumed that the stem-loop structure 1 serves a potential processing site. The 1.2-kb mRNA-d, which was detected only by the hybridization with dnaJ probe, could be a monocistronic mRNA initiated at S3. We have recently reported that the production of DnaJ protein was not completely inhibited by the disruption of the dnaK gene by insertion of the foreign DNA (Hanawa et al 1999). This result could be due to the presence of 1.2-kb transcript.
Analysis of the nucleotide sequence suggested that orf35 and orf29 are part of the dnaK operon, because both are not separated by large noncoding regions and because no apparent promoters are observed. However, transcripts encompassing all 6 genes or demonstrating read through from hrcA to orf29 could not be demonstrated. Even signals suggesting a monocistronic mRNA from each gene were not detected, when the probes specific to orf35 or orf29 genes were used for the hybridization (data not shown). In B. subtilis, the full-length transcript for hrcA-grpE-dnaK-dnaJ-orf35-orf28-orf50 was not detected by Northern blot analysis (Wetzstein et al 1992). Homuth et al (1997) were able to detect the 8-kb transcript extending all 8 genes by Northern blot analysis using riboprobes instead of oligonucleotides for probe. Our failure to demonstrate the RNA species with full length during Northern analysis may be based on its low abundance and/or rapid posttranscriptional processing. Some of the Northern blots shown here exhibit hybridization signals at the 2 positions where the 16S and 23S rRNAs migrate. These bands might occur because of unspecific hybridization or the trapping of degradation products by the rRNAs.
Results from sequence analysis of the dnaK locus revealed the presence of a negative cis-acting CIRCE element (Fig 2) that differed by only 1 bp from the consensus CIRCE sequence (Hecker et al 1996) between the promoter Pl and the initiation codon of hrcA. Furthermore, the transcriptional analysis revealed a significant increase in the amount of the transcript initiated at S1 on heat induction. We found no convincing evidence for an additional CIRCE-like element in the locus. These findings, together with the identification of the gene encoding HrcA homologue (Fig 2), suggest that the L. monocytogenes dnaK operon is regulated, at least in part, in a manner similar to class I heat shock genes in B. subtilis (for review, see Hecker et al 1996). The CIRCE regulon is known to be under negative control of hrcA. Only the promoter P1 becomes active after temperature upshift, probably as a result of inactivation of the HrcA repressor. In addition to the promoter P1, a second internal promoter, P2, was identified between dnaK and dnaJ (Fig 2, Fig 5) and most probably is regulated by the vegetative sigma subunit. The genomic organization of the dnaK operon of B. subtilis (Homuth et al 1997), C. acetobutylicum (Behrens et al 1993), and S. aureus (Ohta et al 1994) is mostly comparable to that of L. monocytogenes. However, in C. acetobutylicum and S. aureus, there is no vegetative promoter located between dnaK and dnaJ.
In conclusion, the dnaK operon of L. monocytogenes consists of 6 genes and at least 4 distinct transcripts are involved in the expression.
REFERENCES
- Ballard SA, Go M, Segers RPAM, Alder B. Molecular analysis of the dnaK locus of Leptospira interrogans serovar Copenhageni. Gene. 1998;216:1–29. doi: 10.1016/s0378-1119(98)00329-1.0378-1119(1998)216<0001:MAOTDL>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Behrens S, Naberhaus F, Bahl H. Cloning, nucleotide sequence and structural analysis of the Clostridium acetobutylicum dnaJ gene. FEMS Microbiol Lett. 1993;114:53–60. doi: 10.1016/0378-1097(93)90141-n.0378-1097(1993)114<0053:CNSASA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Bishop DK, Hinrichs DJ. Adoptive transfer of immunity to Listeria monocytogenes: the influence of in vitro stimulation on lymphocyte subset requirements. J Immunol. 1987;139:2005–2009.0022-1767(1987)139<2005:ATOITL>2.0.CO;2 [PubMed] [Google Scholar]
- Chang ACY, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the p15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978.0021-9193(1978)134<1141:CACOAM>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabri GA, Sanger JM, Portnoy DA, Southwick FS. Listeria monocytogenes moves rapidly through the host-cell cytoplasm by inducing directional actin assembly. Proc Natl Acad Sci USA. 1990;87:6068–6072. doi: 10.1073/pnas.87.16.6068.0027-8424(1990)087<6068:LMMRTT>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton TJ, Sheaman CA, Gasson MJ. Cloning and sequence analysis of the dnaK gene region of Lactococcus lactis subsp. lactis. J Gen Microbiol. 1993;139:3253–3263. doi: 10.1099/00221287-139-12-3253.0022-1287(1993)139<3253:CASAOT>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Gaillard JL, Berche P, Mounier J, Richard S, Sanssonett P. In vitro model of penetration and intracellular growth of Listeria monocytogenes in human enterocyte like line Caco-2. Infect Immunol. 1987;52:2822–2829. doi: 10.1128/iai.55.11.2822-2829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C, Ang D, Liberek K, and Zylicz M 1990 Properties of the Escherichia coli heat shock proteins and their role in bacteriophage lambda growth. In: Stress Proteins in Biology and Medicine, ed Morimoto R, Tissières A, Georgopoulos C. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 191–221. [Google Scholar]
- Graves MC, Rabinowitz JC. In vitro and in vivo transcription of the Clostridium pasteurianium ferredoxin gene. J Biol Chem. 1986;261:11409–11415.0021-9258(1986)261<11409:IVAIVT>2.0.CO;2 [PubMed] [Google Scholar]
- Hanawa T, Fukuda M, Kawakami H, Hirano H, Kamiya S, Yamamoto T. The Listeria monocytogenes DnaK chaperone is required for stress tolerance and efficient phagocytosis with macrophages. Cell Stress Chap. 1999;4:118–128.1355-8145(1999)004<0118:TLMDCI>2.0.CO;2 [PMC free article] [PubMed] [Google Scholar]
- Hanawa T, Yamamoto T, Kamiya S. Listeria monocytogenes can grow in macrophages without the aid of proteins induced by environmental stresses. Infect Immun. 1995;63:4595–4599. doi: 10.1128/iai.63.12.4595-4599.1995.0019-9567(1995)063<4595:LMCGIM>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x.0950-382X(1996)019<0417:HSAGSR>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Homuth G, Masuda S, Mogk A, Kobayashi Y, Schumann W. The dnaK operon of Bacillus subtilis is heptacistronic. J Bacteriol. 1997;179:1153–1164. doi: 10.1128/jb.179.4.1153-1164.1997.0021-9193(1997)179<1153:TDOOBS>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman GC, Penders JE, Burne RA. Transcriptional analysis of the Streptococcus mutans hrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol Microbiol. 1997;25:329–341. doi: 10.1046/j.1365-2958.1997.4671835.x.0950-382X(1997)025<0329:TAOTSM>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Johnson K, Charles I, Dougan G, et al. The role of a stress-response protein in Salmonella typhimurium virulence. Mol Microbiol. 1991;5:401–407. doi: 10.1111/j.1365-2958.1991.tb02122.x.0950-382X(1991)005<0401:TROASR>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Köhler S, Teyssier J, Cloeckaert A, Rouot B, Liautard JP. Participation of the molecular chaperone DnaK in intracellular growth of Brucella suis within U937-derived phagocytes. Mol Microbiol. 1996;20:701–712. doi: 10.1111/j.1365-2958.1996.tb02510.x.0950-382X(1996)020<0701:POTMCD>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Narberhaus F, Giebeler K, Bahl H. Molecular characterization of the dnaK gene region of Clostridium acetobutylicum, including grpE, dnaJ, and a new heat shock gene. J Bacteriol. 1992;174:3290–3299. doi: 10.1128/jb.174.10.3290-3299.1992.0021-9193(1992)174<3290:MCOTDG>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T, Saito K, Kuroda M, Honda K, Hirata H, Hayashi H. Molecular cloning of two new heat shock genes related to the hsp70 genes in Staphylococcus aureus. J Bacteriol. 1994;176:4779–4783. doi: 10.1128/jb.176.15.4779-4783.1994.0021-9193(1994)176<4779:MCOTNH>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips RW, Elzer PH, Roop RM II.. A Brucella melitensis high temperature-requirement A (htrA) deletion mutant demonstrates a stress response defective phenotype in vitro and transient attenuation in the BALB/c mouse model. Microb Pathog. 1995;19:277–284.0882-4010(1995)019<0277:ABMHTR>2.0.CO;2 [PubMed] [Google Scholar]
- Portnoy DA, Jacks PS, Hinrichs DJ. Role of hemolysin for the intracellular growth of Listeria monocytogenes. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459.0022-1007(1988)167<1459:ROHFTI>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquette C, Riplo MT, Pellegri E, Bolla JM, Tascon RI, Vazquez-Boland JA, Berche P. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol Microbiol. 1996;21:977–987. doi: 10.1046/j.1365-2958.1996.641432.x.0950-382X(1996)021<0977:IOACAR>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, and Maniatis T 1989 Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Vanet A, Plumbridge JA, Alix J. Cotranscription of two genes necessary for ribosomal protein L11 methylation (prmA) and pantothenate transport (panF) in Escherichia coli K-12. J Bacteriol. 1993;175:7478–7188. doi: 10.1128/jb.175.22.7178-7188.1993.0021-9193(1993)175<7478:COTGNF>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzstein M, Völker U, Dedio J, et al. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J Bacteriol. 1992;174:3300–3310. doi: 10.1128/jb.174.10.3300-3310.1992.0021-9193(1992)174<3300:CSAMAO>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Hanawa T, Ogata S, Kamiya S. Identification and characterization of the Yersinia enterocolitica gsrA gene, which protectively responds to intracellular stress induced by macrophage phagocytosis and to extracellular environmental stress. Infect Immun. 1996;64:2980–2987. doi: 10.1128/iai.64.8.2980-2987.1996.0019-9567(1996)064<2980:IACOTY>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9.0378-1119(1985)033<0103:IMPCVA>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541.0066-4227(1993)047<0321:ROTHSR>2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- Zuber U, Schumann W. CIRCE: a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994.0021-9193(1994)176<1359:CANHSE>2.0.CO;2 [DOI] [PMC free article] [PubMed] [Google Scholar]