Abstract
V(D)J recombination is the process that generates the diversity among T cell receptors and is one of three mechanisms that contribute to the diversity of antibodies in the vertebrate immune system. The mechanism requires precise cutting of the DNA at segment boundaries followed by rejoining of particular pairs of the resulting termini. The imprecision of aspects of the joining reaction contributes significantly to increasing the variability of the resulting functional genes. Signal sequences target DNA recombination and must participate in a highly ordered protein–DNA complex in order to limit recombination to appropriate partners. Two proteins, RAG1 and RAG2, together form the nuclease that cleaves the DNA at the border of the signal sequences. Additional roles of these proteins in organizing the reaction complex for subsequent steps are explored.
OVERVIEW OF SEQUENCE MODIFICATION PROCESSES
The adaptive immune system of vertebrate animals achieves its protective goals largely through the recognition of target antigens by antigen-binding proteins. These antigen-binding proteins belong to two families: the T cell receptors (TCRs), expressed by and retained on the surface of T cells, and the immunoglobulins (Igs), expressed by B cells and maintained as both cell surface receptors and secreted circulating proteins (antibodies). The overall structures of TCRs and Igs are conserved and are, in both cases, largely composed of repeated domains of the so-called immunoglobulin fold. One special property distinguishes these molecules from any other proteins in vertebrates. In addition to the conserved domains, these molecules also contain loops of peptide near their N-termini that fold into an antigen-binding pocket. The primary sequence of these peptide loops endow each pocket with selective specificity for binding to a particular antigen. In general, a given receptor molecule is very specific in binding to a target. To allow recognition of many antigens, the immune system creates a large population of receptor molecules that differ in the primary sequence of their antigen-binding regions. This population is called the ‘immunologic repertoire’ and is formed by a process of diversification and selection quite different from the way other gene products are produced in our cells.
For most genes two copies per locus is sufficient. Regulation is largely concerned with turning these genes on or off at the right time and in the right cells. Even given the additional complexity introduced by alternative processing pathways, the number of different gene products that are produced from one gene cannot exceed a small number. However, the number of specificities needed by TCR or Ig molecules vastly exceeds this. The total diversity anticipated in a human is estimated at around 1014 for Ig and about a whopping 1018 for TCR molecules [see Janeway et al (1), figure 4.34]. Clearly, with only approximately 105 genes in our entire genome, this kind of diversity cannot be achieved simply by inheriting genes preformed for each specificity. Furthermore, inheriting preformed genes suffers from the additional difficulty of a lack of plasticity. For example, the entire species would be continually at risk from variant viruses that were not recognized by an inherited repertoire. In actuality, when new viral variants do arise, we usually respond to them with new antigen receptors.
Both of these challenges are met by clever DNA sequence modification strategies that allow the immune system to generate a large, expressed repertoire from a much more limited, inherited germline set of sequences. T and B cells each use a site-specific DNA recombination mechanism called V(D)J recombination to cut and rejoin segments of DNA that, when assembled, encode the N-terminal variable portion of the TCR or Ig molecules. This is the portion of the antigen receptors that binds antigen. Elaborating upon the mechanism that coordinates this recombination will occupy most of this review. In addition, two other mechanisms, unique to B cells, further diversify the Ig gene products. One is called ‘Class’ or ‘Isotype’ switching, which is an independent (region targeted) recombination pathway that, upon induction, replaces one gene segment encoding a constant region for another. This allows a B cell (or its descendants) to utilize the same antigen-binding specificity in the context of different constant regions with important immunologic consequences. The mechanism of this reaction is yet to be fully determined, but two recent reviews describe intriguing links between this pathway and mismatch repair, non-homologous end joining (NHEJ) and mRNA processing (2,3). The third diversity-generating pathway present in B cells is termed ‘Somatic Hypermutation’ in which seemingly random single base changes are directed at high frequency into the gene segments encoding the antigen-binding pocket. The consequence is a proliferation of sequence changes that alter the specificity or affinity of an Ig already selected by antigen. If the resulting mutated version of the Ig displays a greater affinity for antigen, the new version of the gene is preferentially selected. Again, the mechanism is still uncertain but candidate error-prone or mutagenic reactions have been suggested (4–6), perhaps also associated with double-strand DNA breaks (7,8).
V(D)J RECOMBINATION TARGETS
Much of the diversity in both TCR and Ig proteins is generated by the combinatorial recombination at the DNA level of inherited gene segments. Both TCR and Ig function as heterodimers, and each polypeptide is comprised of a variable region [assembled by V(D)J recombination] and a constant region (joined to the variable region by conventional mRNA splicing). The variable portion of one polypeptide in each heterodimer (Ig heavy chain and TCR β or δ chains) is assembled from three elements named ‘V’, ‘D’ and ‘J’ segments. The partner polypeptide (one of the Ig light chains κ or λ, or TCR α or γ) is assembled from V and J segments only. In mice and humans these segments are usually inherited as tandem clusters of related elements (Fig. 1A). The organization has evolved and is quite different among phylogenetically older vertebrates such as sharks. A detailed discussion of the origin of this recombination system and the evolution of the chromosomal organization of the various rearranging loci has recently appeared (9 and references therein, 10).
Figure 1.
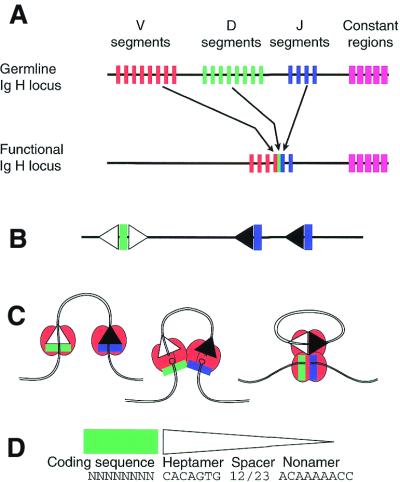
1 The V(D)J recombination reaction scheme. (A) The colored bars represent the tandemly-arranged clusters of coding DNA segments at the Ig heavy chain locus of the mouse. There are actually several hundred V segments, many of which are pseudogenes. One of each of the V, D and J elements are joined to form the functional coding region that encodes the variable portion of the molecule, as shown. Junctional diversity is introduced at each event (see text). The constant regions do not participate in V(D)J joining. (B) Each of the coding segments is associated with an RSS, which specifies the recombination cleavage site. White triangles represent 12-RSS, black triangles represent 23-RSS. Since D elements participate in both upstream and downstream recombination events, they require two RSSs. (C) The conventional reaction course involves RAG proteins (red, stoichiometry not implied) associating at the RSS. Cooperative cleavage generates blunt signal ends on the RSS and hairpinned intermediates on the coding ends. Coding ends are processed and the four ends are then rejoined, signal-containing ends to each other, and similarly for coding ends (see text). (D) The sequence of the coding end is not highly constrained, although some sequences at the cleavage site work better than others. The consensus RSS (triangle) is composed of a conserved heptamer, a spacer of either 12 or 23 nt in length and a conserved nonamer. Some variation in each of these motifs can be tolerated.
A brief discussion of the reaction steps in V(D)J recombination is necessary to introduce the key concepts, but these have been reviewed extensively. The reader is referred elsewhere for a comprehensive introduction to the process from the perspective of the substrate loci and the recombinant products (11,12). The key terminology, elaborated in Figure 1, is that the various ‘coding’ segments (drawn as colored vertical bars) are physically cut and joined together. The cuts occur in two steps, first by nicking a particular DNA strand adjacent to the heptamer, followed by cleavage of the second strand to yield a terminal hairpin on the coding DNA. The ligation of two coding segments forms a ‘coding joint’, which is deliberately imprecise. Small deletions, random insertions and occasional small palindromic additions are common at these junctions and arise through the processing of the intermediate hairpin end shown in Figure 1C. The junctional diversity greatly increases the yield of different receptor molecules, which otherwise would be limited to the multiplicative product of the number of segments. The DNA ligation step is dependent on proteins already known to participate in the repair of other DNA double-strand breaks, the so-called NHEJ pathway (reviewed in 13–19).
The cleavage is directed to a precise location by the presence of recombination signal sequences (RSSs). These are represented by triangles in Figure 1. The RSS is composed of conserved heptamer and nonamer elements separated by an intervening spacer of fixed length. The consensus sequence is shown in Figure 1D, but variations are frequent, and deviation from an optimal sequence may modulate the efficiency with which particular sites are used (20), perhaps through the intervention of additional protein factors (21). The RSS elements have evolved over time (22). There are two classes of RSS, which are distinguished by the length of their spacer regions. Spacer length centers on 12 or 23 nt and the resulting signals are referred to as 12-RSS or 23-RSS (Fig. 1, white and black triangles, respectively). The existence of the two classes helps solve a potential pitfall in the recombination system. While it is desirable to allow recombination between, for example, any one V segment and any one D segment, it would serve no useful purpose to recombine two V segments with each other. This is avoided by organizing the RSS elements such that only one length class is used within a cluster of segments. In the case of the mouse Ig heavy chain locus, each of the V regions is associated with 23-RSS elements. The D regions in this locus are flanked on both sides by 12-RSS elements, thus allowing recombination to occur both upstream and downstream. Finally, the J segments of this locus utilize purely 23-RSS signals (Fig. 1B). The mechanism of recombination incorporates a ‘12/23 rule’ (23), which specifies that recombination be permitted only between segments of complementary RSS length. This allows V to D and D to J recombination but neatly prevents the undesired occurrences. D segments contribute only short lengths of coding information to the assembled polypeptide, typically from two to nine amino acids. True devotees will appreciate the totally different configuration of RSS elements at the mouse TCR δ locus. The D elements there, in contrast, are flanked by one 12-RSS and one 23-RSS, permitting D to D recombination and resulting in both VDJ and VDDJ products. Both result in functional molecules [see Lewis (11), figure 1]. The molecular implementation of the 12/23 rule is not fully determined, but will be addressed later.
While the joining of coding sequences, as portrayed in Figure 1C, creates the assembled receptor gene, it is important for several reasons that the DNA cleaved from the coding sequence, which carries the RSS elements, also be rejoined. This DNA is shown as the circular product in Figure 1C. The cleavage reaction leaves blunt double-strand breaks at the border of the heptamer (24,25), and these are typically joined to each other in precise heptamer to heptamer juxtaposition. These so-called ‘signal joints’ also use the NHEJ pathway, but are distinguishable from coding joints. Signal joints only rarely lose or gain bases and do not require the protein DNA-PKcs (26,27) for their formation. This probably reflects the additional processing demanded by the coding ends. Forming signal joints serves two apparent purposes. First, it prevents the undesirable integration of these DNA ends into other locations as studied in vitro (28–34), and indirectly, in vivo (35,36). The second need for signal joint formation reflects other topologies of the reaction. In the typical rendering of the reaction, as shown in Figure 1C, the opposing orientation of the RSS elements creates a deletion circle. This is the most common configuration in natural loci. However, functional recombination does occur among gene segments in which the participating RSS elements are in direct orientation. Under this circumstance, rearrangement demands inversion of the DNA between the RSS elements rather than deletion, and signal joints are required for chromosomal integrity.
THE RECOMBINASE, RAG1 AND RAG2
The molecular biology of V(D)J recombination experienced a revolution with the isolation of the two genes RAG1 and RAG2 (37,38). The name is derived from the acronym of ‘recombination activating genes’ and they remain the only identified tissue-specific genes required for V(D)J recombination. Extracts containing the encoded proteins, and later, purified versions of these two proteins, are able to cut DNA containing RSSs (39,40). Truncated versions of both RAG proteins are sufficient to catalyze the complete reaction yet proved to be more tractable for expression and purification (41–44).
Biochemical analysis of the recombination pathway is subject to the same fate as the proverbial blind investigators describing an elephant. Each assay only recognizes part of the total. Table 1 lists the many functions to which the recombinase should contribute. Not every step need be an explicit function of the RAG proteins. The table is divided into two categories, reactions operating on one DNA substrate and therefore only requiring one set of interactions, and reactions that demand intelligence of the other DNA and therefore require a more elaborate complex. These steps will be addressed in the discussion that follows.
Table 1. Activities associated with the RAG proteins.
| Steps possible on a low order
complex |
Steps requiring a higher order complex |
| Individual RSS binding | Enforcement of the 12/23 rule |
| Nicking | Coordinated cleavage at two RSSs |
| Hairpin formation | Potential isomerization of the four-end complex, to direct pairing of ends for joining |
| Retention of coding DNA after cleavage | Potential coordination of coding end joining‚ with signal end joining |
| Hairpin opening/processing | Prevention of translocation |
| Recruitment of repair proteins | |
| Obscuring of cut ends from damage sensors |
RAG1 primary structure
The mouse RAG1 protein, as a reference sequence, contains 1040 residues (119 kDa). A previous sequence alignment of five species (41) revealed a central core (residues 384–1008) with a higher degree of conservation than the surrounding sequence. In Table 2, the amino acid alignment has been updated to include more species over the region numbering 354–411 of the mouse sequence. This segment spans the N-terminal border of the RAG1 core to show how striking the evolutionary conservation is between sequences within and outside the core. In this PSI-BLAST (45) alignment, certain residues show absolute conservation and are listed on the top line. For example, the left half of Table 2 shows complete preservation of two cysteine and two histidine residues that form part of a zinc-finger motif (46,47). This motif is believed to participate in RAG1 dimerization, but it does not appear essential for this function since the RAG1 core region dimerizes on its own (48,49). Except for the direct metal-binding amino acids, the N-terminal region shows a tolerance for sequence variation, including insertions, when compared to the mouse sequence. The conservation of sequence becomes almost absolute upon entering the core region and this property extends to the C-terminal end of the core, mouse residue 1008, with only a few islands of variation. The core region of RAG1 is sufficient (along with RAG2) to perform all the known enzymatic functions, and to catalyze the complete recombination of test substrates in cells. The core region alone also localizes to the nucleus and has a short half-life, as does its full-length RAG1 (41). However, the N-terminal third of the protein clearly has functional significance. Restoring parts of this region to the core has increased the efficiency of the complete recombination reaction using test substrates (50–52).
Table 2. Alignment of mouse RAG1 spanning the beginning of the core regiona.
aOnly differences from the mouse sequence are shown. Dashes represent gaps introduced where insertions are present in some sequences. The mouse core region is shown in blue.
bThe first line shows positions absolutely conserved in all species.
cGI# is the National Library of Medicine (USA) GenePep sequence identifier.
dCommon names for some species and bibliographic references.
Naturally arising human RAG1 or RAG2 mutations fall into two classes. Null mutants of either protein result in complete immunodeficiency, but other mutations, which seem to retain low levels of activity, lead to the immunodeficiency called Omenn syndrome (53). The recent finding that some patients demonstrate complete failure of Ig rearrangements but retain some level of TCR rearrangements (54–57) suggests that there may be additional regulatory functions to the RAG proteins that are not identical in the different cell environments of B and T cells, despite the fundamental enzymatic similarity of the recombination reactions.
A basic sequence beginning 389-GGRPR can be found near the start of the core region (Table 2). Deletion of the region from residues 389–486 has been shown to interfere with DNA binding. Thus, this region is commonly termed the NBD (for nonamer-binding domain) although only indirect evidence strictly links these amino acids with that DNA contact (34,58,59). One might anticipate several other parts of the molecule also contribute to DNA binding. Site-directed mutagenesis has identified three conserved acidic amino acids, D600, D708 (60–62) and E962 (60), that are each essential for the nuclease activity. This triad and some evolutionary considerations support the proposal that the RAG proteins share a mechanism with bacterial transposases and HIV integrase (34 and references therein). One would expect that if these amino acids form the catalytic active site, they also must achieve close contact to the DNA. In support of these additional DNA interactions, mutations near the D600 site (substitutions and a deletion in residues 606–611) affect the target specificity of recombination (63–65). Direct contact between RAG1 and DNA has also now been assigned to an N- and C-terminal region of the core protein (66), which will be further discussed below.
RAG2 primary structure
The mouse RAG2 protein is 527 residues in length and, like the RAG1 protein, possesses a core region (1–382) that is sufficient to evoke the complete recombination of test substrates in cells (43,44). However, in contrast to RAG1, the core does not display the same coincidence of sequence conservation with enzymatic function. The C-terminal quarter of the molecule includes a striking acidic region and, as is the case for the N-terminal region of RAG1, may contribute a regulatory role to the reaction. It is reported that a certain stage in B cell rearrangement, the joining of heavy chain V region to the already assembled DJ segment, requires the C-terminal region of RAG2 (67). The authors of this last study suggest a role of RAG2 in governing access to particular loci. Mutations in RAG2 can also lead to Omenn immunodeficiency syndrome and the location of these mutations has been interpreted to support the prediction of a structural model for the RAG2 core (53,68–70).
Single RSS interactions
The direct demonstration that the two RAG proteins together were capable of cleaving a DNA target at the border of the heptamer (40) (Fig. 1D) settled for many the question of whether double-strand breaks in the DNA were a reaction intermediate. Since then, there has been progress in analyzing the interactions between the proteins and substrate. It is now clear that the two proteins cooperate in binding, and both are required for each step of cleavage. RAG1 protein alone exhibits DNA binding with some intrinsic affinity for the nonamer versus random DNA (58,71–73). There is a difference in reports of whether RAG2 alone binds DNA. Mo et al. (73) detected DNA binding by RAG2 alone under different binding conditions and with reduced concentration of non-specific competitor than used by other investigators. All agree that together both proteins exhibit a higher affinity, specificity and stability in assembling a complex on DNA containing an RSS. It is worth noting, however, that the specificity of these two proteins binding to the target over competitor appears to be ∼10–20-fold. This would not be sufficient to prevent frequent recombination activity at undesired locations, such as sequences resembling an RSS in unrelated genes (74).
Studies using oligonucleotide cleavage assays and either mutating the RSS or modifying the DNA structure with single-stranded regions (75,76) indicate that, in addition to base-specific sequence recognition, there is a structural component that favors unpairing of the DNA at the border of the coding region and the heptamer. This is where the hairpin DNA structure forms during cleavage. Distortion of the helical geometry of the DNA at this position in a complex containing both proteins was also detected by footprinting assays (72,77). This distortion might be accompanied by bends in the helical axis, and, in fact, such bends were detected (78,79). Additional footprinting and crosslinking studies provide further details of the interactions of each protein with the DNA. RAG1 was found to make direct contacts at both the nonamer and the heptamer (the latter only in the presence of RAG2) using crosslinking strategies (49,73,80). Footprinting RSS substrates and modification/protection strategies definitely indicate contacts by the proteins at the nonamer and heptamer, and suggest protection over the spacer of the 12-RSS. This last point is subtly contrasted by our own work using crosslinking assays (79), which indicated a much reduced interaction between the RAG proteins in the spacer region. We also detected enhanced binding of the HMG1 protein (which bends DNA and binds more stably at sites that are already distorted) at the 3′ side of the heptamer in the 12-RSS and within the spacer of the 23-RSS. We suggest that the DNA is severely bent between the heptamer and nonamer with the spacer regions looping away from the RAG proteins. This allows similar protein–DNA interactions to occur at the heptamer and nonamer on both types of RSS, with the difference in sequence length accommodated by the size of the loop (Fig. 3). This view is consistent with the previous observation that the length of the spacer appears to demand an integral number of full helical rotations but can tolerate an extra full rotation to 34 bp (76,81).
Figure 3.
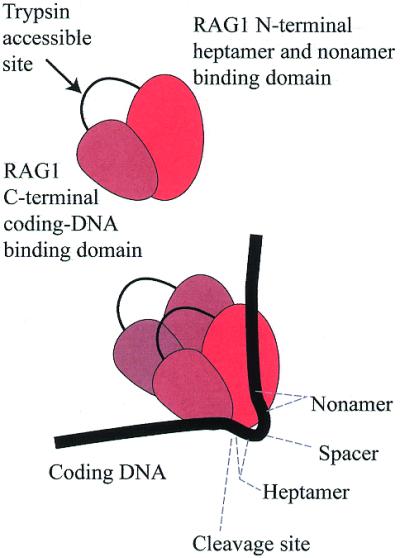
The RAG1 protein and its association to DNA. Top, the RAG1 core region has a trypsin-accessible site, which may divide it into two functional domains. Bottom, the RAG1 protein functions as a dimer, in association with RAG2 (not shown). The N-terminal region of RAG1 associates with the RSS conserved motifs. The C-terminal region associates with the coding DNA. A loop of DNA can be imagined (labeled spacer) that differs in length between the two RSS classes. The cis–trans relationships of protein and DNA are still not well-defined (see text).
Using crosslinking strategies, RAG2 was detected at several nucleotides that also contacted RAG1 (79,80). This supports the idea that RAG2 does make independent DNA contacts, although there still exists the formal possibility that RAG2 is positioned on the DNA purely by tethering through RAG1.
As mentioned above, the protein HMG1 was used in certain binding studies. HMG proteins are abundant and ubiquitous nuclear proteins found throughout eukaryotic evolution (82), and which play a structural role in chromatin. The closely-related proteins HMG1 and HMG2 were each found to stimulate the cleavage reaction with various substrates (83–85). While its effect is more pronounced on 23-RSS substrates, the presence of HMG1 stabilizes the binding and cleavage of both classes of RSS (78,79). One laboratory (78) showed a direct interaction between the HMG proteins (1 or 2) and RAG1, with a relatively modest dissociation constant of 10–5 M. Our laboratory also noted this interaction in pull-down assays (unpublished results).
Coding DNA interactions
Joining is not coupled directly to the cleavage reaction, but rather follows additional processing of the coding ends. In theory, signal ends could be joined instantly to each other, but this also does not necessarily occur (86). Once the coding DNA is detached from the adjacent heptamer, there is no longer any feature that identifies this sequence. In model recombination reactions, the coding DNA can be completely replaced by an experimentally-selected sequence (11). There are consequences to some choices, and these will be discussed further below. Rampant chromosomal breakage could occur if the broken DNA ends were to escape from each other, so the retention of coding ends in a RAG protein complex after cleavage satisfies a conceptual need. The first evidence of an interaction between RAG1 and the coding DNA was the indirect observation that mutants of RAG1 altered the efficiency of recombination as a function of the coding sequence (63,64). Subsequent crosslinking studies directly detected contacts to both RAG1 and, more weakly, to RAG2 using an iodinated base analog at the –1 position (relative to the cut site) (80), or at the –2 position, but not the –4 position, using an azido-bearing adduct attached to the phosphate backbone (79). Consistent data were also obtained independently by crosslinking and immunoprecipitation studies (49). This contact is sufficient to prevent dissociation of the coding end. A gel-shift of the complex following cleavage in vitro still retains the cleaved signal and coding DNA (48). Subsequent joining of coding ends in a cell-free assay fails if the DNA is deproteinized following cleavage (87).
Since RAG proteins constitute an endonuclease capable of nicking DNA, and since the protein is already associated with the hairpinned coding end, it is enticing to picture this activity being used a second time to open the hairpin. The purified proteins are capable of this activity (88,89) and can also act as endonucleases on 3′ flap structures (90). This does not rule out the possibility that other DNA repair pathways, such as that involving MRE-11, could also perform this function in vivo (91,92), perhaps as alternative pathways. An important issue in this regard is the desire for the processing of the coding end to create a variety of products including deletion, palindromic insertion and random insertion through cooperation with terminal deoxynucleotidyl transferase.
The sequence of the coding flank DNA, while not engaging in any base-specific interactions, may influence the efficiency and outcome of the recombination reaction in several ways. This effect was first detected in comparison to the efficiency of recombination of plasmid substrates (93). As has been addressed above, distortion leading to single-stranded bubbles at the coding end/heptamer border may precede the nicking step. Certain coding sequences, originally identified as ‘good flanks’ with respect to recombination by mutants of RAG1 (63,64), are likely to promote this process, and these were found to be better substrates for the cleavage reaction at single sites in vitro (75,76). This issue was revisited very recently (65), to show that substrates containing unpaired coding flanks rescue hairpin formation using the mutant RAG1 proteins. These particular effects on hairpin formation may not be as pronounced when measured by concerted cleavage (81), and careful work seems to distinguish effects at nicking from the subsequent second cleavage step associated with hairpin formation (94,95). This will be discussed further.
The coding end sequence seems to have additional influence at later processing steps. This could affect hairpin opening, exonucleolytic polishing of the opened hairpin and pairing of the coding ends (which may be sensitive to microhomologies) (96–98). Therefore, the designation of coding ends as good or bad may vary considerably based on the assay employed. As mentioned previously, the joining of coding ends is at some stage dependent upon the activity of DNA-PKcs. An interesting connection between signal joint formation and coding end processing was observed in pre B cells and fibroblasts (99) carrying the murine scid mutation, which results in a C-terminal truncation of that kinase (100,101). With certain homopolymeric coding sequences adjacent to both RSSs, scid cells showed a reduction in the abundance of signal joints, arguing that the two processes are linked. Conclusions about the joining reactions depend, of course, on the cleavage efficiency remaining constant across the experiment.
Theorists would support the existence of a four-end complex that retains signal ends as well as coding ends based on the observation that these ends occasionally pair indiscriminately, in violation of the usual rules (11,102,103). This implies that all four ends remain in physical proximity for some time after cleavage.
Coordinated cleavage at two RSSs
The reader has been spared certain details about reaction conditions. The previous in vitro cleavage reactions, performed on single RSS substrates, proceed to double-strand breaks in reactions using Mn+2 as divalent cation (39). This ion seems to relax a constraint in the nuclease that exists when cleavage is undertaken in the presence of the more physiologic ion Mg+2 (104). The constraint is not absolute in activity on single RSS substrates. Nevertheless, when both 12- and 23-RSSs are present, double cleavage coordinated at both sites is obtained (105,106) in Mg+2, especially when supplemented by either HMG1 or HMG2. A detailed review of the role of metal ions in nucleic acid chemistry is available (107), and this topic has been also addressed specifically in V(D)J recombination (34). The demonstration that cleavage at two sites can be concerted under any condition profoundly alters our understanding of the reaction pathway. It means that a protein–DNA complex must assemble at the RSS elements, but the complex must wait for permission to cleave until a partner that satisfies the 12/23 rule is identified. Formation of this complex may be a rate-limiting step (108). Bacterial transposition (109–111), retroviral integration (112) and bacteriophage λ (which assembles distinct complexes under different conditions; 113) also exhibit analogous behaviors, though not always requiring distinguishable ends. In principle, this coordination of cleavage is a very good thing. It prevents chromosomal breakage until the partner for rejoining is already identified, thereby minimizing the duration of broken intermediates. It also provides an increase in specificity since isolated RSS-like sequences may be less likely to result in chromosome breaks. The initial observation that optimal concerted cleavage required both classes of RSS (105,106) showed a few-fold preference over cleavage with only one RSS available. Reality seems to be more complicated, providing uncharted territory for future investigation. Several investigators find that complete cleavage can be detected using substrates that only provide one RSS (81,104,114). The first cleavage step, that of nicking one strand 5′ to the heptamer, may in fact occur in the absence of any requirement for interaction with a second RSS. One recent study demonstrated this by inhibiting potential like-substrate pairing (12/12 or 23/23) by immobilizing the oligonucleotides. Kinetic analysis of cleavage of these substrates was consistent with this interpretation (95). It appears that it is actually the second step in the cleavage reaction, the formation of the hairpin, which is most sensitive to correct pairing (115). This study contends that in a strict sense, pairing can occur equally well among the various combinations of RSS substrates, but that only the 12/23 pairing promotes the reaction. One caveat might be a concern that the short tethered substrates used may impose unnatural constraints on the mechanism, though substrates with the same spacing seem to function in cells (116).
Protein complexes including RAG2
The first evidence that RAG2 participated in some manner in assisting or stabilizing RAG1-binding to DNA was acquired through a transcriptional reporter assay (58). Subsequent gel mobility shift experiments showed that one or more protein–DNA complexes that were formed were dependent on the presence of RAG2 in solution (71,72,117). Indirect assays using resistance to nucleases (118) or accessibility to terminal deoxynucleotidyl transferase (119) suggested similar conclusions. We excised the gel mobility shift bands that formed on 12- or 23-RSS-containing oligonucleotides, and showed that each contained RAG1 and RAG2 in stoichiometric equivalence (73). A study using the additive contribution of multiple copies of the maltose-binding domain to distinguish the mobility of each subunit (49) found RAG1 plus RAG2-containing complexes in both a 2:1 and 2:2 ratio. Bailin et al. (48) showed that RAG1, in solution and in the absence of DNA, existed as a protein dimer, but when isolated in the presence of RAG2, existed as a 2:2 tetramer. Bound to DNA, RAG1 in the absence of RAG2 formed an array of an increasing number of dimers, but in the presence of RAG2, formed two discrete species. In our hands, these represented the RAG1 dimer (only) and the tetramer comprised of RAG1 dimer plus two RAG2 molecules. Ferguson plot analysis suggests that this tetramer of proteins assembles on one DNA target (48). Gel filtration analysis of RAG2 showed monomer and higher forms in solution, but we are not convinced that the higher forms represent meaningful interactions. A current consistent picture is a tight RAG1 dimer with each partner associating independently with RAG2. RAG2 also either caps a surface on RAG1 or changes RAG1 conformation in a manner that prevents multimerization of the RAG1 dimer along one DNA molecule.
The peptide sequences contributing to the RAG1/RAG2 interface have been addressed in deletion and mutation studies (120,121) and are not entirely in agreement. It is, however, impossible at this time to exclude the possibility that deletions perturb the global protein architecture in a manner that would interfere with a distant protein interaction. The later study identifies one residue in RAG2 which, when mutated, interferes with complex formation with RAG1 as measured by coimmunoprecipitation and gel shift assays.
RAG protein complex architecture
The tetramer of RAG1 and RAG2 can be viewed as the fundamental active enzyme unit in the recombination reaction. As suggested by Bailin et al. (48), one or two of these tetramers may participate in binding to the two RSSs. Very recent work (122) provides some additional insight by using mixed dimers of RAG1 in which one partner is defective in DNA binding (through replacement of 10 consecutive residues in the NBD with alanines) and the other is defective in nuclease activity (by single alanine substitution of individual critical acidic residues). The mixtures of mutations show that within the mixed RAG1 plus RAG2 complex, one RAG1 does the binding while the other provides catalysis for cleavage. New in this study is the evidence that all three acidic residues are provided from the same polypeptide, reducing the dimensions of cis–trans relationships within the dimers.
Figure 1C is drawn with the implicit assumption that a synaptic structure is constructed in two steps: first by assembling parallel complexes on the two RSSs, and second by docking these together to make a larger protein–DNA complex. If each RSS interacts with a RAG1 dimer (previously referred to as a single tetramer of RAG1 and RAG2) then the synaptic complex would represent the dimer of dimers of RAG1, plus the associated RAG2 proteins (Fig. 2A). That model would be driven forward by raising the concentration of single-RSS protein-bound intermediates. This is not the only possibility. If a complex assembling on one RSS only occupies one-half of the binding sites within the dimer of RAG1, then a second DNA interaction could occur on that same complex. This would, in essence, require the first protein–DNA complex to sample additional DNA sequences and recruit the second target sequentially into the complex (Fig. 2B). Under the simplest of binding interactions, this process might be expected to be inhibited by high protein concentrations since the free sites would be saturated with RAG complexes that might interfere with successful scanning. This pitfall might be averted by positing a mechanism to hand off the DNA from one complex to another (Fig. 2C). Distinguishing between one dimer of RAG1 or two in the synaptic complex is currently under investigation.
Figure 2.
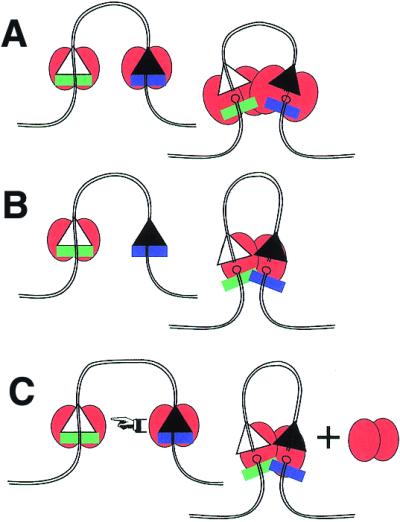
Three paths to the synaptic complex. (A) Docking model. A separate complex, containing a dimer of RAG1 plus associated RAG2 (not pictured), first assembles on each RSS. These then dock with each other, and if the 12/23 rule is satisfied, allow concerted cleavage of both DNA targets. (B) Sequential binding model. The complex of RAG1 and RAG2 binds one DNA first, then must find a second DNA target, which gets incorporated into the same protein complex. The resulting complex would only contain one dimer of RAG1. (C) Cooperative transfer. Complexes first assemble on separate RSSs, but these cooperate to transfer one DNA into the protein complex of the second, resulting in one dimer of RAG1 associated with DNA plus eviction of the second RAG1 dimer. In each case, there would exist intermediates (not shown) prior to the final cleavages.
The suggestion has already been introduced that a protein complex contacts the RSS at the nonamer and heptamer and, in addition, holds the coding DNA through predominantly sequence-independent contacts. An attractive proposal is that the complex undergoes an isomerization following cleavage, which reorients the DNA ends for subsequent joining and may bias which members of the four-end complex rejoin. This isomerization, when executed correctly, would also retain the broken chromosome pieces in a single complex. One difficulty with this model would arise if all the DNA contacts occurred within a single, rigid, DNA-binding region of the protein. Reorientation of DNA ends under this circumstance would require physical dissociation of the DNA from the protein, with increased risk of escape from the complex altogether. Our recent work (66) relieves one of these difficulties. We find a trypsin-accessible site in the middle of the RAG1 core that may divide it into two separate protein domains. Using the chemical crosslinking approach, we find that the coding DNA is associated with the C-terminal region, while heptamer and nonamer make contact with the N-terminal region (Fig. 3). DNA binding would therefore be divided between two parts of RAG1, and allow reconfiguration by conformational changes without dissociation.
RAG protein contributions to the DNA joining reaction
A minimal model of the joining reaction might suggest that once cleavage had occurred, rejoining of the ends becomes the full responsibility of the NHEJ pathway. Alternatively, the RAG proteins may remain associated with the DNA ends and play an active role in recruiting the repair proteins and directing the fidelity of the reaction. Three recent reports show that the minimal model is not sufficient (123–125). The first two reports, using different systems, appear to reach opposing conclusions regarding the effect of topology (cis versus trans) of the coding DNA at the time of joining. Additional support for a role of the RAG proteins in the latter steps of the reaction is obtained by the recent report that mutant versions of either RAG1 (126) or RAG2 (127) permit the initial cleavage of DNA including hairpin formation at the coding end, but fail at the stage of hairpin opening. If the same nuclease active site is used for each of these steps, as preferred, then these observations may be tied together by requiring a conformational change to expose the hairpin to the nuclease. Mutations that interfere with the conformational change would be consistent with the phenotype.
Broken chromosomes commonly induce apoptosis through p53 signaling. Clearly it benefits our cells to abstain from this response during V(D)J recombination. Nevertheless, interfering with the normal pathway does provoke apoptosis (128). One possibility is that a well-formed complex following cleavage sequesters the ends and prevents activation of any damage sensors in the cell. This simple model is not strictly true, as evidenced by a recent report that can serve as a gateway into the world of how DNA breaks are recognized by the cell (129).
CONCLUSION
The DNA recombination process that assembles V, D and J segments encoding the antigen receptors of our immune system requires a series of well-coordinated steps. At the heart of the reaction is the cleavage of the DNA at the appropriate sequences, but other steps are also necessary to generate junctional diversity and to assure the coordinated joining of the intermediates. These steps are performed by the two RAG proteins in association with other factors, some of which have yet to be identified. Overall, the reaction requires an elaborate machine with precise protein–DNA contacts and protein–protein interactions to handle the intermediates, without allowing them to diffuse away from each other.
Acknowledgments
ACKNOWLEDGEMENTS
This review benefited from discussion with others and especially the reviewers comments, but the author apologizes for any omissions. M.J.S. is a scholar of the Leukemia and Lymphoma Society and is supported by NIH grant AI41711.
References
- 1.Janeway C.A., Travers,P., Walport,M. and Capra,D.J. (1999) Immunobiology, 4th edn. Garland Publishing, New York, NY.
- 2.Kenter A.L. (1999) The liaison of isotype class switch and mismatch repair: an illegitimate affair. J. Exp. Med., 190, 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kinoshita K. and Honjo,T. (2000) Unique and unprecedented recombination mechanisms in class switching. Curr. Opin. Immunol., 12, 195–198. [DOI] [PubMed] [Google Scholar]
- 4.Harris R.S., Kong,Q. and Maizels,N. (1999) Somatic hypermutation and the three R’s: repair, replication and recombination. Mutat. Res., 436, 157–178. [DOI] [PubMed] [Google Scholar]
- 5.Poltoratsky V., Goodman,M.F. and Scharff,M.D. (2000) Error-prone candidates vie for somatic mutation. J. Exp. Med., 192, F27–F30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman M.F. and Tippin,B. (2000) Sloppier copier DNA polymerases involved in genome repair. Curr. Opin. Genet. Dev., 10, 162–168. [DOI] [PubMed] [Google Scholar]
- 7.Papavasiliou F.N. and Schatz,D.G. (2000) Cell-cycle-regulated DNA double-stranded breaks in somatic hypermutation of immunoglobulin genes. Nature, 408, 216–221. [DOI] [PubMed] [Google Scholar]
- 8.Bross L., Fukita,Y., McBlane,F., Demolliere,C., Rajewsky,K. and Jacobs,H. (2000) DNA double-strand breaks in immunoglobulin genes undergoing somatic hypermutation. Immunity, 13, 589–597. [DOI] [PubMed] [Google Scholar]
- 9.Hansen J.D. and McBlane,J.F. (2000) Recombination-activating genes, transposition and the lymphoid-specific combinatorial immune system: a common evolutionary connection. Curr. Top. Microbiol. Immunol., 248, 111–135. [DOI] [PubMed] [Google Scholar]
- 10.Lewis S.M. and Wu,G.E. (2000) The old and the restless. J. Exp. Med., 191, 1631–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewis S.M. (1994) The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv. Immunol., 56, 27–150. [DOI] [PubMed] [Google Scholar]
- 12.Honjo T. and Alt,F.W. (1995) Immunoglobulin Genes, 2nd edn. Academic Press, London.
- 13.Bogue M. and Roth,D.B. (1996) Mechanism of V(D)J recombination. Curr. Opin. Immunol., 8, 175–180. [DOI] [PubMed] [Google Scholar]
- 14.Gellert M. (1997) Recent advances in understanding V(D)J recombination. Adv. Immunol., 64, 39–64. [DOI] [PubMed] [Google Scholar]
- 15.Grawunder U., West,R.B. and Lieber,M.R. (1998) Antigen receptor gene rearrangement. Curr. Opin. Immunol., 10, 172–180. [DOI] [PubMed] [Google Scholar]
- 16.Oettinger M.A. (1999) V(D)J recombination: on the cutting edge. Curr. Opin. Cell Biol., 11, 325–329. [DOI] [PubMed] [Google Scholar]
- 17.Smider V. and Chu,G. (1997) The end-joining reaction in V(D)J recombination. Semin. Immunol., 9, 189–197. [DOI] [PubMed] [Google Scholar]
- 18.Lieber M.R. (1999) The biochemistry and biological significance of nonhomologous DNA end joining: an essential repair process in multicellular eukaryotes. Genes Cells, 4, 77–85. [DOI] [PubMed] [Google Scholar]
- 19.Karran P. (2000) DNA double strand break repair in mammalian cells. Curr. Opin. Genet. Dev., 10, 144–150. [DOI] [PubMed] [Google Scholar]
- 20.Livak F., Burtrum,D.B., Rowen,L., Schatz,D.G. and Petrie,H.T. (2000) Genetic modulation of T cell receptor gene segment usage during somatic recombination. J. Exp. Med., 192, 1191–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bassing C.H., Alt,F.W., Hughes,M.M., D’Auteuil,M., Wehrly,T.D., Woodman,B.B., Gartner,F., White,J.M., Davidson,L. and Sleckman,B.P. (2000) Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature, 405, 583–586. [DOI] [PubMed] [Google Scholar]
- 22.Hassanin A., Golub,R., Lewis,S.M. and Wu,G.E. (2000) Evolution of the recombination signal sequences in the Ig heavy-chain variable region locus of mammals. Proc. Natl Acad. Sci. USA, 97, 11415–11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tonegawa S. (1983) Somatic generation of antibody diversity. Nature, 302, 575–581. [DOI] [PubMed] [Google Scholar]
- 24.Roth D.B., Zhu,C. and Gellert,M. (1993) Characterization of broken DNA molecules associated with V(D)J recombination. Proc. Natl Acad. Sci. USA, 90, 10788–10792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlissel M., Constantinescu,A., Morrow,T., Baxter,M. and Peng,A. (1993) Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent and cell cycle regulated. Genes Dev., 7, 2520–2532. [DOI] [PubMed] [Google Scholar]
- 26.Gao Y., Chaudhuri,J., Zhu,C., Davidson,L., Weaver,D.T. and Alt,F.W. (1998) A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity, 9, 367–376. [DOI] [PubMed] [Google Scholar]
- 27.Taccioli G.E., Amatucci,A.G., Beamish,H J., Gell,D., Xiang,X.H., Torres Arzayus,M.I., Priestley,A., Jackson,S.P., Marshak Rothstein,A., Jeggo,P.A. and Herrera,V.L. (1998) Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity, 9, 355–366. [DOI] [PubMed] [Google Scholar]
- 28.Melek M., Gellert,M. and van Gent,D.C. (1998) Rejoining of DNA by the RAG1 and RAG2 proteins. Science, 280, 301–303. [DOI] [PubMed] [Google Scholar]
- 29.Hiom K., Melek,M. and Gellert,M. (1998) DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell, 94, 463–470. [DOI] [PubMed] [Google Scholar]
- 30.Agrawal A., Eastman,Q.M. and Schatz,D.G. (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature, 394, 744–751. [DOI] [PubMed] [Google Scholar]
- 31.Melek M. and Gellert,M. (2000) RAG1/2-mediated resolution of transposition intermediates: two pathways and possible consequences. Cell, 101, 625–633. [DOI] [PubMed] [Google Scholar]
- 32.Plasterk R. (1998) V(D)J recombination. Ragtime jumping. Nature, 394, 718–719. [DOI] [PubMed] [Google Scholar]
- 33.Roth D.B. and Craig,N.L. (1998) VDJ recombination: a transposase goes to work. Cell, 94, 411–414. [DOI] [PubMed] [Google Scholar]
- 34.Fugmann S.D., Lee,A.I., Shockett,P.E., Villey,I.J. and Schatz,D.G. (2000) The RAG proteins and V(D)J recombination: complexes, ends and transposition. Annu. Rev. Immunol., 18, 495–527. [DOI] [PubMed] [Google Scholar]
- 35.Lew S., Franco,D. and Chang,Y. (2000) Activation of V(D)J recombination induces the formation of interlocus joints and hybrid joints in scid pre-B-cell lines. Mol. Cell. Biol., 20, 7170–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams C.J., Grandal,I., Vesprini,D.J., Wojtyra,U., Danska,J.S. and Guidos,C.J. (2001) Irradiation promotes V(D)J joining and RAG-dependent neoplastic transformation in SCID T-cell precursors. Mol. Cell. Biol., 21, 400–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schatz D.G., Oettinger,M.A. and Baltimore,D. (1989) The V(D)J recombination activating gene, RAG-1. Cell, 59, 1035–1048. [DOI] [PubMed] [Google Scholar]
- 38.Oettinger M.A., Schatz,D.G., Gorka,C. and Baltimore,D. (1990) RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science, 248, 1517–1523. [DOI] [PubMed] [Google Scholar]
- 39.van Gent D.C., McBlane,J.F., Ramsden,D.A., Sadofsky,M.J., Hesse,J.E. and Gellert,M. (1995) Initiation of V(D)J recombination in a cell-free system. Cell, 81, 925–934. [DOI] [PubMed] [Google Scholar]
- 40.McBlane J.F., van Gent,D.C., Ramsden,D.A., Romeo,C., Cuomo,C.A., Gellert,M. and Oettinger,M.A. (1995) Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell, 83, 387–395. [DOI] [PubMed] [Google Scholar]
- 41.Sadofsky M.J., Hesse,J.E., McBlane,J.F. and Gellert,M. (1993) Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res., 21, 5644–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silver D.P., Spanopoulou,E., Mulligan,R.C. and Baltimore,D. (1993) Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc. Natl Acad. Sci. USA, 90, 6100–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadofsky M.J., Hesse,J.E. and Gellert,M. (1994) Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res., 22, 1805–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuomo C.A. and Oettinger,M.A. (1994) Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res., 22, 1810–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodgers K.K., Bu,Z., Fleming,K.G., Schatz,D.G., Engelman,D.M. and Coleman,J.E. (1996) A zinc-binding domain involved in the dimerization of RAG1. J. Mol. Biol., 260, 70–84. [DOI] [PubMed] [Google Scholar]
- 47.Bellon S.F., Rodgers,K.K., Schatz,D.G., Coleman,J.E. and Steitz,T.A. (1997) Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nature Struct. Biol., 4, 586–591. [DOI] [PubMed] [Google Scholar]
- 48.Bailin T., Mo,X. and Sadofsky,M.J. (1999) A RAG1 and RAG2 tetramer complex is active in cleavage in V(D)J recombination. Mol. Cell. Biol., 19, 4664–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson P.C. and Desiderio,S. (1999) RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol., 19, 3674–3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roman C.A., Cherry,S.R. and Baltimore,D. (1997) Complementation of V(D)J recombination deficiency in RAG-1(–/–) B cells reveals a requirement for novel elements in the N-terminus of RAG-1. Immunity, 7, 13–24. [DOI] [PubMed] [Google Scholar]
- 51.McMahan C.J., Difilippantonio,M.J., Rao,N., Spanopoulou,E. and Schatz,D.G. (1997) A basic motif in the N-terminal region of RAG1 enhances V(D)J recombination activity. Mol. Cell. Biol., 4544–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steen S.B., Han,J.O., Mundy,C., Oettinger,M.A. and Roth,D.B. (1999) Roles of the ‘dispensable’ portions of RAG-1 and RAG-2 in V(D)J recombination. Mol. Cell. Biol., 19, 3010–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Villa A., Santagata,S., Bozzi,F., Giliani,S., Frattini,A., Imberti,L., Gatta,L.B., Ochs,H.D., Schwarz,K., Notarangelo,L.D., Vezzoni,P. and Spanopoulou,E. (1998) Partial V(D)J recombination activity leads to Omenn syndrome. Cell, 93, 885–896. [DOI] [PubMed] [Google Scholar]
- 54.Signorini S., Imberti,L., Pirovano,S., Villa,A., Facchetti,F., Ungari,M., Bozzi,F., Albertini,A., Ugazio,A.G., Vezzoni,P. and Notarangelo,L.D. (1999) Intrathymic restriction and peripheral expansion of the T-cell repertoire in Omenn syndrome. Blood, 94, 3468–3478. [PubMed] [Google Scholar]
- 55.Notarangelo L.D., Villa,A. and Schwarz,K. (1999) RAG and RAG defects. Curr. Opin. Immunol., 11, 435–442. [DOI] [PubMed] [Google Scholar]
- 56.Santagata S., Gomez,C.A., Sobacchi,C., Bozzi,F., Abinun,M., Pasic,S., Cortes,P., Vezzoni,P. and Villa,A. (2000) N-terminal RAG1 frameshift mutations in Omenn’s syndrome: internal methionine usage leads to partial V(D)J recombination activity and reveals a fundamental role in vivo for the N-terminal domains. Proc. Natl Acad. Sci. USA, 97, 14572–14577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noordzij J.G., Verkaik,N.S., Hartwig,N.G., de Groot,R., van Gent,D.C. and van Dongen,J.J. (2000) N-terminal truncated human RAG1 proteins can direct T-cell receptor but not immunoglobulin gene rearrangements. Blood, 96, 203–209. [PubMed] [Google Scholar]
- 58.Difilippantonio M.J., McMahan,C.J., Eastman,Q.M., Spanopoulou,E. and Schatz,D.G. (1996) RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell, 87, 253–262. [DOI] [PubMed] [Google Scholar]
- 59.Spanopoulou E., Zaitseva,F., Wang,F., Santagata,S., Baltimore,D. and Panayotou,G. (1996) The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell, 87, 263–276. [DOI] [PubMed] [Google Scholar]
- 60.Landree M.A., Wibbenmeyer,J.A. and Roth,D.B. (1999) Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev., 13, 3059–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim D.R., Dai,Y., Mundy,C.L., Yang,W. and Oettinger,M.A. (1999) Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev., 13, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fugmann S.D., Villey,I.J., Ptaszek,L.M. and Schatz,D.G. (2000) Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell, 5, 97–107. [DOI] [PubMed] [Google Scholar]
- 63.Sadofsky M.J., Hesse,J.E., van Gent,D.C. and Gellert,M. (1995) RAG-1 mutations that affect the target specificity of V(D)J recombination: a possible direct role of RAG-1 in site recognition. Genes Dev., 9, 2193–2199. [DOI] [PubMed] [Google Scholar]
- 64.Roman C.A. and Baltimore,D. (1996) Genetic evidence that the RAG1 protein directly participates in V(D)J recombination through substrate recognition. Proc. Natl Acad. Sci. USA, 93, 2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kale S.B., Landree,M.A. and Roth,D.B. (2001) Conditional RAG-1 mutants block the hairpin formation step of V(D)J recombination. Mol. Cell. Biol., 21, 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mo X., Bailin,T. and Sadofsky,M.J. (2001) A C-terminal region of RAG1 contacts the coding DNA during V(D)J recombination. Mol. Cell. Biol. 21, 2038–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirch S.A., Rathbun,G.A. and Oettinger,M.A. (1998) Dual role of RAG2 in V(D)J recombination: catalysis and regulation of ordered Ig gene assembly. EMBO J., 17, 4881–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Callebaut I. and Mornon,J.P. (1998) The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell. Mol. Life Sci., 54, 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Corneo B., Moshous,D., Callebaut,I., de Chasseval,R., Fischer,A. and de Villartay,J.P. (2000) Three-dimensional clustering of human RAG2 gene mutations in severe combined immune deficiency. J. Biol. Chem., 275, 12672–12675. [DOI] [PubMed] [Google Scholar]
- 70.Gomez C.A., Ptaszek,L.M., Villa,A., Bozzi,F., Sobacchi,C., Brooks,E.G., Notarangelo,L.D., Spanopoulou,E., Pan,Z.Q., Vezzoni,P., Cortes,P. and Santagata,S. (2000) Mutations in conserved regions of the predicted RAG2 kelch repeats block initiation of V(D)J recombination and result in primary immunodeficiencies. Mol. Cell. Biol., 20, 5653–5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li W., Swanson,P. and Desiderio,S. (1997) RAG-1 and RAG-2-dependent assembly of functional complexes with V(D)J recombination substrates in solution. Mol. Cell. Biol., 17, 6932–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Akamatsu Y. and Oettinger,M.A. (1998) Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol. Cell. Biol., 18, 4670–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mo X., Bailin,T. and Sadofsky,M.J. (1999) RAG1 and RAG2 cooperate in specific binding to the recombination signal sequence in vitro. J. Biol. Chem., 274, 7025–7032. [DOI] [PubMed] [Google Scholar]
- 74.Lewis S.M., Agard,E., Suh,S. and Czyzyk,L. (1997) Cryptic signals and the fidelity of V(D)J joining. Mol. Cell. Biol., 17, 3125–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cuomo C.A., Mundy,C.L. and Oettinger,M.A. (1996) DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol., 16, 5683–5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramsden D.A., McBlane,J.F., van Gent,D.C. and Gellert,M. (1996) Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J., 15, 3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 77.Swanson P.C. and Desiderio,S. (1998) V(D)J recombination signal recognition: distinct, overlapping DNA–protein contacts in complexes containing RAG1 with and without RAG2. Immunity, 9, 115–125. [DOI] [PubMed] [Google Scholar]
- 78.Aidinis V., Bonaldi,T., Beltrame,M., Santagata,S., Bianchi,M.E. and Spanopoulou,E. (1999) The RAG1 homeodomain recruits HMG1 and HMG2 to facilitate recombination signal sequence binding and to enhance the intrinsic DNA-bending activity of RAG1-RAG2. Mol. Cell. Biol., 19, 6532–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mo X., Bailin,T., Noggle,S. and Sadofsky,M.J. (2000) A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res., 28, 1228–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Eastman Q.M., Villey,I.J. and Schatz,D.G. (1999) Detection of RAG protein–V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol. Cell. Biol., 19, 3788–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim D.R. and Oettinger,M.A. (1998) Functional analysis of coordinated cleavage in V(D)J recombination. Mol. Cell. Biol., 18, 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bustin M. and Reeves,R. (1996) High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog. Nucleic Acid Res. Mol. Biol., 54, 35–100. [DOI] [PubMed] [Google Scholar]
- 83.van Gent D.C., Hiom,K., Paull,T.T. and Gellert,M. (1997) Stimulation of V(D)J cleavage by high mobility group proteins. EMBO J., 16, 2665–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sawchuk D.J., Weis-Garcia,F., Malik,S., Besmer,E., Bustin,M., Nussenzweig,M.C. and Cortes,P. (1997) V(D)J recombination: modulation of RAG1 and RAG2 cleavage activity on 12/23 substrates by whole cell extract and DNA-bending proteins. J. Exp. Med., 185, 2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoshida T., Tsuboi,A., Ishiguro,K., Nagawa,F. and Sakano,H. (2000) The DNA-bending protein, HMG1, is required for correct cleavage of 23 bp recombination signal sequences by recombination activating gene proteins in vitro. Int. Immunol., 12, 721–729. [DOI] [PubMed] [Google Scholar]
- 86.Ramsden D.A. and Gellert,M. (1995) Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev., 9, 2409–2420. [DOI] [PubMed] [Google Scholar]
- 87.Ramsden D.A., Paull,T.T. and Gellert,M. (1997) Cell-free V(D)J recombination. Nature, 388, 488–491. [DOI] [PubMed] [Google Scholar]
- 88.Besmer E., Mansilla-Soto,J., Cassard,S., Sawchuk,D.J., Brown,G., Sadofsky,M., Lewis,S.M., Nussenzweig,M.C. and Cortes,P. (1998) Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol. Cell, 2, 817–828. [DOI] [PubMed] [Google Scholar]
- 89.Shockett P.E. and Schatz,D.G. (1999) DNA hairpin opening mediated by the RAG1 and RAG2 proteins. Mol. Cell. Biol., 19, 4159–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Santagata S., Besmer,E., Villa,A., Bozzi,F., Allingham,J.S., Sobacchi,C., Haniford,D.B., Vezzoni,P., Nussenzweig,M.C., Pan,Z.Q. and Cortes,P. (1999) The RAG1/RAG2 complex constitutes a 3′ flap endonuclease: implications for junctional diversity in V(D)J and transpositional recombination. Mol. Cell, 4, 935–947. [DOI] [PubMed] [Google Scholar]
- 91.Paull T.T. and Gellert,M. (1998) The 3′ to 5′ exonuclease activity of MRE11 facilitates repair of DNA double-strand breaks. Mol. Cell, 1, 969–979. [DOI] [PubMed] [Google Scholar]
- 92.Paull T.T. and Gellert,M. (2000) A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc. Natl Acad. Sci. USA, 97, 6409–6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gerstein R.M. and Lieber,M.R. (1993) Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev., 7, 1459–1469. [DOI] [PubMed] [Google Scholar]
- 94.Yu K. and Lieber,M.R. (1999) Mechanistic basis for coding end sequence effects in the initiation of V(D)J recombination. Mol. Cell. Biol., 19, 8094–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu K. and Lieber,M.R. (2000) The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol., 20, 7914–7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meier J.T. and Lewis,S.M. (1993) P nucleotides in V(D)J recombination: a fine-structure analysis. Mol. Cell. Biol., 13, 1078–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nadel B. and Feeney,A.J. (1997) Nucleotide deletion and P addition in V(D)J recombination: a determinant role of the coding-end sequence. Mol. Cell. Biol., 17, 3768–3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ezekiel U.R., Sun,T., Bozek,G. and Storb,U. (1997) The composition of coding joints formed in V(D)J recombination is strongly affected by the nucleotide sequence of the coding ends and their relationship to the recombination signal sequences. Mol. Cell. Biol., 17, 4191–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun T., Ezekiel,U.R., Erskine,L., Agulo,R., Bozek,G., Roth,D. and Storb,U. (1999) Signal joint formation is inhibited in murine scid preB cells and fibroblasts in substrates with homopolymeric coding ends. Mol. Immunol., 36, 551–558. [DOI] [PubMed] [Google Scholar]
- 100.Blunt T., Finnie,N.J., Taccioli,G.E., Smith,G.C., Demengeot,J., Gottlieb,T.M., Mizuta,R., Varghese,A.J., Alt,F.W., Jeggo,P.A. and Jackson,S.P. (1995) Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell, 80, 813–823. [DOI] [PubMed] [Google Scholar]
- 101.Kirchgessner C.U., Patil,C.K., Evans,J.W., Cuomo,C.A., Fried,L.M., Carter,T., Oettinger,M.A. and Brown,J.M. (1995) DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science, 267, 1178–1183. [DOI] [PubMed] [Google Scholar]
- 102.Lewis S.M., Hesse,J.E., Mizuuchi,K. and Gellert,M. (1988) Novel strand exchanges in V(D)J recombination. Cell, 55, 1099–1107. [DOI] [PubMed] [Google Scholar]
- 103.Lewis S.M. and Hesse,J.E. (1991) Cutting and closing without recombination in V(D)J joining. EMBO J., 10, 3631–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Santagata S., Aidinis,V. and Spanopoulou,E. (1998) The effect of Me2+ cofactors at the initial stages of V(D)J recombination. J. Biol. Chem., 273, 16325–16331. [DOI] [PubMed] [Google Scholar]
- 105.van Gent D.C., Ramsden,D.A. and Gellert,M. (1996) The RAG1 and RAG2 proteins establish the 12/23-rule in V(D)J recombination. Cell, 85, 107–113. [DOI] [PubMed] [Google Scholar]
- 106.Eastman Q.M., Leu,T.M.J. and Schatz,D.G. (1996) Initiation of V(D)J recombination in vitro obeying the 12/23-rule. Nature, 380, 85–88. [DOI] [PubMed] [Google Scholar]
- 107.Cowan J.A. (1998) Metal activation of enzymes in nucleic acid biochemistry. Chem. Rev., 98, 1067–1087. [DOI] [PubMed] [Google Scholar]
- 108.Hiom K. and Gellert,M. (1998) Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell, 1, 1011–1019. [DOI] [PubMed] [Google Scholar]
- 109.Mizuuchi K. (1992) Transpositional recombination: mechanistic insights from studies of µ and other elements. Annu. Rev. Biochem., 61, 1011–1051. [DOI] [PubMed] [Google Scholar]
- 110.Savilahti H. and Mizuuchi,K. (1996) µ transpositional recombination: donor DNA cleavage and strand transfer in trans by the µ transposase. Cell, 85, 271–280. [DOI] [PubMed] [Google Scholar]
- 111.Aldaz H., Schuster,E. and Baker,T.A. (1996) The interwoven architecture of the µ transposase couples DNA synapsis to catalysis. Cell, 85, 257–269. [DOI] [PubMed] [Google Scholar]
- 112.Wei S.Q., Mizuuchi,K. and Craigie,R. (1997) A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J., 16, 7511–7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bankhead T. and Segall,A.M. (2000) Characterization of a mutation of bacteriophage λ integrase. Putative role in core binding and strand exchange for a conserved residue. J. Biol. Chem., 275, 36949–36956. [DOI] [PubMed] [Google Scholar]
- 114.Steen S.B., Gomelsky,L., Speidel,S.L. and Roth,D.B. (1997) Initiation of V(D)J recombination in vivo: role of recombination signal sequences in formation of single and paired double-strand breaks. EMBO J., 16, 2656–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.West R.B. and Lieber,M.R. (1998) The RAG–HMG1 complex enforces the 12/23 rule of V(D)J recombination specifically at the double-hairpin formation step. Mol. Cell. Biol., 18, 6408–6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sheehan K.M. and Lieber,M.R. (1993) V(D)J recombination: signal and coding joint resolution are uncoupled and depend on parallel synapsis of the sites. Mol. Cell. Biol., 13, 1363–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hiom K. and Gellert,M. (1997) A stable RAG1–RAG2–DNA complex that is active in V(D)J cleavage. Cell, 88, 65–72. [DOI] [PubMed] [Google Scholar]
- 118.Agrawal A. and Schatz,D.G. (1997) RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell, 89, 43–53. [DOI] [PubMed] [Google Scholar]
- 119.Grawunder U. and Lieber,M.R. (1997) A complex of RAG-1 and RAG-2 proteins persists on DNA after single-strand cleavage at V(D)J recombination signal sequences. Nucleic Acids Res., 25, 1375–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.McMahan C.J., Sadofsky,M.J. and Schatz,D.G. (1997) Definition of a large region of RAG1 that is important for coimmunoprecipitation of RAG2. J. Immunol., 158, 2202–2210. [PubMed] [Google Scholar]
- 121.Aidinis V., Dias,D.C., Gomez,C.A., Bhattacharyya,D., Spanopoulou,E. and Santagata,S. (2000) Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J. Immunol., 164, 5826–5832. [DOI] [PubMed] [Google Scholar]
- 122.Swanson P.C. (2001) The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol., 21, 449–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Han J.O., Steen,S.B. and Roth,D.B. (1999) Intermolecular V(D)J recombination is prohibited specifically at the joining step. Mol. Cell, 3, 331–338. [DOI] [PubMed] [Google Scholar]
- 124.Tevelev A. and Schatz,D.G. (2000) Intermolecular V(D)J recombination. J. Biol. Chem., 275, 8341–8348. [DOI] [PubMed] [Google Scholar]
- 125.Agard E.A. and Lewis,S.M. (2000) Postcleavage sequence specificity in V(D)J recombination. Mol. Cell. Biol., 5032–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schultz H.Y., Landree,M.A., Qiu,J., Kale,S.B. and Roth,D.B. (2001) Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol. Cell, 7, 65–75. [DOI] [PubMed] [Google Scholar]
- 127.Qiu J., Kale,S.B., Schultz,H.Y. and Roth,D.B. (2001) Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol. Cell, 7, 77–87. [DOI] [PubMed] [Google Scholar]
- 128.Guidos C.J., Williams,C.J., Grandal,I., Knowles,G., Huang,M.T. and Danska,J.S. (1996) V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev., 10, 2038–2054. [DOI] [PubMed] [Google Scholar]
- 129.Chen H.T., Bhandoola,A., Difilippantonio,M.J., Zhu,J., Brown,M.J., Tai,X., Rogakou,E.P., Brotz,T.M., Bonner,W.M., Ried,T. and Nussenzweig,A. (2000) Response to RAG-mediated V(D)J cleavage by NBS1 and γ-H2AX. Science, 290, 1962–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fuschiotti P., Harindranath,N., Mage,R.G., McCormack,W.T., Dhanarajan,P. and Roux,K.H. (1993) Recombination activating genes-1 and -2 of the rabbit: cloning and characterization of germline and expressed genes. Mol. Immunol., 30, 1021–1032. [DOI] [PubMed] [Google Scholar]
- 131.Miller R.D. and Rosenberg,G.H. (1997) Recombination activating gene-1 of the opossum Monodelphis domestica. Immunogenetics, 45, 341–342. [DOI] [PubMed] [Google Scholar]
- 132.Groth J.G. and Barrowclough,G.F. (1999) Basal divergences in birds and the phylogenetic utility of the nuclear RAG-1 gene. Mol. Phylogenet. Evol., 12, 115–123. [DOI] [PubMed] [Google Scholar]
- 133.Greenhalgh P., Olesen,C. and Steiner,L.A. (1993) Characterization and expression of recombination activating genes (RAG-1 and RAG-2) in Xenopus laevis. J. Immunol., 151, 3100–3110. [PubMed] [Google Scholar]
- 134.Willett C.E., Cherry,J.J. and Steiner,L.A. (1997) Characterization and expression of the recombination activating genes (rag1 and rag2) of zebrafish. Immunogenetics, 45, 394–404. [DOI] [PubMed] [Google Scholar]
- 135.Hansen J.D. and Kaattari,S.L. (1995) The recombination activation gene 1 (RAG1) of rainbow trout (Oncorhynchus mykiss): cloning, expression and phylogenetic analysis. Immunogenetics, 42, 188–195. [DOI] [PubMed] [Google Scholar]
- 136.Peixoto B.R., Mikawa,Y. and Brenner,S. (2000) Characterization of the recombinase activating gene-1 and 2 locus in the Japanese pufferfish, Fugu rubripes. Gene, 246, 275–283. [DOI] [PubMed] [Google Scholar]
- 137.Bernstein R.M., Schluter,S.F., Bernstein,H. and Marchalonis,J.J. (1996) Primordial emergence of the recombination activating gene 1 (RAG1): sequence of the complete shark gene indicates homology to microbial integrases. Proc. Natl Acad. Sci. USA, 93, 9454–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]