Abstract
Synapses are specialized cell-cell contacts that mediate communication between neurons. Most excitatory synapses in the brain are housed on dendritic spines, small actin-rich protrusions extending from dendrites. During development and in response to environmental stimuli, spines undergo marked changes in shape and number thought to underlie processes like learning and memory. Improper spine development, in contrast, likely impedes information processing in the brain, since spine abnormalities are associated with numerous brain disorders. Elucidating the mechanisms that regulate the formation and plasticity of spines and their resident synapses is therefore crucial to our understanding of cognition and disease. Rho-family GTPases, key regulators of the actin cytoskeleton, play essential roles in orchestrating the development and remodeling of spines and synapses. Precise spatio-temporal regulation of Rho GTPase activity is critical for their function, since aberrant Rho GTPase signaling can cause spine and synapse defects as well as cognitive impairments. Rho GTPases are activated by guanine nucleotide exchange factors (GEFs) and inhibited by GTPase-activating proteins (GAPs). We propose that Rho-family GEFs and GAPs provide the spatiotemporal regulation and signaling specificity necessary for proper Rho GTPase function based on the following features they possess: (i) existence of multiple GEFs and GAPs per Rho GTPase, (ii) developmentally regulated expression, (iii) discrete localization, (iv) ability to bind to and organize specific signaling networks, and (v) tightly regulated activity, perhaps involving GEF/GAP interactions. Recent studies describe several Rho-family GEFs and GAPs that uniquely contribute to spinogenesis and synaptogenesis. Here, we highlight several of these proteins and discuss how they occupy distinct biochemical niches critical for synaptic development.
1. Introduction: the formation and remodeling of excitatory synapses
The human brain is composed of approximately 100 billion neurons that process and transmit information in the form of electric signals. Communication between neurons occurs at specialized sites of contact called synapses. The majority of excitatory synapses on principal neurons in the brain are located on the tips of dendritic spines, small protrusions on the surface of dendrites (Fig. 1). In recent years, it has become clear that spines are dynamic structures that undergo rapid remodeling important for synapse formation, function and plasticity (Bhatt et al., 2009; Bourne and Harris, 2008). During early postnatal development, dendritic protrusions first appear as long, thin, highly motile filopodia, which can initiate synaptic contacts with nearby axons (Dailey and Smith, 1996; Fiala et al., 1998; Papa et al., 1995; Ziv and Smith, 1996). As development proceeds, dendritic filopodia are replaced by (or mature into) more stable mushroom-shaped spines, which are either maintained into adulthood or eliminated (Engert and Bonhoeffer, 1999; Fiala et al., 1998; Lippman and Dunaevsky, 2005; Ziv and Smith, 1996). Following development, spines continue to remodel in response to a variety of physiological stimuli (Engert and Bonhoeffer, 1999; Lendvai et al., 2000; Maletic-Savatic et al., 1999; Toni et al., 1999). For example, synaptic activity that induces long-term potentiation (LTP), a long-lasting enhancement of synaptic strength, promotes spine enlargement and new spine formation (Matsuzaki et al., 2004), whereas activity that induces long-term depression (LTD), a persistent weakening of synaptic strength, causes spine shrinkage or retraction (Zhou et al., 2004). This synaptic remodeling is thought to be important for neural circuit plasticity associated with learning and memory (Yuste and Majewska, 2001). Since spine structure and synaptic function are intimately related (Kasai et al., 2003), it is easy to imagine that improper spine morphogenesis might result in impaired information processing in the brain. Indeed, spine abnormalities are associated with numerous neurodevelopmental, neuropsychiatric, and neurodegenerative disorders (Fiala et al., 2002; Newey et al., 2005). It is therefore essential to understand the mechanisms that control the development and remodeling of spiny synapses under normal and pathological conditions.
Figure 1. Dendritic spines are the primary sites of excitatory synapses in the brain.
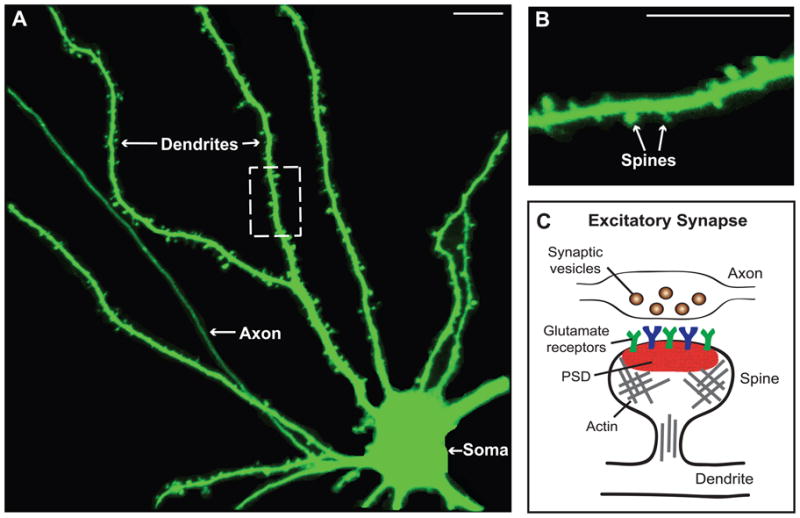
A. Shown is an example of a rat hippocampal neuron expressing green fluorescent protein (GFP). Neurons possess a soma, an axon and branched dendrites containing dendritic spines. B. Image shows an enlargement of the dashed box pictured in A, which provides a clearer view of spines. C. Schematic of an excitatory synapse, which forms between a dendritic spine and a presynaptic bouton on an axon. The postsynaptic density (PSD), which contains glutamate receptors, scaffolding proteins and other signaling molecules, is located on the spine head. Spines are also enriched in actin filaments. Scale bar: 10 νm
Spines are highly enriched in filamentous actin (F-actin), and their ability to change shape depends on the rapid remodeling of the spine actin cytoskeleton (Cingolani and Goda, 2008; Honkura et al., 2008). It is therefore not surprising that Rho GTPases, a subfamily of small GTP-binding proteins known for their ability to control actin cytoskeletal dyanamics, have emerged as key regulators of spine morphogenesis (Fig. 2) (Govek et al., 2005). The best-studied Rho GTPase family members are RhoA, Rac1 and Cdc42. In neurons, Rac1 and Cdc42 promote the formation, growth and maintenance of spines, whereas RhoA induces spine retraction and loss (Newey et al., 2005). Interestingly, mutations in a number of genes involved in Rho GTPase signaling have been linked to non-syndromic mental retardation, an intellectual disability (ID) associated with spine anomalies (Newey et al., 2005; Ramakers, 2002). This apparent correlation between altered Rho GTPase signaling, spine abnormalities, and mental retardation suggests that precise Rho GTPase signaling is important for proper circuit development and normal cognitive function.
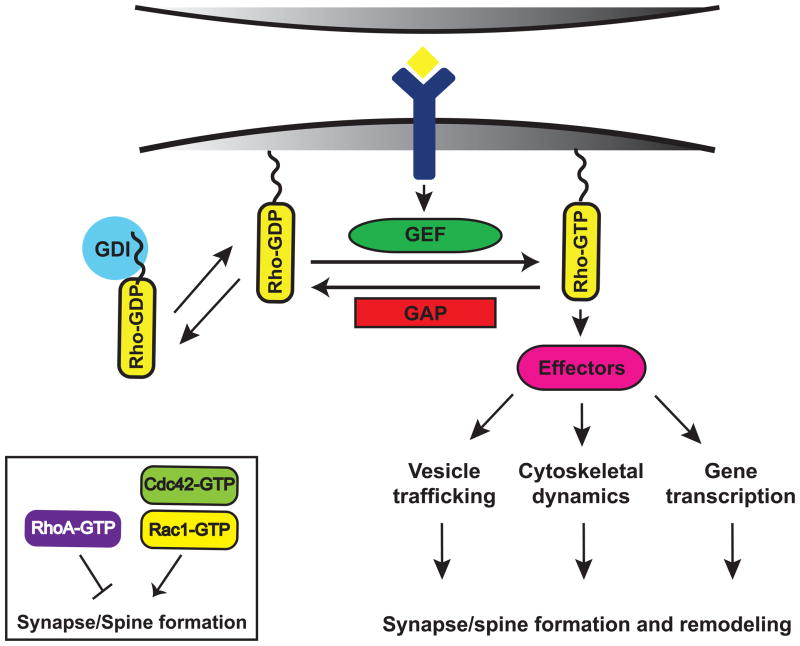
Figure 2. Model of Rho GTPase signaling at synapses.
Rho GTPases function as binary switches by cycling between an active, GTP-bound form and an inactive, GDP-bound form. Rho GTPase activity is tightly regulated in space and time by three different classes of regulatory proteins: guanine nucleotide exchange factors (GEFs), GTP-activating proteins (GAPs), and guanine nucleotide dissociation inhibitors (GDIs). In their active state, Rho GTPases interact with downstream effectors that regulate a variety of cellular processes, which ultimately contribute to spine morphogenesis and excitatory synapse development. The Rho GTPases Rac and Cdc42 promote the formation and growth of synapses and spines, whereas RhoA inhibits synapse development.
Rho GTPases regulate spine morphogenesis and synapse development by functioning as molecular switches, cycling between an active GTP-bound state and an inactive GDP-bound state (Fig. 2). In their active conformation, Rho GTPases interact with specific effector molecules, which induce downstream signaling pathways that control a diverse array of biological processes including actin cytoskeletal reorganization, microtubule dynamics, gene transcription, and membrane trafficking (Govek et al., 2005). Precise spatio-temporal control of Rho GTPase signaling is orchestrated by guanine nucleotide exchange factors (GEFs), which activate Rho GTPases by catalyzing GDP/GTP exchange (Schmidt and Hall, 2002) and GTPase-activating proteins (GAPs), which inhibit Rho GTPases by enhancing their intrinsic GTPase activities (Bernards and Settleman, 2004). Guanine nucleotide dissociation inhibitors (GDIs) also regulate Rho GTPases by preventing GDP/GTP exchange and sequestering inactive Rho GTPases in the cytoplasm (DerMardirossian and Bokoch, 2005). In addition to controlling Rho GTPase activity, GEFs and GAPs contribute to Rho GTPase signaling specificity by interacting with particular upstream receptors and downstream effectors (Buchsbaum et al., 2002, 2003; Jaffe et al., 2005; Tolias et al., 2005; Tolias et al., 2007; Zhang and Macara, 2008). Recent studies have identified a number of Rho-family GEFs and GAPs that play important roles in spine morphogenesis and synapse development (Kiraly et al., 2010). Determining how these GEFs and GAPs are regulated and the distinct functions they serve at synapses will be essential for developing a more mechanistic understanding of synapse formation and remodeling.
2. Regulation of Rho GTPase signaling pathways at synapses
Rho GTPases regulate a variety of neurodevelopmental processes including neuronal migration, axon growth and guidance, dendritic arborization and synaptogenesis (Govek et al., 2005; Linseman and Loucks, 2008). Rho GTPases control these diverse processes by functioning at distinct locations within neurons at different developmental stages and in response to a variety of extracellular signals. The ability of Rho GTPases to act at different times and places in response to different stimuli and generate distinct cellular outcomes suggests a level of specificity that cannot reside in the GTPases alone. If, however, signaling specificity was provided by Rho GTPase regulatory proteins, we would expect these proteins to have some or all of the following features: (i) more than one GEF and/or GAP should exist for each Rho GTPase, (ii) their expression should be developmentally regulated, (iii) they should have discrete localizations within cells, (iv) they should have the capacity to organize specific Rho GTPase signaling networks, i.e. by binding to effectors and/or other signaling molecules, and (v) GEFs and GAPs should have tightly regulated activity and potentially interact with each other, at least indirectly. These features would allow a single Rho GTPase to specifically regulate multiple signaling pathways in the same cell and at the same time in a way that changes with development and is tightly regulated spatially and temporally. Do Rho GTPase GEFs and GAPs possess these features?
A general overview of Rho-family regulatory proteins indicates that they do possess characteristics that would enable them to provide specificity to Rho GTPase signaling (Moon and Zheng, 2003; Rossman et al., 2005; Schmidt and Hall, 2002). First, Rho-family GEFs and GAPs both outnumber Rho GTPases by at least three- to four-fold. Second, their expression levels appear to be developmentally regulated. Third, GEFs and GAPs display varied subcellular localization profiles. Fourth, GEFs and GAPs are typically large proteins that possess multiple signaling domains that enable them to receive diverse upstream inputs as well as recruit downstream components of Rho GTPase-regulated pathways—they are both signal integrators and scaffolds (Fig. 3). Finally, the activities of Rho GTPase GEFs and GAPs are tightly regulated and emerging evidence suggests that they work closely together to control Rho GTPase activation. All of these features suggest that Rho GTPase regulatory proteins are capable of spatially and temporally regulating specific GTPase-mediated signaling pathways. Thus, while at first glance it seems that different Rho-family GEFs or GAPs have identical roles, their chief similarity is their ability to activate or inhibit specific Rho GTPases. The context of that activation or deactivation—the signal(s) to which it responds, the downstream mechanisms, and ultimate outcomes, exhibit striking diversity.
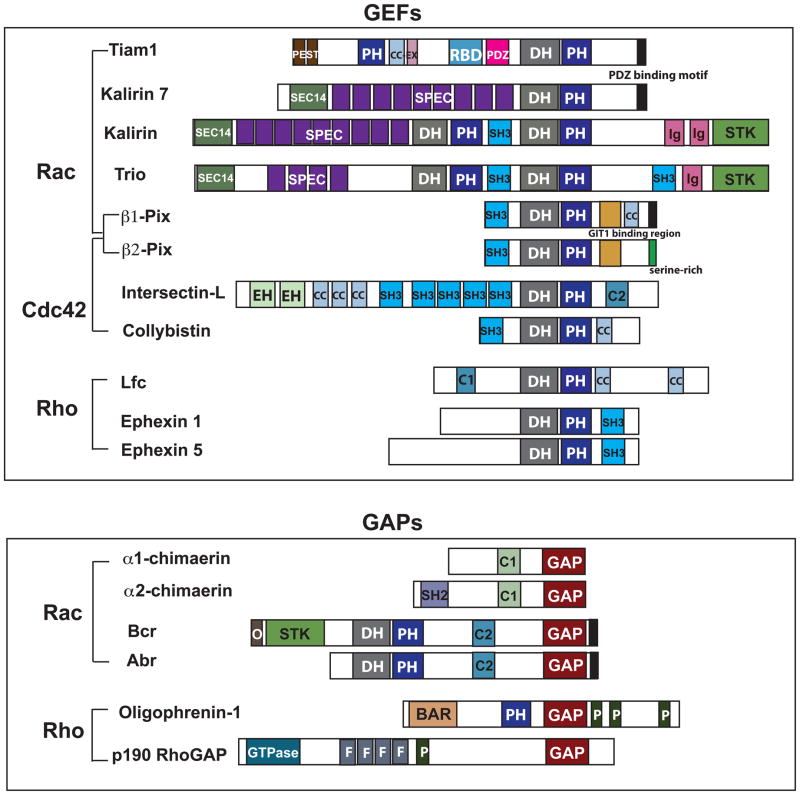
Figure 3. Domain structures of synaptic Rho-family GEFs and GAPs.
Shown are the domain structures of proteins mentioned in this review. The following abbreviations are used in this figure: PH: pleckstrin homology domain, CC-Ex: coiled coil-extended region, RBD: Ras-binding domain, PDZ: domain in PSD-95, Dlg, and ZO-1/2, DH: Dbl homology domain, SEC14: domain in phosphatidylinositol transfer protein Sec14, SPEC: spectrin-like repeats, SH3: Src homology 3 domain, EH: Eps15 homology domain, C2: protein kinase C conserved region 2, C1: protein kinase C conserved region 1, BAR: Bin/Amphiphysin/Rvs domain, and P: proline-rich regions.
In this review, we will highlight a number of Rho-family GEFs and GAPs that are involved in synaptogenesis. We will discuss how these Rho GTPase regulatory proteins contribute to the spatiotemporal regulation and signaling specificity of Rho GTPases by examining their localization, developmental regulation, and network of protein interactions. In this way, we will argue that each Rho GTPase regulatory protein serves a unique role in the formation of synapses in the central nervous system (CNS). In order to disucuss their unique function at synapses, we have focused on better characterized GEFs and GAPs. Reports of additional Rho-family regulatory proteins involved in synaptogenesis exist, however, adequate information is not currently available in the literature for us to delineate a specific role for them at this time.
3. Rho-family GEFs involved in excitatory synaptogenesis
3.1. Rac/Cdc42-GEFs at excitatory synapses
3.1.1 Kalirin-7
The Rac-GEF Kalirin-7 has emerged as a key regulator of spine dynamics (Penzes and Jones, 2008). Kalirin-7 is produced by the KALRN gene, which generates several Kalirin isoforms via alternative splicing (Johnson et al., 2000). Kalirin-7 is the most abundant splice variant of the KALRN gene in the adult brain, and its expression during development correlates with synaptogenesis (Johnson et al., 2000; Penzes et al., 2000). Like most Rho family GEFs, Kalirin-7 possesses multiple domains, including a lipid-binding Sec14p domain, nine spectrin-like repeats, a Rac-GEF domain consisting of a tandem Dbl homology (DH)-pleckstrin homology (PH) domain, and a C-terminal PDZ binding motif (Fig. 3). Kalirin-7 localizes to dendritic spines and is enriched in the postsynaptic density (PSD), where it interacts with numerous PDZ domain-containing proteins, including PSD-95, SAP-102, and SAP-97 (Penzes et al., 2000; Penzes et al., 2001). These adapter proteins link Kalirin-7 with a variety of receptor subtypes and signaling molecules at the PSD, placing Kalirin-7 in a position to integrate diverse signals that regulate different aspects of spine morphogenesis (Penzes and Jones, 2008).
The ability of Kalirin-7 to promote spine formation has been demonstrated in cultured cortical and hippocampal pyramidal neurons as well as in inhibitory aspiny interneurons, where overexpression of Kalirin-7 induces the formation of spiny synapses (Ma et al., 2003; Ma et al., 2008; Penzes et al., 2003; Penzes et al., 2001). Conversely, reduction of Kalirin-7 expression in cultured neurons by antisense RNA or RNA interference (RNAi) results in decreased spine density and loss of excitatory synapses (Ma et al., 2003; Xie et al., 2007). In vivo support for the role of Kalirin-7 in synapse formation and function has been provided by mice lacking Kalirin-7 (Kalirin-7 knockout mice) and mice lacking all Kalirin isoforms (KALRN knockout mice), both of which exhibit decreased spine density in the cortex and specific cognitive deficits (Cahill et al., 2009; Ma et al., 2008; Xie et al., 2010). Interestingly, early stages of excitatory synapse development appear to proceed normally in these mice, suggesting that Kalirin-7, which is primarily expressed late in development and in the adult, may play a more essential role in spine maturation and/or maintenance (Cahill et al., 2009; Ma et al., 2008). Furthermore, reductions in Rac-GTP levels and spine density were only observed in the frontal cortex but not the hippocampus of adult KALRN knockout mice (Cahill et al., 2009). Other Rac-GEFs such as Tiam1 and -PIX, which retain higher levels of expression in the adult hippocampus compared to the cortex, might partially compensate for the absence of Kalirin-7 in the hippocampus (Penzes et al., 2008).
Kalirin-7 regulates spine dynamics downstream of a variety of synaptic receptors (Fig. 4). For instance, Kalirin-7 function is necessary for EphB-induced spine remodeling (Penzes et al., 2003). EphB receptors belong to a large family of Eph receptor tyrosine kinases that play key roles in controlling spine morphogenesis and synapse development and plasticity (Klein, 2004; Yamaguchi and Pasquale, 2004; Klein, 2009). In response to stimulation by their membrane-bound ephrin ligands, EphB receptors promote the formation and maturation of spines (Ethell et al., 2001; Henkemeyer et al., 2003; Kayser et al., 2006; Kayser et al., 2008; Penzes et al., 2003), whereas EphA receptors induce spine retraction (Fu et al., 2007; Murai et al., 2003). The ability of EphB receptors to stimulate spine morphogenesis and synapse maturation requires the activities of several Rho family GEFs (Irie and Yamaguchi, 2002; Penzes et al., 2003; Tolias et al., 2007). Kalirin-7 was shown to play a role in EphB receptor signaling by the demonstration that EphB receptor activation results in the phosphorylation and clustering of Kalirin-7 (Penzes et al., 2003). Furthermore, expression of a GEF-dead Kalirin-7 mutant in cultured hippocampal neurons blocks spine development induced by ephrinB1 activation of EphB receptors (Penzes et al., 2003). Taken together, these results suggest that Kalirin-7 plays an important role in EphB-mediated spine morphogenesis.
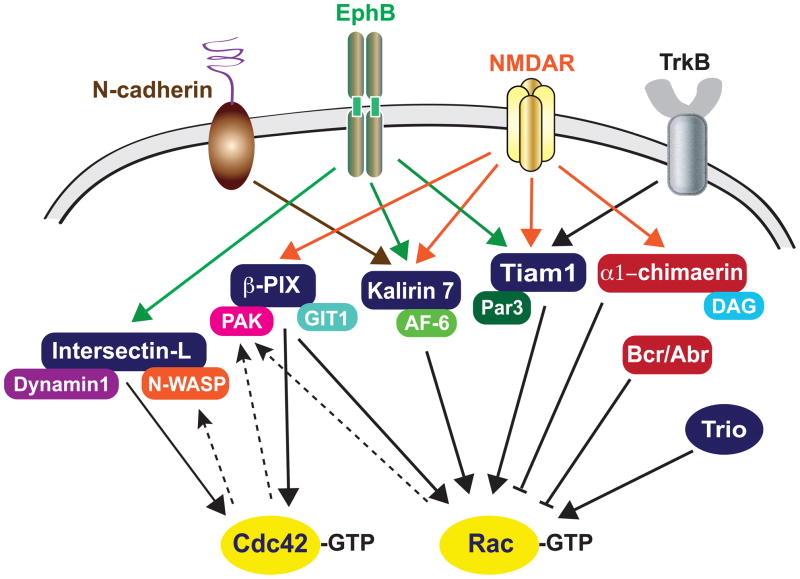
Figure 4. Regulation of Rac/Cdc42 signaling at synapses.
The Rac/Cdc42-GEFs Tiam1, Kalirin7, β-PIX, Intersectin-L and Trio promote Rac/Cdc42 activation, whereas the Rac-GAPs α1-chimaerin, Bcr and Abr inhibit Rac activation at synapses. Although these GEFs and GAPs are regulated by common upstream receptors, e.g. NMDA receptors, EphB, TrkB and N-cadherins, the unique role of each regulator likely arises from the networks of interacting proteins that are unique to each GEF or GAP and/or differential upstream regulation of these molecules in a temporal and spatial manner. GEFs are in blue and GAPs are in red, and dashed lines represent positive feedback loops.
As mentioned previously, neuronal activity exerts profound effects on the development and structural remodeling of dendritic spines (Engert and Bonhoeffer, 1999; Lendvai et al., 2000; Maletic-Savatic et al., 1999; Toni et al., 1999). These activity-induced effects are mediated by the NMDA-type glutamate receptor, a calcium-permeable ion channel that plays a central role in synaptic plasticity and learning and memory formation (Cull-Candy et al., 2001; MacDonald et al., 2006; Yashiro and Philpot, 2008). Several Rac-specific GEFs have been implicated in activity-dependent signaling pathways that regulate the formation and remodeling of spines (Fig. 4) (Saneyoshi et al., 2008; Tolias et al., 2005; Xie et al., 2007). In the case of Kalirin-7, NMDA receptor activation induces a CaMKII-dependent phosphorylation of Kalirin-7 on residue threonine 95, which was suggested to enhance its GEF activity in cultured neurons (Xie et al., 2007). Overexpression of a Kalirin-7 non-phosphorylatable T95A mutant blocked NMDA receptor-dependent spine enlargement but not spine formation, suggesting that Kalirin-7 phosphorylation is necessary for activity-dependent increases in spine area (Xie et al., 2007). Kalirin-7 also interacts with the GluR1 subunit of AMPA receptors, and RNAi knockdown of Kalirin-7 or deletion of all Kalirin isoforms prevented the NMDA receptor-induced spine enlargement and spine delivery of AMPA receptor typically observed in control cortical neuron cultures (Xie et al., 2010; Xie et al., 2007). Kalirin-7 knockout mice, however, have normal levels of GluR1 in purified PSDs, indicating that Kalirin-7 may not be essential for AMPA receptor localization in vivo (Ma et al., 2008).
In addition to acting downstream of EphB and NMDA receptors, Kalirin-7 also regulates spine morphogenesis induced by N-cadherin clustering (Fig. 4). N-cadherin is a trans-synaptic adhesion molecule that plays an important role in regulating the morphology of dendritic spines and the formation, function, and plasticity of synapses (Bozdagi et al., 2000; Bozdagi et al., 2004; Okamura et al., 2004; Takeichi, 2007; Tang et al., 1998; Togashi et al., 2002). N-cadherin interacts with Kalirin-7 via the scaffolding protein AF-6 (Xie et al., 2008). Stimulation of cortical neuronal cultures with clustered N-cadherin results in an increase in spine size, which was blocked by RNAi knockdown of all Kalirin isoforms (Xie et al., 2008). These results suggest that N-cadherin-mediated spine enlargement requires Kalirin.
As described in more detail below, Kalirin-7 is not the only Rac-GEF to localize to excitatory synapses and participate in EphB and NMDA receptor signaling pathways. Interestingly, however, its role is not compensated in the cortex by these other GEFs when Kalirin-7 function is lost. This observation suggests that Kalirin-7 plays a unique role at synapses in the cortex. One clue for determining Kalirin-7’s distinct function at synapses comes from examining its differential expression. In contrast to other Rac-GEFs that are highly expressed during development, Kalirin-7 expression in the cortex and hippocampus begins around postnatal day (P) 10–15 and is retained at high levels throughout adulthood (Penzes et al., 2008). Kalirin-7 may therefore play a more dominant role in spine maturation and maintenance rather than early spine formation. We propose that Kalirin-7 serves as a signal integrator that incorporates inputs from a variety of synaptic receptors late in development and induces Rac GTPase signaling pathways that favor spine maturation and remodeling. Its interaction with multiple PDZ domain-containing proteins may provide additional spatial specificity, possibly giving Kalirin-7-activated Rac access to a unique pool of actin within spines.
3.1.2. Tiam1
The Rac-specific GEF Tiam1 (T-lymphoma invasion and metastasis 1) also functions as a critical mediator of spine development (Tolias et al., 2005; Tolias et al., 2007; Zhang and Macara, 2006). Tiam1 is a large multi-domain protein that consists of a myristoylation site, two PEST sequences, an N-terminal PH domain flanked by a coiled-coiled and an extended region (PHn-CC-Ex), a Ras-binding domain (RBD), a PDZ domain, and the characteristic DH-PH Rac-GEF domain (Fig. 3). The ability of Tiam1 to induce Rac-dependent actin remodeling and cell shape change requires both its Rac-GEF enzymatic activity and its ability to translocate to the plasma membrane, which is mediated by its PHn-CC-Ex domain (Mertens et al., 2003). In Drosophila, the Tiam1 homolog Still life (SIF) localizes to synaptic terminals and plays a role in axonal extension and synaptic development (Sone et al., 1997; Sone et al., 2000). In mammals, Tiam1 is expressed at high levels in the developing brain, and its expression remains high in restricted adult brain regions including the hippocampus, the olfactory bulb and the cerebellum (Ehler et al., 1997). Over the years, Tiam1 has been implicated in several neurodevelopmental processes including neuronal migration, neurite outgrowth, and axon specification (Kawauchi et al., 2003; Kunda et al., 2001; Leeuwen et al., 1997). In addition, Tiam1 is present in dendrites and spines and localizes to the PSD (Tolias et al., 2005). RNAi knockdown of Tiam1 expression in primary hippocampal and cortical neurons results in significant reductions in dendritic arborization and spine and synapse density, suggesting that Tiam1 is necessary for proper dendritic growth as well as spine and synapse development (Tolias et al., 2005; Tolias et al., 2007; Zhang and Macara, 2006).
Like Kalirin-7, Tiam1 functions in a number of signaling pathways (Fig. 4). For instance, Tiam1 interacts with the NMDA receptor and is phosphorylated in a calcium-dependent manner following NMDA receptor stimulation, leading to Rac activation (Tolias et al., 2005). Calcium-dependent Tiam1 phosphorylation may be mediated by CaMKII, which can directly phosphorylate Tiam1 and enhance its GEF activity approximately 2-fold (Fleming et al., 1999). In control cortical neurons, stimulation of the NMDA receptor leads to an increase in spine density (Tolias et al., 2005). Notably, RNAi knockdown of Tiam1 prevents this increase in spine density, suggesting that Tiam1 is required for NMDA receptor-dependent spine formation (Tolias et al., 2005). Furthermore, reduced Tiam1 expression blocks the NMDA receptor-stimulated phosphorylation and activation of AKT, a serine/threonine kinase that controls cell growth and actin cytoskeletal remodeling (Rodgers and Theibert, 2002; Tolias et al., 2005). Together, these results suggest that Tiam1 helps link the NMDA receptor to activity-induced intracellular signaling pathways that regulate spine morphogenesis.
In addition to regulating NMDA receptor-induced spine development, Tiam1 mediates EphB receptor-dependent spine morphogenesis (Tolias et al., 2007). Via its PHn-CC-Ex domain, Tiam1 specifically interacts with the EphB2 tyrosine kinase receptor in a kinase-dependent manner (Tolias et al., 2007). Stimulation of EphB receptors with ephrinB induces the recruitment of Tiam1 to sites of new synaptic contacts and results in the phosphorylation of Tiam1 at tyrosine 829, promoting Rac1 activation (Miyamoto et al., 2006; Tolias et al., 2007). Importantly, disruption of Tiam1 function with RNAi knockdown or a dominant-negative Tiam1 mutant blocks ephrinB-induced spine formation in hippocampal neurons (Tolias et al., 2007). Taken together, these results suggest that EphB receptors regulate spine development in part by recruiting, phosphorylating and activating Tiam1, which leads to Rac-dependent actin remodeling required for spine formation.
Finally, Tiam1 has been shown to cooperate with the partition-defective (PAR) protein PAR-3 in regulating spine morphogenesis (Zhang and Macara, 2006). PAR-3 is a member of the evolutionary conserved PAR polarity complex, consisting of PAR-3, PAR-6, and atypical protein kinase C (aPKCζ). The PAR complex PAR-3 controls many aspects of cell polarity, including asymmetric cell division, directional cell migration, epithelial apical-basal polarity, axon specification and synaptogenesis (Arimura and Kaibuchi, 2007; Goldstein and Macara, 2007). Accumulating evidence indicates that Tiam1 and the closely related Tiam2/STEF are essential components of the PAR complex (Chen and Macara, 2005; Gerard et al., 2007; Mertens et al., 2005; Nishimura et al., 2005; Pegtel et al., 2007; Zhang and Macara, 2006). PAR-3 recruits Tiam1 or Tiam2 to the PAR complex, where they help to establish cell polarity by activating Rac and aPKCζ, resulting in polarized actin and microtubule reorganization (Mertens et al., 2006). In the regulation of hippocampal dendritic spine morphogenesis, PAR-3 has been suggested to spatially restrict Tiam1 to spines, thereby preventing inappropriate Rac activation elsewhere (Zhang and Macara, 2006).
Tiam1, like Kalirin-7, interacts with many proteins and participates in a number of signaling pathways. Yet, several aspects of Tiam1’s biology make it unique among synaptic Rac-GEFs. First, it interacts with the PAR complex, a crucial determinant of cell polarity. This interaction between Tiam1 and the PAR complex is thought to restrict Tiam1 activity in a polarized manner (Zhang and Macara, 2006), which could result in the modulation of a specific subpool of actin within the spine. Second, Tiam1 activates AKT, providing a link between synaptic receptors (NMDA receptor, EphB) and signaling pathways that control cell growth and cytoskeletal rearrangement (Tolias et al., 2005). Third, Tiam1 interacts with GTP-bound Ras through its Ras-binding domain, and may therefore function as a Ras effector that mediates Rac-dependent synaptic remodeling in response to Ras activation (Lambert et al., 2002; Malliri et al., 2002). Finally, in addition to its role in dendritic development, Tiam1 regulates axonal differentiation and growth, and may therefore also contribute to presynaptic development, as is the case for its drosophila homolog SIF (Kunda et al., 2001; Nishimura et al., 2005; Sone et al., 1997; Sone et al., 2000).
3.1.3. β–PIX (Arhgef7)
The Pak-interacting exchange factor β-PIX has also been identified as a regulator of spine morphogenesis and synapse formation (Parnas et al., 2001; Zhang et al., 2003; Zhang et al., 2005). β-PIX is a multidomain protein comprised of an SH3 domain, a DH-PH Rac/Cdc42 GEF domain, a GIT1-binding region, and a proline-rich region (Fig. 3) (Rosenberger and Kutsche, 2006). In the brain, two major β-PIX isoforms are expressed that differ only at their C-termini; β1-PIX possesses a coiled-coil dimerization domain and a PDZ-binding motif, whereas β2-PIX contains a serine-rich region (Koh et al., 2001). These two β-PIX isoforms are present at high levels during development, and they continue to be expressed in the adult in restricted brain regions such as the hippocampus and cerebellum (Kim et al., 2000). β-PIX was originally identified based on its ability to interact via its SH3 domain with the serine/threonine kinase Pak, a major downstream effector of Rac and Cdc42 (Bagrodia et al., 1998; Manser et al., 1998). By binding to Pak and activating Rac and/or Cdc42, β-PIX helps to coordinate Rac/Cdc42-dependent Pak activation, which promotes actin cytoskeletal remodeling (Manabe et al., 2002; Zhang et al., 2005). Interestingly, activated Pak can also phosphorylate β-PIX on residue threonine 526, increasing β-PIX’s membrane localization and enhancing its GEF activity (Shin et al., 2002). This result suggests that the β-PIX/Rac/Cdc42/Pak signaling cascade is regulated by a positive feedback loop.
In hippocampal neurons, β-PIX localizes to synapses and is targeted to the PSD through its interaction with the synaptic scaffolding protein GIT1 (G protein-coupled receptor kinase-interacting protein 1) (Collins et al., 2006; Zhang et al., 2003; Zhang et al., 2005). GIT1 is a ubiquitously expressed Arf-GAP that forms a signaling complex with β-PIX and Pak (Manabe et al., 2002). Disruption of GIT function results in β-PIX mislocalization and a decrease in spine and synapse density, suggesting that GIT1 regulates spine morphogenesis and synapse formation by recruiting β-PIX to synapses and restricting Rac activation (Zhang et al., 2003; Zhang et al., 2005). Localized Rac activation, in turn, induces Pak activation and Pak-mediated myosin light chain (MLC) phosphorylation, which are both required for proper spine formation (Zhang et al., 2005). By interacting with additional pre- and post-synaptic proteins including Shank, Piccolo and liprin-α, β-PIX and GIT1 appear to regulate active zone cytoskeletal matrix organization and AMPA receptor targeting (Kim et al., 2003; Ko et al., 2003; Park et al., 2003). Consistent with these findings, the Drosophila homolog of β-PIX, dPIX, regulates post-synaptic structure and protein localization at the Drosophila glutamatergic neuromuscular junctions (Parnas et al., 2001).
Like Kalirin-7 and Tiam1, β-PIX functions downstream of synaptic receptors (Fig. 4). For instance, NMDA receptor stimulation induces the activation of calmodulin-dependent kinase kinase (CaMKK) and CaMKI, which form a multiprotein complex with β-PIX and GIT1 in spines (Saneyoshi et al., 2008). CaMKI-mediated phosphorylation of serine 516 in β-PIX enhances its GEF activity, resulting in the activation of Rac. Blocking this pathway in the hippocampus with pharmacological inhibitors, dominant-negative constructs or RNAi reduces spine formation and miniature excitatory postsynaptic current (mEPSC) frequency, which can be rescued by constitutively active Pak (Saneyoshi et al., 2008). These results suggest a role for β-PIX in calcium-calmodulin signaling cascades that regulate spine morphogenesis. Furthermore, GIT1 was also recently implicated in spine morphogenesis induced by ephrinB reverse signaling (Segura et al., 2007). EphrinB activation results in the phosphorylation of GIT1 on tyrosine 392, creating a docking site for the adaptor protein Grb4, which binds to the tail of activated ephrinB. Disrupting the ephrinB-Grb4-GIT1 complex blocks reverse signaling and impairs normal spine morphogenesis and synapse formation (Saneyoshi et al., 2008).
Though it shares some similarities with Kalirin-7 and Tiam1, β-PIX has several unique features that may explain why these other Rac-GEFs are unable to fully compensate for its loss. Particularly striking is its interactions with Pak and GIT1 and its putative positive feedback loop. These features suggest both a temporally explosive all-or-none activation of its GEF activity and tight regulation of its signal in space.
3.1.4. Intersectin-L (Itsn1)
Intersectin is a multidomian scaffolding protein that is best known for its role in regulating endocytosis in non-neuronal cells and in synaptic vesicle recycling at the neuromuscular junction (NMJ) in Drosophila and C. elegans (Koh et al., 2004; Marie et al., 2004; Rose et al., 2007; Wang et al., 2008). Intersectin possesses two N-terminal Eps15 homology domains, several coiled-coil domains, and five SH3 domains (Yamabhai et al., 1998) (Fig. 3). In vertebrates, the Intersectin gene is subject to alternative splicing, resulting in the generation of a longer isoform (Intersectin-L) that is expressed exclusively in neurons (Pucharcos et al., 1999). In addition to the domains it shares with the shorter, ubiquitously expressed Intersectin-S isoform, Intersectin-L also contains a C-terminal C2 domain and a DH-PH domain with GEF activity specific for Cdc42 (Hussain et al., 2001; Snyder et al., 2002). Not only does Intersectin-L activate Cdc42, it also interacts with a number of actin regulatory proteins, suggesting a role in actin cytoskeletal regulation (Hussain et al., 2001). Indeed, Intersectin-L can induce filopodia formation when overexpressed in fibroblast cells (Hussain et al., 2001). By controlling both actin remodeling and the recruitment of endocytic proteins such as dynamin1 and synaptojanin1 (Koh et al., 2004; Marie et al., 2004), Intersectin-L may function to couple membrane trafficking with actin cytoskeletal remodeling.
Full length Intersectin-L exhibits little GEF activity and appears to be maintained in an auto-inhibited conformation by an intramolecular interaction between its SH3 and DH domains, which blocks Cdc42 binding (Zamanian and Kelly, 2003). N-WASP (neuronal Wiskott-Aldrich syndrome protein), a critical regulator of Arp2/3-mediated actin polymerization, interacts directly with the SH3 domains of Intersectin-L and relieves this autoinhibition, resulting in Cdc42 activation (Hussain et al., 2001). The EphB2 receptor also interacts with Intersectin-L and stimulates its GEF activity in cooperation with N-WASP (Irie and Yamaguchi, 2002). Cdc42 activation by Intersectin-L then leads to N-WASP activation and the stimulation of actin polymerization (Hussain et al., 2001). Importantly, in hippocampal neurons Intersectin-L co-localizes with dendritic spine F-actin, and RNAi knockdown of Intersectin-L or disruption of the interaction between Intersectin-L and N-WASP perturbs spine development, resulting in an increase in filopodia-like protrusions and a decrease in mushroom-shaped spines (Irie and Yamaguchi, 2002; Nishimura et al., 2006; Thomas et al., 2009). Taken together, these results suggest that Intersectin-L plays an important role in spine maturation by activating Cdc42 and N-WASP, leading to actin cytoskeletal remodeling and possibly membrane trafficking (Fig. 4). Though it is likely that other Cdc42 GEFs participate in synaptogenesis, L-intersectin’s potential coupling of actin dynamics and membrane trafficking and its distinctive relationship with N-WASP provide important clues to its unique role at synapses. Given the plethora of protein interaction domains that L-intersectin contains, identification of additional binding partners will be required to elucidate its particular function.
3.2 RhoA-GEFs at excitatory synapses
3.2.1. Lfc (GEF-H1, Arhgef2)
Rho-specific GEFs such as Lfc have also been implicated in the formation and structural remodeling of synapses (Kang et al., 2009; Ryan et al., 2005). Lfc is a Rho-GEF that is highly expressed in the brain (Ryan et al., 2005). Its domain structure includes a C1 domain, a DH-PH domain and a coiled-coil region (Fig 3). Under basal conditions, Lfc is mainly restricted to the dendritic shafts in cultured hippocampal neurons, likely through its association with microtubules (Glaven et al., 1999; Ren et al., 1998; Ryan et al., 2005). NMDA receptor stimulation results in the rapid translocation of Lfc into dendritic spines (Muly et al., 2008; Ryan et al., 2005). In spines, Lfc is present in the PSD (Collins et al., 2006; Kang et al., 2009), where it interacts with AMPA receptors (Kang et al., 2009) as well as with Spinophilin and Neurabin, two homologous proteins that associate with F-actin (Fig. 5) (Ryan et al., 2005). The recruitment of Lfc into spines likely results in RhoA activation, leading to actin cytoskeletal remodeling and alterations in spine morphology. Indeed, overexpression of Lfc in cultured hippocampal neurons was shown to reduce spine length and size (Ryan et al., 2005), whereas blocking Lfc function with dominant-negative mutants or RNAi results in an increase in spine size and density (Kang et al., 2009; Ryan et al., 2005). Furthermore, AMPA receptor-dependent spine remodeling appears to require Lfc (Kang et al., 2009). Treatment of hippocampal neurons with an AMPA receptor antagonist normally causes a decrease in spine density as well as an increase in RhoA activation. These effects were ablated in neurons in which Lfc function was blocked (Kang et al., 2009). Together, these results indicate that Lfc plays a critical role in RhoA-mediated spine retraction. It is interesting to speculate that Lfc is normally restricted to dendritic shafts to allow for new spine growth during early synapse development. Furthermore, following NMDA receptor activation, Lfc-mediated RhoA activation may be intentionally time-delayed from the onset of an activity-induced signal due to the need of Lfc to translocate from the dendritic shaft into spines. In spines, Lfc’s specificity would be further enhanced by its interaction with AMPA receptors and specific F-actin binding proteins.
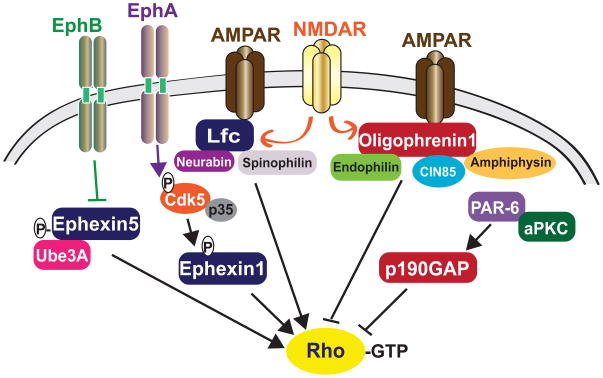
Figure 5. Regulation of RhoA signaling at synapses.
The RhoA-GEFs Lfc, Ephexin1 and Ephexin5 activate RhoA activity, whereas the RhoA-GAPs Oligophrenin1 and p190 RhoGAP inhibit RhoA activity. These regulatory proteins are controlled by upstream receptors, such as NMDA receptors, AMPA receptors, and Eph receptors. As is the case for Rac1/Cdc42 GEFs and GAPs, RhoA regulatory proteins likely achieve their specific functions through a unique set of interactions with downstream effectors and other proteins. GEFs are in blue and GAPs are in red.
3.2.2. Ephexin1/5 (Ngef/Vsm-RhoGEF)
Ephexins are another family of Rho-GEFs implicated in synapse regulation (Frank et al., 2009; Fu et al., 2007; Margolis et al.; Shi et al., 2010). The Ephexin family consists of five members (Ephexin1 through Ephexin5), each encoded by a different gene. Ephexin family members share the same overall structure, including a single DH-PH domain and a C-terminal SH3 domain, but possess unique N-terminal regions (Fig. 3) (Sahin et al., 2005). Of the five family members, only Ephexin1 and Ephexin5 are highly expressed in the developing nervous system (Sahin et al., 2005). Ephexin1 was originally identified based on its ability to interact with the cytoplasmic domain of the EphA4 receptor (Shamah et al., 2001). When overexpressed in cells, Ephexin1 can activate RhoA, Cdc42 and Rac, but its GEF activity towards RhoA appears to be specifically enhanced following EphA4-induced tyrosine phosphorylation (Sahin et al., 2005; Shamah et al., 2001). Ephexin1 is best known for its role in mediating ephrinA-stimulated growth cone collapse (Sahin et al., 2005; Shamah et al., 2001). Recently, however, Ephexin1 has also been implicated in regulating different aspects of synapse development and function. For instance, EphA4-mediated spine retraction in the hippocampus has been shown to require both Ephexin1 and the serine/threonine kinase Cdk5 (Fu et al., 2007). EphA4 receptor activation causes the recruitment of Cdk5 to EphA4, resulting in Cdk5 phosphorylation and activation (Fu et al., 2007). Activated EphA4 and Cdk5 then induce the recruitment and phosphorylation of Ephexin1, leading to RhoA activation and reduced spine density (Fig 5) (Fu et al., 2007). In addition to mediating EphA4-induced spine retraction, Ephexin1 also plays an essential role in regulating presynaptic homeostatic signaling in Drosophila (Frank et al., 2009) and the structural maturation and neurotransmission of NMJs in mice (Shi et al., 2010), which will be discussed in more detail below.
Recently, Ephexin5 was also identified as a negative regulator of excitatory synapse development (Margolis et al., 2010). RNAi knockdown or genetic ablation of Ephexin5 increases dendritic spine and synapse density in hippocampal neurons, whereas overexpression of Ephexin5 decreases excitatory synapse number by selectively activating RhoA (Margolis et al., 2010). Ephexin5 preferentially binds to EphB2, and activation of EphB2 triggers the phosphorylation, ubiquitination, and degradation of Ephexin5, resulting in EphB-dependent excitatory synapse development (Margolis et al., 2010). The degradation of Ephexin5 is mediated by the ubiquitin ligase Ube3A (Margolis et al., 2010), which is mutated in the human cognitive disorder Angelman syndrome and is duplicated in some forms of Autism Spectrum Disorders (ASDs) (Dan, 2009). These findings suggest that Ephexin5 restricts synapse formation by activating RhoA, and that this suppression is relieved by EphB receptor activation during synapse development (Fig. 5). Disruption of EphB-mediated Ephexin5 degradation in the absence of Ube3A function may contribute to the cognitive and synaptic defects associated with Angelman syndrome.
4. Rho-family GEF involved in inhibitory synapse development
Although much of the research in this field has been directed toward the formation of excitatory synapses, Rho GTPase regulatory proteins also function in the formation of other types of synapses. In this section, we will summarize briefly the specific functions of a Rho-family GEF known to participate in the formation of inhibitory synapses.
4.1. Collybistin (Arhgef9)
Collybistin, encoded by rodent ARHGEF9, is by far the best-characterized Rho-family GEF involved in the formation of inhibitory synapses. Collybistin contains an N-terminal SH3 domain, a single DH-PH domain with Cdc42 GEF activity, and a C-terminal coiled-coil domain (Fig. 3). Collybistin knockout mice exhibit a striking loss of inhibitory synapses, both morphologically and functionally (Jedlicka et al., 2009; Papadopoulos et al., 2008; Papadopoulos et al., 2007). These defects are accompanied by increased anxiety and reduced spatial learning (Papadopoulos et al., 2007). Collybistin promotes the formation of inhibitory synapses by clustering gephryn, a protein scaffold that, in turn, clusters glycine and GABA receptors (Harvey et al., 2004; Kins et al., 2000). Collybistin expression is upregulated between embryonic day (E) 7–11, when the postmitotic neurons in which it is primarily expressed begin to arise (Kneussel et al., 2001). In addition to its critical interaction with gephyrin, collybistin’s N-terminal domain interacts with neuroligin-2 (Poulopoulos et al., 2009). This interaction, without which full length collybistin cannot cluster gephyrin, is crucial for the formation of inhibitory synapses in vivo (Poulopoulos et al., 2009).
Interestingly, a question remains as to whether or not collybistin functions as a GEF in the formation of inhibitory synapses. The human homologue of collybistin, h-PEM2, functions in vitro as a GEF for Cdc42, but not for Rac1 or RhoA (Reid et al., 1999). Using a number of collybistin point mutants that are unable to activate Cdc42, Reddy-Alla et al. (Reddy-Alla et al., 2010) showed that collybistin’s Cdc42 GEF activity and, indeed, Cdc42, are dispensable for clustering gephyrin, and, presumably, for inhibitory synapse formation. These results suggest the possibility that collybistin’s GEF activity towards Cdc42 is not required for inhibitory synapse formation. However, it remains to be seen whether the synapses that form with mutant collybistin or in the absence of Cdc42 are functional. It is also not clear whether collybistin activates other Rho-family GTPases, and, if so, whether or not the collybistin mutants used by Reddy-Alla et al. are inactive toward these GTPases. Further investigation will clarify these issues. In sum, collybistin occupies a pivotal location in the protein interactome linking neuroligin-2, which can detect juxtacellular signals, to gephyrin, the scaffold on which inhibitory synapses are built. This location implies a role for collybistin in linking signals to inhibitory synapse formation, though it is presently unclear whether its GEF function is required.
5. Rho-family GEFs involved in neuromuscular junction development
In this section, we will discuss two Rho-family GEFs that play a role in NMJ development. As above, we will consider the regulation and connectivity of these proteins in order to propose a specific niche for each.
5.1. Trio
Although the Rho-family GEF Trio is best known for its role in axon guidance (Bateman and Van Vactor, 2001), recent studies of the Trio homolog UNC-73 in C. elegans have indicated a potential role in synaptogenesis as well. Trio and UNC-73 are large proteins closely related to Kalirin that possess numerous domains including a Sec14p domain, multiple spectrin-like repeats, an SH3 domain, and two DH-PH Rho GEF domains (RhoGEF-1 specific for Rac1/RhoG and RhoGEF-2 specific for RhoA) (Fig. 3). Full length mammalian Trio, containing an additional SH3 domain, an Ig/fibronectin-like domain (Ig/FN) and a kinase domain, is expressed in skeletal muscle, whereas shorter forms of Trio lacking these domains are enriched in the brain (McPherson et al., 2005). Mice lacking Trio exhibit skeletal muscle defects and aberrant organization of several brain regions (O’Brien et al., 2000). In Drosophila and C. elegans, Trio/UNC-73 functions in axon guidance by activating Rac (via its RhoGEF-1 domain), resulting in actin cytoskeletal reorganization necessary for growth cone guidance and outgrowth (Awasaki et al., 2000; Bateman et al., 2000; Forsthoefel et al., 2005; Newsome et al., 2000; Steven et al., 1998; Vanderzalm et al., 2009). Interestingly, mutations restricted to RhoGEF-2 domain of UNC-73 cause early stage lethality and musculature and neurotramission defects in the absence of axon guidance defects, suggesting additional non-guidance functions for UNC-73 that require the RhoGEF-2 domain (Steven et al., 2005). UNC-73 was also independently shown to be necessary for proper extension of the muscle arm, a step involved in the formation of the NMJ (Alexander et al., 2009). Again, UNC-73’s GEF activity was required for this function, though this study implicated UNC-73’s RhoGEF-1 domain (Alexander et al., 2009). Taken together, these data suggest that UNC-73/Trio functions as a GEF in NMJ formation, with the RhoGEF-1 domain being of primary importance. UNC-73 also signals through its RhoGEF-2 domain to regulate musculature and synaptic neurotransmission (Steven et al., 2005), though the relevance of this to NJM formation is not known.
Defining a specific role for UNC-73/Trio in NMJ formation is not yet possible. Though full length mammalian Trio is present in muscle, UNC-73 lacks an equivalent serine/threonine kinase-domain, suggesting that this activity is not required for the functions described here. It is known that UNC-73 localizes to the ends of the muscle arm, a postsynaptic intermediate in NMJ formation (Alexander et al., 2009). There it colocalizes with the transmembrane receptor UNC-40/Dcc, which functions upstream of UNC-73 in NMJ formation (Alexander et al., 2009). UNC-73/Trio also regulates diaphanous-related formins presynaptically in NMJ formation (Pawson et al., 2008). However, the signals that control UNC-73/Trio activity in NMJ formation are not known, nor are whether its actions are due to its ability to activate Rac, RhoG (Estrach et al., 2002), neither, or both. The actin-regulatory WAVE complex functions with UNC-40/73 in NMJ formation, suggesting a role for Rac in this process (Alexander et al., 2009). Axon guidance studies have identified addition UNC-73/Trio-interacting proteins that may also function in NMJ formation, including CRML-1 (Vanderzalm et al., 2009), an inhibitor of Trio’s GEF activity, and the Drosophila netrin receptor Frazzled (Forsthoefel et al., 2005).
5.2. Ephexin1 (Ngef)
In addition to regulating hippocampal excitatory synapse elimination (described above), Ephexin1 participates in the maturation of NMJs. Ephexin-1 knockout mice have weak forelimbs and perform poorly in the rotarod test (Shi et al., 2010). NMJ transmission is decreased in these animals, and severe morphological defects in NMJ are apparent, though contacts between nerve and muscle do form (Shi et al., 2010). Postsynaptic (muscular) clusters of Acetylcholine (Ach) receptors normally undergo a marked rearrangement from an oval morphology to a more complex “pretzel-like” morphology after development of the NMJ. This fails to occur in the Ephexin-1 knockout animals (Shi et al., 2010). Addition of functional Ephexin-1 or constitutively active RhoA allows this transformation to proceed and NMJ development to reach completion (Shi et al., 2010), suggesting a role for Ephexin-1-mediated RhoA activation in controlling the distribution of Ach receptors in developing muscle.
While it is not yet demonstrated what the signal upstream of Ephexin-1 in NMJ maturation is, Shi et al. (Shi et al., 2010) speculate that it comes from EphA4. As in growth cone collapse and spine elimination, phosphorylation of ephexin-1 on tyrosine-87 is required for its activation (Fu et al., 2007; Shi et al., 2010). This result suggests that Ephexin-1 is a multifunctional GEF that ties ephrin-A/EphA signals to a variety of neuronal phenomena through RhoA-mediated cytoskeletal changes. It will be interesting to determine whether Cdk5 is also a common theme in Ephexin-1 signaling and whether Ephexin-1 has other partners whose identity can help us to better understand its precise role in these differing phenomena.
6. Rho-family GAPs involved in excitatory synaptogenesis
Less is known about the role of Rho GTPase GAPs in synaptogenesis, since historically they have been viewed as signal terminators with a secondary role in comparison with the Rho GTPase GEFs, which activate Rho GTPases in response to diverse extracellular stimuli (Tcherkezian and Lamarche-Vane, 2007). It is becoming increasingly clear, however, that GAPs are also highly regulated proteins that play essential roles in controlling specific Rho GTPase-mediated cellular processes such as synaptogenesis. Here we will discuss the function and regulatory mechanisms of several Rho GTPase GAPs that have recently been implicated in synapse development, again highlighting the specific roles that they play.
6.1. Rac/Cdc42-GAPs at excitatory synapses
6.1.1. α 1-chimaerin
The diacyglyerol (DAG)-binding protein α1-chimaerin was one of the first Rho-family GAPs implicated in regulating dendritic spine morphogenesis (Buttery et al., 2006; Van de Ven et al., 2005). α1-chimaerin belongs to the chimaerin family of Rho GAPs that also includes α2-, β1- and β2-chimaerin. These chimaerin isoforms are generated as alternatively spliced products from the α- and β-chimaerin genes. All four chimaerin proteins possess a C1 phorbol ester- and DAG-binding domain and a Rho GAP domain that specifically inhibits Rac (Ahmed et al., 1993; Caloca et al., 2003; Diekmann et al., 1991). α2- and β2-chimaerin also have an additional N-terminal SH2 (Src Homology 2) domain capable of binding to phosphotyrosine-containing residues (Leung et al., 1994) (Fig. 3). Unlike the other chimaerin isoforms, α1-chimaerin is exclusively expressed in the brain, and its expression is upregulated during synaptogenesis in response to neuronal activity (Buttery et al., 2006; Diaz et al., 2002; Lim et al., 1992).
In hippocampal neurons, α1-chimaerin is present in dendrites and spines (Van de Ven et al., 2005). Overexpression of α1-chimaerin causes loss of spines and a simplification of the dendritic arbor that requires both the DAG-binding site and the Rac-GAP activity of α1-chimaerin (Buttery et al., 2006; Van de Ven et al., 2005). In contrast, overexpression of α2-chimaerin induces process outgrowth rather than dendritic pruning, suggesting a differential role for the SH2 domain in α2-chimaerin (Buttery et al., 2006). To further investigate the role of α1-chimaerin in regulating dendritic morphogenesis, two independent groups performed RNAi experiments in hippocampal neurons, with slightly different results. Van de Ven et al. (Van de Ven et al., 2005) reported that α1-chimaerin suppression increases spine density, whereas Buttery et al. (Buttery et al., 2006) demonstrated that α1-chimaerin silencing leads to excess outgrowth from spine heads and dendrites, resulting in a greater number of atypical spines and filopodia-like protrusions. Taken together, these results suggest that -chimaerin plays an important role in restricting dendritic protrusions in hippocampal neurons by inhibiting Rac signaling.
One of the distinguishing features of chimaerin proteins is their C1 domain, which binds to phorbol esters and DAG and regulates the recruitment of chimaerins to the plasma membrane (Ahmed et al., 1990; Caloca et al., 2001). In hippocampal neurons, stimulation of phospholipase Cβ (PLCβ)-coupled cell surface receptors that induce DAG production results in the rapid translocation of α1-chimaerin to the plasma membrane (Buttery et al., 2006). The ability of α1-chimaerin to bind DAG is not only necessary for this translocation but also for the pruning activity of α1-chimaerin (Buttery et al., 2006). α1-chimaerin also binds to the NR2A subunit of NMDA receptor in a phorbol ester-dependent manner, and this interaction is similarly required for α1-chimaerin’s ability to regulate spine density (Van de Ven et al., 2005). DAG signaling may therefore initiate α1-chimaerin recruitment to synaptic NMDA receptors, resulting in local inactivation of Rac1 and the pruning of dendritic protrusions (Fig. 4). Since α1-chimaerin continues to be expressed in adult neurons (Lim et al., 1992), it is possible that α1-chimaerin also contributes to the ongoing remodeling of synapses in response to environmental stimuli and synaptic activity, but verification of this possibility awaits further investigation.
6.1.2. Bcr/Abr
Recently, another family of Rac-GAPs consisting of Bcr (Breakpoint cluster region) and Abr (active Bcr-related) has been implicated in excitatory synapse regulation in the hippocampus (Oh et al., 2010). Bcr is best known for its involvement in Philadelphia chromosome-positive chronic myelogenous leukemia (Groffen and Heisterkamp, 1997). However, Bcr and Abr are highly expressed in the CNS (Fioretos et al., 1995; Tan et al., 1993), and mutant mice lacking both proteins exhibit cerebellar developmental defects, indicating a role for these proteins in nervous system development (Kaartinen et al., 2001). Bcr and Abr both contain a Rho GAP domain (Fig. 3) and function as potent inhibitors of Rac in vitro and in vivo (Cho et al., 2007; Chuang et al., 1995; Diekmann et al., 1991; Kaartinen et al., 2001; Tan et al., 1993; Voncken et al., 1995). In addition to their GAP domains, Bcr and Abr possess a number of additional signaling domains including a C2 domain, a PDZ-binding motif, and a DH-PH GEF domain that modestly activates Cdc42 and RhoA (Korus et al., 2002; Malmberg et al., 2004; Oh et al., 2010; Radziwill et al., 2003; Rizo and Sudhof, 1998; Sahay et al., 2008). Bcr also contains an N-terminal coiled-coil oligomerization domain (McWhirter et al., 1993) and a serine/threonine protein kinase domain that phosphorylates the adaptor proteins AF-6 and 14-3-3 (Fig. 3) (Li and Smithgall, 1996; Maru and Witte, 1991; Radziwill et al., 2003).
Despite the fact that Bcr and Abr are abundantly expressed in the brain, relatively little is known about their function in neurons. In a recent report, Bcr and Abr were shown to localize to excitatory synapses and directly interact with PSD-95 (Oh et al., 2010). Mice lacking Bcr or Abr exhibit a decrease in the maintenance, but not the induction, of LTP, and display impaired spatial learning and object recognition (Oh et al., 2010). Bcr and Abr knockout mice also show a small increase in hippocampal neuron spine density compared to wild-type mice, but no difference in synaptic transmission (Oh et al., 2010). These results indicate that Bcr and Abr play important roles in regulating synaptic plasticity and learning and memory, and suggest that excessive Rac activity hinders synaptic and cognitive function. Since Bcr and Abr have been demonstrated to compensate for each other’s functions in vivo (Cho et al., 2007; Kaartinen et al., 2001; Kaartinen et al., 2002), it will be interesting to determine if mice lacking both proteins have more significant defects in synapse development or function.
6.2. RhoA-GAPs at excitatory synapses
6.2.1. Oligophrenin-1
The Rho-GAP Oligophrenin-1 has also emerged as an important regulator of synapse development (Govek et al., 2004; Khelfaoui et al., 2007; Khelfaoui et al., 2009; Nadif Kasri et al., 2009). Oligophrenin-1 is encoded by OPHN1, which was first identified as an X-linked mental retardation gene (Billuart et al., 1998). It is abundantly expressed in the nervous system during development and at later stages in highly plastic brain regions, such as the olfactory bulb and hippocampus (Fauchereau et al., 2003). Oligophrenin-1 possesses several domains including an N-terminal membrane deforming Bin/Amphiphysin/Rvs (BAR) domain, a lipid binding PH domain, a Rho GAP domain that negatively regulates RhoA, Rac1 and Cdc42, and three proline-rich regions at its C-terminus (Billuart et al., 1998; Fauchereau et al., 2003; Govek et al., 2004; Khelfaoui et al., 2009) (Fig. 3).
Oligophrenin-1 is localized at pre- and postsynaptic sites in hippocampal neurons and has been implicated in regulating spine morphogenesis (Govek et al., 2004). Knocking down Oligophrenin-1 in CA1 neurons from rat hippocampal slices significantly reduces the length of spines (Govek et al., 2004). This phenotype was largely rescued by inhibiting Rho-kinase activity using the pharmacological inhibitor Y-27632, suggesting that the RhoA/Rho-kinase signaling pathway mediates the effect of Oligophrenin-1 knockdown on spine length. These results indicate that Oligophrenin-1 normally maintains spine length by repressing the RhoA/Rho-kinase pathway. Suppression of Oligophrenin-1 expression would therefore relieve RhoA inhibition, resulting in Rho-kinase activation and actin cytoskeletal remodeling that promotes spine shortening. Consistent with this idea, Oligophrenin-1 mutant mice display spine abnormalities and altered pre-synaptic function as well as behavioral, social, and cognitive impairments (Khelfaoui et al., 2007).
Recently, Nadif Kasri et al. (Nadif Kasri et al., 2009) demonstrated that overexpression of Oligophrenin-1 in hippocampal neurons selectively enhances AMPA receptor-mediated synaptic transmission and increases spine size (Nadif Kasri et al., 2009). Conversely, they showed that reduction of Oligophrenin-1 expression inhibits AMPA receptor- and NMDA receptor-mediated currents and hinders synaptic maturation, LTP and structural plasticity (Nadif Kasri et al., 2009). These defects were rescued by reintroduction of wild-type Oligophrenin but not a mutant that lacks Rho-GAP activity, suggesting that Oligophrenin-1 controls AMPA receptor-mediated transmission and spine structure by repressing RhoA activity (Nadif Kasri et al., 2009). Interestingly, Oligophrenin-1 was also shown to translocate into spines in response to neuronal activity and associate with AMPA receptor complexes (Nadif Kasri et al., 2009). Using a peptide derived from the C-terminus of the AMPA receptor GluR2 subunit that blocks AMPA receptor endocytosis (Ahmadian et al., 2004), Nadif Kasri et al. (Nadif Kasri et al., 2009) demonstrated that Oligophrenin-1 regulates synaptic function and structure by stabilizing synaptic AMPA receptors.
Khelfaoui et al. (Khelfaoui et al., 2009) have also recently provided evidence indicating a role for Oligophrenin-1 in regulating AMPA receptor trafficking in the hippocampus. They showed that Oligophrenin-1 is recruited to endocytic sites by interacting with three SH3 domain-containing adaptor proteins involved in clathrin-mediated endocytosis: amphiphysins, endophilins and CIN85 (Khelfaoui et al., 2009). Furthermore, disruption of OPHN1 in mice was shown to reduce endocytosis of synaptic vesicles at presynaptic sites and AMPA receptor internalization at postsynaptic sites, resulting in a significant impairment in NMDA receptor-dependent LTD (Khelfaoui et al., 2009). Importantly, pharmacological inhibition of the RhoA/Rho-kinase pathway fully rescued the endocytosis and LTD defects caused by loss of Oligophrenin-1, indicating that Oligophrenin-1 normally regulates these processes by repressing RhoA/Rho-kinase signaling. Additionally, Nakano-Kobaya et al. (Nakano-Kobayashi et al., 2009) demonstrated that Oligophrenin-1 controls synaptic vesicle cycling by forming a complex with the endocytic regulatory protein endophilin A1. Taken together, these results indicate that Oligophrenin-1 plays a critical role in regulating excitatory synapse morphogenesis and function by inhibiting RhoA-dependent signaling pathways that control both actin cytoskeletal remodeling and membrane trafficking at synapses.
6.2.2. p190 RhoGAP
p190 RhoGAP was first identified as a p120 RasGAP-interacting protein in Src-transformed cells (Ellis et al., 1990). p190 RhoGAP contains an N-terminal GTP-binding domain, four consecutive FF protein-interaction domains, several proline-rich regions, and a C-terminal GAP domain with specific activity towards RhoA (Ridley et al., 1993) (Fig. 3). p190 RhoGAP is a major substrate of the tyrosine kinase Src in the brain, and phosphorylation of p190 by Src enhances its interaction with p120 RasGAP and inhibits its RhoA-GAP and GTP-binding activities (Roof et al., 2000). p190 RhoGAP is highly expressed in the developing nervous system, and mice lacking functional p190 RhoGAP exhibit neural developmental defects including abnormalities in forebrain hemisphere fusion and neural tube closure (Brouns et al., 2000). p190 RhoGAP mutant mice also display aberrations in axon outgrowth, guidance and fasciculation, suggesting a role for p190 RhoGAP in axonal development (Brouns et al., 2001). This role appears to be conserved in Drosophila, since RNAi knockdown of p190 RhoGAP expression in Drosophila causes retraction of axonal branches by up-regulating RhoA signaling, resulting in actin/myosin contractility (Billuart et al., 2001).
In addition to regulating axonal development, p190 RhoGAP localizes to spines and promotes hippocampal neuron dendritic spine maturation and synapse and dendrite stability by inhibiting RhoA activity during late postnatal development (Sfakianos et al., 2007; Zhang and Macara, 2008). The function of p190 RhoGAP at synapses is regulated by the tyrosine kinase Arg, which phosphorylates p190 RhoGAP and promotes its binding to p120 RasGAP, resulting in p190 membrane recruitment and RhoA inactivation (Bradley et al., 2006; Sfakianos et al., 2007). Mice lacking Arg display reduced p190RhoGAP phosphorylation, increased RhoA activity, progressive loss of synapses and dendritic branches, and deficits in a hippocampal-dependent novel object recognition task (Sfakianos et al., 2007). p190 RhoGAP mutations enhance the effects of arg mutations on dendritic arborization, whereas mutations in the Rho-kinase ROCKII suppress the dendritic regression phenotype observed in the Arg knockout mice (Sfakianos et al., 2007). These results indicate that Arg and p190 RhoGAP function together in hippocampal neurons during synaptic refinement to promote spine maturation and synapse and dendrite stability by inhibiting RhoA activity.
p190 RhoGAP has also been implicated in regulating spine morphogenesis in cooperation with members of the PAR-3/PAR-6/aPKC polarity complex in the hippocampus (Zhang and Macara, 2008) (Fig. 5). As mentioned previously, the polarity protein PAR-3 controls spine development by spatially restricting the Rac-GEF Tiam1 to dendritic spines, resulting in local Rac1 activation (Zhang and Macara, 2006). Surprisingly, this ability of PAR-3 to regulate spine morphogenesis does not appear to require the other members of the PAR complex (Zhang and Macara, 2006). Instead, PAR-6 and aPKC were shown to regulate spine formation and maintenance by inhibiting RhoA activity via p190 RhoGAP (Zhang and Macara, 2008). Specifically, a dominant-negative RhoA mutant and a Rho-kinase inhibitor were demonstrated to rescue the loss of spine formation caused by silencing PAR-6 expression, whereas a GAP-deficient p190 mutant inhibited the increase in spine density caused by PAR-6 overexpression (Zhang and Macara, 2008). RNAi knockdown of p190 RhoGAP was also shown to block PAR-6-induced RhoA inactivation, suggesting that PAR-6 inhibits RhoA activity through p190 RhoGAP. The mechanism by which PAR-6/aPKC regulates p190 RhoGAP activity remain to be determined, although it is possible that aPKC activates p190 by direct phosphorylation (Brouns et al., 2000). Taken together, these results indicate that PAR-3 and PAR-6/aPKC regulate hippocampal neuron spine morphogenesis by controlling different aspects of Rho GTPase signaling; PAR-3 promotes spine formation by inducing local Rac activation, whereas PAR-6/aPKC controls spine development by suppressing RhoA activity.
In addition to regulating synaptic structure, 190 RhoGAP has been implicated in fear memory formation in the lateral amygdala (LA) through its interaction with a Grb2-mediated molecular complex (Lamprecht et al., 2002). Grb2 is an SH2/SH3 domain-containing adaptor protein that forms multi-protein complexes with other signaling molecules, resulting in the propagation of intracellular signals (Buday, 1999). Following fear conditioning, Grb2 forms a complex with the tyrosine phosphorylated proteins p190 RhoGAP, RasGAP and Shc in the LA (Lamprecht et al., 2002). Since tyrosine phosphorylation of p190 RhoGAP by Src inhibits its GAP activity towards RhoA (Roof et al., 2000), the appearance of tyrosine phosphorylated p190 RhoGAP following fear conditioning suggests that RhoA activation may be involved in fear conditioning. Indeed, inhibition of the Rho/ROCK pathway by microinjection of the ROCK inhibitor Y-27632 into the LA impairs long-term but not short-term fear conditioning (Lamprecht et al., 2002). Given that p190 RhoGAP regulates spine morphogenesis, these data provide a possible link between structural remodeling of synapses mediated by Rho GTPase signaling and long-term memory formation. As is the case for the other GAPs that we have examined, not enough is known about p190 RhoGAP to fully appreciate the nature of its unique role in synaptogenesis, though its many protein-interaction domains and association with Src, Arg and PAR6 provide ample clues.
7. Concluding remarks
As key regulators of cytoskeletal dynamics, Rho GTPases control many different aspects of nervous system development, including neuronal migration, axon growth and guidance, dendrite arborization, synapse formation, and spine morphogenesis (Govek et al., 2005). How do Rho GTPases regulate such diverse cellular processes? Rho GTPases require both precise spatio-temporal regulation of their activities and the activation of particular downstream pathways in order to dynamically control these distinct processes in response to extracellular cues (Pertz, 2010). Rho-family GEFs and GAPs likely provide this signaling specificity. As described in this review, Rho GTPase regulatory proteins typically contain an assortment of functional domains in addition to their GEF or GAP domains. These signaling domains enable Rho GEFs and GAPs to precisely control Rho GTPase activity in space and time and couple it to specific upstream receptors and downstream effector molecules by acting as scaffolding proteins (Moon and Zheng, 2003; Rossman et al., 2005; Schmidt and Hall, 2002). Localized Rho GTPase signaling then induces specific actin and microtubule remodeling that is required for the different neurodevelopmental processes.
In the last ten years, a number of Rho-family GEFs and GAPs have been identified that play critical roles in different aspects of synapse development and plasticity (Kiraly et al., 2010). In general, Rac- and Cdc42-GEFs promote the formation and/or maturation of synapses and spines, whereas Rho-GEFs induce spine shrinkage and synapse elimination. Likewise, Rac-GAPs restrict dendritic protrusions and synapse formation, whereas Rho-GAPs prevent spine/synapse retraction. Since Rho-family GEFs or GAPs with a common GTPase target appear to act in a similar manner at synapses, it is surprising that multiple Rho regulatory proteins have been found to function in the same signaling pathway. For example, the Rac-GEFs Kalirin-7 and Tiam1 have both been shown to regulate synapse development downstream of EphB and NMDA receptors (Penzes et al., 2003; Tolias et al., 2005; Tolias et al., 2007; Xie et al., 2007). Why would more than one Rac-GEF be required to mediate the effects of a particular receptor? It is possible that Kalirin-7 and Tiam1 function at different times during development, in distinct brain regions, and/or in different types of neurons or synapses. Alternatively, since Kalirin-7 and Tiam1 associate with distinct multi-protein complexes via their unique protein-interaction domains, they may have non-overlapping roles in regulating different aspects of Rac signaling at the same synapse. The specific roles of the individual Rho-family regulatory proteins and the mechanistic details of how they act in concert to regulate synapse development is currently unclear, and would benefit from further comparative analysis.
To achieve precise spatio-temporal control of Rho GTPase signaling that is required for proper synapse development and plasticity, the actions of Rho-family GEFs and GAPs likely need to be coordinately regulated. This possibility has been demonstrated by the finding that members of the PAR polarity complex regulate spine morphogenesis by modulating the activities of a Rac-GEF and a Rho-GAP. The polarity protein PAR-3 spatially restricts Tiam1 and induces local Rac activation at spines (Zhang and Macara, 2006), whereas PAR-6 induces p190 RhoGAP-mediated RhoA inhibition (Zhang and Macara, 2008). Since Rac and RhoA have opposite effects on spine morphogenesis, by enhancing Rac activity and suppressing RhoA activity, members of the PAR polarity complex appear to tilt the balance between Rac and RhoA towards Rac-dependent spine formation. It is also possible that Rho-family GEFs and GAPs with specificity for the same Rho GTPase function together to precisely regulate Rho GTPase activity in space and time. Further investigation is required to understand how Rho regulatory proteins act in concert to direct synapse development and plasticity.
In this review, we have focused our attention on Rac-, RhoA- and Cdc42-specific regulatory proteins that play critical roles in synapse formation and remodeling. While regulation of Rac, RhoA and Cdc42 signaling is clearly important for proper synapse development, it is unlikely that these GTPases are the only Rho family members functioning at synapses. Indeed, the novel Rho GTPase Rnd1 has been shown to promote the elongation and maturation of spines by inhibiting RhoA activity (Ishikawa et al., 2003). Currently, more than 20 mammalian Rho-family GTPases have been identified, however little is known about the functions of most of these Rho GTPase family members in neurons. Based on their ability to regulate cytoskeletal dynamics and cell morphology, it will be important to investigate whether any additional Rho GTPases play a role in synapse development and/or plasticity.
Given the essential roles that Rho GTPases play in regulating spine morphogenesis and the strong association between spine abnormalities and mental retardation, it is not surprising that mutations in a number of genes involved in Rho GTPase signaling have been found to cause mental retardation in humans (Govek et al., 2005; Ramakers, 2002). These genes include Rho GTPase GEFs (ARHGEF6, FGD1), GAPs (OPHN1, OCRL1, MEGAP) and downstream signaling molecules (PAK3, FMR1) (Allen et al., 1998; Attree et al., 1992; Billuart et al., 1998; Billuart and Chelly, 2003; Endris et al., 2002; Kutsche et al., 2000; Lebel et al., 2002; Soderling et al., 2003). The connection between altered Rho GTPase signaling, spine abnormalities, and mental retardation suggests that proper Rho GTPase signaling is important for normal cognitive development. In vivo studies in mice lacking Rho GTPases and their regulatory proteins will help to clarify the individual roles these proteins play in synapse development and plasticity and provide insight into how disruptions in Rho GTPase signaling could give rise to cognitive disorders such as mental retardation.
Research Highlights.
Proper synapse development and plasticity are essential for normal cognitive function.
Rho GTPases, key regulators of the actin cytoskeleton, control the formation and structural remodeling of synapses.
Precise spatio-temporal regulation of Rho GTPase signaling is critical for their function.
Rho GTPase regulatory proteins direct synapse development and plasticity by controlling the spatio-temporal regulation and signaling specificity of Rho GTPases.
Acknowledgments
Work in the authors’ laboratory was generously supported by the National Institutes of Health, the Department of Defense, and Mission Connect.
Abbreviation List
- GEF
Guanine nucleotide exchange factor
- GAP
GTPase-activating protein
- LTP
Long-term potentiation
- LTD
Long-term depression
- F-actin
filamentous actin
- CNS
central nervous system
- PH
pleckstrin homology domain
- CC-Ex
coiled coil-extended region
- RBD
Ras-binding domain
- PDZ
PSD-95, Dlg, and ZO-1/2 domain
- DH
Dbl homology domain
- SEC14
Sec14p homology domain
- SPEC
spectrin-like repeats
- SH3
Src homology 3 domain
- EH
Eps15 homology domain
- C1
protein kinase C conserved region 1
- C2
protein kinase C conserved region 2
- BAR
Bin/Amphiphysin/Rvs domain
- P
proline-rich regions
- PSD
post-synaptic density
- NMDA receptor
N-methyl-D-aspartate receptor
- AMPA receptor
α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid
- PAR
partition-defective protein
- aPKCζ
atypical protein kinase C
- Pak
p21-activated kinase
- CaMKK
calmodulin-dependent kinase kinase
- mEPSC
miniature excitatory postsynaptic current
- N-WASP
neuronal Wiskott-Aldrich syndrome protein
- Cdk5
cyclin dependent kinase 5
- ASDs
Autism Spectrum Disorders
- NMJ
neuromuscular junction
- GABA receptor
gamma-amino butyric acid receptor
- Ach receptor
acetylcholine receptor
- DAG
diacyglyerol
- Bcr
breakpoint cluster region
- Abr
active Bcr-related
- CIN85
c-Cbl-interacting protein of 85 kDa
- Arg
Abl-related gene
- LA
lateral amygdala
- Grb2
growth factor receptor-bound protein 2
- ROCK
Rho-associated coil-containing protein kinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmadian G, Ju W, Liu L, Wyszynski M, Lee SH, Dunah AW, Taghibiglou C, Wang Y, Lu J, Wong TP, Sheng M, Wang YT. Tyrosine phosphorylation of GluR2 is required for insulin-stimulated AMPA receptor endocytosis and LTD. Embo J. 2004;23:1040–1050. doi: 10.1038/sj.emboj.7600126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Kozma R, Monfries C, Hall C, Lim HH, Smith P, Lim L. Human brain n-chimaerin cDNA encodes a novel phorbol ester receptor. Biochem J. 1990;272:767–773. doi: 10.1042/bj2720767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Lee J, Kozma R, Best A, Monfries C, Lim L. A novel functional target for tumor-promoting phorbol esters and lysophosphatidic acid. The p21rac-GTPase activating protein n-chimaerin. J Biol Chem. 1993;268:10709–10712. [PubMed] [Google Scholar]
- Alexander M, Chan KK, Byrne AB, Selman G, Lee T, Ono J, Wong E, Puckrin R, Dixon SJ, Roy PJ. An UNC-40 pathway directs postsynaptic membrane extension in Caenorhabditis elegans. Development. 2009;136:911–922. doi: 10.1242/dev.030759. [DOI] [PubMed] [Google Scholar]
- Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, Mulley JC, Walsh CA. PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- Arimura N, Kaibuchi K. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat Rev Neurosci. 2007;8:194–205. doi: 10.1038/nrn2056. [DOI] [PubMed] [Google Scholar]
- Attree O, Olivos IM, Okabe I, Bailey LC, Nelson DL, Lewis RA, McInnes RR, Nussbaum RL. The Lowe’s oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature. 1992;358:239–242. doi: 10.1038/358239a0. [DOI] [PubMed] [Google Scholar]
- Awasaki T, Saito M, Sone M, Suzuki E, Sakai R, Ito K, Hama C. The Drosophila trio plays an essential role in patterning of axons by regulating their directional extension. Neuron. 2000;26:119–131. doi: 10.1016/s0896-6273(00)81143-5. [DOI] [PubMed] [Google Scholar]
- Bagrodia S, Taylor SJ, Jordon KA, Van AL, Cerione RA. A novel regulator of p21-activated kinases. J Biol Chem. 1998;273:23633–23636. doi: 10.1074/jbc.273.37.23633. [DOI] [PubMed] [Google Scholar]
- Bateman J, Shu H, Van Vactor D. The guanine nucleotide exchange factor trio mediates axonal development in the Drosophila embryo. Neuron. 2000;26:93–106. doi: 10.1016/s0896-6273(00)81141-1. [DOI] [PubMed] [Google Scholar]
- Bateman J, Van Vactor D. The Trio family of guanine-nucleotide-exchange factors: regulators of axon guidance. J Cell Sci. 2001;114:1973–1980. doi: 10.1242/jcs.114.11.1973. [DOI] [PubMed] [Google Scholar]
- Bernards A, Settleman J. GAP control: regulating the regulators of small GTPases. Trends Cell Biol. 2004;14:377–385. doi: 10.1016/j.tcb.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Bhatt DH, Zhang S, Gan WB. Dendritic spine dynamics. Annu Rev Physiol. 2009;71:261–282. doi: 10.1146/annurev.physiol.010908.163140. [DOI] [PubMed] [Google Scholar]
- Billuart P, Bienvenu T, Ronce N, des Portes V, Vinet MC, Zemni R, Roest Crollius H, Carrie A, Fauchereau F, Cherry M, Briault S, Hamel B, Fryns JP, Beldjord C, Kahn A, Moraine C, Chelly J. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;392:923–926. doi: 10.1038/31940. [DOI] [PubMed] [Google Scholar]
- Billuart P, Chelly J. From fragile X mental retardation protein to Rac1 GTPase: new insights from Fly CYFIP. Neuron. 2003;38:843–845. doi: 10.1016/s0896-6273(03)00362-3. [DOI] [PubMed] [Google Scholar]
- Billuart P, Winter CG, Maresh A, Zhao X, Luo L. Regulating axon branch stability: the role of p190 RhoGAP in repressing a retraction signaling pathway. Cell. 2001;107:195–207. doi: 10.1016/s0092-8674(01)00522-0. [DOI] [PubMed] [Google Scholar]
- Bourne JN, Harris KM. Balancing structure and function at hippocampal dendritic spines. Annu Rev Neurosci. 2008;31:47–67. doi: 10.1146/annurev.neuro.31.060407.125646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Shan W, Tanaka H, Benson DL, Huntley GW. Increasing numbers of synaptic puncta during late-phase LTP: N-cadherin is synthesized, recruited to synaptic sites, and required for potentiation. Neuron. 2000;28:245–259. doi: 10.1016/s0896-6273(00)00100-8. [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Valcin M, Poskanzer K, Tanaka H, Benson DL. Temporally distinct demands for classic cadherins in synapse formation and maturation. Mol Cell Neurosci. 2004;27:509–521. doi: 10.1016/j.mcn.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley WD, Hernandez SE, Settleman J, Koleske AJ. Integrin signaling through Arg activates p190RhoGAP by promoting its binding to p120RasGAP and recruitment to the membrane. Mol Biol Cell. 2006;17:4827–4836. doi: 10.1091/mbc.E06-02-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns MR, Matheson SF, Hu KQ, Delalle I, Caviness VS, Silver J, Bronson RT, Settleman J. The adhesion signaling molecule p190 RhoGAP is required for morphogenetic processes in neural development. Development. 2000;127:4891–4903. doi: 10.1242/dev.127.22.4891. [DOI] [PubMed] [Google Scholar]
- Brouns MR, Matheson SF, Settleman J. p190 RhoGAP is the principal Src substrate in brain and regulates axon outgrowth, guidance and fasciculation. Nat Cell Biol. 2001;3:361–367. doi: 10.1038/35070042. [DOI] [PubMed] [Google Scholar]
- Buchsbaum RJ, Connolly BA, Feig LA. Interaction of Rac exchange factors Tiam1 and Ras-GRF1 with a scaffold for the p38 mitogen-activated protein kinase cascade. Mol Cell Biol. 2002;22:4073–4085. doi: 10.1128/MCB.22.12.4073-4085.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchsbaum RJ, Connolly BA, Feig LA. Regulation of p70 S6 kinase by complex formation between the Rac guanine nucleotide exchange factor (Rac-GEF) Tiam1 and the scaffold spinophilin. J Biol Chem. 2003;278:18833–18841. doi: 10.1074/jbc.M207876200. [DOI] [PubMed] [Google Scholar]
- Buday L. Membrane-targeting of signalling molecules by SH2/SH3 domain-containing adaptor proteins. Biochim Biophys Acta. 1999;1422:187–204. doi: 10.1016/s0304-4157(99)00005-2. [DOI] [PubMed] [Google Scholar]
- Buttery P, Beg AA, Chih B, Broder A, Mason CA, Scheiffele P. The diacylglycerol-binding protein alpha1-chimaerin regulates dendritic morphology. Proc Natl Acad Sci U S A. 2006;103:1924–1929. doi: 10.1073/pnas.0510655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill ME, Xie Z, Day M, Photowala H, Barbolina MV, Miller CA, Weiss C, Radulovic J, Sweatt JD, Disterhoft JF, Surmeier DJ, Penzes P. Kalirin regulates cortical spine morphogenesis and disease-related behavioral phenotypes. Proc Natl Acad Sci U S A. 2009;106:13058–13063. doi: 10.1073/pnas.0904636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caloca MJ, Wang H, Delemos A, Wang S, Kazanietz MG. Phorbol esters and related analogs regulate the subcellular localization of beta 2-chimaerin, a non-protein kinase C phorbol ester receptor. J Biol Chem. 2001;276:18303–18312. doi: 10.1074/jbc.M011368200. [DOI] [PubMed] [Google Scholar]
- Caloca MJ, Wang H, Kazanietz MG. Characterization of the Rac-GAP (Rac-GTPase-activating protein) activity of beta2-chimaerin, a ‘non-protein kinase C’ phorbol ester receptor. Biochem J. 2003;375:313–321. doi: 10.1042/BJ20030727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- Cho YJ, Cunnick JM, Yi SJ, Kaartinen V, Groffen J, Heisterkamp N. Abr and Bcr, two homologous Rac GTPase-activating proteins, control multiple cellular functions of murine macrophages. Mol Cell Biol. 2007;27:899–911. doi: 10.1128/MCB.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TH, Xu X, Kaartinen V, Heisterkamp N, Groffen J, Bokoch GM. Abr and Bcr are multifunctional regulators of the Rho GTP-binding protein family. Proc Natl Acad Sci U S A. 1995;92:10282–10286. doi: 10.1073/pnas.92.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci. 2008;9:344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J Neurochem. 2006;97(Suppl 1):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Dailey ME, Smith SJ. The dynamics of dendritic structure in developing hippocampal slices. J Neurosci. 1996;16:2983–2994. doi: 10.1523/JNEUROSCI.16-09-02983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan B. Angelman syndrome: current understanding and research prospects. Epilepsia. 2009;50:2331–2339. doi: 10.1111/j.1528-1167.2009.02311.x. [DOI] [PubMed] [Google Scholar]
- DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Diaz E, Ge Y, Yang YH, Loh KC, Serafini TA, Okazaki Y, Hayashizaki Y, Speed TP, Ngai J, Scheiffele P. Molecular analysis of gene expression in the developing pontocerebellar projection system. Neuron. 2002;36:417–434. doi: 10.1016/s0896-6273(02)01016-4. [DOI] [PubMed] [Google Scholar]
- Diekmann D, Brill S, Garrett MD, Totty N, Hsuan J, Monfries C, Hall C, Lim L, Hall A. Bcr encodes a GTPase-activating protein for p21rac. Nature. 1991;351:400–402. doi: 10.1038/351400a0. [DOI] [PubMed] [Google Scholar]
- Ehler E, van LF, Collard JG, Salinas PC. Expression of Tiam-1 in the developing brain suggests a role for the Tiam-1-Rac signaling pathway in cell migration and neurite outgrowth. Mol Cell Neurosci. 1997;9:1–12. doi: 10.1006/mcne.1997.0602. [DOI] [PubMed] [Google Scholar]
- Ellis C, Moran M, McCormick F, Pawson T. Phosphorylation of GAP and GAP-associated proteins by transforming and mitogenic tyrosine kinases. Nature. 1990;343:377–381. doi: 10.1038/343377a0. [DOI] [PubMed] [Google Scholar]
- Endris V, Wogatzky B, Leimer U, Bartsch D, Zatyka M, Latif F, Maher ER, Tariverdian G, Kirsch S, Karch D, Rappold GA. The novel Rho-GTPase activating gene MEGAP/srGAP3 has a putative role in severe mental retardation. Proc Natl Acad Sci U S A. 2002;99:11754–11759. doi: 10.1073/pnas.162241099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engert F, Bonhoeffer T. Dendritic spine changes associated with hippocampal long-term synaptic plasticity. Nature. 1999;399:66–70. doi: 10.1038/19978. [DOI] [PubMed] [Google Scholar]
- Estrach S, Schmidt S, Diriong S, Penna A, Blangy A, Fort P, Debant A. The Human Rho-GEF trio and its target GTPase RhoG are involved in the NGF pathway, leading to neurite outgrowth. Curr Biol. 2002;12:307–312. doi: 10.1016/s0960-9822(02)00658-9. [DOI] [PubMed] [Google Scholar]
- Ethell IM, Irie F, Kalo MS, Couchman JR, Pasquale EB, Yamaguchi Y. EphB/syndecan-2 signaling in dendritic spine morphogenesis. Neuron. 2001;31:1001–1013. doi: 10.1016/s0896-6273(01)00440-8. [DOI] [PubMed] [Google Scholar]
- Fauchereau F, Herbrand U, Chafey P, Eberth A, Koulakoff A, Vinet MC, Ahmadian MR, Chelly J, Billuart P. The RhoGAP activity of OPHN1, a new F-actin-binding protein, is negatively controlled by its amino-terminal domain. Mol Cell Neurosci. 2003;23:574–586. doi: 10.1016/s1044-7431(03)00078-2. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Allwardt B, Harris KM. Dendritic spines do not split during hippocampal LTP or maturation. Nat Neurosci. 2002;5:297–298. doi: 10.1038/nn830. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Feinberg M, Popov V, Harris KM. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci. 1998;18:8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fioretos T, Voncken JW, Baram TZ, Kamme F, Groffen J, Heisterkamp N. Regional localization and developmental expression of the BCR gene in rodent brain. Cell Mol Biol Res. 1995;41:97–102. [PMC free article] [PubMed] [Google Scholar]
- Fleming IN, Elliott CM, Buchanan FG, Downes CP, Exton JH. Ca2+/calmodulin-dependent protein kinase II regulates Tiam1 by reversible protein phosphorylation. J Biol Chem. 1999;274:12753–12758. doi: 10.1074/jbc.274.18.12753. [DOI] [PubMed] [Google Scholar]
- Forsthoefel DJ, Liebl EC, Kolodziej PA, Seeger MA. The Abelson tyrosine kinase, the Trio GEF and Enabled interact with the Netrin receptor Frazzled in Drosophila. Development. 2005;132:1983–1994. doi: 10.1242/dev.01736. [DOI] [PubMed] [Google Scholar]
- Frank CA, Pielage J, Davis GW. A presynaptic homeostatic signaling system composed of the Eph receptor, ephexin, Cdc42, and CaV2.1 calcium channels. Neuron. 2009;61:556–569. doi: 10.1016/j.neuron.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu WY, Chen Y, Sahin M, Zhao XS, Shi L, Bikoff JB, Lai KO, Yung WH, Fu AK, Greenberg ME, Ip NY. Cdk5 regulates EphA4-mediated dendritic spine retraction through an ephexin1-dependent mechanism. Nat Neurosci. 2007;10:67–76. doi: 10.1038/nn1811. [DOI] [PubMed] [Google Scholar]
- Gerard A, Mertens AE, van der Kammen RA, Collard JG. The Par polarity complex regulates Rap1- and chemokine-induced T cell polarization. J Cell Biol. 2007;176:863–875. doi: 10.1083/jcb.200608161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaven JA, Whitehead I, Bagrodia S, Kay R, Cerione RA. The dbl-related protein, lfc, localizes to microtubules and mediates the activation of rac signaling pathways in cells [In Process Citation] J Biol Chem. 1999;274:2279–2285. doi: 10.1074/jbc.274.4.2279. [DOI] [PubMed] [Google Scholar]
- Goldstein B, Macara IG. The PAR proteins: fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Akerman CJ, Cross JR, Van der Veken L, Van Aelst L. The X-linked mental retardation protein oligophrenin-1 is required for dendritic spine morphogenesis. Nat Neurosci. 2004;7:364–372. doi: 10.1038/nn1210. [DOI] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Groffen J, Heisterkamp N. The chimeric BCR-ABL gene. Baillieres Clin Haematol. 1997;10:187–201. doi: 10.1016/s0950-3536(97)80002-9. [DOI] [PubMed] [Google Scholar]
- Harvey K, Duguid IC, Alldred MJ, Beatty SE, Ward H, Keep NH, Lingenfelter SE, Pearce BR, Lundgren J, Owen MJ, Smart TG, Luscher B, Rees MI, Harvey RJ. The GDP-GTP exchange factor collybistin: an essential determinant of neuronal gephyrin clustering. J Neurosci. 2004;24:5816–5826. doi: 10.1523/JNEUROSCI.1184-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkemeyer M, Itkis OS, Ngo M, Hickmott PW, Ethell IM. Multiple EphB receptor tyrosine kinases shape dendritic spines in the hippocampus. J Cell Biol. 2003;163:1313–1326. doi: 10.1083/jcb.200306033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Hussain NK, Jenna S, Glogauer M, Quinn CC, Wasiak S, Guipponi M, Antonarakis SE, Kay BK, Stossel TP, Lamarche-Vane N, McPherson PS. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat Cell Biol. 2001;3:927–932. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- Irie F, Yamaguchi Y. EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat Neurosci. 2002;5:1117–1118. doi: 10.1038/nn964. [DOI] [PubMed] [Google Scholar]
- Ishikawa Y, Katoh H, Negishi M. A role of Rnd1 GTPase in dendritic spine formation in hippocampal neurons. J Neurosci. 2003;23:11065–11072. doi: 10.1523/JNEUROSCI.23-35-11065.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe AB, Hall A, Schmidt A. Association of CNK1 with Rho guanine nucleotide exchange factors controls signaling specificity downstream of Rho. Curr Biol. 2005;15:405–412. doi: 10.1016/j.cub.2004.12.082. [DOI] [PubMed] [Google Scholar]
- Jedlicka P, Papadopoulos T, Deller T, Betz H, Schwarzacher SW. Increased network excitability and impaired induction of long-term potentiation in the dentate gyrus of collybistin-deficient mice in vivo. Mol Cell Neurosci. 2009;41:94–100. doi: 10.1016/j.mcn.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Johnson RC, Penzes P, Eipper BA, Mains RE. Isoforms of kalirin, a neuronal Dbl family member, generated through use of different 5′- and 3′-ends along with an internal translational initiation site. J Biol Chem. 2000;275:19324–19333. doi: 10.1074/jbc.M000676200. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Gonzalez-Gomez I, Voncken JW, Haataja L, Faure E, Nagy A, Groffen J, Heisterkamp N. Abnormal function of astroglia lacking Abr and Bcr RacGAPs. Development. 2001;128:4217–4227. doi: 10.1242/dev.128.21.4217. [DOI] [PubMed] [Google Scholar]
- Kaartinen V, Nagy A, Gonzalez-Gomez I, Groffen J, Heisterkamp N. Vestibular dysgenesis in mice lacking Abr and Bcr Cdc42/RacGAPs. Dev Dyn. 2002;223:517–525. doi: 10.1002/dvdy.10071. [DOI] [PubMed] [Google Scholar]
- Kang MG, Guo Y, Huganir RL. AMPA receptor and GEF-H1/Lfc complex regulates dendritic spine development through RhoA signaling cascade. Proc Natl Acad Sci U S A. 2009;106:3549–3554. doi: 10.1073/pnas.0812861106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Matsuzaki M, Noguchi J, Yasumatsu N, Nakahara H. Structure-stability-function relationships of dendritic spines. Trends Neurosci. 2003;26:360–368. doi: 10.1016/S0166-2236(03)00162-0. [DOI] [PubMed] [Google Scholar]
- Kaufmann WE, Moser HW. Dendritic anomalies in disorders associated with mental retardation. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. Embo J. 2003;22:4190–4201. doi: 10.1093/emboj/cdg413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser MS, McClelland AC, Hughes EG, Dalva MB. Intracellular and trans-synaptic regulation of glutamatergic synaptogenesis by EphB receptors. J Neurosci. 2006;26:12152–12164. doi: 10.1523/JNEUROSCI.3072-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser MS, Nolt MJ, Dalva MB. EphB receptors couple dendritic filopodia motility to synapse formation. Neuron. 2008;59:56–69. doi: 10.1016/j.neuron.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelfaoui M, Denis C, van Galen E, de Bock F, Schmitt A, Houbron C, Morice E, Giros B, Ramakers G, Fagni L, Chelly J, Nosten-Bertrand M, Billuart P. Loss of X-linked mental retardation gene oligophrenin1 in mice impairs spatial memory and leads to ventricular enlargement and dendritic spine immaturity. J Neurosci. 2007;27:9439–9450. doi: 10.1523/JNEUROSCI.2029-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelfaoui M, Pavlowsky A, Powell AD, Valnegri P, Cheong KW, Blandin Y, Passafaro M, Jefferys JG, Chelly J, Billuart P. Inhibition of RhoA pathway rescues the endocytosis defects in Oligophrenin1 mouse model of mental retardation. Hum Mol Genet. 2009;18:2575–2583. doi: 10.1093/hmg/ddp189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim T, Lee D, Park SH, Kim H, Park D. Molecular cloning of neuronally expressed mouse betaPix isoforms. Biochem Biophys Res Commun. 2000;272:721–725. doi: 10.1006/bbrc.2000.2845. [DOI] [PubMed] [Google Scholar]
- Kim S, Ko J, Shin H, Lee JR, Lim C, Han JH, Altrock WD, Garner CC, Gundelfinger ED, Premont RT, Kaang BK, Kim E. The GIT family of proteins forms multimers and associates with the presynaptic cytomatrix protein Piccolo. J Biol Chem. 2003;278:6291–6300. doi: 10.1074/jbc.M212287200. [DOI] [PubMed] [Google Scholar]
- Kins S, Betz H, Kirsch J. Collybistin, a newly identified brain-specific GEF, induces submembrane clustering of gephyrin. Nat Neurosci. 2000;3:22–29. doi: 10.1038/71096. [DOI] [PubMed] [Google Scholar]
- Kiraly DD, Eipper-Mains JE, Mains RE, Eipper BA. Synaptic plasticity, a symphony in GEF. ACS Chem Neurosci. 2010;1:348–365. doi: 10.1021/cn100012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. Eph/ephrin signaling in morphogenesis, neural development and plasticity. Curr Opin Cell Biol. 2004;16:580–589. doi: 10.1016/j.ceb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Klein R. Bidirectional modulation of synaptic functions by Eph/ephrin signaling. Nat Neurosci. 2009;12:15–20. doi: 10.1038/nn.2231. [DOI] [PubMed] [Google Scholar]
- Kneussel M, Engelkamp D, Betz H. Distribution of transcripts for the brain-specific GDP/GTP exchange factor collybistin in the developing mouse brain. Eur J Neurosci. 2001;13:487–492. doi: 10.1046/j.0953-816x.2000.01411.x. [DOI] [PubMed] [Google Scholar]
- Ko J, Kim S, Valtschanoff JG, Shin H, Lee JR, Sheng M, Premont RT, Weinberg RJ, Kim E. Interaction between liprin-alpha and GIT1 is required for AMPA receptor targeting. J Neurosci. 2003;23:1667–1677. doi: 10.1523/JNEUROSCI.23-05-01667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CG, Manser E, Zhao ZS, Ng CP, Lim L. Beta1PIX, the PAK-interacting exchange factor, requires localization via a coiled-coil region to promote microvillus-like structures and membrane ruffles. J Cell Sci. 2001;114:4239–4251. doi: 10.1242/jcs.114.23.4239. [DOI] [PubMed] [Google Scholar]
- Koh TW, Verstreken P, Bellen HJ. Dap160/intersectin acts as a stabilizing scaffold required for synaptic development and vesicle endocytosis. Neuron. 2004;43:193–205. doi: 10.1016/j.neuron.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Korus M, Mahon GM, Cheng L, Whitehead IP. p38 MAPK-mediated activation of NF-kappaB by the RhoGEF domain of Bcr. Oncogene. 2002;21:4601–4612. doi: 10.1038/sj.onc.1205678. [DOI] [PubMed] [Google Scholar]
- Kunda P, Paglini G, Quiroga S, Kosik K, Caceres A. Evidence for the involvement of Tiam1 in axon formation. J Neurosci. 2001;21:2361–2372. doi: 10.1523/JNEUROSCI.21-07-02361.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, Moraine C, Ropers HH, Hamel BC, van Bokhoven H, Gal A. Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat Genet. 2000;26:247–250. doi: 10.1038/80002. [DOI] [PubMed] [Google Scholar]
- Lambert JM, Lambert QT, Reuther GW, Malliri A, Siderovski DP, Sondek J, Collard JG, Der CJ. Tiam1 mediates Ras activation of Rac by a PI(3)K-independent mechanism. Nat Cell Biol. 2002;4:621–625. doi: 10.1038/ncb833. [DOI] [PubMed] [Google Scholar]
- Lamprecht R, Farb CR, LeDoux JE. Fear memory formation involves p190 RhoGAP and ROCK proteins through a GRB2-mediated complex. Neuron. 2002;36:727–738. doi: 10.1016/s0896-6273(02)01047-4. [DOI] [PubMed] [Google Scholar]
- Lebel RR, May M, Pouls S, Lubs HA, Stevenson RE, Schwartz CE. Non-syndromic X-linked mental retardation associated with a missense mutation (P312L) in the FGD1 gene. Clin Genet. 2002;61:139–145. doi: 10.1034/j.1399-0004.2002.610209.x. [DOI] [PubMed] [Google Scholar]
- Leeuwen FN, Kain HE, Kammen RA, Michiels F, Kranenburg OW, Collard JG. The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J Cell Biol. 1997;139:797–807. doi: 10.1083/jcb.139.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvai B, Stern EA, Chen B, Svoboda K. Experience-dependent plasticity of dendritic spines in the developing rat barrel cortex in vivo. Nature. 2000;404:876–881. doi: 10.1038/35009107. [DOI] [PubMed] [Google Scholar]
- Leung T, How BE, Manser E, Lim L. Cerebellar beta 2-chimaerin, a GTPase-activating protein for p21 ras- related rac is specifically expressed in granule cells and has a unique N-terminal SH2 domain. J Biol Chem. 1994;269:12888–12892. [PubMed] [Google Scholar]
- Li J, Smithgall TE. Co-expression with BCR induces activation of the FES tyrosine kinase and phosphorylation of specific N-terminal BCR tyrosine residues. J Biol Chem. 1996;271:32930–32936. doi: 10.1074/jbc.271.51.32930. [DOI] [PubMed] [Google Scholar]
- Lim HH, Michael GJ, Smith P, Lim L, Hall C. Developmental regulation and neuronal expression of the mRNA of rat n-chimaerin, a p21rac GAP:cDNA sequence. Biochem J. 1992;287 (Pt 2):415–422. doi: 10.1042/bj2870415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linseman DA, Loucks FA. Diverse roles of Rho family GTPases in neuronal development, survival, and death. Front Biosci. 2008;13:657–676. doi: 10.2741/2710. [DOI] [PubMed] [Google Scholar]
- Lippman J, Dunaevsky A. Dendritic spine morphogenesis and plasticity. J Neurobiol. 2005;64:47–57. doi: 10.1002/neu.20149. [DOI] [PubMed] [Google Scholar]
- Ma XM, Huang J, Wang Y, Eipper BA, Mains RE. Kalirin, a multifunctional Rho guanine nucleotide exchange factor, is necessary for maintenance of hippocampal pyramidal neuron dendrites and dendritic spines. J Neurosci. 2003;23:10593–10603. doi: 10.1523/JNEUROSCI.23-33-10593.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XM, Kiraly DD, Gaier ED, Wang Y, Kim EJ, Levine ES, Eipper BA, Mains RE. Kalirin-7 is required for synaptic structure and function. J Neurosci. 2008;28:12368–12382. doi: 10.1523/JNEUROSCI.4269-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JF, Jackson MF, Beazely MA. Hippocampal long-term synaptic plasticity and signal amplification of NMDA receptors. Crit Rev Neurobiol. 2006;18:71–84. doi: 10.1615/critrevneurobiol.v18.i1-2.80. [DOI] [PubMed] [Google Scholar]
- Maletic-Savatic M, Malinow R, Svoboda K. Rapid dendritic morphogenesis in CA1 hippocampal dendrites induced by synaptic activity. Science. 1999;283:1923–1927. doi: 10.1126/science.283.5409.1923. [DOI] [PubMed] [Google Scholar]
- Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867–871. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- Malmberg EK, Andersson CX, Gentzsch M, Chen JH, Mengos A, Cui L, Hansson GC, Riordan JR. Bcr (breakpoint cluster region) protein binds to PDZ-domains of scaffold protein PDZK1 and vesicle coat protein Mint3. J Cell Sci. 2004;117:5535–5541. doi: 10.1242/jcs.01472. [DOI] [PubMed] [Google Scholar]
- Manabe R, Kovalenko M, Webb DJ, Horwitz AR. GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J Cell Sci. 2002;115:1497–1510. doi: 10.1242/jcs.115.7.1497. [DOI] [PubMed] [Google Scholar]
- Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- Margolis SS, Salogiannis J, Lipton DM, Mandel-Brehm C, Wills ZP, Mardinly AR, Hu L, Greer PL, Bikoff JB, Ho HY, Soskis MJ, Sahin M, Greenberg ME. EphB-Mediated Degradation of the RhoA GEF Ephexin5 Relieves a Developmental Brake on Excitatory Synapse Formation. Cell. 2010;143:442–455. doi: 10.1016/j.cell.2010.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Sweeney ST, Poskanzer KE, Roos J, Kelly RB, Davis GW. Dap160/intersectin scaffolds the periactive zone to achieve high-fidelity endocytosis and normal synaptic growth. Neuron. 2004;43:207–219. doi: 10.1016/j.neuron.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Maru Y, Witte ON. The BCR gene encodes a novel serine/threonine kinase activity within a single exon. Cell. 1991;67:459–468. doi: 10.1016/0092-8674(91)90521-y. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson CE, Eipper BA, Mains RE. Multiple novel isoforms of Trio are expressed in the developing rat brain. Gene. 2005;347:125–135. doi: 10.1016/j.gene.2004.12.028. [DOI] [PubMed] [Google Scholar]
- McWhirter JR, Galasso DL, Wang JY. A coiled-coil oligomerization domain of Bcr is essential for the transforming function of Bcr-Abl oncoproteins. Mol Cell Biol. 1993;13:7587–7595. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens AE, Pegtel DM, Collard JG. Tiam1 takes PARt in cell polarity. Trends Cell Biol. 2006;16:308–316. doi: 10.1016/j.tcb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Mertens AE, Roovers RC, Collard JG. Regulation of Tiam1-Rac signalling. FEBS Lett. 2003;546:11–16. doi: 10.1016/s0014-5793(03)00435-6. [DOI] [PubMed] [Google Scholar]
- Mertens AE, Rygiel TP, Olivo C, van der Kammen R, Collard JG. The Rac activator Tiam1 controls tight junction biogenesis in keratinocytes through binding to and activation of the Par polarity complex. J Cell Biol. 2005;170:1029–1037. doi: 10.1083/jcb.200502129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Yamauchi J, Tanoue A, Wu C, Mobley WC. TrkB binds and tyrosine-phosphorylates Tiam1, leading to activation of Rac1 and induction of changes in cellular morphology. Proc Natl Acad Sci U S A. 2006;103:10444–10449. doi: 10.1073/pnas.0603914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon SY, Zheng Y. Rho GTPase-activating proteins in cell regulation. Trends Cell Biol. 2003;13:13–22. doi: 10.1016/s0962-8924(02)00004-1. [DOI] [PubMed] [Google Scholar]
- Muly EC, Nairn AC, Greengard P, Rainnie DG. Subcellular distribution of the Rho-GEF Lfc in primate prefrontal cortex: effect of neuronal activation. J Comp Neurol. 2008;508:927–939. doi: 10.1002/cne.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai KK, Nguyen LN, Irie F, Yamaguchi Y, Pasquale EB. Control of hippocampal dendritic spine morphology through ephrin-A3/EphA4 signaling. Nat Neurosci. 2003;6:153–160. doi: 10.1038/nn994. [DOI] [PubMed] [Google Scholar]
- Nadif Kasri N, Nakano-Kobayashi A, Malinow R, Li B, Van Aelst L. The Rho-linked mental retardation protein oligophrenin-1 controls synapse maturation and plasticity by stabilizing AMPA receptors. Genes Dev. 2009;23:1289–1302. doi: 10.1101/gad.1783809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano-Kobayashi A, Kasri NN, Newey SE, Van Aelst L. The Rho-linked mental retardation protein OPHN1 controls synaptic vesicle endocytosis via endophilin A1. Curr Biol. 2009;19:1133–1139. doi: 10.1016/j.cub.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newey SE, Velamoor V, Govek EE, Van Aelst L. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol. 2005;64:58–74. doi: 10.1002/neu.20153. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Schmidt S, Dietzl G, Keleman K, Asling B, Debant A, Dickson BJ. Trio combines with dock to regulate Pak activity during photoreceptor axon pathfinding in Drosophila. Cell. 2000;101:283–294. doi: 10.1016/s0092-8674(00)80838-7. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Yamaguchi T, Kato K, Yoshizawa M, Nabeshima Y, Ohno S, Hoshino M, Kaibuchi K. PAR-6-PAR-3 mediates Cdc42-induced Rac activation through the Rac GEFs STEF/Tiam1. Nat Cell Biol. 2005;7:270–277. doi: 10.1038/ncb1227. [DOI] [PubMed] [Google Scholar]
- Nishimura T, Yamaguchi T, Tokunaga A, Hara A, Hamaguchi T, Kato K, Iwamatsu A, Okano H, Kaibuchi K. Role of numb in dendritic spine development with a Cdc42 GEF intersectin and EphB2. Mol Biol Cell. 2006;17:1273–1285. doi: 10.1091/mbc.E05-07-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien SP, Seipel K, Medley QG, Bronson R, Segal R, Streuli M. Skeletal muscle deformity and neuronal disorder in Trio exchange factor-deficient mouse embryos. Proc Natl Acad Sci U S A. 2000;97:12074–12078. doi: 10.1073/pnas.97.22.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D, Han S, Seo J, Lee JR, Choi J, Groffen J, Kim K, Cho YS, Choi HS, Shin H, Woo J, Won H, Park SK, Kim SY, Jo J, Whitcomb DJ, Cho K, Kim H, Bae YC, Heisterkamp N, Choi SY, Kim E. Regulation of synaptic Rac1 activity, long-term potentiation maintenance, and learning and memory by BCR and ABR Rac GTPase-activating proteins. J Neurosci. 2010;30:14134–14144. doi: 10.1523/JNEUROSCI.1711-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Tanaka H, Yagita Y, Saeki Y, Taguchi A, Hiraoka Y, Zeng LH, Colman DR, Miki N. Cadherin activity is required for activity-induced spine remodeling. J Cell Biol. 2004;167:961–972. doi: 10.1083/jcb.200406030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa M, Bundman MC, Greenberger V, Segal M. Morphological analysis of dendritic spine development in primary cultures of hippocampal neurons. J Neurosci. 1995;15:1–11. doi: 10.1523/JNEUROSCI.15-01-00001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos T, Eulenburg V, Reddy-Alla S, Mansuy IM, Li Y, Betz H. Collybistin is required for both the formation and maintenance of GABAergic postsynapses in the hippocampus. Mol Cell Neurosci. 2008;39:161–169. doi: 10.1016/j.mcn.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Papadopoulos T, Korte M, Eulenburg V, Kubota H, Retiounskaia M, Harvey RJ, Harvey K, O’Sullivan GA, Laube B, Hulsmann S, Geiger JR, Betz H. Impaired GABAergic transmission and altered hippocampal synaptic plasticity in collybistin-deficient mice. Embo J. 2007;26:3888–3899. doi: 10.1038/sj.emboj.7601819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Na M, Choi J, Kim S, Lee JR, Yoon J, Park D, Sheng M, Kim E. The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42. J Biol Chem. 2003;278:19220–19229. doi: 10.1074/jbc.M301052200. [DOI] [PubMed] [Google Scholar]
- Parnas D, Haghighi AP, Fetter RD, Kim SW, Goodman CS. Regulation of postsynaptic structure and protein localization by the Rho-type guanine nucleotide exchange factor dPix. Neuron. 2001;32:415–424. doi: 10.1016/s0896-6273(01)00485-8. [DOI] [PubMed] [Google Scholar]
- Pawson C, Eaton BA, Davis GW. Formin-dependent synaptic growth: evidence that Dlar signals via Diaphanous to modulate synaptic actin and dynamic pioneer microtubules. J Neurosci. 2008;28:11111–11123. doi: 10.1523/JNEUROSCI.0833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel DM, Ellenbroek SI, Mertens AE, van der Kammen RA, de Rooij J, Collard JG. The Par-Tiam1 complex controls persistent migration by stabilizing microtubule-dependent front-rear polarity. Curr Biol. 2007;17:1623–1634. doi: 10.1016/j.cub.2007.08.035. [DOI] [PubMed] [Google Scholar]
- Penzes P, Beeser A, Chernoff J, Schiller MR, Eipper BA, Mains RE, Huganir RL. Rapid induction of dendritic spine morphogenesis by trans-synaptic ephrinB-EphB receptor activation of the Rho-GEF kalirin. Neuron. 2003;37:263–274. doi: 10.1016/s0896-6273(02)01168-6. [DOI] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, Srivastava DP. Convergent CaMK and RacGEF signals control dendritic structure and function. Trends Cell Biol. 2008;18:405–413. doi: 10.1016/j.tcb.2008.07.002. [DOI] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Alam MR, Kambampati V, Mains RE, Eipper BA. An isoform of kalirin, a brain-specific GDP/GTP exchange factor, is enriched in the postsynaptic density fraction. J Biol Chem. 2000;275:6395–6403. doi: 10.1074/jbc.275.9.6395. [DOI] [PubMed] [Google Scholar]
- Penzes P, Johnson RC, Sattler R, Zhang X, Huganir RL, Kambampati V, Mains RE, Eipper BA. The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron. 2001;29:229–242. doi: 10.1016/s0896-6273(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Penzes P, Jones KA. Dendritic spine dynamics--a key role for kalirin-7. Trends Neurosci. 2008;31:419–427. doi: 10.1016/j.tins.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O. Spatio-temporal Rho GTPase signaling - where are we now? J Cell Sci. 2010;123:1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- Poulopoulos A, Aramuni G, Meyer G, Soykan T, Hoon M, Papadopoulos T, Zhang M, Paarmann I, Fuchs C, Harvey K, Jedlicka P, Schwarzacher SW, Betz H, Harvey RJ, Brose N, Zhang W, Varoqueaux F. Neuroligin 2 drives postsynaptic assembly at perisomatic inhibitory synapses through gephyrin and collybistin. Neuron. 2009;63:628–642. doi: 10.1016/j.neuron.2009.08.023. [DOI] [PubMed] [Google Scholar]
- Pucharcos C, Fuentes JJ, Casas C, de la Luna S, Alcantara S, Arbones ML, Soriano E, Estivill X, Pritchard M. Alu-splice cloning of human Intersectin (ITSN), a putative multivalent binding protein expressed in proliferating and differentiating neurons and overexpressed in Down syndrome. Eur J Hum Genet. 1999;7:704–712. doi: 10.1038/sj.ejhg.5200356. [DOI] [PubMed] [Google Scholar]
- Radziwill G, Erdmann RA, Margelisch U, Moelling K. The Bcr kinase downregulates Ras signaling by phosphorylating AF-6 and binding to its PDZ domain. Mol Cell Biol. 2003;23:4663–4672. doi: 10.1128/MCB.23.13.4663-4672.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers GJ. Rho proteins, mental retardation and the cellular basis of cognition. Trends Neurosci. 2002;25:191–199. doi: 10.1016/s0166-2236(00)02118-4. [DOI] [PubMed] [Google Scholar]
- Reddy-Alla S, Schmitt B, Birkenfeld J, Eulenburg V, Dutertre S, Bohringer C, Gotz M, Betz H, Papadopoulos T. PH-domain-driven targeting of collybistin but not Cdc42 activation is required for synaptic gephyrin clustering. Eur J Neurosci. 2010;31:1173–1184. doi: 10.1111/j.1460-9568.2010.07149.x. [DOI] [PubMed] [Google Scholar]
- Reid T, Bathoorn A, Ahmadian MR, Collard JG. Identification and characterization of hPEM-2, a guanine nucleotide exchange factor specific for Cdc42. J Biol Chem. 1999;274:33587–33593. doi: 10.1074/jbc.274.47.33587. [DOI] [PubMed] [Google Scholar]
- Ren Y, Li R, Zheng Y, Busch H. Cloning and characterization of GEF-H1, a microtubule-associated guanine nucleotide exchange factor for Rac and Rho GTPases. J Biol Chem. 1998;273:34954–34960. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Self AJ, Kasmi F, Paterson HF, Hall A, Marshall CJ, Ellis C. rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. Embo J. 1993;12:5151–5160. doi: 10.1002/j.1460-2075.1993.tb06210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J, Sudhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- Rodgers EE, Theibert AB. Functions of PI 3-kinase in development of the nervous system. Int J Dev Neurosci. 2002;20:187–197. doi: 10.1016/s0736-5748(02)00047-3. [DOI] [PubMed] [Google Scholar]
- Roof RW, Dukes BD, Chang JH, Parsons SJ. Phosphorylation of the p190 RhoGAP N-terminal domain by c-Src results in a loss of GTP binding activity. Febs Lett. 2000;472:117–121. doi: 10.1016/s0014-5793(00)01439-3. [DOI] [PubMed] [Google Scholar]
- Rose S, Malabarba MG, Krag C, Schultz A, Tsushima H, Di Fiore PP, Salcini AE. Caenorhabditis elegans intersectin: a synaptic protein regulating neurotransmission. Mol Biol Cell. 2007;18:5091–5099. doi: 10.1091/mbc.E07-05-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger G, Kutsche K. AlphaPIX and betaPIX and their role in focal adhesion formation. European journal of cell biology. 2006;85:265–274. doi: 10.1016/j.ejcb.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Ryan XP, Alldritt J, Svenningsson P, Allen PB, Wu GY, Nairn AC, Greengard P. The Rho-specific GEF Lfc interacts with neurabin and spinophilin to regulate dendritic spine morphology. Neuron. 2005;47:85–100. doi: 10.1016/j.neuron.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Sahay S, Pannucci NL, Mahon GM, Rodriguez PL, Megjugorac NJ, Kostenko EV, Ozer HL, Whitehead IP. The RhoGEF domain of p210 Bcr-Abl activates RhoA and is required for transformation. Oncogene. 2008;27:2064–2071. doi: 10.1038/sj.onc.1210841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin M, Greer PL, Lin MZ, Poucher H, Eberhart J, Schmidt S, Wright TM, Shamah SM, O’Connell S, Cowan CW, Hu L, Goldberg JL, Debant A, Corfas G, Krull CE, Greenberg ME. Eph-dependent tyrosine phosphorylation of ephexin1 modulates growth cone collapse. Neuron. 2005;46:191–204. doi: 10.1016/j.neuron.2005.01.030. [DOI] [PubMed] [Google Scholar]
- Saneyoshi T, Wayman G, Fortin D, Davare M, Hoshi N, Nozaki N, Natsume T, Soderling TR. Activity-dependent synaptogenesis: regulation by a CaM-kinase kinase/CaM-kinase I/betaPIX signaling complex. Neuron. 2008;57:94–107. doi: 10.1016/j.neuron.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Segura I, Essmann CL, Weinges S, Acker-Palmer A. Grb4 and GIT1 transduce ephrinB reverse signals modulating spine morphogenesis and synapse formation. Nat Neurosci. 2007;10:301–310. doi: 10.1038/nn1858. [DOI] [PubMed] [Google Scholar]
- Sfakianos MK, Eisman A, Gourley SL, Bradley WD, Scheetz AJ, Settleman J, Taylor JR, Greer CA, Williamson A, Koleske AJ. Inhibition of Rho via Arg and p190RhoGAP in the postnatal mouse hippocampus regulates dendritic spine maturation, synapse and dendrite stability, and behavior. J Neurosci. 2007;27:10982–10992. doi: 10.1523/JNEUROSCI.0793-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamah SM, Lin MZ, Goldberg JL, Estrach S, Sahin M, Hu L, Bazalakova M, Neve RL, Corfas G, Debant A, Greenberg ME. EphA receptors regulate growth cone dynamics through the novel guanine nucleotide exchange factor ephexin. Cell. 2001;105:233–244. doi: 10.1016/s0092-8674(01)00314-2. [DOI] [PubMed] [Google Scholar]
- Shi L, Butt B, Ip FC, Dai Y, Jiang L, Yung WH, Greenberg ME, Fu AK, Ip NY. Ephexin1 is required for structural maturation and neurotransmission at the neuromuscular junction. Neuron. 2010;65:204–216. doi: 10.1016/j.neuron.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin EY, Shin KS, Lee CS, Woo KN, Quan SH, Soung NK, Kim YG, Cha CI, Kim SR, Park D, Bokoch GM, Kim EG. Phosphorylation of p85 beta PIX, a Rac/Cdc42-specific guanine nucleotide exchange factor, via the Ras/ERK/PAK2 pathway is required for basic fibroblast growth factor-induced neurite outgrowth. J Biol Chem. 2002;277:44417–44430. doi: 10.1074/jbc.M203754200. [DOI] [PubMed] [Google Scholar]
- Snyder JT, Worthylake DK, Rossman KL, Betts L, Pruitt WM, Siderovski DP, Der CJ, Sondek J. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat Struct Biol. 2002;9:468–475. doi: 10.1038/nsb796. [DOI] [PubMed] [Google Scholar]
- Soderling SH, Langeberg LK, Soderling JA, Davee SM, Simerly R, Raber J, Scott JD. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc Natl Acad Sci U S A. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M, Hoshino M, Suzuki E, Kuroda S, Kaibuchi K, Nakagoshi H, Saigo K, Nabeshima Y, Hama C. Still life, a protein in synaptic terminals of Drosophila homologous to GDP-GTP exchangers [published erratum appears in Science 1997 Mar 7;275(5305):1405] Science. 1997;275:543–547. doi: 10.1126/science.275.5299.543. [DOI] [PubMed] [Google Scholar]
- Sone M, Suzuki E, Hoshino M, Hou D, Kuromi H, Fukata M, Kuroda S, Kaibuchi K, Nabeshima Y, Hama C. Synaptic development is controlled in the periactive zones of Drosophila synapses. Development. 2000;127:4157–4168. doi: 10.1242/dev.127.19.4157. [DOI] [PubMed] [Google Scholar]
- Steven R, Kubiseski TJ, Zheng H, Kulkarni S, Mancillas J, Ruiz Morales A, Hogue CW, Pawson T, Culotti J. UNC-73 activates the Rac GTPase and is required for cell and growth cone migrations in C. elegans. Cell. 1998;92:785–795. doi: 10.1016/s0092-8674(00)81406-3. [DOI] [PubMed] [Google Scholar]
- Steven R, Zhang L, Culotti J, Pawson T. The UNC-73/Trio RhoGEF-2 domain is required in separate isoforms for the regulation of pharynx pumping and normal neurotransmission in C. elegans. Genes Dev. 2005;19:2016–2029. doi: 10.1101/gad.1319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. The cadherin superfamily in neuronal connections and interactions. Nat Rev Neurosci. 2007;8:11–20. doi: 10.1038/nrn2043. [DOI] [PubMed] [Google Scholar]
- Tan EC, Leung T, Manser E, Lim L. The human active breakpoint cluster region-related gene encodes a brain protein with homology to guanine nucleotide exchange proteins and GTPase-activating proteins. J Biol Chem. 1993;268:27291–27298. [PubMed] [Google Scholar]
- Tang L, Hung CP, Schuman EM. A role for the cadherin family of cell adhesion molecules in hippocampal long-term potentiation. Neuron. 1998;20:1165–1175. doi: 10.1016/s0896-6273(00)80497-3. [DOI] [PubMed] [Google Scholar]
- Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biol Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- Thomas S, Ritter B, Verbich D, Sanson C, Bourbonniere L, McKinney RA, McPherson PS. Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J Biol Chem. 2009;284:12410–12419. doi: 10.1074/jbc.M809746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- Tolias KF, Bikoff JB, Burette A, Paradis S, Harrar D, Tavazoie S, Weinberg RJ, Greenberg ME. The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron. 2005;45:525–538. doi: 10.1016/j.neuron.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Tolias KF, Bikoff JB, Kane CG, Tolias CS, Hu L, Greenberg ME. The Rac1 guanine nucleotide exchange factor Tiam1 mediates EphB receptor-dependent dendritic spine development. Proc Natl Acad Sci U S A. 2007;104:7265–7270. doi: 10.1073/pnas.0702044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toni N, Buchs PA, Nikonenko I, Bron CR, Muller D. LTP promotes formation of multiple spine synapses between a single axon terminal and a dendrite. Nature. 1999;402:421–425. doi: 10.1038/46574. [DOI] [PubMed] [Google Scholar]
- Van de Ven TJ, VanDongen HM, VanDongen AM. The nonkinase phorbol ester receptor alpha 1-chimerin binds the NMDA receptor NR2A subunit and regulates dendritic spine density. J Neurosci. 2005;25:9488–9496. doi: 10.1523/JNEUROSCI.2450-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderzalm PJ, Pandey A, Hurwitz ME, Bloom L, Horvitz HR, Garriga G. C. elegans CARMIL negatively regulates UNC-73/Trio function during neuronal development. Development. 2009;136:1201–1210. doi: 10.1242/dev.026666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken JW, van Schaick H, Kaartinen V, Deemer K, Coates T, Landing B, Pattengale P, Dorseuil O, Bokoch GM, Groffen J, et al. Increased neutrophil respiratory burst in bcr-null mutants. Cell. 1995;80:719–728. doi: 10.1016/0092-8674(95)90350-x. [DOI] [PubMed] [Google Scholar]
- Wang W, Bouhours M, Gracheva EO, Liao EH, Xu K, Sengar AS, Xin X, Roder J, Boone C, Richmond JE, Zhen M, Egan SE. ITSN-1 controls vesicle recycling at the neuromuscular junction and functions in parallel with DAB-1. Traffic. 2008;9:742–754. doi: 10.1111/j.1600-0854.2008.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Cahill ME, Penzes P. Kalirin loss results in cortical morphological alterations. Mol Cell Neurosci. 2010;43:81–89. doi: 10.1016/j.mcn.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Photowala H, Cahill ME, Srivastava DP, Woolfrey KM, Shum CY, Huganir RL, Penzes P. Coordination of synaptic adhesion with dendritic spine remodeling by AF-6 and kalirin-7. J Neurosci. 2008;28:6079–6091. doi: 10.1523/JNEUROSCI.1170-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Srivastava DP, Photowala H, Kai L, Cahill ME, Woolfrey KM, Shum CY, Surmeier DJ, Penzes P. Kalirin-7 controls activity-dependent structural and functional plasticity of dendritic spines. Neuron. 2007;56:640–656. doi: 10.1016/j.neuron.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamabhai M, Hoffman NG, Hardison NL, McPherson PS, Castagnoli L, Cesareni G, Kay BK. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem. 1998;273:31401–31407. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Pasquale EB. Eph receptors in the adult brain. Curr Opin Neurobiol. 2004;14:288–296. doi: 10.1016/j.conb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Philpot BD. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology. 2008;55:1081–1094. doi: 10.1016/j.neuropharm.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuste R, Majewska A. On the function of dendritic spines. Neuroscientist. 2001;7:387–395. doi: 10.1177/107385840100700508. [DOI] [PubMed] [Google Scholar]
- Zamanian JL, Kelly RB. Intersectin 1L guanine nucleotide exchange activity is regulated by adjacent src homology 3 domains that are also involved in endocytosis. Mol Biol Cell. 2003;14:1624–1637. doi: 10.1091/mbc.E02-08-0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The polarity protein PAR-3 and TIAM1 cooperate in dendritic spine morphogenesis. Nat Cell Biol. 2006;8:227–237. doi: 10.1038/ncb1368. [DOI] [PubMed] [Google Scholar]
- Zhang H, Macara IG. The PAR-6 polarity protein regulates dendritic spine morphogenesis through p190 RhoGAP and the Rho GTPase. Dev Cell. 2008;14:216–226. doi: 10.1016/j.devcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Horwitz AF. Synapse formation is regulated by the signaling adaptor GIT1. J Cell Biol. 2003;161:131–142. doi: 10.1083/jcb.200211002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J Neurosci. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, Poo MM. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Ziv NE, Smith SJ. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron. 1996;17:91–102. doi: 10.1016/s0896-6273(00)80283-4. [DOI] [PubMed] [Google Scholar]