Abstract
Computational systems biology is empowering the study of drug action. Studies on biological effects of chemical compounds have increased in scale and accessibility, allowing integration with other large-scale experimental data types. Here, we review computational approaches for elucidating the mechanisms of both intended and undesirable effects of drugs, with the collective potential to change the nature of drug discovery and pharmacological therapy.
Keywords: Computation, Computer Modeling, Drug Action, Genetics, Yeast Genetics
Introduction
Empowered by advances in genomic research, target-based drug discovery promises greater throughput with systematic knowledge-driven approaches. Target-based drug discovery starts by identifying genes for which perturbation of activity can yield a desirable phenotype and seeks to discover or engineer compounds that interact with these genes' products selectively to alter the disease state or symptoms in a positive way.
Here, we look at an important problem in target-based drug discovery: identifying the molecular targets of chemical compounds. Being able to recognize the cellular entities that a compound interacts with helps identify potential therapeutic applications and undesirable effects and facilitates development of binding assays for identifying alternative compounds with greater specificity. High attrition rate is well recognized as a major hurdle in the drug discovery pipeline (1–3), suggesting a general insufficiency in the understanding of drug effects in the initial stages. Even drugs with well established mechanisms can have less understood secondary targets. Such “off-targets” can lead to unexpected side effects and, in extreme cases, severe adverse reactions in individuals with certain genotypes (4).
Advances in biotechnology in recent years made it possible to study genome-wide responses to genetic and chemical perturbations in vivo (5–12). These studies generated unbiased phenotypic data that capture drug response on a systems level. The proliferation of public databases documenting structural, chemical, and biological activities of chemical compounds also contributes to a diverse pool of accessible data for studying drug effects (13, 14). Increasingly rich data sources have outgrown the ability of humans to reason over the data in its entirety. However, computational biology offers to reveal novel connections between chemical compounds, biological entities, and phenotypes. Here, we will review computational approaches in systems biology that seek to understand both intended and undesired effects of known drugs and novel chemical compounds.
Chemical-Genomic Expression Profiling
DNA microarrays offer the power to characterize genome-wide changes in gene expression and have had a profound influence on molecular biology (15). By using expression profiling to characterize cell response to genetic and chemical perturbations, associations between genes and drugs can be identified.
Comparative Analysis
In 1998, Marton et al. (16) hypothesized that, because an ideal inhibitory drug inhibits the activities of its protein target specifically and completely, null mutants in the corresponding gene should “phenocopy” wild-type cells treated with the drug. To provide support for this hypothesis, the authors compared the genome-wide gene expression profiles of wild-type Saccharomyces cerevisiae cells exposed to FK506 and cyclosporin A with that of a null mutant of calcineurin, a protein phosphatase inactivated (indirectly) by both drugs. They reported that the gene expression patterns of cells treated with either compound exhibited significant correlation with those of the mutants. In contrast, no correlation was observed between the expression profiles of drug-treated cells and mutants deleted for a randomly selected gene. Similarly, cells treated with 3-aminotriazole exhibited gene expression patterns that highly correlated with those of null mutants in the HIS3 gene, which encodes one of the targets of 3-aminotriazole. Anticipating that drugs inhibiting different molecular targets may yield similar expression patterns, the authors proposed a “Decoder” strategy to identify the real drug target given a set of candidate targets. Based on the authors' hypothesis, one expects drug treatment of null mutants in the target gene to exhibit little expression fluctuation relative to the mutant strain. Conversely, drug treatment of null mutants in a non-target should yield expression patterns similar to drug treatment of the wild-type strain. The authors demonstrated that this strategy can distinguish cna and fpr1 strains, for which deleted genes are involved in the drug action of FK506, from a control mutant not expected to be inhibited by the drug. Iterative use of the strategy to identify secondary targets was also proposed. Cells treated with higher doses of FK506 exhibited significant expression changes in some genes that showed no significant expression fluctuation in cna and fpr1 mutants. A similar off-target expression subpattern based on this set of genes is found in gcn4 mutants, indicating that Gcn4 is a possible secondary target of FK506. Treatment of gcn4 mutants with FK506 did not lead to significant expression changes in the majority of this subset of genes relative to gcn4 mutants, suggesting that Gcn4 is implicated in a secondary FK506 response pathway.
Similar concepts were revisited at a larger scale by Hughes et al. (5), who described the “Rosetta” Compendium of 300 genome-wide expression profiles of gene mutants or drug treatments versus mock-treated cultures grown in otherwise similar conditions. In addition to 276 null mutants of viable genes, 11 tetracycline-repressible alleles of essential genes were included. Two-dimensional hierarchical clustering of the profiles revealed many biologically relevant clusters. For several of the 13 experiments that involved treatment of cells with well characterized drugs, drug treatment clustered with mutants implicated in a targeted pathway. Examples include lovastatin/hmg2, itraconazole/erg11, cycloheximide/yef3, hydroxyurea/rnr1, and tunicamycin/gas1. A novel association was found between ERG2 and the topical anesthetic dyclonine and was confirmed in independent experiments.
Lamb et al. (17) used a similar approach to study drug mechanisms in mammalian species. The authors profiled the mRNA expression of ∼22,000 genes in cultured human cells from several cancer cell lines subjected to the treatment of 164 small molecules, including Food and Drug Administration-approved drugs. A “query signature” can be searched against these expression data (referred to as the “Connectivity Map”) to look for interesting connections with these compounds. The query signature is a list of genes labeled with binary values indicating increase or decrease of expression in a biologically interesting condition such as chemical or genetic perturbations. A rank-based enrichment analysis similar to that proposed in Ref. 18 is used to compute the “connectivity score” between the signature and each drug in the collection. Connectivity scores identified the uncharacterized drug gedunin as an HSP90 inhibitor based on connectivity to geldanamycin and its analogs. A conceptual advance relative to the Rosetta Compendium was identification of compounds with an expression effect opposite that of a reference profile. For example, the anti-estrogens fulvestrant, tamoxifen, and raloxifene showed negative connectivity scores to 17β-estradiol.
Network Reconstruction
Deletions of genes downstream of a drug target pathway can yield expression patterns similar to null mutants of the target, hindering target identification via comparative analyses. Gardner et al. (19) proposed a computational approach to alleviate this problem. Adapting a technique from a branch of engineering known as system identification, the authors proposed network identification by multiple regression (NIR),3 which reconstructs an approximated causal network between genes in the drug-affected pathway from steady-state gene expression patterns exhibited from various perturbations. Expression is modeled as a weighted sum of changes due to external perturbations and abundances of regulatory genes. Weights are estimated using linear regression. The concept was tested on a nine-gene subnetwork in Escherichia coli. Perturbations were delivered by inducing overexpression of gene(s) on a plasmid, and expression was measured using quantitative PCR. By applying the model to expression data of cells with two genes simultaneously overexpressed, perturbed genes were accurately identified. Given expression data generated from cells treated with mitomycin C, the model identified known target recA but also flagged another operon (umuDC).
Although NIR effectively identified primary drug targets from expression data of a small set of genes, it was not clear if it would work with genome-scale expression data from microarrays. di Bernardo et al. (20) proposed mode-of-action by network identification (MNI) and applied it to 515 whole genome expression profiles to identify drug targets in S. cerevisiae, including profiles from the Rosetta Compendium. MNI is similar to NIR; however, instead of requiring single gene perturbations to train the network model, MNI can use generic conditions such as drug treatment and environmental stress as inputs. A recursive approach similar to expectation maximization is used to estimate perturbations of specific genes from steady-state expression profiles. MNI correctly identified perturbed genes in 8 of 11 tetracycline-regulatable alleles from the Rosetta Compendium. The algorithm also identified genes involved in target-associated pathways for 7 of 9 drugs with known targets.
Chemical-Genomic Genetic Profiling
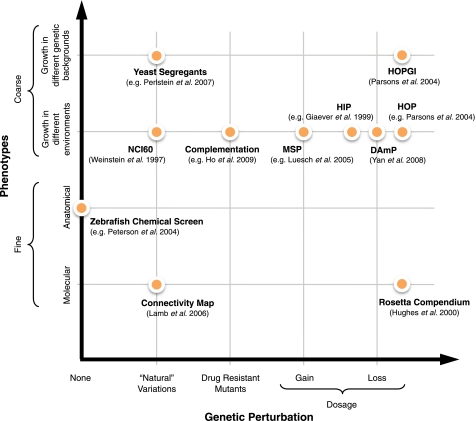
Phenotypic assays of cells under simultaneous genetic and chemical perturbations allow systematic assessment of the effect of genetic changes on drug response. Fig. 1 summarizes chemical-genetic approaches used to study drug/target relationships.
FIGURE 1.
Chemical-genetic approaches for studying drug/target relationships. HIP, haploinsufficiency profiling; HOP, homozygous deletion profiling; HOPGI, HOP/genetic interactions analysis; MSP, multicopy suppression profiling.
Drug Response of Genetic Variants
In the late 1980s, researchers at the National Cancer Institute (NCI) developed a drug screen to systematically characterize inhibitory effects of chemical compounds on the growth of ∼60 distinct human tumor cell lines. Most compounds were tested at five concentrations to determine the GI50, the concentration at which cell growth is inhibited by 50% (21). Using these GI50 profiles for each compound and a database associating molecular targets or modulators of activity in the 60 cell lines, Weinstein et al. (22) related molecular targets to drugs. The database included small-scale studies as well as protein expression measured en masse using two-dimensional PAGE. By computing correlation of drug/target pairs for 3989 compounds and 113 targets, the authors found clusters of similar and mechanistically related compounds (e.g. Taxol analogs, thiosemicarbazones, and the closely related clinical agents cisplatin and carboplatin). In another example, the authors identified a cluster of 155 compounds exhibiting highly negative correlation with targets related to Pgp/Mdr-1 activity and significantly enriched for Mdr-1 substrates. The approach was used to predict compounds that are particularly active against mutant p53 human tumor cells.
Brem and Kruglyak (23) studied the properties of quantitative trait locus (QTL) detection from genome-wide gene expression patterns of 104 segregants from the mating of two genetically diverse yeast strains, lab strain BY4716 and wild-strain RM11-1a. Perlstein et al. (10) treated these segregants with 100 small molecule compounds and measured their growth at multiple time points. Hierarchical clustering of the growth data revealed clusters of compounds that induce similar physiological effects but lack significant structural similarity. Linkage analysis identified 124 unique linkages between genetic markers and loci. Seven of eight QTL “hot spots” associated with multiple compounds are at loci reported to affect the abundance of multiple transcripts and therefore may contain pleiotropic regulators. Secondary assays supported the associations of two loci with distinct compounds.
A major limitation of the approach is its dependence on informative genetic variation between parental strains. In the study, Perlstein et al. (10) reported that five loci corresponding to known targets are undetected in the QTLs, as no variation exists in these genes between parental strains. For example, two drugs used in the experiment with known molecular targets (cycloheximide/RPL41a and RPL41b (24) and haloperidol/ERG2) were not associated with their targets. Also, associated loci often contain many genes, making it difficult to pinpoint the gene responsible for the association.
Gene Dosage Assays
Phenotypic abnormality arising from function loss of one gene copy in a diploid cell, or haploinsufficiency, is not common in wild-type organisms (25, 26). However, Giaever et al. (11) systematically identified many S. cerevisiae genes that exhibit haploinsufficiency under certain conditions, such as in the presence of drugs that target their gene products. For example, heterozygous deletion strains of six known drug targets exhibited haploinsufficiency in the presence of the drug that targeted them. The authors explored the feasibility of using this phenomenon to study drug/target interactions at a larger scale by growing 233 heterozygous deletion strains competitively in the presence of tunicamycin. The strains are tagged with molecular barcodes, facilitating quantification of relative growth rates in a mixed population of competing cells. Drug-induced haploinsufficiency was observed at three loci, one of which encodes the known target of tunicamycin, ALG7. This exciting finding spurred a series of chemical-genomic studies at increasing scales.
Lum et al. (6) screened ∼3500 heterozygous yeast strains in 78 compounds and reported that many known drug targets displayed significant drug-induced haploinsufficiency. Parsons et al. (7) conducted a genome-wide drug sensitivity screen on ∼4700 viable haploid deletion strains in S. cerevisiae for 12 compounds. Employing genetic interactions previously reported using synthetic genetic array analysis (27) as well as genetic interactions selectively tested based on known targets and pathways of the drugs, the authors performed two-dimensional hierarchical clustering of the combined set of chemical-genetic and genetic interaction profiles. Three known drug/target pairs were found to cluster together: fluconazole with ERG1, benomyl with TUB2, and cyclosporine A and FK506 with CNB1. They followed this up with a larger study in 2006 that encompassed 82 compounds and ∼5000 haploid deletion strains (8). Cluster analyses using two-dimensional hierarchical clustering and probabilistic sparse matrix factorization analysis (28) revealed compounds with related drug effects such as actin-binding agents and microtubule poisons. The similarity between the haploid deletion profiles of the breast cancer drug tamoxifen and amiodarone suggested a common effect of calcium homeostasis disruption that was novel to tamoxifen. Independent assays provided evidence that treatment with both drugs activated the Ca2+/calcineurin/Crz1 signaling pathway. Hillenmeyer et al. (9) conducted the largest drug sensitivity screening to date, testing each of ∼5000 haploid and ∼6000 heterozygous diploid deletion strains of S. cerevisiae against 178 and 354 unique conditions, respectively. These conditions included both environmental and chemical perturbations.
Yan et al. (29) explored “decreased abundance by mRNA perturbation” (DAmP) alleles, wherein an mRNA-destabilizing cassette is introduced into the 3′-untranslated region of a gene as an alternative to heterozygous gene deletions to capture drug effects. An earlier study of 20 DAmP alleles in yeast reported mRNA levels in these strains to be ∼5–50% that of wild-type levels (30), which is lower than the typical ∼50% reduction in heterozygous deletion strains and therefore more likely to yield detectable phenotypes. To efficiently barcode large collections of yeast strains, the authors built a set of “Barcoders,” donor strains with unique molecular barcodes that can be transferred to any yeast collection en masse (29), and used them to tag DAmP strains of 958 essential yeast genes. Relative to wild-type controls, DAmP alleles exhibited greater sensitivity to drug treatment compared with heterozygous deletion strains for most drugs.
The preceding approaches reduced gene dosage to find the drug target gene, which might be expected to exhibit increased drug sensitivity. A complementary genetic approach is to seek genes for which increased gene dosage increases drug resistance, e.g. multicopy suppression of drug sensitivity (31, 32). Meijer et al. (33) employed a similar approach to identify genes mediating anti-estrogen resistance in human breast cancer cells. The cancer cells were infected with retroviral cDNA libraries derived from human brain and placenta and mouse embryo. PCR amplification from the integrated construct revealed genes that enabled proliferation in the presence of 4-hydroxytamoxifen. Thus, seven known genes and one putative gene were identified in this screen, with the cDNA of the latter being the most abundant. Subsequent analysis confirmed the gene to be responsible for tamoxifen resistance and designated it as BCAR4 (breast cancer anti-estrogen resistance 4). Hoon et al. (12) integrated haploinsufficiency and homozygous profiling with multicopy suppression to study known and novel compounds. Using heterozygous deletion strains for essential genes and homozygous deletion strains for nonessential genes, the authors found that multicopy suppression allowed them to distinguish target from non-target genes among those genes exhibiting haploinsufficiency for the drugs rapamycin, methotrexate, fluconazole, and calyculin A.
Complementation Assays
Ho et al. (34) explored another large-scale chemical-genetic approach that diverges from conventional gene dosage-altering assays. To identify targets of a drug, drug-resistant mutants were isolated. Where mutants are recessive and localized to one gene, introducing a wild-type copy of the mutated gene should restore drug sensitivity. To apply this approach at large-scale, the authors constructed a library of barcoded plasmids, each harboring a distinct yeast ORF (collectively covering ∼90% of the genome). Plasmids for each gene were crossed into each drug-resistant mutant strain using synthetic genetic array technology (27). Potential drug targets were identified among barcoded plasmids that were highly depleted upon drug treatment. The assay revealed known drug mechanisms for cycloheximide and rapamycin. The sordarin-associated gene EFT2 was also identified using a yeast strain with higher mutation rates. The novel target MVD1 was isolated for the drugs theopalauamide and stichloroside, and secondary analysis suggested that the drugs are likely to act on the ergosterol pathway modulated by Mvd1 activity.
Complex Phenotypes
Although cell growth is the primary phenotype used in genetic and chemical perturbation studies, more complex and specific phenotypes (e.g. cell morphology traits) can be quantified with microscopy and image analysis (35–37). These genetic approaches can also be extended to mammalian organisms using cell lines and gene silencing with siRNA. Loo et al. (37) conducted image-based profiling of HeLa cells under the treatment of 100 compounds. Neumann et al. (38) performed genome-wide phenotypic profiling of HeLa cells silenced for each of ∼21,000 human protein-coding genes through transfection with RNAi using time-lapse microscopy and computational image processing to identify mitosis-related morphological features. Among metazoans, zebrafish is highly amenable to whole organism large-scale genetic or chemical screens and serves as a faithful model of a variety of human diseases (39). A screen for compounds suppressing the zebrafish mutation gridlock, which models aortic “narrowing” (coarctation), led to the discovery of two structurally related compounds that can ameliorate the disease phenotype (40). The nematode Caenorhabditis elegans is another good model for some human pathways and is amenable to large-scale genetics studies in vivo (41). To circumvent its extensive physical and enzymatic xenobiotic defenses, Burns et al. (42) developed a structure-based computational model that predicts bioavailability of small molecules in the worm. The model can be used to preselect suitable compounds prior to screening and is expected to accelerate future screening efforts.
Drug/Protein Association Networks
Large-scale efforts in chemical and pharmacological research have made available vast amounts of chemical-related information in structured formats that are amenable to computational integration and analysis (43). This opens up new opportunities for studying drug effects using systems level computational methods.
Yamanishi et al. (44) constructed a bipartite interaction network between compounds and targets for four classes of human drug targets (enzymes, ion channels, G protein-coupled receptors, and nuclear receptors) and designed a Euclidean space matrix representation for the graph based on Gaussian functions. They refer to this matrix as the “pharmacological feature space.” To predict novel interactions for a protein or a compound that is not within the network, two similarity measures are used: sequence similarity between the protein and targets in the network based on sequence alignment (45) and structural similarity between compounds and drugs in the network based on SIMCOMP (46). The authors used a kernel regression model to represent mapping between structure/sequence similarities of a new entity (compound/protein) and existing entities in the network to its predicted vector in the Euclidean space matrix. Weight parameters in the model are derived from the existing network by minimizing a loss of function. Given a new protein or compound, the trained model can then be used to predict its drug/gene interactions with entities in the network by mapping the structure/sequence similarities back into the pharmacological feature space. He et al. (47) proposed a similar approach using different feature representations. The frequency of 28 functional chemical groups is used as a descriptor for compounds (48), whereas protein targets are represented by a set of features based on the concept of “pseudo amino acid composition” (49). These features include amino acid compositions as well as summary statistics of the distribution and transition frequency between different states in the protein sequence for each of six biochemical and physiochemical properties. A nearest neighbor approach is used to predict novel drug/target relationships from existing ones.
Keiser et al. (50) proposed the “similarity ensemble approach,” which groups proteins based on the chemical similarity of the ligands with which they interact. Using data extracted from the MDL drug reports, the authors compiled a set of 246 protein targets, each annotated with hundreds of drug-like ligands. A similarity score between two protein targets is computed based on the Tanimoto coefficients between all possible pairs of ligands such that one ligand is in each target set. Among protein targets with significant similarity to ligands associated with methadone, the authors found μ-opioid receptors known to be targeted by methadone. They also found M3 muscarinic receptor antagonists, suggesting an off-target relationship subsequently confirmed by binding assay. Applying this technique to >12,000 PubChem compounds associated 30 compounds with unexpected drug effects. The authors selected two of these, emetine/α2-adrenergic receptors and loperamide/neurokinin-2 receptors, and established significant binding affinities for these off-targets in direct binding and function assays. The same technique was used to identify new molecular targets for 3665 compounds, which included 878 Food and Drug Administration-approved drugs (51). The authors inspected 184 predictions in detail and tested 30 of them with radioligand competition assays, of which 23 yielded low inhibition constants (Ki < 15 μm).
Campillos et al. (52) described the identification of secondary targets for existing drugs based on the similarity of their side effects. Terms describing side effects were extracted from drug package inserts of 746 human-marketed drugs and mapped onto the UMLS® (Unified Medical Language System) ontology. Similarity between each drug pair was computed by the number of common side effects, corrected for dependences between related terms. Comparing side effect annotations with a reference set of 502 drugs with known drug/target relations from the MATADOR (53), DrugBank (14), and the Psychoactive Drug Screening Program (PDSP) Ki (54) Databases, the authors found an inverse logarithmic correlation between the frequency of a side effect term and the likelihood that drugs with the term share a common protein target. Benchmarking against the reference set, the authors found the p values of the side effect similarity score between drug pairs to correlate very well with target sharing. Using a combined scoring method incorporating both two-dimensional Tanimoto chemical similarity scores and side effect similarity, 2903 pairs of drugs were estimated to share drug targets with a probability of >25%. The authors tested some of the findings that were most surprising (little chemical similarity, not known to share targets, and not known to be in the same therapeutic category). Among the drugs predicted to share targets with the anti-ulcer drug rabeprazole are two other nervous system drugs: fluoxetine and pergolide. Binding assays revealed that like fluoxetine and pergolide, rabeprazole targets the dopamine receptor D3, which may explain some of its reported side effects. Among 20 drug pairs tested, 13 have at least one predicted target validated by in vitro binding assays.
As more complex organisms are studied, future screens will inevitably assay phenotypes of greater variety and complexity, creating new opportunities and challenges for computational analysis. Creative aggregation and manipulation of diverse data such as chemical structure, genomic sequence, ontologies, and unstructured text data should provide novel ways to understand drug mechanisms from systematically collected data.
This is the fifth article in the Thematic Minireview Series on Computational Systems Biology. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012
- NIR
- network identification by multiple regression
- MNI
- mode-of-action by network identification
- QTL
- quantitative trait locus
- DAmP
- decreased abundance by mRNA perturbation.
REFERENCES
- 1. Kola I., Landis J. (2004) Nat. Rev. Drug. Discov. 3, 711–715 [DOI] [PubMed] [Google Scholar]
- 2. Sams-Dodd F. (2005) Drug Discov. Today 10, 139–147 [DOI] [PubMed] [Google Scholar]
- 3. Brown D. (2007) Drug Discov. Today 12, 1007–1012 [DOI] [PubMed] [Google Scholar]
- 4. Tatonetti N. P., Liu T., Altman R. B. (2009) Genome Biol. 10, 238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hughes T. R., Marton M. J., Jones A. R., Roberts C. J., Stoughton R., Armour C. D., Bennett H. A., Coffey E., Dai H., He Y. D., Kidd M. J., King A. M., Meyer M. R., Slade D., Lum P. Y., Stepaniants S. B., Shoemaker D. D., Gachotte D., Chakraburtty K., Simon J., Bard M., Friend S. H. (2000) Cell. 102, 109–126 [DOI] [PubMed] [Google Scholar]
- 6. Lum P. Y., Armour C. D., Stepaniants S. B., Cavet G., Wolf M. K., Butler J. S., Hinshaw J. C., Garnier P., Prestwich G. D., Leonardson A., Garrett-Engele P., Rush C. M., Bard M., Schimmack G., Phillips J. W., Roberts C. J., Shoemaker D. D. (2004) Cell. 116, 121–137 [DOI] [PubMed] [Google Scholar]
- 7. Parsons A. B., Brost R. L., Ding H., Li Z., Zhang C., Sheikh B., Brown G. W., Kane P. M., Hughes T. R., Boone C. (2004) Nat. Biotechnol. 22, 62–69 [DOI] [PubMed] [Google Scholar]
- 8. Parsons A. B., Lopez A., Givoni I. E., Williams D. E., Gray C. A., Porter J., Chua G., Sopko R., Brost R. L., Ho C. H., Wang J., Ketela T., Brenner C., Brill J. A., Fernandez G. E., Lorenz T. C., Payne G. S., Ishihara S., Ohya Y., Andrews B., Hughes T. R., Frey B. J., Graham T. R., Andersen R. J., Boone C. (2006) Cell. 126, 611–625 [DOI] [PubMed] [Google Scholar]
- 9. Hillenmeyer M. E., Fung E., Wildenhain J., Pierce S. E., Hoon S., Lee W., Proctor M., St Onge R. P., Tyers M., Koller D., Altman R. B., Davis R. W., Nislow C., Giaever G. (2008) Science 320, 362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perlstein E. O., Ruderfer D. M., Roberts D. C., Schreiber S. L., Kruglyak L. (2007) Nat. Genet. 39, 496–502 [DOI] [PubMed] [Google Scholar]
- 11. Giaever G., Shoemaker D. D., Jones T. W., Liang H., Winzeler E. A., Astromoff A., Davis R. W. (1999) Nat. Genet. 21, 278–283 [DOI] [PubMed] [Google Scholar]
- 12. Hoon S., Smith A. M., Wallace I. M., Suresh S., Miranda M., Fung E., Proctor M., Shokat K. M., Zhang C., Davis R. W., Giaever G., St Onge R. P., Nislow C. (2008) Nat. Chem. Biol. 4, 498–506 [DOI] [PubMed] [Google Scholar]
- 13. Kuhn M., von Mering C., Campillos M., Jensen L. J., Bork P. (2008) Nucleic Acids Res. 36, D684–D688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wishart D. S., Knox C., Guo A. C., Shrivastava S., Hassanali M., Stothard P., Chang Z., Woolsey J. (2006) Nucleic Acids Res. 34, D668–D672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lenoir T., Giannella E. (2006) J. Biomed. Discov. Collab. 1, 11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marton M. J., DeRisi J. L., Bennett H. A., Iyer V. R., Meyer M. R., Roberts C. J., Stoughton R., Burchard J., Slade D., Dai H., Bassett D. E., Jr., Hartwell L. H., Brown P. O., Friend S. H. (1998) Nat. Med. 4, 1293–1301 [DOI] [PubMed] [Google Scholar]
- 17. Lamb J., Crawford E. D., Peck D., Modell J. W., Blat I. C., Wrobel M. J., Lerner J., Brunet J. P., Subramanian A., Ross K. N., Reich M., Hieronymus H., Wei G., Armstrong S. A., Haggarty S. J., Clemons P. A., Wei R., Carr S. A., Lander E. S., Golub T. R. (2006) Science 313, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 18. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gardner T. S., di Bernardo D., Lorenz D., Collins J. J. (2003) Science 301, 102–105 [DOI] [PubMed] [Google Scholar]
- 20. di Bernardo D., Thompson M. J., Gardner T. S., Chobot S. E., Eastwood E. L., Wojtovich A. P., Elliott S. J., Schaus S. E., Collins J. J. (2005) Nat. Biotechnol. 23, 377–383 [DOI] [PubMed] [Google Scholar]
- 21. Shoemaker R. H. (2006) Nat. Rev. Cancer. 6, 813–823 [DOI] [PubMed] [Google Scholar]
- 22. Weinstein J. N., Myers T. G., O'Connor P. M., Friend S. H., Fornace A. J., Jr., Kohn K. W., Fojo T., Bates S. E., Rubinstein L. V., Anderson N. L., Buolamwini J. K., van Osdol W. W., Monks A. P., Scudiero D. A., Sausville E. A., Zaharevitz D. W., Bunow B., Viswanadhan V. N., Johnson G. S., Wittes R. E., Paull K. D. (1997) Science 275, 343–349 [DOI] [PubMed] [Google Scholar]
- 23. Brem R. B., Kruglyak L. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 1572–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stöcklein W., Piepersberg W. (1980) Antimicrob. Agents Chemother. 18, 863–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lindsley D. L., Sandler L., Baker B. S., Carpenter A. T., Denell R. E., Hall J. C., Jacobs P. A., Miklos G. L., Davis B. K., Gethmann R. C., Hardy R. W., Steven A. H., Miller M., Nozawa H., Parry D. M., Gould-Somero M. (1972) Genetics 71, 157–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fisher E., Scambler P. (1994) Nat. Genet. 7, 5–7 [DOI] [PubMed] [Google Scholar]
- 27. Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., Andrews B., Tyers M., Boone C. (2001) Science 294, 2364–2368 [DOI] [PubMed] [Google Scholar]
- 28. Dueck D., Morris Q. D., Frey B. J. (2005) Bioinformatics 21, i144–i151 [DOI] [PubMed] [Google Scholar]
- 29. Yan Z., Costanzo M., Heisler L. E., Paw J., Kaper F., Andrews B. J., Boone C., Giaever G., Nislow C. (2008) Nat. Methods 5, 719–725 [DOI] [PubMed] [Google Scholar]
- 30. Schuldiner M., Collins S. R., Thompson N. J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J. F., Weissman J. S., Krogan N. J. (2005) Cell 123, 507–519 [DOI] [PubMed] [Google Scholar]
- 31. Luesch H., Wu T. Y., Ren P., Gray N. S., Schultz P. G., Supek F. (2005) Chem. Biol. 12, 55–63 [DOI] [PubMed] [Google Scholar]
- 32. Butcher R. A., Bhullar B. S., Perlstein E. O., Marsischky G., LaBaer J., Schreiber S. L. (2006) Nat. Chem. Biol. 2, 103–109 [DOI] [PubMed] [Google Scholar]
- 33. Meijer D., van Agthoven T., Bosma P. T., Nooter K., Dorssers L. C. (2006) Mol. Cancer Res. 4, 379–386 [DOI] [PubMed] [Google Scholar]
- 34. Ho C. H., Magtanong L., Barker S. L., Gresham D., Nishimura S., Natarajan P., Koh J. L., Porter J., Gray C. A., Andersen R. J., Giaever G., Nislow C., Andrews B., Botstein D., Graham T. R., Yoshida M., Boone C. (2009) Nat. Biotechnol. 27, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ohya Y., Sese J., Yukawa M., Sano F., Nakatani Y., Saito T. L., Saka A., Fukuda T., Ishihara S., Oka S., Suzuki G., Watanabe M., Hirata A., Ohtani M., Sawai H., Fraysse N., Latgé J. P., François J. M., Aebi M., Tanaka S., Muramatsu S., Araki H., Sonoike K., Nogami S., Morishita S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 19015–19020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Neumann B., Held M., Liebel U., Erfle H., Rogers P., Pepperkok R., Ellenberg J. (2006) Nat. Methods 3, 385–390 [DOI] [PubMed] [Google Scholar]
- 37. Loo L. H., Wu L. F., Altschuler S. J. (2007) Nat. Methods 4, 445–453 [DOI] [PubMed] [Google Scholar]
- 38. Neumann B., Walter T., Hériché J. K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebel U., Cetin C., Sieckmann F., Pau G., Kabbe R., Wünsche A., Satagopam V., Schmitz M. H., Chapuis C., Gerlich D. W., Schneider R., Eils R., Huber W., Peters J. M., Hyman A. A., Durbin R., Pepperkok R., Ellenberg J. (2010) Nature 464, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lieschke G. J., Currie P. D. (2007) Nat. Rev. Genet. 8, 353–367 [DOI] [PubMed] [Google Scholar]
- 40. Peterson R. T., Shaw S. Y., Peterson T. A., Milan D. J., Zhong T. P., Schreiber S. L., MacRae C. A., Fishman M. C. (2004) Nat. Biotechnol. 22, 595–599 [DOI] [PubMed] [Google Scholar]
- 41. Kaletta T., Hengartner M. O. (2006) Nat. Rev. Drug. Discov. 5, 387–398 [DOI] [PubMed] [Google Scholar]
- 42. Burns A. R., Wallace I. M., Wildenhain J., Tyers M., Giaever G., Bader G. D., Nislow C., Cutler S. R., Roy P. J. (2010) Nat. Chem. Biol. 6, 549–557 [DOI] [PubMed] [Google Scholar]
- 43. Paolini G. V., Shapland R. H., van Hoorn W. P., Mason J. S., Hopkins A. L. (2006) Nat. Biotechnol. 24, 805–815 [DOI] [PubMed] [Google Scholar]
- 44. Yamanishi Y., Araki M., Gutteridge A., Honda W., Kanehisa M. (2008) Bioinformatics 24, i232–i240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith T. F., Waterman M. S. (1981) J. Mol. Biol. 147, 195–197 [DOI] [PubMed] [Google Scholar]
- 46. Hattori M., Okuno Y., Goto S., Kanehisa M. (2003) J. Am. Chem. Soc. 125, 11853–11865 [DOI] [PubMed] [Google Scholar]
- 47. He Z., Zhang J., Shi X. H., Hu L. L., Kong X., Cai Y. D., Chou K. C. (2010) PLoS ONE 5, e9603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chou K. C., Cai Y. D. (2002) J. Biol. Chem. 277, 45765–45769 [DOI] [PubMed] [Google Scholar]
- 49. Chou K. C. (2001) Proteins Struct. Funct. Genet. 43, 246–255 [DOI] [PubMed] [Google Scholar]
- 50. Keiser M. J., Roth B. L., Armbruster B. N., Ernsberger P., Irwin J. J., Shoichet B. K. (2007) Nat. Biotechnol. 25, 197–206 [DOI] [PubMed] [Google Scholar]
- 51. Keiser M. J., Setola V., Irwin J. J., Laggner C., Abbas A. I., Hufeisen S. J., Jensen N. H., Kuijer M. B., Matos R. C., Tran T. B., Whaley R., Glennon R. A., Hert J., Thomas K. L., Edwards D. D., Shoichet B. K., Roth B. L. (2009) Nature 462, 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Campillos M., Kuhn M., Gavin A. C., Jensen L. J., Bork P. (2008) Science 321, 263–266 [DOI] [PubMed] [Google Scholar]
- 53. Günther S., Kuhn M., Dunkel M., Campillos M., Senger C., Petsalaki E., Ahmed J., Urdiales E. G., Gewiess A., Jensen L. J., Schneider R., Skoblo R., Russell R. B., Bourne P. E., Bork P., Preissner R. (2008) Nucleic Acids Res. 36, D919–D922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Roth B. L., Lopez E., Patel S., Kroeze W. K. (2000) Neuroscientist 6, 252–262 [Google Scholar]