Abstract
Life scientists today cannot hope to read everything relevant to their research. Emerging text-mining tools can help by identifying topics and distilling statements from books and articles with increased accuracy. Researchers often organize these statements into ontologies, consistent systems of reality claims. Like scientific thinking and interchange, however, text-mined information (even when accurately captured) is complex, redundant, sometimes incoherent, and often contradictory: it is rooted in a mixture of only partially consistent ontologies. We review work that models scientific reason and suggest how computational reasoning across ontologies and the broader distribution of textual statements can assess the certainty of statements and the process by which statements become certain. With the emergence of digitized data regarding networks of scientific authorship, institutions, and resources, we explore the possibility of accounting for social dependences and cultural biases in reasoning models. Computational reasoning is starting to fill out ontologies and flag internal inconsistencies in several areas of bioscience. In the not too distant future, scientists may be able to use statements and rich models of the processes that produced them to identify underexplored areas, resurrect forgotten findings and ideas, deconvolute the spaghetti of underlying ontologies, and synthesize novel knowledge and hypotheses.
Keywords: Biophysics, Computation, Computer Modeling, Drug Design, Epigenetics, Computational Biology, Information Cascade, Sociology of Science, Text Mining
Introduction
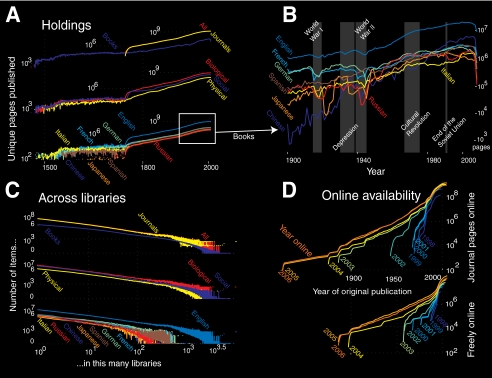
A vast and rapidly growing volume of text traces the succession of findings and ideas that constitute modern science. Extrapolating from global library data, we estimate the world hosts at least a trillion scholarly pages. An incomplete inventory (Fig. 1), divided into biological, social, and physical sciences, contains 400, 200, and 65 billion pages, respectively (see supplemental data). From the Western invention of the printing press in 1453, scientific knowledge has grown, increasingly become published in English, and shifted from books to journals (Fig. 1A). Published knowledge has accumulated fastest in eras of peace and prosperity; it grows much more slowly in turmoil (Fig. 1B). Until recently, access to this knowledge required going to a library. The complete collection of science is, however, distributed so widely across libraries that to assemble all knowledge on any broad topic would require lifelong travel (Fig. 1C). More than one-quarter of the world's basic and applied science books appear in less than ten libraries. Google and the Google Books settlement, which finalized in November 2009, is beginning to reverse this trend by making millions of these books available for search and reading through the Internet. This follows the massive migration of scientific journals online over the past decade (Fig. 1D).
FIGURE 1.
A, estimated number of distinct pages from the Online Computer Library Center WorldCat Database of books and journals in 71,000 libraries across 121 countries, split by manuscripts and journals, broad subject area, and the most common eight languages from 1450 to present. B, manuscript pages, by language, mapped against major historical events of the 20th century. C, distribution of volumes across libraries: number of volumes plotted against the number of Online Computer Library Center libraries in which each are held, split by manuscript and serials, subject, and language. All distributions feature a spiking tail, suggesting a core collection of books that appears in nearly all libraries. D, growth in the number and publication age of journal pages available via the Internet and freely on the Internet (without institutional subscription) from 1998 to 2006.
With the emergence of new journals that are digital at publication and novel ways of expressing findings and hypotheses in science tweets, blogs, and online databases (and with more scientists producing science than ever before), researchers can catalogue only a vanishing fraction of what is relevant to their work by traditional reading and note taking. In response, scientists in many fields have begun to use computation not only to search and browse scientific texts (1) but also to read and reason about them (2). Numerous obstacles remain, but the possibility of enlisting computation in discovery as well as analysis has inspired a growing body of knowledge and tools whose use and development have been nowhere more active than in the molecular life sciences.
Processing Natural Language
The process by which text is refigured into standardized machine-readable representations of meaning is often called semantic analysis. The expressive richness and ambiguity of natural language, however, make automatically extracting statements from scientific text a formidable challenge. For example, consider the difficulty involved in extracting all information from the text “NCOA3 in turn acylates histones, which makes downstream DNA more accessible.” As a result, information extraction (IE),3 a robust approach to semantic analysis, currently avoids attempting to process every phrase in text (e.g. by simply extracting “NCOA3 acetylates histones”). IE assumes a relatively simple fixed template of expected information. Researchers then fill semantic slots with information from text through a series of steps.
First, what is to be extracted is narrowed using a supplied lexicon of semantically classified terms (e.g. genes, enzymes, cofactors, small molecules) (3). The researcher then identifies these classes in text, a step called named entity recognition, using deterministic rules or computational techniques that statistically learn from human-coded data. The accuracy of the best named entity recognition in some domains rivals that of human coders at >90% (4). The researcher then assembles these mentions into basic and then complex noun, verb, and prepositional phrases. Finally, semantic entities and events are recognized, inserted into the template, and merged if they are determined to share a referent. Many biomedical research teams in recent years have extracted protein-protein interactions from text using an IE approach and filling a simple template that involves two or more proteins, an interaction, and occasionally evidence associated with the claim. Following from our previous example, NCOA3 acetylates histones as determined with high resolution mass spectrometry.
A related but more involved method of robust semantic analysis builds on formal grammars that model how the mind generates language. Several exist, including transformational, relational, dependency, construction, and categorial grammars (5). Transformational or constituency grammars (TGs) are the most commonly used and comprise a set of symbols representing constituent words and phrases and the rules by which they are substituted to create language. Parsing a sentence, given a TG, boils down to reconstructing the substitution steps that most likely generated the sentence. Context-free grammars are currently the TG most commonly implemented as they offer the most affordable compromise between expressive power and computational cost. A syntactic context-free grammar renders a sentence through symbols that signify structures like noun and verb phrases, which are each, in turn, substituted for word classes like nouns, verbs, adjectives, and prepositions, which are ultimately replaced with words like “bright” or “phosphorylate.”
Similarly, a semantic grammar operates with symbols that stand for semantic categories such as enzyme, substrate, or enzymatic reaction. The semantic grammar constitutes rules that include selection restrictions, which limit the ways in which entities can appear in meaningful statements within the domain (5). Semantic grammars in science rely on the notion that scientific domains are characterized by specalized scientific sublanguages, which can be characterized by finite sets of rules (6). In robust semantic grammars, one typically mixes syntactic and semantic rules and may implement them deterministically or probabilistically. In a probabilistic grammar, each rule is associated with a probability that can be used to infer the most likely genesis or parse of the sentence. Collectively, these approaches to robust semantic analysis are used extensively to extract meaningful statements in computer science, business intelligence, biology, and medicine. They have been particularly fruitful in fields at the interface of chemistry and biology, where the fundamental importance of molecules and reactions has driven more linguistic conventions than many other areas of science and scholarship. These approaches are also beginning to enter many other natural and social sciences.
Assembling Ontologies
If computation could distill articles into a database of statements, it would still be too large for a researcher to browse. Consider a scientist who is interested in the relationship between human gene p53, which regulates the cell cycle, and cancer, a cell proliferation disorder, and who finds 36,589 relevant articles in PubMed. Alternately, imagine a biochemical researcher interested in all published statements of the form “molecule A is a substrate for enzyme B.” The path to computational reasoning commonly involves aggregating statements from text into singular declarations ordered into a consistent system or ontology. Modern scientific ontologies derive from two traditions. In philosophy, ontology (historically, a branch of metaphysics) is the study of existence. For modern science, this translates into a specification of empirical entities and their organization into statements of fact that highlight essential qualities, parts, and relationships. Ontologies emerged in computer science as an approach to knowledge representation in the early 1990s (7), preceded by practical classification systems like the 19th century Dewey Decimal System and the International Classification of Diseases. First used in artificial intelligence, ontologies now permeate software engineering and database theory, where consistent content schemas facilitate the interoperability of data stores. They are also becoming an integral part of the World Wide Web. In 2004, the World Wide Web Consortium endorsed the OWL Web Ontology Language, which supports the formal semantics required by ontologies, as the centerpiece of its Semantic Web Framework.
Modern scientific ontologies, often classified into light- and heavyweight, contain a controlled vocabulary of concepts and relationships that link them. Lightweight ontologies comprise terminologies or simple taxonomies with little or no information about the entities or relationships. For example, all known enzymes organized into a list or a simple taxonomy (e.g. a kinase is an enzyme) would serve as a valuable data resource. In contrast, heavyweight ontologies like Cyc, a massive schema of common sense knowledge, or the Foundational Model of Anatomy add formal axioms and constraints to characterize entities and relationships distinctive to the domain. A formal axiom in biology might specify that genes encode proteins, but proteins cannot encode genes. Light and heavyweight ontologies may draw upon reference or upper level ontologies to characterize their parts: abstract entities and processes as in Cyc or concrete elements like human bones within the Foundational Model of Anatomy. They then subclass these elements and link them with domain-specific information. The most frequently cited ontology in science, the Gene Ontology (GO), is a structurally lightweight taxonomy that comprises 22,000 entities biologists use to characterize gene products (8). GO statements are concept-relation-concept triplets like “oxidative phosphorylation is a metabolic process” and “photosynthesis, light harvesting is part of photosynthesis, light reaction.” Knowledge bases of text-mined statements similarly draw upon ontologies for the lexicon used to extract entities and relations from text, but duplication and contradiction are permitted as they are in articles (9).
After computer and information science, ontologies are most used in biomedicine but also increasingly in astronomy and diverse areas of engineering, government, and business. Recall the recent ontological debate over whether Pluto is a planet, an asteroid, or a dwarf planet, the controversial appellation eventually contrived by the XXVIth General Assembly of the International Astronomical Union in Prague 2006. Among applied ontologists, there is broad agreement that ontologies should primarily be understood as precise data structures to facilitate sharing and reuse, a kind of object-oriented content.4 Ontologists are divided, however, over whether to promote one ontology that enables/constrains the interoperation of all others or to let a thousand flowers bloom and encourage a wide range customized to scientific usage. Mark Musen, coeditor of Applied Ontology and author of the popular ontology editor Protégé, wrote, “So much of scientific knowledge is not absolute—it is constructed—it is context-dependent. Ontologies can provide an impression of certainty that may not always be appropriate.” Consider, for example, the multiple coexisting definitions of gene concurrently used in biology: a unit of inheritance, a chunk of DNA, and a template for a group of proteins. Others believe ontologies are and should be built only on “settled parts of science.”5 By creating one ontological compendium or one description of existence, however, scientists necessarily preclude others.
Many ontology communities routinely update changes in their systems, and recent work in artificial intelligence is beginning to assist ontology evolution by automatically comparing ontologies and designing repair plans that split functions and add arguments to make them commensurable.6 Some ontologies are also beginning to incorporate uncertainty.7 These amendments suggest an emerging interpretation of ontologies in science, not simply as truth statements or data-sharing structures but as representations of mental constructs through which we organize our growing understanding about the world.
Computational Reasoning
The culminating step of computational reasoning involves building a reasoner or agent that infers new knowledge from the existing statements of an ontology. Many reasoners use variants of unambiguous proposition or first-order predicate logic to make inferences and prove theorems. Automated theorem proving has advanced in recent years and is used intensively in circuit and software design, aerospace, and related industries where verifying a particular operation under all possible conditions is critical. The WolframAlpha Computational Knowledge Engine takes a similar approach to computational question answering by using rules to assemble systematic knowledge across a variety of domains and fit it with algorithms to produce on-demand analysis. Another approach takes computational inference still further by using extracted published knowledge to condition models of chemical and biological agents and then enables these agents to interact to simulate and predict higher level cellular, tissue, and organism outcomes.
Scientific disagreement, alternative views of the world, and uncertainty become most consequential in the reasoning process. For example, are three amino acids in sequence a “small molecule” or a “macromolecule” (a very short protein). Crisp logical reasoners have been unleashed on biomedical ontologies to extend them, for example, by associating genes with biochemical pathways (10).8 Logical approaches have been most successful, however, not in generating new knowledge but in flagging internal contradictions for repair (11).9 Because of contradictions and self-references, many of the most used ontologies (like the SNOMED medical ontology and GO) are insufficiently restricted to be expressed with protocols like OWL and cannot take advantage of many of the reasoners available for consistent systems. This has suggested to others the need to incorporate error into the reasoning process.
Early approaches to probabilistic logic dealt with the challenge of inducing support for a proposition from evidence. Belief networks combine proposition probabilities to model uncertainty within a broader domain.10 Belief networks include nodes, which represent variables, interconnected with arcs, which signify probabilistic influences. In biology, a node might constitute the active or inactive state of a gene, and an arc the probability that the gene is active given the state of the protein that regulates it. The strength of influence between nodes is propagated along the graph via forward conditional probability. Bayesian networks are the most commonly implemented form of belief network. They use Bayesian conditioning as the basis for probability updating and so emphasize the relationship between assumptions and the accuracy of evidence in assessment (12). Consider the use of Bayesian networks to diagnose illness. Given some set of observed symptoms, one can compute probabilities along arcs of the network to compare the likelihood of each possible disease (13). Several implementations of Bayesian networks have been developed to handle particular types of inference: dynamic networks for dynamic systems, causal networks for causal processes, and influence diagrams that add values and choices to the belief network to optimize decision making (14).
Probabilistic reasoners allow researchers to relax the assumption that statements within an ontology or associated knowledge base are certain and universal.11 This strategy reduces the precision of conclusions but can reduce the influence of isolated mistakes and make computational inference possible even in the presence of contradiction. In developing ontologies from literature, however, researchers called curators often excise repetitive, contradictory, and incommensurable statements. They recognize that article statements are unequal, but rather than rank their certainty, curators have tended to censor “uncertain” ones. This is changing. Gene function annotation using the GO now allows for the distinction between experimental findings and structural inference.9
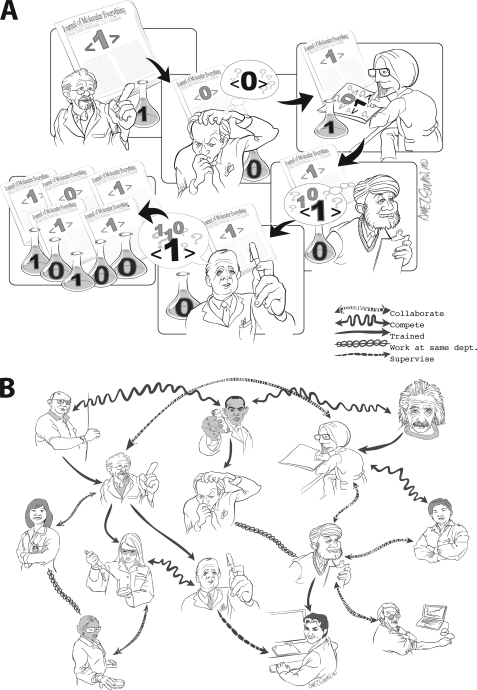
A few researchers have begun to use probabilistic reasoners on the complete collection of statements extracted from literature. Retaining uncertain statements has allowed them to explore how statements become certain. For example, two recent papers explicitly model the process by which statements and citations in molecular biomedical articles respond to each other in information cascades (15, 16). An information cascade is a chain of collective reasoning that degenerates into repetition (17–19). Fig. 2A illustrates this process by showing a sequence of experiments about the same phenomenon (e.g. NCOA3 acetylates histones). The first researcher to investigate the relationship interprets his experiment directly. The second interprets his experiment taking both his own research and the previously published interpretation into account. The third scientist accounts for still more published history and proportionally discounts her findings. This may not appear irrational to the individual scientist. She is acting like a Bayesian statistician: the more prior collective knowledge, the more that knowledge should influence her interpretation. The process is not collectively rational, however, because the sequence of prior published experiments were not independent. The first had much more influence on the resolution than the last.
FIGURE 2.
A, hypothetical temporal sequence of experimental findings (1 and 0 in the beakers) and published articles (1 and 0 in the papers) (15). Early findings are reflected accurately in publications, whereas scientists' interpretation of later findings incorporate the history of publication into account. B, the broader social network in which the scientists in A live. The positive correlation between social ties in B and the propositional agreement in A suggests that communication induces accord.
A recent investigation into the claim that β-amyloid, a protein concentrated in the brains of Alzheimer patients, is produced by and injures skeletal muscle in patients with inclusion body myositis illustrates this process and its potential costs (16). The relationship was first published in five 1992 and 1993 articles produced by two research groups. Before 1996, six articles critical of the claim were also published, two from the same laboratory that had produced four of the five original supportive articles. This is consistent with the hypothesized “Proteus phenomena,” whereby early findings are subjected to contradiction (21). Nevertheless, 242 new analyses and reviews that explored the relationship came out before 2008, and all but one disproportionally cited the positive articles and amplified the claim within the community. Some reviews even made the claim firmer and more general than it had been in its initial context. As a consequence of this cascade of support, interpretation of new experiments neither doubted the claim nor explored other possible roles of β-amyloid. A 2010 paper takes a wider view and shows how β-amyloid may beneficially function as an antimicrobial peptide in the innate immune system (20). This insight raises the possibility that Alzheimer disease is infectious and suggests novel treatment strategies. Regardless of this new claim's efficacy, the information cascade surrounding the deleterious effect of β-amyloid almost certainly prolonged experimental consideration of its possible beneficial role in Alzheimer disease and immunity.
Building on this work could allow analysts to identify the scope of convergence and divergence processes in biology, chemistry, and related fields. Other work has highlighted how research communities are much more aware of some findings than others: those appearing in their own journals over others published about identical topics (22). Ultimately, reasoning across the distribution of untidy statements could allow us to examine the dynamic nature of research attention in the sciences.
Incorporating Social Structure and Culture
The complete distribution of published research statements (even perfectly parsed and analyzed) would still ignore dependences between statements induced by communication. To produce research, scientists engage in multiparty conversations that span university hallways, workshops, conferences, and libraries. Research on the social production of science has begun to trace these linkages using article bylines and acknowledgments to understand how authors and resources organize around research problems into teams (23), networks (24), institutions, and regions (25). In large article collections, author identification has been a challenge, but recent approaches that use many article features are accurate at >98% even for large article collections like PubMed (26).
Improved models of scientific production could allow reasoning models to statistically deconvolute social, geographic, and funding-related dependences among published statements (27). For example, offline discussions about “the best” techniques for inferring protein interactions will likely constrain the range of methods deployed. The projection of social, geographic, and funding networks onto the network of published statements exposes correlations, common repetitions likely induced by the communication of ideas. Fig. 2B illustrates this correlation between personal relationships and scientific interpretations. A finding corroborated by articles from three laboratories with no known association from distant locales using different methods is much more persuasive than one repeated by a Ph.D. advisor and his students. Computational models conditioned on social dependences might weight independent statements with greater confidence.
Incorporating social structure could also improve computational prediction. In a classic analysis, Don Swanson observed that the community studying Raynaud disease noted blood viscosity as a common symptom and that the socially disconnected nutrition community published how dietary fish oil reduced blood viscosity. Swanson hypothesized that fish oil could be beneficial for Raynaud disease patients, and it was found successful in an independent randomized clinical trial (28). Where scientific elements (e.g. blood viscosity) cross the social boundaries between communities, co-occurring elements, problems, and solutions from one domain can be connected with those in another. This approach is the scientific equivalent of market arbitrage: accelerated by computation, it could facilitate “conversations” between contemporary and historical or orphan ideas that were underappreciated in the scientific context of their debut (29).
Beyond social relations, scientists sort into fields with differing knowledge cultures: methods of reasoning, evidentiary standards, and styles of articulation (30). Textual clues provide insight into the patterns that distinguish these cultures. A meta-analysis of oncology articles found that methods sections in those citing industry support were systematically more vague than in articles citing only government funding (31). Articles and patents, even for the same discovery, are also quite different, with the article emphasizing continuity with previous research and the patent highlighting distinction. Uneven contact between related fields leads to concepts from one domain having imperfect analogs in another. The gene of genetics (the basic unit of heredity) historically had an emphasis and theoretical function distinct from the gene of biochemistry (an encoding segment of DNA), but the two have become mangled with contact. When scientists move between fields or draw from multiple domains, meanings intermix. Established terms attract new meanings over time as the context and concerns of research shift (32). In this way, fields, communities, and even individuals within papers host multiple clustered but distinct symbolic systems: their texts draw concepts from a mixture of ontologies.
Following this logic, reasoners could be trained to account for cultural bias. By computationally classifying published statements and estimating the likelihood that each class will enter scientific discourse, reasoners could reweight certainty in underrepresented claims. For example, the results of successful experiments appear in print much more frequently than those of unsuccessful ones. This approach could quantify that likelihood and begin to correct for it. It might also enable prediction by suggesting that logically possible but never published negative statements (e.g. the human gene MIR96 does not increase susceptibility to heart disease) are more likely than unpublished positive ones in densely crowded research areas, but less so in sparse ones between disciplines where many questions remain unasked.
Reasoners that account for the sociocultural dispersion of statements could enable us to recover the mixture of cognitive ontologies that gave rise to them. This could highlight (and subject to testing) higher level ontological disputes in science that lie above the level of most ad hoc hypotheses. Theoretical progress is often associated with the reconciliation of ontologies from multiple theories. Consider the merger of evolution and genetics that precipitated modern evolutionary theory (33) or the common impulse in physics to generate a unified theory. Separating, formalizing, and explicitly comparing the conceptual ontologies that give rise to statements may create novel opportunities to blend ontologies for theoretical experimentation and improvement.6
Conclusion
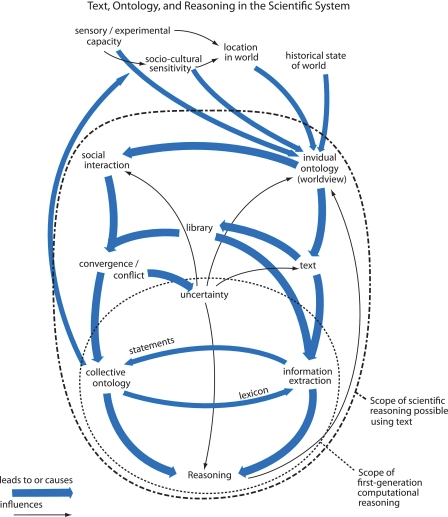
The rapidly increasing volume and electronic availability of published science can seem overwhelming to the modern bioscientist. It also poses a unique opportunity. Recent advances in natural language processing, ontology construction, and reasoning models are being brought together by scientists to computationally read and reason. As analysts begin to extract more of the richness from texts, computational reasoners may become capable of modeling certainty and generating predictions based on the full range of factors scientists have always considered, including the sociocultural processes through which science is produced. Fig. 3 diagrams relationships between published science, ontologies, and reasoning in the scientific system. It points to this expansion of features that researchers are beginning to engage in their models of science and their tools to analyze and advance it.
FIGURE 3.
Elements, context, and processes involved in scientific reasoning. Arrows represent causes or influences. The figure emphasizes the scope of first-generation computational reasoning and the emerging second-generation reasoning we describe in this minireview.
Many obstacles remain, including the need for better models of the production of language and science, more efficient algorithms, and faster computation. Nevertheless, computational extraction and reasoning with ontologies have already begun to help scientists overcome some of the limitations of working within a distinct community. By expanding the set of useful documents through text mining, scientists enlarge the distribution from which they can sample ideas and extend the length and potential of their inferences. Reasoning models have allowed scientists to complete paradigms, like working out the genetic details of hypothesized biopathways. They are also beginning to enable ontology comparison, which could flag opportunities for theoretical recombination that punctuate scientific advance (see supplemental data).
Acknowledgments
We thank all those who agreed to be interviewed for this project (as listed in Footnotes 3 and 4) and for helpful comments on the manuscript and also John Goldsmith and Ian Foster. We are especially grateful to the Online Computer Library Center for allowing us to use WorldCat data, Thomson Reuters for allowing us to use Citation data, and Fulltext Sources Online (FSO) for granting us access to online access data for the analysis in Fig. 1.
This is the sixth article in the Thematic Minireview Series on Computational Systems Biology. This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012

The on-line version of this article (available at http://www.jbc.org) contains supplemental data, Fig. S1, and additional references.
M. Musen, P. Karp, B. Smith, J. Blake, L. Hunter, L. Hirschman, B. Carpenter, and J. Shrager, personal communications.
B. Smith and P. D. Karp, personal communications.
A. Bundy, paper presented at the Fall Symposium for the Association for the Advancement of Artificial Intelligence, Athens, GA, November 5, 2006.
P. C. G. Costa, K. B. Laskey, and K. J. Laskey, paper presented at the Proceedings of the Second ISWC Workshop on Uncertainty Reasoning in the Semantic Web: Frontiers in Artificial Intelligence and Applications (FLOIS), Arlington, VA, November 7–9, 2006.
S. Eker et al., paper presented at the Proceedings of the Pacific Symposium on Biocomputing, 2002.
J. Blake, personal communication.
The mathematics of belief networks are closely related to those of game theory, where winning a game parallels the verification of a scientific conclusion, and the uncertainty of competitive moves maps to uncertainty about scientific evidence and assumptions.
This approach can be captured by second-order logic in the principle of bivalence but cannot exist in first-order statements that demand universal relationships.
- IE
- information extraction
- TG
- transformational grammar.
REFERENCES
- 1. Renear A. H., Palmer C. L. (2009) Science 325, 828–832 [DOI] [PubMed] [Google Scholar]
- 2. Leach S. M., Tipney H., Feng W., Baumgartner W. A., Kasliwal P., Schuyler R. P., Williams T., Spritz R. A., Hunter L. (2009) PLoS Comput. Biol. 5, e1000215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hirschman L., Morgan A. A., Yeh A. S. (2002) J. Biomed. Inform. 35, 247–259 [DOI] [PubMed] [Google Scholar]
- 4. Morgan A. L., Lu Z., Wang X., Cohen A. M., Fluck J., Ruch P., Divoli A., Fundel K., Leaman R., Hakenberg J., Sun C., Liu H., Torres R., Krauthammer M., Lau W. W., Liu H., Hsu C. N., Schuemie M., Cohen K. B., Hirschman L. (2008) Genome Biol. 9, Suppl. 2, S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jurafsky D., Martin J. H. (2000) Speech and Language Processing: An Introduction to Natural Language Processing, Computational Linguistics, and Speech Recognition, Prentice Hall, Upper Saddle River, NJ [Google Scholar]
- 6. Harris Z. S. (1982) in Sublanguage: Studies of Language in Restricted Semantic Domains (Kittredge R., Lehrberger J. eds) pp. 231–236, Walter de Gruyter & Co., Berlin [Google Scholar]
- 7. Gruber T. R. (1993) Knowledge Acquisition 5, 199–220 [Google Scholar]
- 8. Bodenreider O. (2008) Yearb. Med. Inform. 47, 67–79 [PMC free article] [PubMed] [Google Scholar]
- 9. Friedman C., Kra P., Yu H., Krauthammer M., Rzhetsky A. (2001) Bioinformatics 17, S74–S82 [DOI] [PubMed] [Google Scholar]
- 10. Green M. L., Karp P. D. (2006) Nucleic Acids Res. 34, 3687–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horrocks I., Voronkov A. (2006) in Foundations of Information and Knowledge Systems: 4th International Symposium, FolKS 2006, Budapest, Hungary, February 14–17, 2006, Proceedings (Dix J., Hegner S. J. eds) Vol. 3861, p. 201–218, Springer, New York [Google Scholar]
- 12. Pearl J. (1988) Probabalistic Reasoning in Intelligent Systems: Networks of Plausible Inference, p. xix, Morgan Kaufmann Publishers, San Mateo, CA [Google Scholar]
- 13. Shortliffe E. H., Buchanan B. (1975) Math. Biosci. 23, 351–379 [Google Scholar]
- 14. Pearl J. (2000) Causality: Models, Reasoning, and Inference, Cambridge University Press, Cambridge [Google Scholar]
- 15. Rzhetsky A., Iossifov I., Loh J. M., White K. P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4940–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Greenberg S. A. (2009) BMJ 339, b2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Banerjee A. V. (1992) Q. J. Econ. 107, 797–817 [Google Scholar]
- 18. Bikhchandani S., Hirshleifer D., Welch I. (1992) J. Polit. Econ. 100, 992–1026 [Google Scholar]
- 19. Sunstein C. R. (2006) Infotopia: How Many Minds Produce Knowledge?, Oxford University Press, New York [Google Scholar]
- 20. Soscia S. J., Soscia S. J., Kirby J. E., Washicosky K. J., Tucker S. M., Ingelsson M., Hyman B., Burton M. A., Goldstein L. E., Duong S., Tanzi R. E., Moir R. D. (2010) PLoS ONE 5, e9505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ioannidis J. P., Trikalinos T. A. (2005) J. Clin. Epidemiol. 58, 543–549 [DOI] [PubMed] [Google Scholar]
- 22. Cokol M., Iossifov I., Weinreb C., Rzhetsky A. (2005) Nat. Biotechnol. 23, 1243–1247 [DOI] [PubMed] [Google Scholar]
- 23. Guimerà R., Uzzi B., Spiro J., Amaral L. A. (2005) Science 308, 697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Newman M. E. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5200–5205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones B. F., Wuchty S., Uzzi B. (2008) Science 322, 1259–1262 [DOI] [PubMed] [Google Scholar]
- 26. Smalheiser N. R., Torvik V. I. (2009) Annu. Rev. Inform. Sci. Technol. 43, 287–313 [Google Scholar]
- 27. Heckerman D., Chickering D. M., Meek C., Rounthwaite R., Kadie C. (2001) J. Mach. Learn. Res. 1, 49–75 [Google Scholar]
- 28. Swanson D. R. (1990) Bull. Med. Libr. Assoc. 78, 29–37 [PMC free article] [PubMed] [Google Scholar]
- 29. Yetisgen-Yildiz M., Pratt W. (2006) J. Biomed. Inform. 39, 600–611 [DOI] [PubMed] [Google Scholar]
- 30. Knorr-Cetina K. (1999) Epistemic Cultures: How the Sciences Make Knowledge, p. xiii, Harvard University Press, Cambridge, MA [Google Scholar]
- 31. Knox K. S., Adams J. R., Djulbegovic B., Stinson T. J., Tomor C., Bennet C. L. (2000) Ann. Oncol. 11, 1591–1595 [DOI] [PubMed] [Google Scholar]
- 32. Feder M. E. (2007) J. Exp. Biol. 210, 1653–1660 [DOI] [PubMed] [Google Scholar]
- 33. Fisher R. A. (1930) The Genetical Theory of Natural Selection, Clarendon Press, Oxford [Google Scholar]