Abstract
Genetic evidence has implicated both Mdm2 and MdmX as essential in negative regulation of p53. However, the exact role of MdmX in this Mdm2-dependent protein degradation is not well understood. Most, if not all, previous Mdm2 studies used GST-Mdm2 fusion proteins in the in vitro assays. Here, we show that the p53 polyubiquitination activity of GST-Mdm2 is conferred by the GST tag and non-GST-tagged Mdm2 only catalyzes monoubiquitination of p53 even at extremely high concentrations. We further demonstrate that MdmX is a potent activator of Mdm2, facilitating dose-dependent p53 polyubiquitination. This activation process requires the RING domains of both MdmX and Mdm2 proteins. The polyubiquitination activity of Mdm2/MdmX is Mdm2-dependent. Unlike Mdm2 or MdmX overexpression alone, co-overexpression of MdmX and Mdm2 consistently triggered p53 degradation in cells. Moreover, cellular polyubiquitination of p53 was only observable in the cytoplasm where both Mdm2 and MdmX are readily detectable. Importantly, RNAi knockdown of MdmX increased levels of endogenous p53 accompanied by reduced p53 polyubiquitination. In conclusion, our work has resolved a major confusion in the field derived from using GST-Mdm2 and demonstrated that MdmX is the cellular activator that converts Mdm2 from a monoubiquitination E3 ligase to a polyubiquitination E3 ligase toward p53. Together, our findings provide a biochemical basis for the requirement of both Mdm2 and MdmX in the dynamic regulation of p53 stability.
Keywords: E3 Ubiquitin Ligase, Enzymes, p53, Protein Degradation, Protein Stability, Mdm2, MdmX, Monoubiquitination, Polyubiquitination, Tumor Suppressor
Introduction
p53 is a major tumor suppressor protein whose gene mutation or functional inactivation occurs at high frequency in many cancer types (1). p53 is also a stress-responsive protein; it is dormant under normal physiological conditions and activated when cells are exposed to various types of stress and aberrant oncogenic stimuli (2). Upon activation, p53 elicits either cytostatic and/or proapoptotic consequences depending on the cell type and signaling context. Years of studies have revealed multiple mechanisms operating a tight and dynamic regulation of cellular p53 activity. These mechanisms protect cells from accidental p53 activation and ensure proper unleashing of its tumor suppressor activity when needed (3). Such dynamic regulation determines the following properties of p53: its protein abundance, subcellular localization, posttranslational modifications, and binding partners (4). These properties subsequently dictate the proper rapidity, intensity, and duration of p53 activation, as well as its preference for individual transcriptional targets, thus determining the eventual biological outcomes.
The key molecular device in the p53 regulatory network is the p53/Mdm2 feedback loop. Recent advances in the field provided insights into the molecular details of this feedback loop. Importantly, this central regulatory process is further controlled by a number of other critical molecules such as HAUSP, MdmX, and the tumor suppressor ARF (4, 5).
Mdm2 was discovered as the first p53 binding protein and an inhibitor of the transcriptional activity of p53 (6). Subsequently, it was demonstrated by mouse genetics that Mdm2 is an essential negative regulator of p53 activity because Mdm2 knock-out in mice causes embryonic lethality, which can be completely rescued by simultaneous deletion of the p53 gene (7). It is now well established that Mdm2 inhibits p53 through two actions: one is to mask the transcriptional activity of p53 via direct protein interaction and another is to promote p53 proteasomal degradation through its ubiquitin ligase activity, the latter of which provides a very efficient way to down-regulate cellular p53 levels and sets p53 a half-life of ∼20–30 min in normal cells (8–12). Moreover, this Mdm2-mediated ubiquitination process was found to be mutually exclusive with p300/CBP-mediated acetylation at the same lysine residues of p53 (13). Mdm2 belongs to the RING domain E3 ligase family whose enzymatic activity relies on the RING domain (14). A recent mouse genetic study indicated that the RING domain of Mdm2 is indeed essential for its inhibitory effects on p53 in mice (15).
MdmX (or Mdm4) is another p53 binding protein with structural similarity to Mdm2. The p53 binding and RING domains of Mdm2 and MdmX share high levels of homology (16). MdmX and Mdm2 can also bind to each other through their RING domains (17). Mouse genetic studies showed that MdmX is a physiological p53 inhibitor and is not redundant with Mdm2 (18–21). It appears that MdmX inhibits p53 through Mdm2-dependent and independent mechanisms (19). MdmX is also regulated by DNA damage signaling events, indicating its involvement in the p53 response to DNA damage (22–26). Despite our knowledge about the significant role of MdmX in p53 biology, the molecular mechanisms underlying MdmX action remain poorly defined. For example, opposing reports exist in the literature regarding the E3 ligase activity of MdmX and its effect on p53 ubiquitination (5).
Most if not all previous Mdm2 studies used GST-Mdm2 fusion proteins in the in vitro assays. It has been shown that GST-Mdm2 can mediate monoubiquitination of p53 at low concentrations, while promoting polyubiquitination of p53 at high concentrations (30). However, whether Mdm2 is a true monoubiquitination E3 ligase or a polyubiquitination E3 ligase has not been established (31, 30). Intriguingly, here we report that, unlike GST-tagged Mdm2, non-GST full-length Mdm2 possesses weak E3 ligase activity for p53, mediating only monoubiquitination of the protein in an in vitro-reconstituted system even at high concentrations. Addition of MdmX to the reaction activated Mdm2 and converted Mdm2 to a polyubiquitination E3 ligase for p53. Our biochemical and subsequent cellular studies have (a) led to resolution of a major confusion in the field regarding the E3 ligase activity of Mdm2 derived from using GST-Mdm2 fusion proteins and (b) identified the biochemical role of MdmX as a cellular activator of Mdm2 for polyubiquitination of p53. These findings provide a biochemical basis underlying the requirement of both Mdm2 and MdmX in the dynamic regulation of p53 stability in cells.
EXPERIMENTAL PROCEDURES
Cell Culture, Chemicals, and Treatment
293T, PC3, U2OS, and MCF7 cells were maintained in DMEM supplemented with 10% fetal calf serum (Atlanta Biologicals, Inc.) and antibiotics. HCT116 cells were maintained in McCoy's 5A medium supplemented with 10% fetal calf serum and antibiotics. Transfection was carried out with LipofectamineTM 2000 (Invitrogen). ATP (catalog no. A-7699) and leptomycin B were purchased from Sigma-Aldrich. Ubiquitin and ubiquitin variants were purchased from Boston Biochem, Inc. (Cambridge, MA).
Plasmids
The cloning of human E1 (ubiquitin-activating enzyme) into pFastBacI vector with a His9 tag for baculoviral expression in insect cells has been described previously (27). The open reading frame of Mdm2 (human sequence) and MdmX (mouse sequence) were subcloned by PCR into pFastBacI vector with a His9 tag at the C terminus for baculoviral expression and pcDNA3.1 for mammalian expression. Human p53 was subcloned into pET28a with a His6 tag at the N terminus of p53 for Escherichia coli expression. The RING mutations of Mdm2 (C461A/C464A, L468A) or MdmX (C459S/C462S) were generated by site-directed mutagenesis. Mammalian expression plasmids pcDNA3.1(+)-His-FLAG-MdmX, pcDNA3.0-HA-His-FLAG-MdmX were constructed by PCR cloning. The pFLAG-CMV-Mdm2 was subcloned from pGEX6P-Mdm2 using EcoRI and SmaI sites. All of the plasmids were confirmed by DNA sequencing.
Recombinant Protein Preparation
GST-Mdm2, GST-Mdm2L468A, and UbcH5c were expressed in E. coli and affinity purified with glutathione-Sepharose 4B (Amersham Biosciences). The cleavage of GST from GST-Mdm2 was carried out with PreScission protease (Amersham Biosciences) as described previously (28). Insect cell expressed His-Mdm2-HA, His-FLAG-MdmX, and human His-tagged E1 (human E1) and E. coli expressed His-p53 were subjected to affinity purification with a nickel column as described previously (27). For recombinant His-p53, the eluate from the nickel column was further purified with a 1-ml Q Sepharose column on AKTA Purifier 10 (GE Healthcare) with 20–54% buffer B (20 mm Tris-HCl, pH 7.5, 1 mm DTT, 1 m NaCl), followed by 1-ml a SP Sepharose column eluted with 23–47% buffer B. The SP fractions (F6–8) with a single p53 band were used for in vitro p53 ubiquitination assay.
In Vitro p53 Ubiquitination Assay
The reaction was carried out at 30 °C for 1 h in a volume of 20 μl containing 40 mm Tris (pH 7.5), 2 mm DTT, 5 mm MgCl2, 40 μm wild type ubiquitin, or other ubiquitin variants as indicated, 50 nm hE1, 200 nm UbCH5c, 5 mm ATP, 0.7 μm recombinant p53, and indicated concentrations of purified Mdm2 or MdmX. The reaction was then stopped by adding 10 μl of 4×SDS-PAGE sample buffer and boiled for 5 min. The samples were then resolved by 8% SDS-PAGE, and the ubiquitinated p53 species were detected by Western blotting with p53-specific antibodies (mixture of monoclonal p53 antibody PAb1801 and DO-1 at a 1:1000 dilution for each). When using monoubiquitinated p53 as substrate, in vitro p53 ubiquitination was first carried out using GST-free Mdm2 (200 nm) to generate monoubiquitinated p53. Then, p53, including monoubiquitinated p53, was purified by 50 μl of nickel resins (Qiagen) in 20 mm Tris, pH 7.5, 100 mm NaCl, 10% glycerol, rotating at 4 °C for 1 h. The resins were followed by three washes with 20 mm Tris, pH 7.5, 500 mm NaCl, 10% glycerol; three washes with 20 mm Tris, pH 7.5, 1000 mm NaCl, 10% glycerol; and one wash with 20 mm Tris, pH 7.5, 10 mm NaCl, 10% glycerol. Monoubiquitinated p53 was eluted twice with 50 μl of 20 mm Tris, pH 7.5, 100 mm NaCl, 10% glycerol, 100 mm imidazole by rotating the resins in the elution buffer at 4 °C for 30 min. The eluate served as monoubiquitinated p53 substrates.
Cytosolic and Nuclear Fractionation
Cytosolic and nuclear fractionation was carried out as follows: 0.5–1 × 106 cells from a 6-cm plate or 2 × 106 cells from a 10-cm plate were washed once with 10 ml TBS and pelleted by centrifugation at 500 × g for 5 min. TBS was completely removed after transferring the cells in suspension into Eppendorf tubes followed by a quick spin at 10,000 × g for 1 min. The cell pellets were resuspended in 200–400 μl of hypotonic buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, supplemented with protease inhibitors). The cells were left on ice for 15 min. The cytosolic fraction was released by a 10-s gentle vortex immediately after adding 10% Nonidet P-40 to a final concentration of 0.625%. The cytosolic fraction was recovered by a 30-s centrifugation at 10,000 × g at 4 °C. The nuclear pellets were washed once with 1 ml of buffer A and then the same volume of buffer A containing 1% SDS were added. After boiling the sample for 10 min, the nuclear fraction was recovered by centrifugation for 10 min at 14,000 × g at room temperature.
Gel Filtration
Gel filtration was carried out on a SMART-FPLC system (Amersham Biosciences). A 50-μl mixture of MdmX and Mdm2 containing 1.8 mg/ml of MdmX and 0.36 mg/ml of Mdm2 were loaded on a 2.4-ml Superdex 200 column and resolved in 3% buffer B (20 mm Tris, pH 7.5, 40 mm NaCl, 1 mm DTT) at a flow rate of 50 μl/min. The fractions were collected at a size of 100 μl. Subsequently, 3 μl of each fraction were used for Western analysis of protein elution profiles, and 10 μl were used for in vitro p53 ubiquitination assay.
In Vivo p53 Ubiquitination Assay
HCT116 cells or MCF7 cells were plated at a density of 10 × 106 cells per 6-cm plate. Transfection with Lipofectamine 2000 (Invitrogen, catalog no. 11668-019) was carried out 18 h after cell plating. Each plate was transfected with 0.5 μg His-ubiquitin (Ub)3 and 4 μl of 20 μm control siRNA (Dharmacon, ON_TARGET SMART pool siControl, catalog no. L-001816-10) or siRNA for MdmX (Dharmacon, ON_TARGET SmartPool MDM4, catalog no. L-006536-00). Five identical plates were transfected for each siRNA oligonucleotide. Cells were treated with MG132 at 50 μm for 5 h before cell harvest and cytosolic fractions were prepared as described (“Cytosolic and Nuclear Fractionation”). One ml of urea buffer A (8 m urea, 0.1 m phosphate, 0.01 m Tris, pH 8.0, 15 mm imidazole, 0.2% Triton X-100) was added to the 200-μl cytosolic fractions from five plates. After 1 min of vortexing, the lysates were transferred to tubes with 15-μl Dynabeads slurry (Invitrogen, catalog no. 101.3D, Dynabeads His-Tag Isolation and Purification) prewashed twice with urea buffer A. Then, the lysates were incubated with the beads for 1 h at room temperature by rotation. After two washes with 1 ml of urea buffer A, the beads were further washed twice with buffer B (buffer B: 20 mm Tris, pH 6.3, 25 mm imidazole, 0.2% Triton X-100). The His-tagged proteins were then released by boiling the beads for 10 min in 30 μl of 2× SDS-PAGE sample buffer. Then, the samples were loaded on an 8% SDS-PAGE gel and subjected to Western blotting for p53 with a mixture of D0–1 and PAb1801 antibodies (Santa Cruz Biotechnology, catalog nos. sc-126 and sc-98). The efficiency of MdmX knockdown was confirmed by Western blotting with a rabbit antibody for MdmX (Bethyl Laboratories, Inc., A300-287A). Mdm2 was detected with SMP14 monoclonal antibody (Santa Cruz Biotechnology, catalog no. sc-965). Polyubiquitin was detected with a monoclonal antibody for polyubiquitin (BD Biosciences, catalog no. 550944, clone 6C1.17).
RESULTS
Mdm2 Mediates Multiple Monoubiquitination of p53 in Vitro
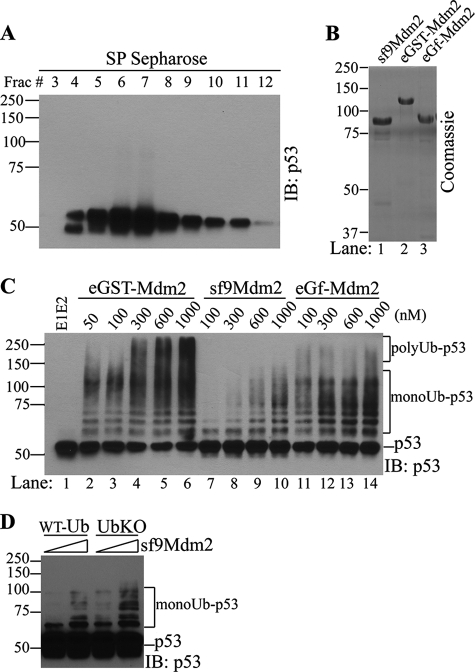
To investigate in vitro ubiquitination of p53, we purified recombinant p53 expressed in E. coli and recombinant Mdm2 expressed in insect cells (Fig. 1, A, p53, and B, sf9Mdm2). Surprisingly, in the ubiquitination assay, the recombinant Mdm2 expressed in insect cells consistently displayed very weak E3 ligase activity toward p53, which is in a sharp contrast to previous results in the literature using GST-tagged Mdm2 as the E3 source. This observation led to a careful comparison between GST-Mdm2 expressed from E. coli (Fig. 1B, eGST-Mdm2) and the non-GST fusion Mdm2 with a short His tag purified from insect cells. As shown in Fig. 1C, GST-Mdm2 expressed in E. coli can catalyze both multiple mono (recognized by seven distinct bands above nonubiquitinated p53 because p53 has seven major ubiquitination sites in its C terminus) and polyubiquitination of p53 (recognized by smearing bands >140 kDa (53 + (7 + 3) × 8.5 kDa per Ub) in a concentration-dependent manner (Fig. 1C, eGST-Mdm2 panel), consistent with previous reports (29). However, His-tagged Mdm2 displayed a significantly reduced E3 ligase activity toward p53 as compared with GST-Mdm2. His-tagged Mdm2 at a high concentration up to 1 μm catalyzed p53 ubiquitination less efficiently than 50 nm GST-Mdm2 (compare Fig. 1C, lane 10 with lane 2). This marked difference in E3 ligase activity of Mdm2 from different expression systems led us to suspect that either the GST tag or certain posttranslational modifications conferred by the insect cell expression system might be responsible. To differentiate between these two possibilities, we cleaved GST from GST-Mdm2 and purified the GST-free Mdm2 (Fig. 1B, lane 3, eGf-Mdm2, which represents E. coli, GST-free Mdm2) and compared its activity in the p53 ubiquitination assay. As shown in Fig. 1C, GST-free Mdm2 from E. coli lost the activity to catalyze p53 polyubiquitination but remained as effective in mediating multiple monoubiquitination of p53 as His-tagged Mdm2 (compare Fig. 1C, lanes 2–6 with lanes 11–14). GST-free Mdm2 generated a ubiquitination pattern reminiscent of that reported by Lai and colleagues (30), who showed multiple monoubiquitination of p53 in vitro when using very low concentrations of GST-Mdm2. These results indicate that the polyubiquitination activity of GST-Mdm2 at high concentrations is conferred by the presence of the GST tag and is not an intrinsic activity associated with Mdm2 protein.
FIGURE 1.
Mdm2 only mediates multiple monoubiquitination of p53 in vitro. A, recombinant p53 used in the in vitro reaction was purified from E. coli by His tag affinity purification followed by SP Sepharose cation-exchange chromatography. p53 Western blots of SP Sepharose fractions (Frac) are shown. A pool of p53 (fractions 8–11) was used as substrates for in vitro assay. B, Coomassie staining of Mdm2 recombinant proteins used in the in vitro assay. sf9Mdm2, HA-Mdm2-His purified from sf9 cells; eGST-Mdm2, GST-Mdm2 purified from E. coli; eGf-Mdm2, GST-free E. coli expressed Mdm2. C and D, in vitro p53 ubiquitination by GST-Mdm2 or GST-free Mdm2 at concentrations indicated. Ubiquitinated p53 was detected by Western blot analysis of p53 after the reaction using different versions of Mdm2 (C) or ubiquitin (D) as indicated. IB, immunoblot; WT-Ub, wild type ubiquitin; UBK0, lysine-less ubiquitin. polyUb-p53, polyubiquitinated p53 (p53-ubiquitin adducts >140 kDa); monoUb-p53, multimonoubiquitinated p53 (seven bands above p53).
To further confirm that insect cell-expressed Mdm2 mediates multiple monoubiquitination but not polyubiquitination of p53, we compared the p53 ubiquitination patterns catalyzed by the insect cell-expressed Mdm2 with either wild type (WT) ubiquitin or a lysine-less ubiquitin variant (UBK0), which prevents ubiquitin chain assembly after conjugation of the first ubiquitin to primary sites on the substrate. The p53 ubiquitination patterns produced by His-tagged Mdm2 at 300 or 900 nm in the presence of WT ubiquitin are identical to those of UBK0 (Fig. 1D), except for differences in band intensities. These results indicate that Mdm2 without a GST tag is only active in mediating multiple monoubiquitination of p53 and is not capable of mediating p53 polyubiquitination on its own.
MdmX Is an Essential Activator of Mdm2 E3 Ligase for p53 Polyubiquitination in Vitro
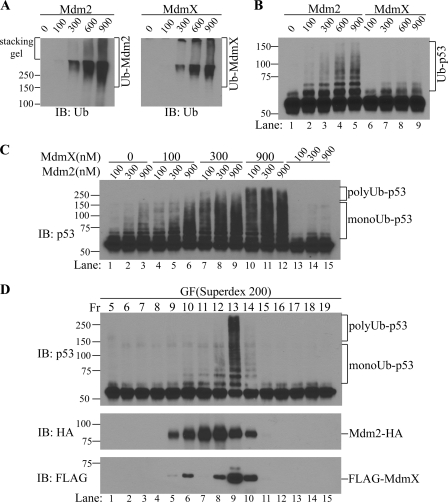
Because GST has dimerization properties (31, 32), we speculated that GST-Mdm2 dimerization might be critical for its artificial, self-sufficient activity to catalyze p53 polyubiquitination. This explanation pointed to the potential existence of other player(s) in cells that function to induce p53 polyubiquitination when heterodimerized with Mdm2. We further hypothesized that MdmX might be such a cellular activator of Mdm2 because MdmX is a binding partner of Mdm2 (17). Therefore, to investigate the potential role of MdmX in Mdm2 activation, we expressed and purified recombinant MdmX protein from insect cells. MdmX protein displayed minor autoubiquitination activity weaker than that of Mdm2 (Fig. 2A). When used alone, only Mdm2 but not MdmX showed E3 ligase activity for p53 (Fig. 2B, compare lanes 2–5 with lanes 6–8). The observation that MdmX is not capable of mediating p53 ubiqutination on its own is consistent with previous reports (33). Strikingly, we found that combination of MdmX and Mdm2 resulted in potent p53 ubiquitination in an MdmX dose-dependent manner (Fig. 2C). In addition, although Mdm2 alone can only mediate multiple monoubiquitination of p53, the presence of MdmX generated a ladder of ubiquitinated p53 species higher than 140 kDa, representing polyubiquitinated p53.
FIGURE 2.
MdmX is an activator of Mdm2 E3 ligase for p53 polyubiquitination. A, autoubiquitination of Mdm2 and MdmX. In vitro ubiquitination assays with Mdm2 or MdmX alone in the absence of p53 were carried out and the ubiquitinated adducts are shown by anti-ubiquitin Western blotting. B, p53 ubiquitination by Mdm2 or MdmX. In vitro p53 ubiquitination assays were carried out with Mdm2 or MdmX alone. Western blotting of p53 is shown. Ub-p53, ubiquitinated p53. C, MdmX activates Mdm2-catalyzed p53 polyubiquitination. In vitro p53 ubiquitination assays were carried out with different concentrations of Mdm2 and/or MdmX as indicated. The numbers indicate the concentrations of each component in nm. Polyubiquitinated p53 (polyUb-p53), and multiple monoubiquitinated p53 (monoUb-p53) are indicated. D, in vitro p53 ubiquitination with fractions of gel filtration (GF) of the Mdm2/MdmX mixture; IB, immunoblot.
To further confirm the requirement for MdmX in Mdm2-mediated p53 polyubiquitination, a gel filtration experiment was carried out using a mixture of recombinant MdmX and Mdm2 proteins; active p53 E3 ligase fractions were resolved under conditions that allow ready differentiation between free Mdm2 and potential Mdm2/MdmX heterodimers. As shown in Fig. 2D, a potent p53 E3 ligase activity was detected in a single fraction, which is the peak fraction of MdmX and subpeak fraction of Mdm2 (Fig. 2D, lane 9, fraction 13). As expected, the fractions containing free Mdm2 (Fig. 2D, lane 7, fraction 11), or Mdm2 with minor amounts of MdmX (Fig. 2D, lanes 8 and 10, fractions 12 and 14) only exhibited weak p53 monoubiquitination activity. Significantly, the presence of MdmX generated a potent E3 ligase activity sufficient to produce a polyubiquitinated p53 ladder that nearly reached the top of the gel. This result corroborated our findings that MdmX is an essential activator of Mdm2 and can convert Mdm2 into a p53 polyubiquitination E3 ligase in vitro. Our result also provides a biochemical explanation for the finding that targeted deletion of the MdmX gene in mice results in p53-dependent embryonic lethality; increased p53 cellular concentrations is a likely cause of this effect (18).
Requirement of RING Domains for Mdm2 Activation by MdmX
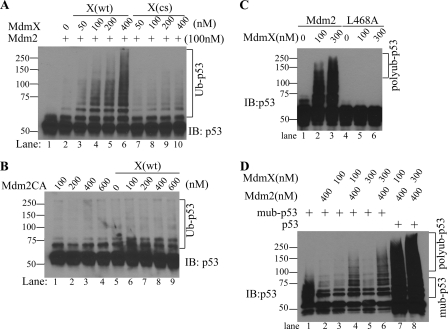
Mdm2 is a member of the RING domain E3 ligase family whose E3 ligase activity relies on its RING domain (14). The role of the RING domain of MdmX is not well defined as MdmX itself does not show E3 ligase activity toward p53 (33). However, it has been shown that MdmX binds to Mdm2 via its RING domain (17). To investigate the potential role of the MdmX RING domain in activation of Mdm2, we generated an MdmX RING mutant (C459S and C462S) and assessed whether this mutant can still activate Mdm2. As shown in Fig. 3A, whereas wild type MdmX potently activated Mdm2 E3 ligase activity for p53 polyubiquitination, the RING domain mutant failed to do so, indicating that the MdmX RING domain is indispensable for its activation of Mdm2 (Fig. 3A, compare lanes 2–6 with lanes 7–10). To confirm that p53 ubiquitination also requires the RING domain of Mdm2, an in vitro p53 ubiquitination experiment using RING domain mutated Mdm2 (Mdm2CA, C461A and C464A) and wild type MdmX was carried out. As the results indicate, the Mdm2 mutant without the intact RING domain completely lost its activity to mediate monoubiquitination of p53 in the absence of MdmX or mediate polyubiquitination of p53 in the presence of MdmX (Fig. 3B). These data indicate that the RING domains of both Mdm2 and MdmX are required for Mdm2-dependent polyubiquitination of p53.
FIGURE 3.
The RING domains and Mdm2 are required for p53 polyubiquitination. A, in vitro p53 ubiquitination by Mdm2 in the presence of either wild type MdmX (X(wt)) or RING domain mutant MdmX (X(cs)) at concentrations indicated. Ubiquitinated p53 was detected by Western blotting of p53 and is indicated by Ub-p53. B, in vitro p53 ubiquitination by RING domain mutant Mdm2 in the presence or absence of wild type MdmX. C, effects of E2 binding mutation of Mdm2 (Mdm2L468A) on MdmX activation of Mdm2 for p53 polyubiquitination. In vitro p53 ubiquitination by GST-free Mdm2 (400 nm) or GST-free Mdm2(L468A) (400 nm) in the presence of MdmX (100 and 300 nm) is shown. Ubiquitinated p53 was detected by Western blotting with a mixture of D0–1 and PAb1801. D, polyubiquitination of monoubiquitinated p53. In vitro p53 ubiquitination was analyzed with monoubiquitinated p53 (lanes 1–6) or p53 (lanes 7 and 8) as substrates. Further ubiquitination of monoubiquitinated p53 was tested by Mdm2 (400 nm) alone, MdmX (100 and 300 nm) or the combination of Mdm2 and MdmX. mub-p53, monoubiquitinated p53; IB, immunoblot.
Dependence of Mdm2 in p53 Polyubiquitination by Mdm2/MdmX
A combination of Mdm2 and MdmX generates a potent polyubiquitination E3 ligase for p53. It is possible that MdmX possesses the polyubiquitination capability after p53 is monoubiquitinated by Mdm2 or when in complex with Mdm2. To identify the catalytic component of Mdm2/MdmX-mediated polyubiquitination of p53, we used a RING domain-intact but enzyme-dead Mdm2 (GST-free Mdm2L468A), which is deficient in E2 interaction (35). This mutant Mdm2 is expected to interact with MdmX through its intact RING domain. As shown in Fig. 3C, although MdmX can dose-dependently activate GST-free wild type Mdm2 for p53 polyubiquitination, it could not stimulate p53 polyubiquitination in the presence of Mdm2L468A (Fig. 3C, compare lanes 2 and 3 with lanes 5–7). These results indicate that Mdm2 is the catalytic component of Mdm2/MdmX that recruits E2 to mediate p53 polyubiquitination. To investigate the potential capability of MdmX in p53 polyubiquitination after p53 is monoubiquitinated by Mdm2, we first carried out in vitro p53 ubiquitination by Mdm2 (GST-free) and then purified the p53, including the monoubiquitinated species for their further ubiquitination by MdmX alone, Mdm2 alone, or their combination. As shown in Fig. 3D, MdmX or Mdm2 alone could not mediate polyubiquitination of monoubiquitinated p53 (Fig. 3D, lanes 2, 3, and 5). Polyubiquitination of monoubiquitinated p53 only happened when both Mdm2 and MdmX were present (Fig. 3D, lanes 4 and 6). Of note, monoubiquitinated p53 is a poorer substrate compared with nonubiquitinated p53 for Mdm2/MdmX-mediated polyubiquitination (Fig. 3D, compare lane 4 with 7 and lane 6 with 8). Together, these results establish that Mdm2 is the catalytic component of Mdm2/MdmX in p53 polyubiquitination, and MdmX functions as an activator component.
p53 as a Competitive Substrate over MdmX by Mdm2/MdmX
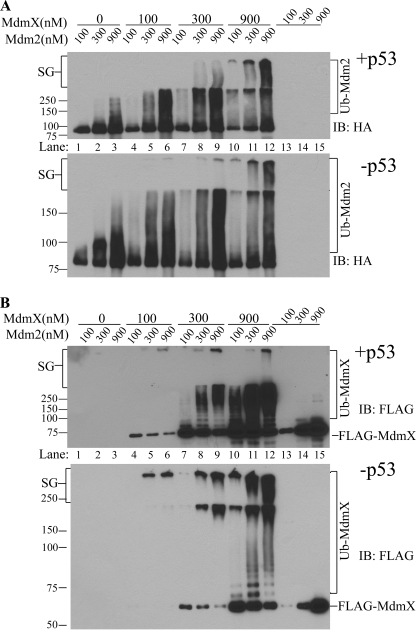
Although MdmX shows little activity for p53 ubiquitination even at extremely high concentrations (Fig. 1B), MdmX indeed possesses some autoubiquitination activity detectable with anti-ubiquitin immunoblotting (Fig. 2A, right panel) but not readily detected with anti-FLAG antibody (recombinant MdmX is FLAG-tagged) (Fig. 4B, lanes 13–15). To determine whether MdmX can be a substrate for Mdm2 because it binds to Mdm2, we investigated MdmX ubiquitination by Mdm2 and the effects of p53 on MdmX ubiquitination and Mdm2 autoubiquitination. As the results indicate, MdmX dose-dependently activated the E3 ligase activity of Mdm2 as measured by Mdm2 autoubiquitnation in this experiment. The Mdm2 autoubiquitination was so robust in the presence of MdmX that a significant portion of the ubiquitinated Mdm2 species remained in the stacking gel. We found that the Mdm2 autoubiquitination process was not altered by the presence or absence of p53 in the system (Fig. 4A, compare +p53, upper panel, with −p53, lower panel). In the same experiment, MdmX became heavily ubiquitinated in an Mdm2 concentration-dependent manner readily detected by anti-FLAG immunoblotting. Indeed, a reduction in non-ubiquitinated FLAG-MdmX (Fig. 4B, FLAG-MdmX) correlated with the increase in ubiquitinated MdmX (Fig. 4B, compare lanes 4–6 between upper and lower panels for FLAG-MdmX and Ub-MdmX). On the other hand, in the absence of Mdm2, MdmX itself showed little autoubiquitination (Fig. 4B, compare lanes 4–6 with lanes 13–15). Importantly, the presence of p53 in the reaction exhibited a significant inhibitory effect on the production of the very high molecular weight ubiquitin conjugates of MdmX (Fig. 4B, compare the stacking gel region between +p53 and −p53). In fact, the reduction in nonubiquitinated MdmX nicely reflected the increase in ubiquitinated MdmX and the effects of p53 on this process. Nearly all MdmX at 100 nm concentration was ubiquitinated in the absence of p53 under the assay conditions used, but the protein was largely protected from being ubiquitinated in the presence of p53 (Fig. 4B, compare lanes 4–6 in upper and lower panels). These results suggest that p53 can out-compete MdmX as a substrate for Mdm2, a mechanism that might have important cellular implications.
FIGURE 4.
MdmX ubiquitination by Mdm2 and effects of p53 on Mdm2 and MdmX ubiquitination. A, effects of p53 on Mdm2 autoubiquitination. In vitro Mdm2 autoubiquitination was analyzed in the presence of different concentrations of MdmX (0–900 nm) and the presence (+p53) or absence of p53 (−p53). Ubiquitinated Mdm2 was detected by Western blotting of HA (recombinant Mdm2 is HA-tagged) and was indicated by Ub-Mdm2. SG, stacking gel. B, effects of p53 on Mdm2-mediated MdmX ubiquitination. In vitro MdmX ubiquitination was analyzed in the presence of different concentrations of Mdm2 (0–900 nm) and the presence (+p53) or absence of p53 (−p53). Ubiquitinated MdmX was detected by Western blotting of FLAG (recombinant MdmX is FLAG-tagged).
Compartmentalization of p53 Ubiquitination in Cells
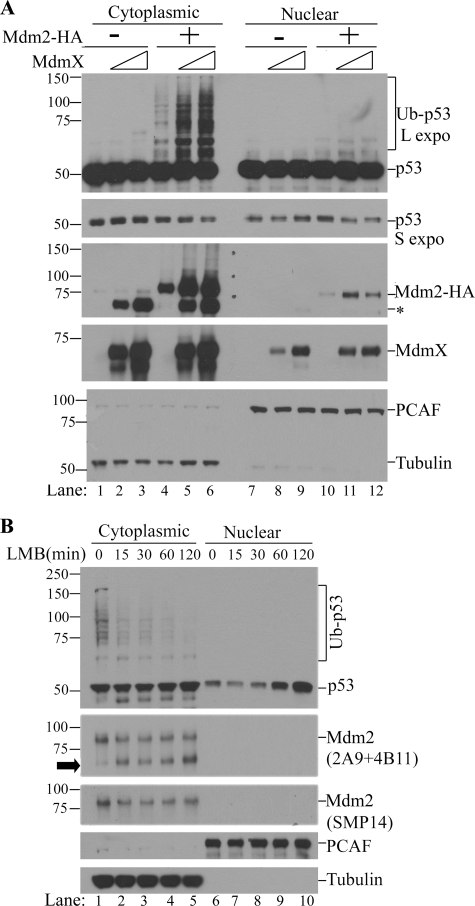
Our in vitro results suggest that in cells, p53 polyubiquitination would only take place in a cellular compartment containing both Mdm2 and MdmX. We chose to use HEK 293T cells (a human embryonic kidney cell line expressing SV40 large T antigen) to test this possibility for the following reasons: (1) p53 expression is relatively high in these cells; (2) p53 protein is distributed almost equally in the nucleus and cytoplasm; and (3) endogenous Mdm2 expression is very low in these cells due to inactivation of the transcription activity of p53 by SV40 large T antigen. We found that when MdmX alone was overexpressed in 293T cells, it caused a modest increase in endogenous Mdm2 levels in the cytoplasm without discernable effects on endogenous p53 levels (Fig. 5A, lanes 1–3, asterisk for endogenous Mdm2). However, when MdmX and Mdm2 were coexpressed, an MdmX dose-dependent increase in the ubiquitination of endogenous p53 was observed mainly in the cytoplasm, which coincided with a decrease in p53 levels in both the cytoplasm and nucleus (Fig. 5A, lanes 4–6, long exposure for Ub-p53, and short exposure for unmodified p53). As expected, endogenous p53 ubiquitination took place in the subcellular compartment where a major fraction of Mdm2 and MdmX co-reside, i.e. in the cytoplasm. To confirm that the cytoplasm is the site of polyubiquitination of endogenous p53, we also used U2OS cells, a human osteosarcoma cell line expressing wild type functional p53. In this experiment, we first used leptomycin B to block the nuclear export of endogenous p53 (and thus allow one-directional transport of p53 to the nucleus). Subsequently, we monitored ubiquitination of p53 and its steady-state levels in both the cytoplasm and nucleus. As shown in Fig. 5B, we found that polyubiquitinated p53 was only observable in the cytoplasm (Fig. 5B, compare lanes 1–5 with lanes 6–10). Interestingly, we found that a decrease in the polyubiquitinated p53 in the cytoplasm precedes the nuclear accumulation of p53 (Fig. 5B, compare lanes 1–5 with lanes 6–10 in p53 and Ub-p53). Furthermore, the decrease in polyubiquitinated p53 after leptomycin B treatment is accompanied by a decrease in full-length Mdm2 due to protein cleavage (Fig. 5B, arrowhead, Mdm2). This result indicates that polyubiquitination of endogenous p53 by endogenous Mdm2 and MdmX takes place mainly in the cytoplasm where MdmX resides.
FIGURE 5.
p53 ubiquitination mainly takes place in the cytoplasm where Mdm2 and MdmX colocalize. A, in vivo ubiquitination of endogenous p53 in 293T cells. 293T cells were transfected with 200 or 400 ng of pcDNA3.0-HA-MdmX DNA per 60-mm plate in the absence (lanes 1–3 and 7–8) or presence of pFLAG-CMV-Mdm2 (400 ng DNA/60-mm plate). Twenty-four hours later, cytoplasmic and nuclear fractions were prepared, and Western blot analysis of p53 (using a mixture of DO-1 and Pab1801 antibodies), Mdm2 (using a mixture of 2A9 and 4B11 antibodies), and MdmX (anti-HA) was performed. PCAF and tubulin were used as loading controls as well as nuclear and cytoplasmic markers, respectively. L expo and S expo indicate long and short exposure, respectively. The asterisk indicates the position of endogenous human Mdm2. B, in vivo ubiquitination of endogenous p53 in U2OS cells. U2OS cells were treated with a nuclear export inhibitor, leptomycin B (LMB; 20 ng/ml) for various times; cytoplasmic and nuclear fractions were prepared and subjected to Western blot analysis. The arrowhead indicates a short fragment of endogenous Mdm2.
Mdm2 and MdmX Cooperation in p53 Degradation in Cells
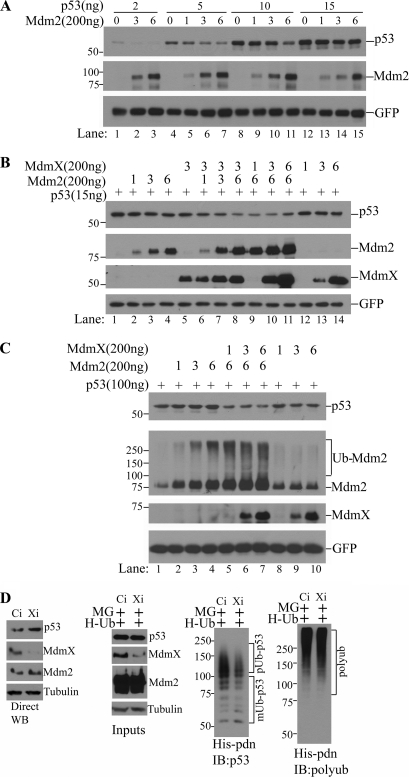
Based on the finding that MdmX is indispensible for the p53 polyubiquitination activity of Mdm2, we hypothesized that both Mdm2 and MdmX are limiting factors for p53 degradation in cells. To test this hypothesis, we chose to use p53-null PC3 cells in our co-transfection experiments to easily monitor the relative ratios of p53, Mdm2, and MdmX. As shown in Fig. 6A, co-transfection of Mdm2 with p53 induced an Mdm2 dose-dependent degradation of transfected p53. These data indicate that endogenous MdmX was sufficient to support Mdm2-mediated p53 polyubiquitination and subsequent proteasomal degradation within the range of ectopically expressed Mdm2 and p53 in this experiment. However, when higher amounts of p53 were ectopically expressed, Mdm2-mediated p53 degradation was apparently less efficient (Fig. 6A, compare lanes 1–3, 4–7, and 8–11 with lanes 12–15, p53), which might imply that endogenous MdmX became insufficient to produce enough fully active Mdm2 E3 ligase to mediate p53 polyubiquitination, i.e. MdmX became a limiting factor for p53 degradation. If this was the case, co-transfection of MdmX under these conditions would resume p53 degradation. As shown in Fig. 6B, when MdmX was co-transfected with Mdm2 and p53, dose-dependent Mdm2-mediated p53 degradation was restored, whereas MdmX transfection alone did not have any effect (Fig. 6B, compare lanes 1–4 with lanes 5–8, 9–11, and 12–14). Remarkably, even when ectopic p53 expression reached such a high level that coexpression of Mdm2 failed to induce any detectable degradation of p53 (Fig. 6C, lanes 1–4), transfection of MdmX was still able to restore efficient p53 degradation in an Mdm2-dependent manner (Fig. 6C, compare lanes 1–4 with 5–7). MdmX also consistently induced potent autoubiquitination of Mdm2 (Fig. 6C, compare lanes 1–4 with lanes 5–7, Ub-Mdm2). To extend the findings revealed by in vitro biochemical assays and in vivo overexpression systems and to determine whether MdmX is also a limiting factor for degradation of endogenous p53, we examined the effects of siRNA-mediated knockdown of MdmX in HCT116 cells. We found that knockdown of MdmX increased steady-state levels of endogenous p53 (Fig. 6D, left panel), accompanied by reduced p53 polyubiquitination (Fig. 6D, second panel from right, His-pdn, IB:p53). The equal efficiency of the His tag pulldown of polyubiquitinated proteins was demonstrated by reprobing the same membrane with a polyubiquitin antibody after stripping the p53 signal (Fig. 6D, right panel, His-pdn, IB:polyub). The same phenomena were also observed with MCF7 cells (supplemental Fig. 2). Taken together, it appears that the relative quantities of p53 versus Mdm2 and MdmX determine the rates of p53 degradation in cells and thus the biological function of this crucial cell regulator.
FIGURE 6.
Efficient p53 degradation in cells depends on both Mdm2 and MdmX. A, titration of Mdm2-mediated p53 degradation in vivo. PC3 cells were transfected with increasing amounts of Mdm2 and increasing amounts of p53 in 60-mm plates as indicated. Twenty-four hours later, whole cell lysates were prepared, and the effect of p53 DNA amounts on Mdm2-mediated p53 degradation was examined by Western blot analysis of steady-state levels of p53. B and C, effects of MdmX on Mdm2-mediated p53 degradation. PC3 cells were transfected with indicated amounts of Mdm2 and p53 expressing plasmids in the presence or absence of co-transfection with indicated amounts of MdmX. p53 steady-state levels were shown by Western blotting of p53. GFP was used as a transfection control. The numbers (1, 3, and 6) represent times of 200 ng DNA in corresponding transfections. D, effects of siRNA knockdown of MdmX on endogenous p53 levels and p53 polyubiquitination. HCT116 cells were transfected with either control (Ci) or MdmX (Xi) siRNA oligonucleotides together with a His-ubiquitin expression plasmid (H-Ub). Thirty-six hours after transfection, cells were either left untreated or treated with 50 μm MG132 (MG) for 5 h. Cell lysates from untreated cells were used for p53 analysis by Western blotting (WB; left panel) and cell lysates from MG132-treated cells were used for His-tag pulldown (His-pdn) for in vivo p53 ubiquitination assay (right three panels). Polyubiquitinated p53 (pUb-p53) and monoubiquitinated p53 (mUb-p53) are shown (second panel from right). The equal efficiency of His tag pulldown (His-pdn) of polyubiquitinated proteins was shown by reprobing the same membrane with a polyubiquitin antibody after stripping the p53 signal (right panel).
DISCUSSION
After the initial cloning of MdmX, its critical role in negative regulation of p53 was unambiguously demonstrated by mouse genetics (16, 18–21). Although the molecular cell biology studies have reached a consensus that MdmX can inhibit p53 transcriptional activity through direct physical interaction, the picture of how MdmX functions within the central p53/Mdm2 regulatory loop to control p53 stability remains obscure, partly because opposing effects of MdmX on p53 levels have been described in the literature (5). Thus, the biochemical basis underlying the genetic findings has been missing. Although MdmX is a well established p53 binder and the crystal structure of the RING domains of Mdm2 and MdmX are nearly identical (34), whether MdmX possesses intrinsic E3 ligase activity toward p53 and how it regulates p53 stability remained unsolved.
In this report, using a biochemical reconstitution system, we found that in the absence of MdmX, Mdm2 alone can only mediate monoubiquitination of p53 in vitro (Fig. 1). The monoubiquitination activity of Mdm2 has been reported when using GST-Mdm2 at low concentrations (30). However, this finding was dismissed later by more detailed experiments, showing that p53 ubiquitination by GST-Mdm2 is a concentration-dependent phenomenon: at low concentrations, it mediates multiple monoubiquitination, and at high concentrations, it can also mediate p53 polyubiquitination (29). Our current results indicate that the concentration dependent polyubiquitination of p53 by GST-Mdm2 is delivered artificially, likely through a GST-mediated dimerization process (Fig. 1). Furthermore, we found that MdmX, the physiological heterodimerizing molecule of Mdm2, can convert Mdm2 to a polyubiquitination E3 for p53. This is reminiscent of activation of BRCA1 E3 ligase activity by BARD1 heterodimerization (35). The gel filtration results further supported the potent stimulation of Mdm2 by MdmX through their heterodimerization (Fig. 2D). In this gel filtration experiment, we observed a single activity fraction (Fig. 2D, fraction 13) in which the estimated concentrations of Mdm2 and MdmX are 200 nm and 160 nm, respectively, whereas the two neighboring fractions (fractions 12 and 14) showed no significant E3 ligase activity because the Mdm2 and MdmX concentrations were below the lower limit of the linear range of stimulation (400 nm Mdm2 and 40 nm MdmX for fraction 12; 100 nm Mdm2 and 80 nm of MdmX for fraction 14). Notably, the activating effect of MdmX could also be observed with GST-Mdm2 when low concentrations (10–100 nm) of GST-Mdm2 were used (supplemental Fig. 1), which is consistent with the previous report in which ∼100 nm GST-Mdm2 was used in the in vitro reaction (33). Thus, this report resolved a major problem in the field derived from using GST-Mdm2 in previous studies and uncovered the biochemical role of MdmX as an essential activator of Mdm2 for p53 polyubiquitination. Our work provided the solid biochemical evidence to explain why lack of either Mdm2 or MdmX causes a complete disruption of the essential p53 negative regulatory module and results in p53-dependent embryonic lethality in mice (7, 18). Therefore, the conclusion from this study marks a significant step forward in the field.
Our finding that the RING domains of MdmX and Mdm2 are both required for a fully active p53 polyubiquitination E3 ligase (Fig. 3, A and B) is mirrored in an in vivo observation that a RING domain-mediated interaction between MdmX and Mdm2 is necessary for the E3 ligase activity of Mdm2 using Mdm2/MdmX/p53 triple knock-out mouse embryonic fibroblasts (36). More importantly, we demonstrated that the E3 ligase activity of Mdm2/MdmX for p53 polyubiquitination depends on Mdm2 but not MdmX (Fig. 3, C and D). This defines MdmX as a sole activator for Mdm2 that can convert Mdm2 from a monoubiquitination E3 ligase to a polyubiquitination E3 ligase.
Our biochemical evidence predicts that both Mdm2 and MdmX are limiting factors for degradation of p53 in cells because only polyubiquitinated p53 undergoes proteasomal degradation; this is supported by the results from exogenous p53 in PC3 cells and endogenous p53 in 293T cells and HCT116 cells after either overexpression of Mdm2 and MdmX or siRNA knockdown of endogenous MdmX (Figs. 5 and 6). These findings are in line with previous reports on a cooperative role of MdmX in stimulating Mdm2 E3 ligase activity and p53 degradation in cells (33, 36–39). Our results from 293T and U2OS cells showing the cytoplasmic ubiquitination of p53 where Mdm2 and MdmX are co-localized are consistent with a recent finding that MdmX can tether p53 and Mdm2 for a predominant cytoplasmic localization (40).
We found that MdmX actually plays dual roles in the Mdm2-mediated ubiquitination reaction: as an activator of Mdm2 on one hand and as an Mdm2 substrate on the other hand (Fig. 4). This will add another layer of complexity to the effects of MdmX on the p53/Mdm2 loop. Of note, although MdmX significantly stimulates Mdm2-mediated p53 ubiquitination in vitro (Fig. 2) and p53 degradation in vivo (Fig. 6), the dose-dependent effects of MdmX disappear at higher levels of Mdm2 and constant levels of p53 (Fig. 2C, compare lane 8 with 9 and lane 11 with 12; Fig. 6B, compare lane 10 with 11, and Fig. 6C, lanes 5–7). This outcome may be explained by the dual role of MdmX as both an Mdm2 activator and a substrate. Although p53 is a preferential substrate over MdmX for Mdm2/MdmX E3 activity (Fig. 4), MdmX at higher concentrations can competitively inhibit Mdm2-mediated p53 ubiquitination and degradation. An optimal stoichiometric ratio of p53, Mdm2, and MdmX appears to be required for maximal p53 ubiquitination and degradation in vivo as revealed in this study (Figs. 2 and 6). This might help to explain some early observations in MdmX studies, i.e. MdmX overexpression stabilized p53 and Mdm2 in a number of cell types (41–43).
In contrast to Mdm2, MdmX expression is not regulated transcriptionally after DNA damage (44). On the other hand, posttranslational down-regulation of MdmX that precedes p53 stabilization in response to DNA damage has been observed in several studies. Because MdmX is an essential component for Mdm2-driven p53 polyubiquitnation and degradation, DNA damage triggered removal of MdmX would be consistent with the rapid uncoupling of the p53/Mdm2 regulatory loop, which heralds p53 accumulation and nuclear accumulation in response to DNA damage (23–25). In a more cancer-relevant context, MdmX was shown to be transcriptionally up-regulated by mitogenic signaling (45), suggesting its involvement in procancer inactivation of p53.
In light of the critical role of MdmX in p53 down-regulation and the fact that MdmX expression is regulated by mitogenic signaling (45), targeting MdmX will undoubtedly restore p53 activity in a more robust way for therapeutic treatment of cancer. Indeed, it has been reported that the levels of MdmX dictate the sensitivity of normal and transformed cells to the Mdm2 inhibitor Nutlin-3 (46), and efficient p53 activation and apoptosis can be achieved by simultaneous disruption of binding to Mdm2 and MdmX (47). Therefore, MdmX inhibitors may be promising anti-cancer drugs for targeted therapies that aim to potentiate p53 tumor suppressor activity.
Supplementary Material
Acknowledgment
We thank Dr. Moshe Oren for pGEX GP-Hdm2, pCMV-p53 and pT7-UbcH5c constructs and Dr. Jennifer Black for critical reading of the manuscript.
This work was supported by the American Cancer Society and a Geoffrey Beene Cancer Research Fund award (to X. J.) and by start-up funds from the Roswell Park Cancer Institute (to X. W.) and Elsa U. Pardee Foundation (to X. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- Ub
- ubiquitin.
REFERENCES
- 1. Levine A. J., Oren M. (2009) Nat. Rev. Cancer 9, 749–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oren M. (1999) J. Biol. Chem. 274, 36031–36034 [DOI] [PubMed] [Google Scholar]
- 3. Vousden K. H., Prives C. (2009) Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 4. Kruse J. P., Gu W. (2009) Cell 137, 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marine J. C., Jochemsen A. G. (2005) Biochem. Biophys. Res. Commun. 331, 750–760 [DOI] [PubMed] [Google Scholar]
- 6. Momand J., Zambetti G. P., Olson D. C., George D., Levine A. J. (1992) Cell 69, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 7. Montes de Oca Luna R., Wagner D. S., Lozano G. (1995) Nature 378, 203–206 [DOI] [PubMed] [Google Scholar]
- 8. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 9. Kubbutat M. H., Ludwig R. L., Ashcroft M., Vousden K. H. (1998) Mol. Cell Biol. 18, 5690–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Honda R., Tanaka H., Yasuda H. (1997) FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 11. Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M. (2000) J. Biol. Chem. 275, 8945–8951 [DOI] [PubMed] [Google Scholar]
- 12. Wang X., Michael D., de Murcia G., Oren M. (2002) J. Biol. Chem. 277, 15697–15702 [DOI] [PubMed] [Google Scholar]
- 13. Li M., Luo J., Brooks C. L., Gu W. (2002) J. Biol. Chem. 277, 50607–50611 [DOI] [PubMed] [Google Scholar]
- 14. Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Itahana K., Mao H., Jin A., Itahana Y., Clegg H. V., Lindström M. S., Bhat K. P., Godfrey V. L., Evan G. I., Zhang Y. (2007) Cancer Cell 12, 355–366 [DOI] [PubMed] [Google Scholar]
- 16. Shvarts A., Steegenga W. T., Riteco N., van Laar T., Dekker P., Bazuine M., van Ham R. C., van der Houven van Oordt W., Hateboer G., van der Eb A. J., Jochemsen A. G. (1996) EMBO J. 15, 5349–5357 [PMC free article] [PubMed] [Google Scholar]
- 17. Tanimura S., Ohtsuka S., Mitsui K., Shirouzu K., Yoshimura A., Ohtsubo M. (1999) FEBS Lett. 447, 5–9 [DOI] [PubMed] [Google Scholar]
- 18. Parant J., Chavez-Reyes A., Little N. A., Yan W., Reinke V., Jochemsen A. G., Lozano G. (2001) Nat. Genet. 29, 92–95 [DOI] [PubMed] [Google Scholar]
- 19. Xiong S., Van Pelt C. S., Elizondo-Fraire A. C., Liu G., Lozano G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3226–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiong S., Van Pelt C. S., Elizondo-Fraire A. C., Fernandez-Garcia B., Lozano G. (2007) Circulation 115, 2925–2930 [DOI] [PubMed] [Google Scholar]
- 21. Barboza J. A., Iwakuma T., Terzian T., El-Naggar A. K., Lozano G. (2008) Mol. Cancer Res. 6, 947–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li C., Chen L., Chen J. (2002) Mol. Cell Biol. 22, 7562–7571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen L., Gilkes D. M., Pan Y., Lane W. S., Chen J. (2005) EMBO J. 24, 3411–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Okamoto K., Kashima K., Pereg Y., Ishida M., Yamazaki S., Nota A., Teunisse A., Migliorini D., Kitabayashi I., Marine J. C., Prives C., Shiloh Y., Jochemsen A. G., Taya Y. (2005) Mol. Cell Biol. 25, 9608–9620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pereg Y., Shkedy D., de Graaf P., Meulmeester E., Edelson-Averbukh M., Salek M., Biton S., Teunisse A. F., Lehmann W. D., Jochemsen A. G., Shiloh Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5056–5061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y. V., Wade M., Wong E., Li Y. C., Rodewald L. W., Wahl G. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12365–12370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang X., Trotman L. C., Koppie T., Alimonti A., Chen Z., Gao Z., Wang J., Erdjument-Bromage H., Tempst P., Cordon-Cardo C., Pandolfi P. P., Jiang X. (2007) Cell 128, 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang X., Taplick J., Geva N., Oren M. (2004) FEBS Lett. 561, 195–201 [DOI] [PubMed] [Google Scholar]
- 29. Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. (2003) Science 302, 1972–1975 [DOI] [PubMed] [Google Scholar]
- 30. Lai Z., Ferry K. V., Diamond M. A., Wee K. E., Kim Y. B., Ma J., Yang T., Benfield P. A., Copeland R. A., Auger K. R. (2001) J. Biol. Chem. 276, 31357–31367 [DOI] [PubMed] [Google Scholar]
- 31. Yan H., Lim J. T., Contillo L. G., Krolewski J. J. (1995) Anal. Biochem. 231, 455–458 [DOI] [PubMed] [Google Scholar]
- 32. Maru Y., Afar D. E., Witte O. N., Shibuya M. (1996) J. Biol. Chem. 271, 15353–15357 [DOI] [PubMed] [Google Scholar]
- 33. Linares L. K., Hengstermann A., Ciechanover A., Müller S., Scheffner M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12009–12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Linke K., Mace P. D., Smith C. A., Vaux D. L., Silke J., Day C. L. (2008) Cell Death Differ. 15, 841–848 [DOI] [PubMed] [Google Scholar]
- 35. Hashizume R., Fukuda M., Maeda I., Nishikawa H., Oyake D., Yabuki Y., Ogata H., Ohta T. (2001) J. Biol. Chem. 276, 14537–14540 [DOI] [PubMed] [Google Scholar]
- 36. Kawai H., Lopez-Pajares V., Kim M. M., Wiederschain D., Yuan Z. M. (2007) Cancer Res. 67, 6026–6030 [DOI] [PubMed] [Google Scholar]
- 37. Badciong J. C., Haas A. L. (2002) J. Biol. Chem. 277, 49668–49675 [DOI] [PubMed] [Google Scholar]
- 38. Gu J., Kawai H., Nie L., Kitao H., Wiederschain D., Jochemsen A. G., Parant J., Lozano G., Yuan Z. M. (2002) J. Biol. Chem. 277, 19251–19254 [DOI] [PubMed] [Google Scholar]
- 39. Okamoto K., Taya Y., Nakagama H. (2009) FEBS Lett. 583, 2710–2714 [DOI] [PubMed] [Google Scholar]
- 40. Ohtsubo C., Shiokawa D., Kodama M., Gaiddon C., Nakagama H., Jochemsen A. G., Taya Y., Okamoto K. (2009) Cancer Sci. 100, 1291–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sharp D. A., Kratowicz S. A., Sank M. J., George D. L. (1999) J. Biol. Chem. 274, 38189–38196 [DOI] [PubMed] [Google Scholar]
- 42. Stad R., Little N. A., Xirodimas D. P., Frenk R., van der Eb A. J., Lane D. P., Saville M. K., Jochemsen A. G. (2001) EMBO Rep. 2, 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jackson M. W., Berberich S. J. (2000) Mol. Cell Biol. 20, 1001–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jackson M. W., Berberich S. J. (1999) DNA Cell Biol. 18, 693–700 [DOI] [PubMed] [Google Scholar]
- 45. Gilkes D. M., Pan Y., Coppola D., Yeatman T., Reuther G. W., Chen J. (2008) Mol. Cell Biol. 28, 1999–2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Patton J. T., Mayo L. D., Singhi A. D., Gudkov A. V., Stark G. R., Jackson M. W. (2006) Cancer Res. 66, 3169–3176 [DOI] [PubMed] [Google Scholar]
- 47. Hu B., Gilkes D. M., Chen J. (2007) Cancer Res. 67, 8810–8817 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.