Abstract
Two processes, DNA replication and DNA damage repair, are key to maintaining genomic fidelity. The Dna2 enzyme lies at the heart of both of these processes, acting in conjunction with flap endonuclease 1 and replication protein A in DNA lagging strand replication and with BLM/Sgs1 and MRN/X in double strand break repair. In vitro, Dna2 helicase and flap endo/exonuclease activities require an unblocked 5′ single-stranded DNA end to unwind or cleave DNA. In this study we characterize a Dna2 nuclease activity that does not require, and in fact can create, 5′ single-stranded DNA ends. Both endonuclease and flap endo/exonuclease are abolished by the Dna2-K677R mutation, implicating the same active site in catalysis. In addition, we define a novel ATP-dependent flap endo/exonuclease activity, which is observed only in the presence of Mn2+. The endonuclease is blocked by ATP and is thus experimentally distinguishable from the flap endo/exonuclease function. Thus, Dna2 activities resemble those of RecB and AddAB nucleases even more closely than previously appreciated. This work has important implications for understanding the mechanism of action of Dna2 in multiprotein complexes, where dissection of enzymatic activities and cofactor requirements of individual components contributing to orderly and precise execution of multistep replication/repair processes depends on detailed characterization of each individual activity.
Keywords: DNA Enzymes, DNA Helicase, DNA Repair, DNA Replication, Enzyme Mechanisms, Dna2, Homologous Recombination, Manganese, Okazaki Fragment Processing, RPA
Introduction
The Dna2 protein is an essential enzyme involved in both Okazaki fragment processing and double strand break (DSB)4 repair (1–5). The conserved Dna2 enzymatic activities, single-stranded DNA (ssDNA)-dependent ATPase, 5′-3′ helicase, 5′-3′ endo/exonuclease, 3′-5′ exonuclease, single strand annealing, and strand exchange, function in DNA replication and DSB repair in both the nucleus and mitochondria of yeast and human cells (6–16). Genetic and biochemical studies have shown yeast, Xenopus, and human Dna2 nuclease to function with Sgs1/BLM helicase and the MRX-MRN complex in the 5′ end resection of DSBs and with flap endonuclease 1 (FEN1), replication protein A (RPA), and Ctf4 in lagging strand DNA replication (11, 12, 17–27). Given its key roles in these two important pathways and its multiplicity of biochemical functions, it is of interest to characterize further the unique enzymatic functions and substrate preferences of the Dna2 enzyme that underlie these physiological processes.
The preference of Dna2 for ssDNA flaps, such as 5′-terminated tails generated during strand displacement synthesis by polymerase δ during Okazaki fragment processing, is evident in binding and enzymatic studies (28, 29). The helicase activity of Dna2 requires a free 5′ ssDNA end to unwind DNA, and the flap endo/exonuclease activity requires an unblocked 5′ end to cleave flap structures (30, 31). Although the DNA end is necessary to stimulate the flap nuclease activity, cleavage occurs within the flap up to 6–7 nucleotides from the ssDNA/dsDNA junction making the reaction technically endonuclease (2, 32). This function is termed flap endo/exonuclease in this study consistent with other proteins that have similar modes of action. DNA fork structures or ssDNA flap structures (single-stranded tails at a nick or gap in duplex DNA) compete for Dna2 binding more effectively than nicked duplex or ssDNA gap regions, showing that Dna2 is a structure-specific enzyme (29). This binding is independent of the free end required for nucleolytic cleavage, because Dna2 can bind to a DNA fork or flap when the 5′ end is blocked, even though its nuclease activity cannot cleave the structure (18, 28).
The purpose of the DNA end is of considerable interest in understanding how Dna2 functions in the cell. The Dna2 enzyme is proposed to initially bind to the ssDNA/dsDNA junction and then track down the flap from the free end. Evidence for this model comes from experiments with a blockage in the middle of the flap that results in cleavage between the end and the blockage point but not beyond the block toward the base of the flap (31). One unique role of Dna2 is to remove long flaps that are coated by the ssDNA-binding protein RPA because these structures cannot be removed by FEN1 during Okazaki fragment processing (18, 33, 34). Interestingly, yeast Dna2 can displace RPA from flaps with blocked 5′ ends, even though Dna2 cannot track down the length of the blocked flap (33).
These observations have led us to investigate further the requirement for free ends in Dna2 activity. In this study, we characterize Dna2 endonuclease activity in the absence of DNA ends (32). We show that Dna2 can bind circular ssDNA and cleave it, but that, in contrast to previous results, circular DNA is not an effector of the ATPase activity (35). We also find that the endonuclease and the flap endo/exonuclease are catalyzed by the same active site, but that, nevertheless, the activities are distinguishable based on their cofactor requirements. The presence of this endonuclease function allows the Dna2 enzyme to create free ends for further helicase and flap endo/exonuclease processing, which might be encountered during repair of DSBs with ends modified by proteins or terminating in modified nucleotides.
EXPERIMENTAL PROCEDURES
Materials and Substrates
M13mp18 and ΦX174 phage ssDNA was purchased from New England Biolabs. All synthetic oligonucleotides were purchased from Integrated DNA Technologies. Oligonucleotide sequences are listed in Table 1. [γ-32P]ATP, 10 mCi/ml, was purchased from MP Biomedicals. The 5′ oligonucleotide labeling reactions were performed as described previously with T4 polynucleotide kinase and [γ-32P]ATP (36). For annealing to M13mp18, oligonucleotides were placed in TE (1 mm EDTA and 10 mm Tris-HCl (pH 8.0)), heated to 100 °C for 5 min and slowly cooled to room temperature. Yeast RPA was purified as described (37).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotides | Length | Sequence |
|---|---|---|
| nt | ||
| 5′ Flap | 98 | TTCACGAGATTTACTTATTTCACTGCGGCTACATGATGCATCGTTAGGCGATTCCGCCTAACGATGCATCATGTCGCGAACCCTATTTAGGGTTCGCG |
| 5′ Tail | 74 | TTCACGAGATTTACTTATTTCACTGCGGCTACATGATGCATCGTTAGGCGATTCCGCCTAACGATGCATCATGT |
| H1 (5′ flap) | 42 | AGCTAGCTCTTGATCGTAGACGTTGTAAAACGACGGCCAGTG |
| H2 (annealed) | 24 | GACGTTGTAAAACGACGGCCAGTG |
Dna2 Protein Purification
Human Dna2 was purified as described previously (6). Wild-type and nuclease-dead (K677R) Saccharomyces cerevisiae Dna2 proteins were expressed in yeast grown in 2 liters of minimal medium without uracil supplemented with 2% glycerol, 3% lactic acid, and 3% galactose. Cells were lysed using a CryoMill (Retsch) with continuous liquid nitrogen cooling. Pellets were resuspended in 50 ml of 20 mm Tris-HCl (pH 8.0), 35 mm imidazole, 750 mm NaCl, 5% glycerol (v/v), 1 mm 2-mercaptoethanol, 0.01% Triton X-100, and Complete EDTA-free protease inhibitor mixture (Roche Applied Science). Extracts were clarified by ultracentrifugation at 29,000 rpm for 20 min in a Beckman Ti-45 rotor. The supernatant was mixed with 2 ml of Ni2+-nitrilotriacetic acid-agarose (Qiagen) for 1 h at 4 °C. Resin was collected by gravity flow and washed three times with 20 ml of 20 mm Tris-HCl (pH 8.0), 35 mm imidazole, 5% glycerol, 1 mm 2-mercaptoethanol buffer containing decreasing levels of NaCl (750 mm, 300 mm, and 100 mm NaCl, respectively). Dna2 was eluted in 1-ml fractions of 20 mm Tris-HCl (pH 8.0), 400 mm imidazole, 5% glycerol, 1 mm 2-mercaptoethanol, and 100 mm NaCl. Fractions containing Dna2 were pooled and loaded onto a 1-ml MonoQ FPLC column (GE Healthcare) equilibrated with MonoQ buffer (25 mm Tris-HCl (pH 7.5), 10% glycerol, 100 mm NaCl, 1 mm EDTA). The column was washed with 10 ml of MonoQ buffer and eluted over a 10-ml gradient of 100–600 mm NaCl. Fractions containing Dna2 were pooled and dialyzed into storage buffer containing 25 mm Tris-HCl (pH 7.5), 500 mm NaCl, 25% glycerol, and 1 mm EDTA. Aliquots were stored at −80 °C.
Endonuclease Assay with Circular ssDNA
Nuclease reactions containing 50 fmol of Dna2 protein, 500 ng of M13mp18 or ΦX174 circular ssDNA, and MgCl2, MnCl2, and ATP as noted in the figures and figure legends in 20 μl of reaction buffer (50 mm Tris-HCl (pH 7.5), 25 mm NaCl, 2 mm dithiothreitol (DTT), 0.25 mg/ml bovine serum albumin (BSA)) were incubated at 37 °C for 15 min. Reactions were stopped by the addition 5× buffer (60 mm EDTA, 40% sucrose, 0.6% SDS, 0.25% bromphenol blue, and 0.25% xylene cyanole FF) and electrophoresed on a 1% agarose gel containing ethidium bromide. Reactions including RPA, 0.75 μg or 1.5 μg, were first incubated in reaction buffer (25 mm Tris-HCl (pH 7.5), 25 mm NaCl, 2 mm DTT, 0.25 mg/ml BSA, 1 mm MgCl2 or MnCl2, and 100 ng of M13mp18) minus Dna2 for 10 min at room temperature to facilitate RPA-ssDNA binding. 50 fmol of Dna2 was then added, and reactions were incubated at 37 °C for 15 min.
Electrophoretic Mobility Shift Assay with Competitor
Reactions, mixed on ice, contained 1 pmol of nuclease-dead Dna2-K677R and 20 fmol of 32P-labeled 5′ flap substrate in 20 μl of reaction buffer (25 mm Tris-HCl (pH 7.5), 2 mm DTT, 0.5 mg/ml BSA, 2 mm ATP, 5% glycerol, and 1 mm or 2 mm MgCl2 as indicated). Unlabeled competitors, M13mp18 circular ssDNA, and 5′ flap substrate were added as indicated, and reactions were incubated at 30 °C for 20 min. Samples were electrophoresed on a 12% polyacrylamide gel at 4 °C, 100 V for 3 h with 0.5× TBE running buffer (89 mm Tris, 89 mm boric acid, and 2 mm EDTA), and products were detected by PhosphoImager.
ATPase Assay
ATPase reactions containing 1 pmol of wild-type or nuclease-dead Dna2 protein in 20 μl of reaction buffer (40 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 25 mm NaCl, 1 mm DTT, 0.5 mg/ml BSA, 0.2 mm ATP, 10% glycerol, and 3 μCi of [γ-32P]ATP) were supplemented with 15.625 ng, 62.5 ng. 250 ng, or 1 μg of ssDNA (M13mp18 ssDNA circle and 5′-flap substrate as indicated) and incubated at 30 °C for 1 h. The reactions were stopped by adding EDTA to a final concentration of 4 mm. 0.8 μl of each reaction was spotted onto a polyethyleneimine-cellulose TLC plate (Selecto Scientific) and developed in 0.5 m LiCl, 1 m formic acid solution. Products were detected by PhosphoImager.
Exonuclease Assay with Radiolabeled Substrate
Nuclease reactions containing 50 fmol of Dna2 protein, 20 fmol of radiolabeled substrate, and MgCl2, MnCl2, and ATP as noted in 20 μl of reaction buffer (50 mm Tris-HCl (pH 7.5), 25 mm NaCl, 2 mm DTT, 0.25 mg/ml BSA) were incubated at 37 °C for 15 min. Reactions were stopped by using 2× denaturing termination dye (95% deionized formamide, 10 mm EDTA, 0.1% bromphenol blue, and 0.1% xylene cyanol), and boiled for 5 min. The cleavage products were separated on a 12% sequencing gel (SequaGel; National Diagnostics) and detected by PhosphoImager.
Endonuclease and Helicase Assays with Radiolabeled Substrate
Standard reactions contained 300 fmol of Dna2 or Dna2-K677R and 10 fmol of helicase substrates (32P-labeled H1 (5′ flap) or H2 (fully annealed) oligonucleotides annealed to M13mp18) in 20 μl of reaction buffer (25 mm Tris-HCl (pH 7.5), 25 mm NaCl, 2 mm DTT, 0.25 mg/ml BSA) with MgCl2, MnCl2, and ATP as indicated. Reaction buffers with increasing NaCl contained from 0 to 200 mm NaCl as noted. After incubation at 37 °C for 30 min, reactions were stopped with 5× stop solution (60 mm EDTA, 40% sucrose, 0.6% SDS, 0.25% bromphenol blue, and 0.25% xylene cyanole FF). Reaction products were then separated using 8% native polyacrylamide gels containing 0.1% SDS and detected by PhosphoImager.
For endonuclease reactions with yeast RPA using 32P-labeled H2 oligonucleotide annealed to M13mp18 as the substrate, 2.5 pmol of RPA/reaction was incubated with 10 fmol of the substrate in 1× reaction buffer for 10 min at room temperature prior to the addition of yeast Dna2 protein and 1 mm MgCl2 or 1 mm MnCl2 as indicated.
RESULTS
Dna2 Has Endonuclease Activity
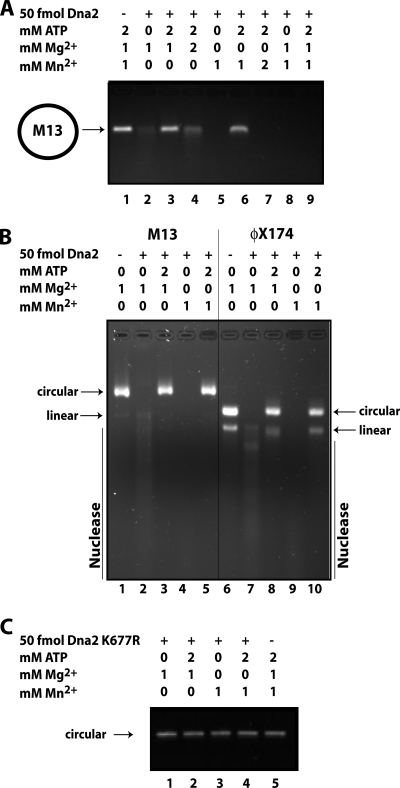
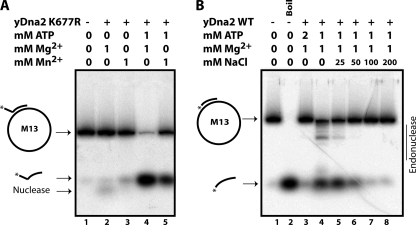
Several recent biochemical studies have demonstrated the requirement for a free 5′ DNA end for Dna2 helicase and flap endo/exonuclease activity on synthetic oligonucleotide substrates (30, 31). However, prior studies showed that Dna2 has nuclease activity on circular ssDNA, i.e. DNA entirely without ends (2, 32). To reconcile these observations, we revisited the endonuclease function of Dna2. We first asked whether yeast Dna2 could cut a long ssDNA substrate without an end. For this, we used the circular ssDNA of phage M13mp18 (M13). Under standard Dna2 nuclease assay conditions, as shown in Fig. 1A, lane 2, the phage DNA was degraded by Dna2. ATP is known to inhibit the flap endo/exonuclease activity of Dna2. As shown in Fig. 1A, lane 3, ATP also protects the circular ssDNA substrate from Dna2 endonuclease activity. A ratio of 1:1 Mg2+:ATP significantly reduces the endonuclease activity of Dna2 (Fig. 1A, lane 4).
FIGURE 1.
Dna2 has endonuclease activity. A, nuclease assays were performed as described under “Experimental Procedures.” Briefly, 50 fmol of yeast Dna2 was incubated with 500 ng of M13mp18 phage ssDNA and MgCl2, MnCl2, and ATP as indicated for 15 min at 37 °C. Reaction products were subjected to electrophoresis on a 1% agarose gel, and DNA was stained with ethidium bromide. B, nuclease reactions were performed as above with M13mp18 and ΦX174 ssDNA as indicated. C, nuclease assay with M13mp18 ssDNA and Dna2-K677R, nuclease-defective, is shown.
Recently, Dna2 has been shown to function in conjunction with Mre11 and Sgs1/Rmi1/Top3 in resection of a DSB. It is not clear whether the Mre11 is performing a preprocessing event, such as preliminary resection, or whether its primary role is in recruiting the Dna2/Sgs1 proteins to the unprocessed break. The nuclease activity of Mre11 is dependent on Mn2+ and is not supported by Mg2+ under most conditions. However, Mn2+, even in the presence of Mg2+, inhibits the DSB resection reaction reconstituted from the human counterparts to the yeast proteins: MRN, BLM, and DNA2, raising the question of which protein in the resection reaction is inhibited by Mn2+ (12). We found that M13mp18 is completely degraded by yeast Dna2 in the presence of Mn2+ alone as a cofactor (Fig. 1A, lane 5). Addition of ATP at a ratio of 2:1 ATP:Mn2+ inhibits the endonuclease activity (Fig. 1A, lane 6), as it does with Mg2+, but at a 1:1 ATP:Mn2+ ratio, ATP is not sufficient to protect the M13 circular DNA (lane 7). Endonuclease activity is somewhat greater with Mn2+ than Mg2+ with a circular substrate. A mixture of Mg2+ and Mn2+ also supported endonuclease activity (Fig. 1A, lane 8) and was not inhibited by a 1:1 ratio of ATP to Mg2+ and Mn2+ (lane 9), unlike the reaction with Mg2+ alone, suggesting that Mn2+ is dominant over Mg2+ in supporting Dna2 endonuclease.
Because Mn2+ does not inhibit, but stimulates Dna2, it is not Dna2 in the Dna2/BLM/MRN reconstituted resection reaction that is inhibited by Mn2+, although the effects of Mn2+ could be different in the presence of the additional proteins. Similar endonuclease properties were observed with ΦX174 circular ssDNA (Fig. 1B). In this case it is clear that the ΦX174 DNA preparation contained a significant fraction of linear as well as circular DNA. Both are degraded by Dna2 in the presence of either Mg2+ or Mn2+. The contamination of the ΦX174 circular DNA preparation with linear DNA, documented by the provider, can explain a previous observation suggesting that the helicase activity of Dna2 could function in the absence of a free end for Dna2 loading (35). No endonuclease is observed with Dna2-K677R, which is endo/exonuclease-defective in the presence of either Mg2+ or Mn2+ (Fig. 1C). We conclude that, unlike Dna2 helicase and flap endo/exonuclease functions, Dna2 can act as an endonuclease in the absence of a free DNA end and that the same active site catalyzes both nucleolytic activities.
Association of Dna2 with ssDNA Does Not Require an End
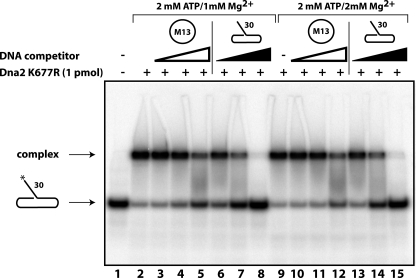
We next investigated whether association of Dna2 with ssDNA required a DNA end. Previous studies have shown that Dna2 can bind to ssDNA flaps with blocked ends, but those substrates contained ssDNA/dsDNA junctions and other secondary structures that are strong binding sites for Dna2 (29). We asked whether circular ssDNA could compete with a radiolabeled flap substrate for Dna2 binding as measured by EMSA. To protect the labeled flap substrate from Dna2 nuclease activity, the nuclease-deficient Dna2-K677R mutant was used. In Fig. 2, lane 2, it is clear that Dna2 binds to the flap substrate. Competition with increasing amounts of unlabeled flap substrate reduces the amount shifted (Fig. 2, lanes 6–8 and 13–15). When unlabeled circular ssDNA was added as a competitor, the amount of flap substrate shifted by Dna2 was also reduced (Fig. 2, lanes 3–5 and 10–12). Therefore, Dna2 does not require a free DNA end or a flap/fork junction to bind ssDNA. As in the case of the flap substrate, the binding to the ssDNA circle was not inhibited by ATP, even though these conditions protect the circle from Dna2 endonuclease degradation (see Fig. 1). We conclude that Dna2 can bind to DNA without ends and that the inhibitory effect of high levels of ATP on nuclease activity is not due to interference with DNA binding.
FIGURE 2.
Association of Dna2 with ssDNA does not require an end. EMSA was performed as described under “Experimental Procedures.” 32P-Labeled 5′ flap substrate was mixed on ice with 1 pmol of Dna2-K677R protein in reaction buffer supplemented with Mg2Cl and ATP as indicated. Lane 1 contains the substrate in the absence of Dna2 protein. Lanes 2 and 9 indicate the electrophoretic mobility shift of the substrate with Dna2 protein in the absence of DNA competitor. Lanes 3–5 and 10–12 show binding of the labeled substrate in the presence of 2.5, 10, and 100 ng of M13mp18 circular ssDNA competitor as noted by white triangles. Lanes 6–8 and 13–15 show binding of the labeled substrate in the presence of 2.5, 10, and 100 ng of unlabeled 5′ flap substrate competitor, as noted by black triangles.
DNA Ends Are Required to Stimulate Dna2 ATPase Activity
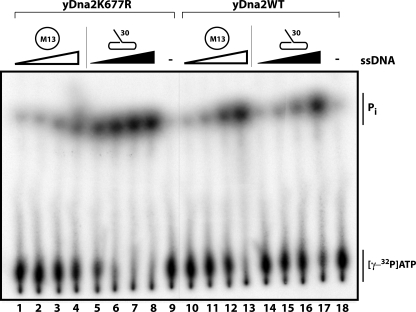
Dna2 is a ssDNA-dependent ATPase. With wild-type Dna2 protein, the circular ssDNA and linear ssDNA promote ATPase activity equally (Fig. 3, lanes 10–13 and 14–17). With the nuclease-dead Dna2-K677R protein, however, only linear DNA serves as an ATPase effector (Fig. 3, lanes 1–4), even though we demonstrated that Dna2-K677R does bind to the circular ssDNA (Fig. 2). We infer that wild-type Dna2 first linearizes the circular DNA, which then serves as a Dna2 cofactor. This verifies that DNA ends, not ssDNA alone, are required to stimulate ATPase activity. This also establishes the fact that the K677R mutation prevents the generation of these ends, confirming that the K677R mutation abolishes both endonuclease and flap endo/exonuclease activities.
FIGURE 3.
DNA ends are required for ATPase activity. ATPase assays were performed as described under “Experimental Procedures.” Reactions in lanes 1–9 were performed with 1 pmol of nuclease-dead Dna2-K677R. Reactions in lanes 10–18 were performed with 1 pmol of wild-type Dna2 protein. Lanes 1–4 and 10–13 were supplemented with 15.625 ng, 62.5 ng, 250 ng, and 1 μg of M13mp18 circular ssDNA as denoted by white triangles. Lanes 5–8 and 14–17 were supplemented with 15.625 ng, 62.5 ng, 250 ng, and 1 μg of 5′ flap substrate as denoted by black triangles. Reactions in lanes 9 and 18 lacked ssDNA.
Endo- and Exonuclease Activities Can Be Separated
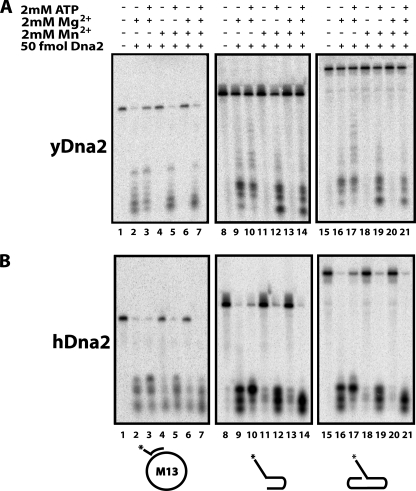
The strong Dna2 endonuclease activity in the presence of Mn2+ prompted us to investigate flap endo/exonuclease activity in the presence of Mn2+. As shown in Fig. 4A, lane 2, yeast Dna2 flap endo/exonuclease, in the presence of Mg2+, removes the 5′ ssDNA flap from 20 fmol of radiolabeled substrate. This activity is reduced in the presence of ATP as evidenced by the remaining uncut substrate (Fig. 4A, lane 3). Mn2+ cannot substitute for Mg2+ in cutting the 5′ flap (Fig. 4A, lane 4). Surprisingly, however, when Mn2+ is supplemented with ATP, the flap endo/exonuclease activity is restored (Fig. 4A, lane 5). This ATP-dependent exonuclease activity in the presence of Mn2+ stands in direct contrast to the Mg2+-dependent endonuclease activity that is inhibited by the presence of ATP.
FIGURE 4.
Endonuclease and exonuclease functions can be separated. Nuclease assays were performed as described under “Experimental Procedures” with 50 fmol of yeast Dna2 (A) or human Dna2 (B) protein for 15 min at 37 °C. The radiolabeled substrates are depicted under the respective panels with an asterisk denoting the position of the label: H1 (5′ flap) annealed to M13 phage ssDNA (lanes 1–7), 5′ tail with dsDNA hairpin (lanes 8–14), and 5′ flap with dsDNA hairpin (lanes 15–21). Addition of 2 mm ATP, 2 mm MgCl2, and 2 mm MnCl2 is denoted by +/− marks.
When Mg2+ and Mn2+ are present in equal concentrations, no flap endo/exonuclease is detected, similar to Mn2+ alone (compare Fig. 4A, lanes 4 and 6). When ATP is added to the reaction with Mg2+ and Mn2+ (Fig. 4A, lane 7), the substrate is cleaved. The inhibition of Mg2+-dependent flap endo/exonuclease activity by Mn2+ is puzzling, considering that both cofactors support robust endonuclease activity alone or together. However, this observation demonstrates that the endonuclease and flap endo/exonuclease functions are experimentally separable, even though they are both abolished with a mutation at the same residue in the nuclease active site. We analyzed three different 5′ flap substrates and found that this Mn2+-dependent behavior was consistent regardless of the substrate structure at the ssDNA/dsDNA junction (Fig. 4A).
Comparison of the reaction products in Fig. 4A, especially between lanes 9 and 12, for the 5′ overhang substrate and lanes 16 and 19 for the 5′ flap substrate adjacent to an upstream oligonucleotide, shows the Mn2+/ATP-stimulated nuclease products to be shorter than those observed with Mg2+ alone. It is unknown whether this is due to lower processivity along the flap or secondary cutting after the initial flap removal with Mn2+ and ATP. However, similar nuclease assays with 3′ labels on the 5′ fork substrate show the final products after both Mg2+ and Mn2+/ATP mediated cutting to be the same length (data not shown). The presence of Mn2+ does not enable Dna2 to cut or degrade linear double-stranded DNA (dsDNA), with or without ATP, consistent with behavior with cofactor Mg2+ (data not shown).
All of these activities, including the ATP-dependent, Mn2+-stimulated flap removal activity, were conserved in the human Dna2 protein (Fig. 4B). As previously reported, human Dna2 does not show nuclease when Mn2+ is substituted for Mg2+ (Fig. 4B, lanes 4 and 6). However, we now show that Mn2+ does exhibit nuclease activity if ATP is added (Fig. 4B, lanes 5 and 7).
Endonuclease Can Create DNA End for Helicase or Exonuclease Activity
It has been recently established that the Dna2 helicase requires a free DNA end to unwind DNA (30). Considering the end-independent endonuclease activity characterized above, we wondered whether Dna2 could create a free end suitable for its helicase activity. We first measured helicase activity using the Dna2-K677R mutant and a 5′ 32P-labeled oligonucleotide with an 18-nucleotide 5′ noncomplementary flap annealed to the M13mp18 circular ssDNA. Helicase products are generated in the presence of ATP with either Mg2+ or Mn2+ but not in the absence of ATP (Fig. 5A, compare lanes 4 and 5 with lanes 2 and 3). Note that the small amount of label migrating ahead of the oligonucleotide in lane 2 is due to minor residual exonuclease seen in Dna2-K677R preparations after extended incubation, but that no endonuclease products, such as are seen in Fig. 5B, lane 4, are apparent. Although not as potent as Mg2+, we find that Mn2+ is a functional cofactor for Dna2 helicase activity.
FIGURE 5.
Endonuclease activity can generate helicase substrate. A, helicase assays were performed as described under “Experimental Procedures” using nuclease-dead Dna2-K677R and 1 mm MgCl2, 1 mm MnCl2, and 2 mm ATP as indicated with radiolabeled H1 (5′ flap) oligonucleotide annealed to M13mp18 ssDNA. Reactions were incubated at 37 °C for 30 min, and products were resolved on nondenaturing PAGE. Lane 1 represents the substrate alone. Helicase reaction products are indicated by the arrow corresponding to the oligonucleotide alone. Products of residual Dna2 flap endo/exonuclease activity in lane 2 are indicated by the nuclease arrow. B, helicase reactions were performed as above using wild-type Dna2 and a radiolabeled H2 (fully annealed) oligonucleotide annealed to M13mp18 ssDNA. Lane 1 contains the substrate alone and lane 2 the substrate after boiling. Helicase products are noted by the arrow corresponding to the labeled oligonucleotide after boiling. Lanes 4–8 contain increasing concentrations of NaCl from 0 to 200 mm as indicated.
In keeping with the demonstration that substrates without a ssDNA flap or with a flap whose 5′ end is blocked by a bulky adduct cannot be unwound by Dna2, when we annealed an oligonucleotide lacking a noncomplementary 5′ flap to M13mp18 and incubated with wild-type Dna2 under conditions that inhibit the endonuclease, 1 mm Mg2+ and 2 mm ATP, the substrate is not unwound (Fig. 5B, lane 3). This is likely because there is no free 5′ ssDNA tail on the oligonucleotide and the circle is protected from degradation by the excess ATP and cannot provide an alternative end. As shown in Fig. 5B, lane 4, when this assay is repeated in conditions that allow some endonuclease activity, 1 mm Mg2+ and 1 mm ATP as established in Fig. 1, both endonuclease products and helicase products are seen. Increasing amounts of NaCl have been shown previously to inhibit the flap endo/exonuclease activity of Dna2 (6, 15, 35). The endonuclease activity was also clearly inhibited by increasing NaCl (Fig. 5B, lanes 5–8), and this resulted in a corresponding reduction of unwound helicase product. These results led to the conclusion that Dna2 endonuclease can create a substrate for its helicase activity.
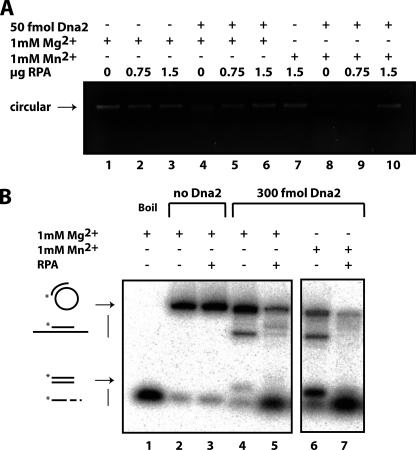
Replication Protein A Inhibits Dna2 Endonuclease Activity
Several studies have shown that Dna2 enzymatic activity is modulated by RPA; 5′ flap endo/exonuclease activity is stimulated whereas 3′ exonuclease activity is inhibited (7, 26, 33). This regulation is thought to give Dna2 specificity to process the correct strand of DSB ends and nascent Okazaki fragments. We were interested to see how RPA affected the Dna2 endonuclease function on the M13mp18 circular ssDNA lacking dsDNA/ssDNA junctions. As shown in Fig. 6A, RPA inhibited endonucleolytic cleavage of M13 DNA by Dna2 protein in the presence of either Mg2+ (Fig. 6A, lanes 4–6) or Mn2+ (Fig. 6A, lanes 8–10). This result is consistent with the inferred inhibition of Dna2 endonuclease by RPA on a substrate comprising a radiolabeled oligonucleotide (52-mer) annealed to ΦX174, a molecule with a ssDNA/dsDNA junction (2). In Fig. 6B, we show a similar experiment using M13 annealed to the shorter H2 (24-mer) oligonucleotide. Dna2 alone cleaves the circular substrate but does not displace the oligonucleotide, as expected (Fig. 6B, lanes 4 and 6). In the presence of RPA, however, we could not assess endonuclease activity, because, surprisingly, in the presence of both RPA and high levels of Dna2 (Fig. 6B, lanes 5 and 7), the entire labeled oligonucleotide was displaced. This displacement is due to yet another form of interaction between RPA and Dna2 because neither RPA (Fig. 6B, lane 3) nor Dna2 alone (Fig. 6B, lanes 4 and 6) displaces the oligonucleotide. Therefore, we conclude that RPA inhibits the endonuclease activity of Dna2 and that RPA and Dna2 can together destabilize partial duplex DNA in the absence of ATP.
FIGURE 6.
Replication protein A inhibits Dna2 endonuclease. A, endonuclease reactions were performed as described under “Experimental Procedures.” Briefly, RPA, 0, 0.75 μg, or 1.5 μg, was incubated with 100 ng of M13mp18 ssDNA in endonuclease buffer containing 1 mm MgCl2 or 1 mm MnCl2 as indicated, for 10 min at room temperature prior to the addition of wild-type Dna2. Reactions were then incubated at 37 °C for 15 min, products were resolved using electrophoresis on a 1% agarose gel, and DNA was stained with ethidium bromide. B, Dna2 and RPA together, but neither alone, can displace short oligonucleotides annealed to M13mp18. 2.5 pmol of RPA was incubated with radiolabeled H2 (fully annealed) oligonucleotide annealed to M13mp18 ssDNA in 1× endonuclease/helicase buffer containing no NaCl for 10 min at room temperature prior to the addition of wild-type Dna2, 1 mm MgCl2, and 1 mm MnCl2 as indicated. Reactions were incubated at 37 °C for 30 min, and products were resolved by nondenaturing PAGE. Lane 1 shows the substrate after boiling to displace the oligonucleotide.
DISCUSSION
In this study, we have established that the Dna2 enzyme exhibits true endonuclease activity. Unlike the well characterized flap endo/exonuclease function and helicase activity, the Dna2 endonuclease does not require a free ssDNA end. Although the endonuclease and flap endo/exonuclease appear to share an active site, assays with the cofactor Mn2+ show that these two functions are distinguishable.
The physiological significance of the endonuclease activity is unclear. The ability to cut ssDNA internally could conceivably result in a DSB at sites of DNA damage or between nascent Okazaki fragments. Therefore, the endonuclease activity must be somehow regulated. In the cell, long stretches of ssDNA would be bound by RPA, and we have shown that RPA inhibits the endonuclease on circular ssDNA in addition to circular ssDNA with an annealed oligonucleotide. Although RPA stimulates the Dna2 flap endo/exonuclease and RPA can be removed from DNA by Dna2 without threading from an end (33), RPA shows inhibitory effects on multiple Dna2 functions in addition to the endonuclease (7, 12, 38). Other structural features or participation of additional proteins in a complex at the replication fork may also modulate the endonuclease activity, like the endonuclease of RecB in the RecBCD complex (39). The need for regulation of this activity, however, should not obscure the fact that endonuclease may, at times, be beneficial because it could allow Dna2 to process intermediate structures into a form compatible with its helicase and flap endo/exonuclease requirements. The helicase function of Dna2 as studied in vitro would imply that it is not important to cells because the substrates are more likely to be cut by the flap endo/exonuclease than unwound. However, genetic experiments show that the helicase activity does play a role in the survival of damage in vivo (4).
Roles for the endonuclease may be hard to discern because mutants are also defective in flap endo/exonuclease activity. The observation that the two activities are distinguishable using the cofactor Mn2+ provides an approach to elucidate the contributions of the endonuclease function to Okazaki fragment processing or DBS resection in vitro. Future studies must also address the role of post-translational modifications and other proteins, especially Sgs1/BLM, in stimulating or inhibiting the endonuclease function of Dna2 on both DNA repair intermediates and Okazaki fragment processing steps.
In reactions with Mn2+, Dna2 exhibits helicase activity and endonuclease activity as with Mg2+. What is surprising is that addition of Mn2+ reveals an ATP-dependent exonuclease activity, like that of the bacterial enzymes RecBCD and AddAB (39–41). The nuclease domain of Dna2 shares some sequence homology with RecB (32). The AddAB enzyme, like RecBCD, is involved in DNA repair and homologous recombination with dsDNA- and ssDNA-dependent ATPase, helicase, and ATP-dependent endo- and exonuclease activities. Dna2 and AddAB have also been proposed to share structural similarities in the nuclease active site and in an iron staple domain spanning the active site (42). Perhaps conformational changes in the nuclease active site of Dna2 while bound to Mn2+ cause Dna2 to behave more like these related enzymes. Weak ATP-dependent nuclease has been seen previously with Dna2 in the presence of Mg2+, but was most likely 3′ exonuclease dependent on ATP-driven helicase activity exposing a 3′ ssDNA tail (32, 35, 43).
Functionally, the use of Mn2+ as a cofactor in vitro will allow Dna2 to be studied under conditions that promote both helicase and flap endo/exonuclease strongly. Use has been made of the ratio of cofactor to ATP in priming the Dna2 enzyme for preferentially observing either helicase or nuclease activity, respectively (31). However, the activity of the two domains is likely to be coupled (35). In fact, genetic experiments show that a mutant with Dna2 helicase activity but not nuclease activity is more sensitive to DSBs than a double helicase and nuclease-dead mutant (19). Understanding how the two domains work in concert with each other and with other enzymes present at sites of DNA replication and repair will be important, however, to understanding the role of Dna2 in genomic stability in a comprehensive fashion.
Although Mn2+ is not as abundant in the cell as Mg2+, several enzymes are manganese-dependent (44–46). Prominent among these is DSB-processing nuclease Mre11. Recently, the purified Mre11-Rad50 complex from bacteriophage T4, which also uses Mn2+ as a divalent cation, was found to exhibit some nuclease activity in Mg2+ when assayed with proteins UvsY (RAD52) and gp32 (RPA) (47). As with Dna2, the products from the Mg2+- and Mn2+-catalyzed reactions were slightly different. Another example of a manganese-dependent repair enzyme is the latent endonuclease of human MutLα, which is critical in human mismatch repair (48).
Dna2 is one of the key players in eukaryotic DSB repair (8, 10, 11, 38). It is important to point out that human Mre11, one of the stimulatory components of the DNA end resection machinery, exhibits strictly Mn2+-dependent endo- and exonuclease activity in vitro (44). Substitution of Mg2+ for Mn2+ in DNA end resection assays did not generate resection products (12). Furthermore, addition of Mn2+ to the Mg2+-driven reactions inhibited resection. It was suggested that this is due to inability of one or more of the proteins in the complex to function in the presence of Mn2+. Based on results presented here, the presence of Mn2+ in the resection reactions should not have an adverse effect with respect to Dna2 activity.
Changes in Dna2 active site conformation, and thus activity, caused by Mn2+ instead of Mg2+ binding in vitro may mimic a change in conformation when bound to other proteins in the cell or when post-translationally modified in vivo. These results may have importance in studying higher order DNA-protein complexes involving Dna2, a snapshot of which is provided by the studies of DSB resection, referred to above, by Nimonkar et al. (12). Detailed biochemical characterization of Dna2 will be important for the design of in vitro reconstitution experiments involving Dna2 in multiprotein complexes and dissection of enzymatic activities and cofactor requirements of individual components contributing to orderly and precise execution of multistep replication/repair processes.
This work was supported, in whole or in part, by National Institutes of Health Grants GM024441 (to R. A. B.) and GM078666 (to J. L. C.). This work was also supported by Army Research Office Grant ARO 09-1-0041 and Ellison Foundation Grant AG-55-2143 (to J. L. C.).
- DSB
- double strand break
- dsDNA
- double-stranded DNA
- FEN1
- flap endonuclease 1
- RPA
- replication protein A
- ssDNA
- single-stranded DNA.
REFERENCES
- 1. Budd M. E., Campbell J. L. (1997) Mol. Cell. Biol. 17, 2136–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bae S. H., Seo Y. S. (2000) J. Biol. Chem. 275, 38022–38031 [DOI] [PubMed] [Google Scholar]
- 3. Budd M. E., Campbell J. L. (2000) Mutat. Res. 459, 173–186 [DOI] [PubMed] [Google Scholar]
- 4. Weitao T., Budd M., Hoopes L. L., Campbell J. L. (2003) J. Biol. Chem. 278, 22513–22522 [DOI] [PubMed] [Google Scholar]
- 5. Budd M. E., Tong A. H., Polaczek P., Peng X., Boone C., Campbell J. L. (2005) PLoS Genet 1, e61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Masuda-Sasa T., Imamura O., Campbell J. L. (2006) Nucleic Acids Res. 34, 1865–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Masuda-Sasa T., Polaczek P., Peng X. P., Chen L., Campbell J. L. (2008) J. Biol. Chem. 283, 24359–24373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G. (2008) Cell 134, 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng L., Zhou M., Guo Z., Lu H., Qian L., Dai H., Qiu J., Yakubovskaya E., Bogenhagen D. F., Demple B., Shen B. (2008) Mol. Cell 32, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liao S., Toczylowski T., Yan H. (2008) Nucleic Acids Res. 36, 6091–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wawrousek K. E., Fortini B. K., Polaczek P., Chen L., Liu Q., Dunphy W. G., Campbell J. L. (2010) Cell Cycle 9, 1156–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nimonkar A. V., Genschel J., Kinoshita E., Polaczek P., Campbell J. L., Wyman C., Modrich P., Kowalczykowski S. C. (2011) Genes Dev. 25, 350–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Duxin J. P., Dao B., Martinsson P., Rajala N., Guittat L., Campbell J. L., Spelbrink J. N., Stewart S. A. (2009) Mol. Cell. Biol. 29, 4274–4282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Copeland W. C., Longley M. J. (2008) Mol. Cell 32, 457–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim J. H., Kim H. D., Ryu G. H., Kim D. H., Hurwitz J., Seo Y. S. (2006) Nucleic Acids Res. 34, 1854–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Masuda-Sasa T., Polaczek P., Campbell J. L. (2006) J. Biol. Chem. 281, 38555–38564 [DOI] [PubMed] [Google Scholar]
- 17. Formosa T., Nittis T. (1999) Genetics 151, 1459–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stewart J. A., Campbell J. L., Bambara R. A. (2009) J. Biol. Chem. 284, 8283–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Budd M. E., Campbell J. L. (2009) PloS One 4, e4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mimitou E. P., Symington L. S. (2009) Trends Biochem. Sci. 34, 264–272 [DOI] [PubMed] [Google Scholar]
- 21. Mimitou E. P., Symington L. S. (2009) DNA Repair 8, 983–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burgers P. M. (2009) J. Biol. Chem. 284, 4041–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stith C. M., Sterling J., Resnick M. A., Gordenin D. A., Burgers P. M. (2008) J. Biol. Chem. 283, 34129–34140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsutsui Y., Morishita T., Natsume T., Yamashita K., Iwasaki H., Yamao F., Shinagawa H. (2005) Curr. Genet 48, 34–43 [DOI] [PubMed] [Google Scholar]
- 25. Garg P., Burgers P. M. (2005) Crit. Rev. Biochem. Mol. Biol. 40, 115–128 [DOI] [PubMed] [Google Scholar]
- 26. Bae K. H., Kim H. S., Bae S. H., Kang H. Y., Brill S., Seo Y. S. (2003) Nucleic Acids Res. 31, 3006–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Niu H., Chung W. H., Zhu Z., Kwon Y., Zhao W., Chi P., Prakash R., Seong C., Liu D., Lu L., Ira G., Sung P. (2010) Nature 467, 108–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stewart J. A., Campbell J. L., Bambara R. A. (2006) J. Biol. Chem. 281, 38565–38572 [DOI] [PubMed] [Google Scholar]
- 29. Stewart J. A., Campbell J. L., Bambara R. A. (2010) Nucleic Acids Res. 38, 920–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Balakrishnan L., Polaczek P., Pokharel S., Campbell J. L., Bambara R. A. (2010) J. Biol. Chem. 285, 38861–38868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kao H. I., Campbell J. L., Bambara R. A. (2004) J. Biol. Chem. 279, 50840–50849 [DOI] [PubMed] [Google Scholar]
- 32. Budd M. E., Choe W., Campbell J. L. (2000) J. Biol. Chem. 275, 16518–16529 [DOI] [PubMed] [Google Scholar]
- 33. Stewart J. A., Miller A. S., Campbell J. L., Bambara R. A. (2008) J. Biol. Chem. 283, 31356–31365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pike J. E., Burgers P. M., Campbell J. L., Bambara R. A. (2009) J. Biol. Chem. 284, 25170–25180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bae S. H., Kim D. W., Kim J., Kim J. H., Kim D. H., Kim H. D., Kang H. Y., Seo Y. S. (2002) J. Biol. Chem. 277, 26632–26641 [DOI] [PubMed] [Google Scholar]
- 36. Kao H. I., Veeraraghavan J., Polaczek P., Campbell J. L., Bambara R. A. (2004) J. Biol. Chem. 279, 15014–15024 [DOI] [PubMed] [Google Scholar]
- 37. Alani E., Thresher R., Griffith J. D., Kolodner R. D. (1992) J. Mol. Biol. 227, 54–71 [DOI] [PubMed] [Google Scholar]
- 38. Cejka P., Cannavo E., Polaczek P., Masuda-Sasa T., Pokharel S., Campbell J. L., Kowalczykowski S. C. (2010) Nature 467, 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sun J. Z., Julin D. A., Hu J. S. (2006) Biochemistry 45, 131–140 [DOI] [PubMed] [Google Scholar]
- 40. Kooistra J., Haijema B. J., Venema G. (1993) Mol. Microbiol. 7, 915–923 [DOI] [PubMed] [Google Scholar]
- 41. Niu H., Raynard S., Sung P. (2009) Genes Dev. 23, 1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yeeles J. T., Cammack R., Dillingham M. S. (2009) J. Biol. Chem. 284, 7746–7755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bae S. H., Choi E., Lee K. H., Park J. S., Lee S. H., Seo Y. S. (1998) J. Biol. Chem. 273, 26880–26890 [DOI] [PubMed] [Google Scholar]
- 44. Paull T. T., Gellert M. (1998) Mol. Cell 1, 969–979 [DOI] [PubMed] [Google Scholar]
- 45. Supek F., Supekova L., Nelson H., Nelson N. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 5105–5110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schneider C., Leung E., Brown J., Tollervey D. (2009) Nucleic Acids Res. 37, 1127–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Herdendorf T. J., Albrecht D. W., Benkovic S. J., Nelson S. W. (2011) J. Biol. Chem. 286, 2382–2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kadyrov F. A., Dzantiev L., Constantin N., Modrich P. (2006) Cell 126, 297–308 [DOI] [PubMed] [Google Scholar]