Abstract
Initiation of eukaryotic genome duplication begins when a six-subunit origin recognition complex (ORC) binds to DNA. However, the mechanism by which this occurs in vivo and the roles played by individual subunits appear to differ significantly among organisms. Previous studies identified a soluble human ORC(2–5) complex in the nucleus, an ORC(1–5) complex bound to chromatin, and an Orc6 protein that binds weakly, if at all, to other ORC subunits. Here we show that stable ORC(1–6) complexes also can be purified from human cell extracts and that Orc6 and Orc1 each contain a single nuclear localization signal that is essential for nuclear localization but not for ORC assembly. The Orc6 nuclear localization signal, which is essential for Orc6 function, is facilitated by phosphorylation at its cyclin-dependent kinase consensus site and by association with Kpna6/1, nuclear transport proteins that did not co-purify with other ORC subunits. These and other results support a model in which Orc6, Orc1, and ORC(2–5) are transported independently to the nucleus where they can either assemble into ORC(1–6) or function individually.
Keywords: Cell Cycle, DNA Enzymes, DNA Replication, Eukaryote, Nuclear Transport, Orc1, Orc2, Orc6, Karyopherin-α, Origin Recognition Complex
Introduction
Genome duplication among the eukarya is a highly conserved process in which either homologues or orthologues of the proteins used by the budding yeast Saccharomyces cerevisiae are found throughout the eukarya and the sequence of events from one species to another is remarkably similar, if not identical (1, 2). Arguably the most critical event in this process is the assembly of prereplication complexes throughout the genome, because it is tightly linked to cell division and highly regulated. Prereplication complex assembly begins when the origin recognition complex (ORC)3 binds to a DNA replication origin where it associates with another initiator protein termed Cdc6 and the helicase loading protein Cdt1 to mount the replicative DNA helicase Mcm(2–7) onto chromosomal DNA. Upon activation by cyclin-dependent kinase CDK2-CcnA and Dbf4-dependent kinase Cdc7, prereplication complexes are converted into preinitiation complexes that begin DNA unwinding and DNA synthesis.
Studies of yeast and flies have led to the generally held view that ORC is a stable complex of six different subunits essential for initiation of DNA replication (1, 3, 4). However, the organization and function of ORC subunits varies markedly among organisms (5). ORC isolated from frog eggs lacks the Orc6 subunit, which is not essential for DNA replication (6). ORC isolated from human cells also lacks the Orc6 subunit (7–10), and only the human ORC(1–5) complex is required to initiate DNA replication in an ORC-depleted frog egg extract (9, 10). Human ORC(2–5) binds to DNA and initiates DNA replication only when associated with Orc1 (9, 10, 26). The most striking difference occurs with the Orc6 subunit. Human ORC assembled from baculovirus-expressed proteins either lacks Orc6 or it is present only in trace amounts (11–14). Nevertheless, interactions between mammalian Orc6 and other ORC subunits have been detected by yeast two-hybrid screens (15), by association of ORC subunits in vitro (10, 11), and by immunoprecipitation of ORC subunits from cell extracts (11). Thus, the interaction of mammalian Orc6 with other ORC subunits appears tenuous.
Although ORC can assemble spontaneously in vitro, the mechanisms by which it assembles and disassembles in vivo appear to play critical roles in cell proliferation and differentiation. For example, Orc1 in yeast, flies, and humans is selectively modified in a cell cycle-dependent manner, thereby providing a mechanism by which ORC activity can be regulated during cell division (16). Mitotic cell cycles and endocycles in plants appear to use different Orc1 subunits (17). ORC appears to disassemble as cells progress from S to M phase (11). Orc6 is required for binding ORC to DNA in flies (19) but not in yeast (20, 21), frogs (6), or humans (Hs) (13, 14). On the other hand, Orc6 is essential for initiation of DNA replication in yeast and flies (19, 22–25) although not in frogs or humans (6, 13, 14). A role for Orc6 in mitosis and cytokinesis also has been reported in flies and humans (19, 26–28), although it is not essential for these events in budding yeast (22, 23). In addition, various ORC subunits appear to play a role in heterochromatin assembly, ribosomal biogenesis, centrosome and kinetochore function, sister chromatin cohesion, and neural dendritic branching (5, 29–31). Collectively, these varied and seemingly contradictory observations suggest that ORC assembly is a dynamic rather than a static process.
To account for these observations, we reasoned that assembly of vertebrate ORCs in vivo may occur through independent pathways to allow assembly of different complexes and to accommodate the various functions currently ascribed to individual subunits. Here we demonstrate that human Orc6 and Orc1 are transported into nuclei independently of the ORC(2–5) core complex. These components can then either be assembled into ORC(1–5), ORC(2–6), and ORC(1–6) complexes or function individually. Such a mechanism would allow the six ORC subunits to elicit the broad spectrum of activities suggested in the literature.
EXPERIMENTAL PROCEDURES
Cells
The human cell lines HeLa (#CCL-2, American Type Culture Collection) and HeLa-RR and their culture conditions have been described (9). These cells were synchronized as they entered S-phase by culturing them in the presence of 4 mm deoxy-Thd for 18 h, then in fresh medium for 12 h, and again with deoxy-Thd for 16 h. Cells were arrested in metaphase by culturing them in the presence of deoxy-Thd for 18 h and then releasing them into 100 ng/ml nocodazole for 8 h. HeLa-RR lines that constitutively expressed either wild-type or mutant Orc6 genes in the absence of continual selection of a genetic marker were constructed as previously described (9).
Recombinant Proteins
A full-length human Orc6 open reading frame was amplified from HeLa cell RNA using reverse transcriptase and PCR to produce a DNA copy that was then cloned into the XhoI/NotI site in pCI (Promega). The cloned human Orc6 sequence was identical to that given under accession number NM_014321 (National Center for Biotechnology Information). Orc6 deletions and amino acid substitutions were generated by PCR amplification. The sequence 198RKRKK202 was converted into AAAVA. The single valine created a PstI site that facilitated cloning. The site-directed mutagenesis kit from Promega was used to clone the −NLS, T195A, and T195E mutants into pCI. All of the primers used in these constructions are available upon request.
Affinity Purification of Proteins
Purification of FH-Orc6 proteins was carried out by a two-step affinity purification protocol, as described previously (9). In brief, FH-Orc6 protein was purified from TK100 extracts of HeLa-RR cells. TK100 was previously termed B100 (20 mm Tris-HCl (pH 7.9), 100 mm KCl, 5 mm MgCl2, 10% glycerol, 0.1% Nonidet P40, 10 μm leupeptin, 1 μm pepstatin, 1 μm PMSF, and 0.5 μg/ml aprotinin). About 500 ng of FLAG-ORC (estimated by Coomassie Blue staining) was incubated with anti-HA resin, and the beads were then washed and eluted with HA peptide (Roche Applied Science) in TK100 buffer. The eluate was fractionated by SDS-PAGE and stained with colloidal Coomassie Blue stain or silver stain (SilverSnap kit, Pierce) to estimate the protein amount, and the amino sequence of the major bands in the stained gel was determined by nano-HPLC/mass spectroscopic analysis (32).
To purify ORC(1–6), HeLa-RR cells were lysed in GN50 (20 mm glycylglycine (pH 9.0), 50 mm NaCl, 5 mm MgCl2, 10% glycerol, 1 mm ATP, 0.1% Nonidet P40, 10 μm leupeptin, 1 μm pepstatin, 1 μm PMSF, and 0.5 μg/ml aprotinin). When FH-Orc6 was used as the affinity tag, ORC(1–6) was purified from the GN50 extract by the two-step affinity purification protocol (9), except that GN buffers were used. When FH-Orc1 was used as the affinity tag, the GN50-extracted HeLa-RR cell pellet was extracted further with GN300 (300 mm NaCl instead of 50 mm NaCl) to maximize recovery of Orc1 protein. To preserve to the greatest degree the existing ORC(1–6) complex, each step was performed at 4 °C: 15-min extraction, 15-min centrifugation at 15,000 × g, 1-h incubation of antibody with antigen, 6 washes each with 20 resin volumes GN50, and a 1-h elution of resin with either FLAG or HA peptide in GN50.
Immunofluorescence
Cytology was done as previously described (9), except that mouse anti-FLAG antibody (Sigma) was diluted 1:1500 in PBS, and rabbit anti-Orc6 antibody was diluted at 1:1200 in PBS. Images were merged using either MetaVue software (Universal Imaging) or Adobe Photoshop.
Western Immunoblotting
Western immunoblotting was done as described previously (9). Proteins were fractionated by SDS-polyacrylamide gel electrophoresis. Proteins were detected with anti-FLAG M2 monoclonal antibody (Sigma), goat anti-karyopherin-α1/6 (1:500, #sc6918, Santa Cruz Biotechnology), goat anti-karyopherin-β (1:500, #sc1863, Santa Cruz Biotechnology), and mouse anti-NPI-1 (1:250, #37-0800, Invitrogen). Mouse anti-NPI-1 is specific for the NPI-1 (nucleoprotein interactor-1, karyopherin-α1) protein. Goat anti-Kpna1/6 detects karyopherin-α1 and karyopherin-α6. Rabbit anti-Orc6 antibody was generated in our laboratory against the entire human Orc6 protein expressed in Escherichia coli. The antigen-antibody complex was detected with HRP-conjugated antibodies (Amersham Biosciences) followed by Super Signal West Dura Extended Duration Substrate kit (Pierce). Goat polyclonal anti-Lamin B (Santa Cruz Biotechnology), anti-Orc1 (Santa Cruz Biotechnology), anti-Orc2 (Santa Cruz Biotechnology), anti-Orc3 (U. S. Biological), and anti-Orc4 (Abcam) were used at a dilution of 1:500 followed by HRP-conjugated rat anti-goat antibody (1:30,000; Pierce).
In Vivo Phosphorylation
Cells were cultured in the presence of radioactive phosphate and then extracted with TK100. FH-Orc6 was affinity-purified from these lysates using anti-FLAG resin, fractionated by SDS-polyacrylamide gel electrophoresis, and the 32P-labeled FH-Orc6 was visualized by autoradiography.
siRNA
Cells were transfected for 5 h with 100 μm siRNA targeted against the 3′-untranslated region of the human Orc6 mRNA (GACUUGACGGCUUUGGGAUtt) or control firefly luciferase GL2 (AACGUACGCGGAAUACUUCGA) using RNAiMAX (Invitrogen) according to the manufacturer's protocol. Cells were transfected again 24 h later, harvested 48 h after the second transfection, and then analyzed by immunoblotting and FACS.
RESULTS
Orc6 Contains a Single Nuclear Localization Signal
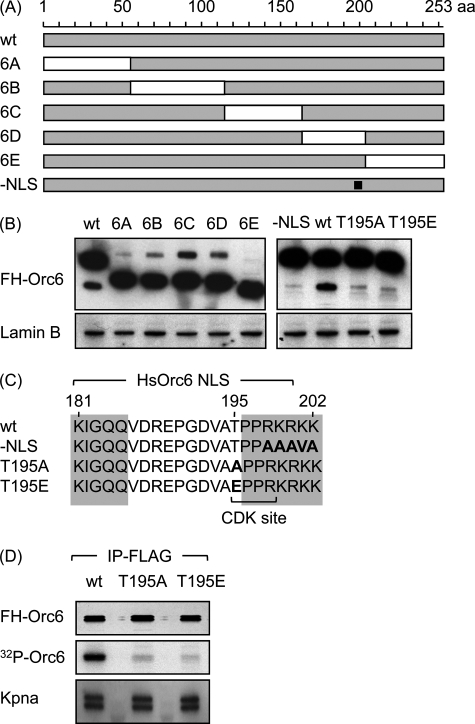
We reasoned that if the human Orc6 protein were transported into the nucleus independently of other ORC proteins, it would require its own nuclear localization signal (NLS). Therefore, to identify a functional NLS in Orc6, deletions were constructed throughout the entire length of the human Orc6 molecule (Fig. 1A). Orc6 proteins from different species were aligned to identify the non-conserved regions where end points for each deletion were less likely to interfere with functional domains. Each protein was tagged at its N terminus with the FLAG-hemagglutinin epitopes (FH) to facilitate its purification, and HeLa cell lines that constitutively expressed one of these proteins were constructed. Subsequent analysis revealed that each recombinant FH-Orc6 mutant was expressed at levels equivalent to that of FH-Orc6 wild-type protein (Fig. 1B). However, whereas deletions 6A, 6B, 6C, and 6E and wild-type Orc6 each localized to the nucleus of interphase cells, deletion 6D (residues 163–204) localized to the cytoplasm regardless of whether it was expressed transiently or constitutively or if it was expressed in monolayers or in suspension cultures of HeLa cells (Fig. 2; data not shown). Thus, only deletion 6D affected nuclear localization.
FIGURE 1.
Orc6 contains a putative monopartite NLS and a single CDK consensus phosphorylation site. A, five deletions were constructed that spanned the entire length of the Orc6 coding sequence (6A (Δ1–55), 6B (Δ55–115), 6C (Δ115–163), 6D (Δ163–204), 6E (Δ204–253)). One mutation was constructed in the putative NLS (−NLS; amino acids 198–202 (squlf]). B, Western immunoblotting using anti-FLAG antibody revealed that constitutive expression of each mutant protein in HeLa-RR cells was comparable with that of wild-type FH-ORC protein. Lamin B protein was used as a loading control. C, alignment of Orc6 amino acid sequences from frog, mouse, rat, cattle, dog, chimpanzee, and human revealed a conserved sequence (shaded) characteristic of a monopartite NLS (PRKRKK) is shown. The −NLS mutant contained AAAVA in place of RKRKK. The T195A Orc6 mutant contained alanine (A) in place of threonine (T) at position 195 to prevent phosphorylation at the only CDK consensus sequence in human Orc6. The T195E mutant contained glutamic acid (E) in place of T to mimic phosphorylation. D, cell proteins were labeled with radioactive phosphate by culturing cells that constitutively expressed the indicated protein for 4 h in phosphate-free DMEM containing [32P]orthophosphate (200 μCi/ml). FH-Orc6 was then immunoprecipitated (IP) with anti-FLAG resin, and the precipitate was fractionated by SDS-PAGE. [32P]Orc6 was visualized by autoradiography, whereas Kpna and FH-Orc6 were visualized by Western immunoblotting.
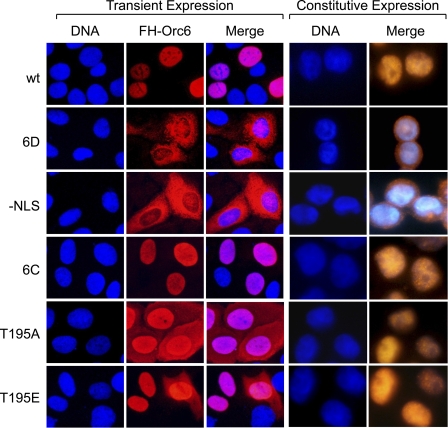
FIGURE 2.
Orc6 contains a single nuclear localization signal. Wild-type FH-Orc6 (wt) and FH-Orc6 deletion mutants 6A, 6B, 6C, and 6E were localized to the nucleus (wt), as was endogenous Orc6 (not shown). Only deletion mutant 6D and the NLS substitution mutant (−NLS) were localized to the cytoplasm. Because the images of Orc6 mutant proteins that localized to the nucleus were indistinguishable from wild-type Orc6, only 6C is shown as an example. Cells were stained with Hoechst to visualize nuclear DNA and with anti-FLAG antibody to visualize FH-Orc6. Transiently expressed Orc6(T195A) was localized both in the nuclear and cytoplasmic compartments. 90% of transiently expressed Orc6(T195E) cells were localized to the nucleus, and 10% was in the cytoplasm. However, constitutively expressed Orc6(T195A) and Orc6(T195E) were localized to the nucleus, as discussed in the text. Transient expression assays were carried out on HeLa cell monolayers. Constitutive expression of Orc6 and other proteins was engineered in HeLa-RR cells that proliferate in suspension.
Comparison of the sequences of Orc6 proteins from seven different vertebrates revealed the presence of an NLS consensus sequence within deletion 6D consisting of KIGLQX11PPRKRKK (Fig. 1C). To determine whether or not this sequence was, in fact, the Orc6 NLS, the basic amino acid sequence RKRKK was replaced with the neutral amino acid sequence AAAVA to generate a NLS-defective mutant (Fig. 1C, FH-Orc6(-NLS)). Expression of FH-Orc6(-NLS) was equivalent to FHOrc6(wt) (Fig. 1B), and Orc6(-NLS), like Orc6(6D), localized to the cytoplasm in both transient and constitutive expression assays (Fig. 2). Therefore, HsOrc6 contains a single NLS within residues 180–202.
Orc6 Phosphorylation Facilitates Nuclear Localization
Orc6 contains a single CDK-dependent consensus phosphorylation site ((S/T)PX(K/R)) at Thr-195 (Fig. 1C), suggesting that phosphorylation at this site may affect nuclear localization. Therefore, two mutants were constructed in which Thr-195 was changed either to alanine (T195A) to prevent phosphorylation at this site or to glutamate (T195E) to mimic phosphorylation at this site. Both mutants carried the FH epitopes at their N terminus, and both were expressed constitutively to levels comparable with wild-type FH-Orc6 (Fig. 1B). When Orc6(T195A) was expressed transiently in HeLa cells, the protein was distributed throughout both nucleus and cytoplasm (Fig. 2), whereas transiently expressed Orc6(T195E) was localized to the nucleus in 90% of the cells (Fig. 2) and distributed throughout both nucleus and cytoplasm in 10% of the cells (data not shown).
To determine whether or not Orc6 is phosphorylated at Thr-195 in vivo, cells expressing either FH-Orc6, FH-Orc6(T195A), or FH-Orc6(T195E) were cultured in the presence of radioactive phosphate, and then FH-Orc6 was affinity-purified from cell lysates using anti-FLAG resin, fractionated by SDS-polyacrylamide gel electrophoresis, and the 32P-labeled FH-Orc6 was visualized by autoradiography. Only wild-type Orc6 was radiolabeled significantly, revealing that Orc6 was indeed phosphorylated in vivo primarily, if not exclusively, at Thr-195 (Fig. 1D).
These results suggest that CDK-dependent phosphorylation of HsOrc6 facilitates its localization to the nucleus. However, when Orc6(T195A) and Orc6(T195E) were expressed constitutively in HeLa cells, both proteins localized to the nucleus (Fig. 2). Therefore, as previously shown for the SV40 large tumor antigen (33), phosphorylation at the NLS can expedite nuclear localization, but it is not essential for nuclear localization. The fact that cellular levels of Orc6(T195A) were severalfold greater during transient expression than during constitutive expression would amplify the facilitating effect of phosphorylation.
Orc6 Is Bound to Karyopherin-α
Proteins that localize to the nucleus generally, if not always, associate with one of the karyopherin-α (Kpna) or karyopherin-β nuclear transport proteins (34, 35). To determine whether or not HsOrc6 in its native environment is bound to a nuclear transport protein, FH-Orc6 was purified by double affinity chromatography from HeLa cells that expressed it constitutively. Previous applications of this strategy demonstrated that constitutively expressed FH-Orc1 and FH-Orc2 proteins modeled their endogenous counterparts (8, 9). They are assembled into ORCs in the nuclei of G1-phase cells where they preferentially bind to DNA replication origins and stimulate DNA replication. Moreover, FH-Orc1, like endogenous Orc1, is selectively degraded during S-phase.
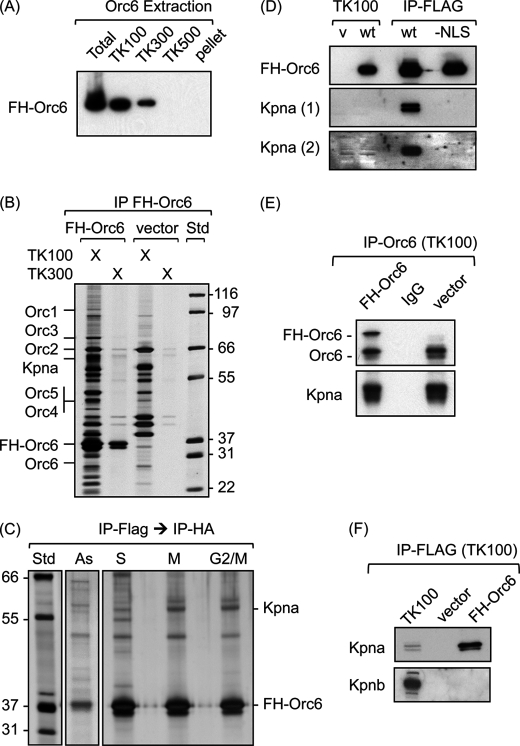
Although all of the FH-Orc6 was localized within the nucleus of all interphase cells stained with anti-FLAG or anti-HA antibodies (see the examples in Fig. 2), most of the FH-Orc6 was solubilized when cells were lysed with TK100, a Tris-buffered nonionic detergent containing 100 mm KCl (Fig. 3A). The remaining FH-Orc6 could be extracted with TK300, the same concoction but with 300 mm KCl instead of 100 mm KCl. The soluble FH-Orc6 was bound to anti-FLAG resin and then recovered by eluting with FLAG peptide (Fig. 3B). HeLa cells that harbored the expression vector alone served as a negative control. To further eliminate proteins that were not associated specifically with FH-Orc6, the FLAG peptide-eluted proteins were purified a second time by binding them to anti-HA resin and then eluting with HA peptide. The resulting Orc6-associated proteins detected in asynchronous populations of cells could be enriched by synchronizing cells either in S-phase with excess thymidine or in metaphase with nocodazole or in G2/M by releasing cells from the thymidine block for 9 h (Fig. 3C). The major bands in the HA eluate from FH-Orc6-expressing cells that were absent from the eluate from control cells were then excised and analyzed by mass spectrometry. Nine peptides were identified from Kpna6 and -5 peptides from Kpna1, two proteins with extensive sequence homology (36). This protein was clearly enriched in all three populations of synchronized cells. None of the Orc6-associated proteins was known to be involved in DNA replication.
FIGURE 3.
Orc6-Kpna complexes were affinity purified from HeLa cells. A, HeLa-RR cells constitutively expressing the FH-Orc6 gene were extracted sequentially with buffer TK containing the indicated concentration of KCl. The TK100 pellet was extracted with TK300, and the TK300 pellet was extracted with TK500. The proteins in each soluble fraction (TK100, TK300, TK500) and the pellet from B500 were then subjected to Western immunoblotting with anti-FLAG antibody. B, FH-Orc6 was purified from TK100 and TK300 fractions using anti-FLAG resin, fractionated by SDS-PAGE, and stained with silver. Positions of Orc1 (100 kDa), Orc3 (82 kDa), Orc2 (66 kDa), Orc4 (50.4 kDa), Orc5 (50.3 kDa), FH-Orc6 (31 kDa), Orc6 (28 kDa), and Kpna (60 kDa) and the Mark 12 protein standards (Std) are indicated. Proteins from HeLa-RR cells transformed by the parent expression vector were analyzed in parallel (vector). IP, immunoprecipitates. C, TK100 extracts were subjected to double affinity purification, first using anti-FLAG resin, then using anti-HA resin. The cells were either asynchronous (As), arrested in S-phase (S), arrested in metaphase (M), or G2/M-phase cells isolated by centrifugal centrifugation (G2 /M). D, FH-Orc6(wt) and FH-Orc6(−NLS) protein was immunoprecipitated with anti-FLAG resin from TK100 extracts of cells that expressed these proteins constitutively. The total immunoprecipitate was fractionated by SDS-PAGE in parallel with 0.3% of the total cell extract from cells transformed by the empty vector (v) and cells expressing FH-Orc6 (wt) and immunoblotted with anti-Kpna antibody. Kpna (2) is a longer exposure of Kpna (1). E, native Orc6 was immunoprecipitated with a rabbit anti-Orc6 polyclonal antibody from TK100 extracts of FH-Orc6-expressing cells and from cells harboring the expression vector alone. The center lane is an immunoprecipitate of vector cells with rabbit IgG. The immunoprecipitates were fractionated by SDS-PAGE and immunoblotted using anti-Orc6 and anti-Kpna antibody. F, FH-Orc6 was immunoprecipitated from a TK100 extract using anti-FLAG resin, fractionated by SDS-PAGE, and immunoblotted with either anti-Kpna or anti-Kpnb antibody.
The existence of an Orc6-Kpna complex was confirmed by affinity purification. FH-Orc6 in a TK100 cell extract was eluted from anti-FLAG resin with FLAG peptide, and the eluate was subjected to Western immunoblotting using two different antibodies that recognize both Kpna1 and Kpna6. Both antibodies detected Kpna associated with Orc6 (Fig. 3D; data not shown). Moreover, Kpna in the purified FH-Orc6 fraction was enriched considerably relative to Kpna in the TK100 extract (Fig. 3D). In contrast to FH-Orc6(wt), Kpna was absent from affinity-purified FH-Orc6(-NLS) (Fig. 3D). This result reflected the fact that Kpna binds to stretches of basic amino acids within the NLS (37), the same residues that were changed to neutral amino acids in the −NLS mutant.
To determine whether or not Kpna associated with native Orc6 as well as FH-Orc6, a rabbit polyclonal antiserum was prepared against the intact Orc6 protein. This antiserum did not recognize any of the other ORC subunits, and it did not recognize Kpna (data not shown). Immunoprecipitation of Orc6 from TK100 lysates using anti-Orc6 antibodies confirmed that Kpna interacted specifically with Orc6 (Fig. 3E); it did not depend either on the presence of FLAG or HA peptides or on overexpression of a recombinant Orc6 protein in HeLa cells. In contrast to Kpna, Kpnb was easily detected in TK100 lysates, but it did not co-purify with FH-Orc6 (Fig. 3F). This was consistent with the fact that Kpnb dissociates rapidly from protein-Kpna complexes immediately after they enter the nucleus (34).
To determine whether or not the Orc6 CDK phosphorylation site was required for Orc6 binding to Kpna, FH-Orc6 wild-type, T195A, and T195E proteins were immunoprecipitated from TK100 lysates of cells that constitutively expressed these proteins. Kpna was associated with Orc6(T195A) and Orc6(T195E) as well as with wild-type Orc6 protein (Fig. 1D), revealing that that Orc6 phosphorylation was not essential for binding Kpna.
Kpna Is Bound to Free Orc6
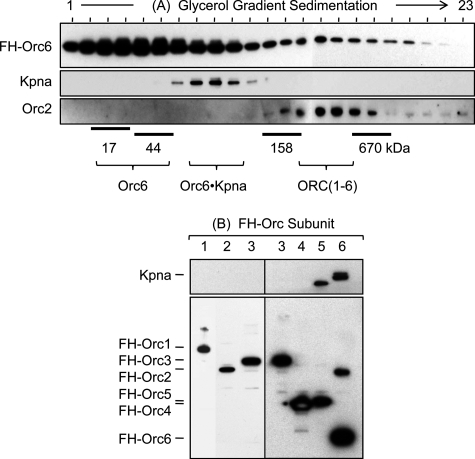
Orc6 is localized to the nucleus where it is presumed to function as a component of ORC. To determine what fraction of the FH-Orc6 was associated with Kpna and what fraction was associated with ORC, FH-Orc6 was immunoprecipitated from a TK100 cell extract using anti-FLAG antibody and then fractionated by glycerol gradient centrifugation (Fig. 4A). By far the largest fraction of FH-Orc6 migrated as a monomer (31 kDa). A smaller fraction migrated as a binary complex with Kpna (91 kDa). The smallest fraction of FH-Orc6 migrated as a complex with other ORC subunits (∼400 kDa). This result is consistent with the formation of an Orc6-Kpna complex in the cytoplasm that is then transported into the nucleus where Kpna is released and Orc6 associates with other ORC subunits.
FIGURE 4.
Kpna was associated only with Orc6 that was not associated with other ORC subunits. A, an anti-FLAG immunoprecipitate was prepared from a TK100 extract of 20 × 108 cells that constitutively expressed FH-Orc6. The immunoprecipitate was then fractionated by glycerol gradient centrifugation (4 ml 10–25% glycerol gradient for 5 h at 55,000 rpm and 10 °C, Beckman Ti-60 rotor), and aliquots from each fraction were subjected to Western immunoblotting using the indicated antibody. B, Orc6 is the only ORC subunit bound to karyopherin-α. TK100 extracts were prepared from HeLa-RR cell lines that constitutively expressed one of the six FH-tagged ORC subunits (1, 2, 3, 4, 5, or 6). Anti-FLAG immunoprecipitates were fractionated by SDS-PAGE and immunoblotted using anti-Kpna (upper panel) and anti-FLAG (lower panel) antibodies. Experiments with the six FH-tagged ORC subunits were done at different times, and therefore, separate gels are presented together.
Only the Orc6 Subunit Is Associated with Kpna
To determine whether or not Kpna facilitates nuclear localization of other ORC subunits, FH-tagged ORC subunits 2, 3, 4, 5, and 6 were was isolated from TK100 lysates of cells that constitutively expressed the protein and then and affinity-eluted from anti-FLAG resin. Lysates were prepared from the same number of cells. FH-Orc1 was purified by the same procedure, but from TK300 lysates. The purified proteins were then fractionated by SDS gel electrophoresis and immunoblotted using either anti-Kpna or anti-FLAG antibodies. Kpna was detected only with Orc6 (Fig. 4B), even when larger amounts of FLAG eluent were analyzed. The anti-Kpna antibody also detected an unidentified 50-kDa protein associated only with FH-Orc5. The goat anti-Kpna1/6 used to detect Kpna was made against a peptide from the C terminus of human Kpna1, a region that is almost identical to the corresponding regions in human Kpna6 and Kpna5, suggesting that the antibody recognizes several Kpna paralogs. Thus, analysis of equivalent amounts of cell lysate detected Kpna associated only with Orc6, suggesting that Kpna selectively transports the Orc6 subunit into the nucleus.
Association between Orc6 and Other ORC Subunits Is Tenuous
Previous studies found that human and frog Orc6 binds weakly if at all to other ORC subunits (see the Introduction). For example, FH-Orc2 purified by double affinity chromatography from TK100 extracts of HeLa cells that express this protein constitutively co-purifies with Orc3, Orc4, and Orc5 but not with Orc6 (9). Orc1 was present only when these cells were extracted with TK300, suggesting that Orc1 is required to bind a stable ORC(2–5) core complex tightly to the chromatin fraction. Therefore, either Orc6 did not form a tight complex with other ORC subunits under these extraction and purification conditions or the amount of Orc6 associated with other ORC subunits was too low to be detected by the colloidal Coomassie stain used to identify protein bands for mass spectrometry analysis.
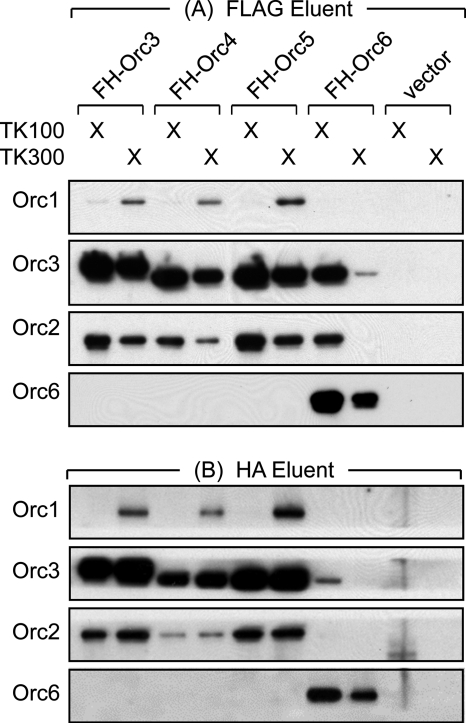
To determine whether or not Orc6 was associated with any of the other ORC subunits, HeLa cells constitutively expressing FH-Orc3, FH-Orc4, FH-Orc5, or FH-Orc6 were lysed in TK100, and the epitope-tagged protein was purified by affinity chromatography on anti-FLAG resin (Fig. 5A). As previously found by affinity purification of FH-Orc2, ORC(2–5) was purified from TK100 lysates, and ORC(1–5) was purified from TK300 lysates. Orc6 did not co-purify with FH-Orc3, FH-Orc4, or FH-Orc5. However, both Orc2 and Orc3 did co-purify with FH-Orc6, revealing that a small fraction of the Orc6 localized within the nuclei was part of a complex consisting of ORC(2–6). Purification of FH-Orc6 from a TK300 lysate was not associated with other ORC subunits.
FIGURE 5.
Orc6 was associated weakly with other ORC subunits in TK100 cell extracts. The indicated FH-ORC subunit was purified in two steps from TK100 and TK300 extracts of 20 × 107 cells constitutively expressing either FH-ORC3, -4, -5, or -6. (A) FH-ORC subunits were first purified by elution from anti-FLAG resin using FLAG peptide. (B) The FLAG eluent was then applied to anti-HA resin and HA-tagged proteins eluted off with HA-peptide. HeLa-RR cells transformed with the empty expression vector served as a control (vector). Each immunoprecipitate (IP) was subjected to Western immunoblotting using the indicated antibody.
To determine whether or not the ORC complexes in the FLAG eluents were stable, they were purified further by affinity chromatography on anti-HA resin (Fig. 5B). As before, complexes consisting of ORC(2–5) and ORC(1–5) were recovered easily and, therefore, were considered stable under these conditions. However, consistent with the results in Fig. 3, B and C, ORC(2–6) was not recovered.
ORC(1–6) Can Be Purified from Cell Lysates
Based on the results described above, we reasoned that the absence of Orc6 from ORC complexes might have been due simply to the absence of optimal conditions for protein-protein interactions among ORC subunits. Indeed, small variations in the relative amounts of salt and detergents in the lysis buffer as well as the speed and efficiency of cell breakage are particularly important in the isolation of proteins that associate with macromolecular structures (38). Therefore, conditions for purification of ORC(1–6) from cell lysates were reinvestigated. These included extensive titrations using MES, PIPES, HEPES, Tris, glycylglycine, or CAPSO buffering agents to cover the range from pH 5 to 10. Both potassium and sodium salts were tested at various concentrations with chloride, acetate, or glutamate as the anion. In addition, consideration was given to stabilizing agents such as glycerol, sucrose, and polyethylene glycol, detergent concentration, and the effect of ATP. The results revealed that an extraction buffer containing glycylglycine (pH 9.0), NaCl, and 10% glycerol allowed purification of ORC(1–6) complexes from human cell extracts.
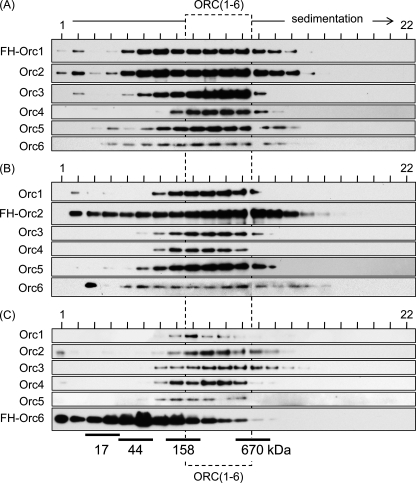
Extracts of HeLa cells that expressed constitutively FH-Orc1, FH-Orc2, or FH-Orc6 were subjected first to affinity chromatography against the FLAG epitope, then to affinity chromatography against the HA epitope, and finally to fractionation by glycerol gradient sedimentation. In each case, an ORC(1–6) complex sedimented between 158 and 670 kDa (Fig. 6). The fact that the apparent molecular weight of ORC complexes containing all six subunits was the same regardless of which of the three ORC subunits were purified suggested the presence of a single protein complex containing all six ORC subunits. Moreover, FH-Orc2 and FH-Orc6 complexes were purified using buffers containing 50 mm NaCl, whereas the FH-Orc1 complex was purified using buffers containing 300 mm NaCl to ensure complete release of FH-Orc1.
FIGURE 6.
Both GN50 and GN300 extracts of HeLa cells contained stable complexes of ORC(1–6). A, FH-Orc1 was purified from a GN300 extract of 20 × 107 cells by two-step affinity chromatography, and 100 μl was layered over a 3.7-ml 10–25% linear glycerol gradient in GN300. The gradient was centrifuged in a Beckman SW60 rotor for 4.5 h at 55,000 rpm (4 °C). B, FH-Orc2 was purified from a GN50 extract of 20 × 107 cells and subjected to glycerol gradient centrifugation in GN50. C, FH-Orc6 was purified as in B.
The apparent molecular weight of the ORC(1–6) detected under these conditions was consistent with the presence of one copy of each subunit. Such a complex would have a molecular weight of 380 kDa. The absence of higher molecular weight aggregates of Orc proteins suggests that the purified complexes were not simply random aggregates but rather a soluble, specific, stable, multimeric complex that survived the 12 h required for two affinity purifications and one glycerol gradient sedimentation.
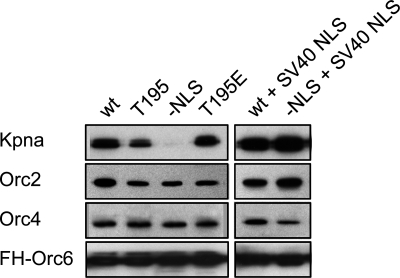
Using conditions developed for stabilizing ORC(1–6), the importance of the Orc6 NLS and the Orc6 CDK consensus phosphorylation site on protein-protein interactions was reexamined. Neither the presence nor absence of phosphorylation at Thr-195 prevented Orc6 from co-purifying with Kpna and other ORC subunits (Fig. 7). Moreover, the Orc6 NLS remained essential for binding Kpna. This conclusion was confirmed by fusing the SV40 large tumor antigen NLS onto FH-Orc6(−NLS), which restored Orc6(−NLS) nuclear localization and binding to Kpna. The SV40 large tumor antigen NLS, like that of Orc6, is mediated by association with Kpna (39).
FIGURE 7.
The Orc6 NLS is required for binding Kpna but not for binding other ORC subunits. The SV40 large tumor antigen NLS (126PKKKRKV132) was fused to the N terminus of both the wt and the −NLS FH-Orc6 proteins. FH-Orc6 was then immunoprecipitated using anti-FLAG resin from extracts of HeLa cells that constitutively expressed the indicated protein. Each immunoprecipitate was fractionated on SDS-PAGE and subjected to Western immunoblotting using anti-Kpna, anti-Orc2, anti-Orc4, and anti-FLAG antibodies.
The Orc6 NLS Is Essential for Orc6 Function
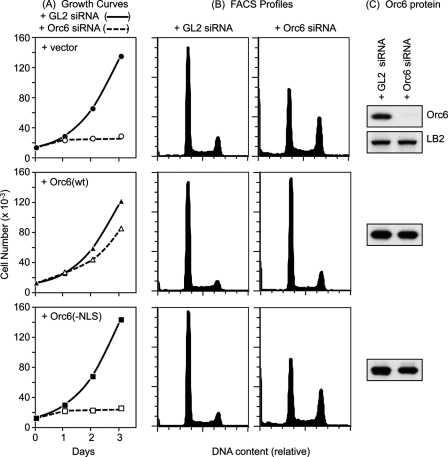
The results described above show that the Orc6 NLS is required to bind Kpna and localize Orc6 to the nucleus but not for association of Orc6 with other ORC subunits. Therefore, to determine whether or not nuclear localization of Orc6 is required for Orc6 function, endogenous Orc6 expression in HeLa cells was suppressed with siRNA targeted against its 3′-nontranslated region so that the effects of constitutively expressed recombinant FH-Orc6 could be evaluated. Under conditions where endogenous Orc6 protein was not detected, cell proliferation ceased with most cells arrested either with a 2N (G1-phase) or 4N (G2- and M-phase) DNA content (Fig. 8, + vector row). When the same experiment was carried out in cells that constitutively expressed wild-type recombinant FH-Orc6 protein, cell proliferation was restored (Fig. 8, + Orc6(wt) row). Thus, recombinant Orc6 can replace of endogenous Orc6, thereby demonstrating that the effects of siOrc6–3′ were specific for the Orc6 gene. In contrast, constitutive expression of recombinant Orc6 lacking a functional NLS did not compensate for loss of endogenous Orc6 (Fig. 8, + Orc6(−NLS) row). Therefore, consistent with previous studies showing that siRNA suppression of human Orc6 expression retards both DNA replication and cytokinesis (28), nuclear localization of Orc6 is essential for cell proliferation.
FIGURE 8.
Nuclear localization of Orc6 is essential for cell proliferation. A, U2OS cell lines that constitutively expressed the indicated protein were transfected twice with siRNA targeted against either the 3′-untranslated region of the endogenous Orc6 gene or against the coding region of the firefly luciferase GL2 gene. The cell number per well was counted each day, and the results were plotted. B, FACS profiles of the samples in panel A. C, at 72 h post-transfection, total cell lysates from the same number of cells were prepared and subjected to Western immunoblotting using anti-Orc6 antibody to reveal the total amount of Orc6 protein (endogenous + recombinant). The lamin B2 (LB2) loading control is shown for the cells that did not express recombinant FH-Orc6.
Orc1 Localizes to the Nucleus Independently of the Other ORC Subunits
Given that Orc1 as well as Orc6 interact weakly with the ORC(2–5) core complex, we considered the possibility that Orc1 also is localized independently to the nucleus. The same strategy used to identify the human Orc2 (9) and Orc6 (Fig. 1) NLS was applied to Orc1. A series of deletions was constructed throughout the Orc1 protein, tagged at their N terminus with FLAG-HA epitopes, and expressed constitutively in HeLa cells (Fig. 9A). The results restricted the NLS to residues 240–288, a region in which previous studies had identified a NLS at residues 259–266 (PGRIKRKV) in transient expression assays (40). Therefore, this sequence was changed from RIKRK to GLEGV, and the resulting recombinant FH-Orc1(−NLS) protein was expressed constitutively in HeLa cells. The result confirmed that PGRIKRKV was required to localize Orc1 to the nucleus; FH-Orc1(−NLS) was distributed throughout both the nucleus and cytoplasm, whereas all other Orc1 variants were localized to the nucleus (Fig. 9C; data not shown). Furthermore, FH-Orc1(wt) and endogenous Orc1 were recovered almost exclusively in the TK300 extract, but about 15% of FH-Orc1(−NLS) was recovered in the TK100 extract.
FIGURE 9.
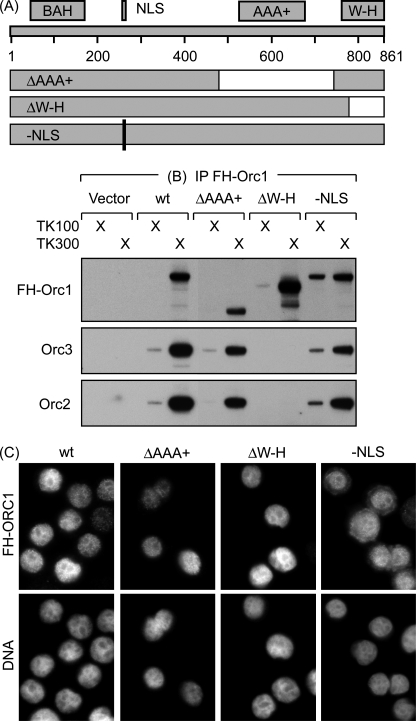
The NLS and ORC binding domain in Orc1 are physically and functionally distinct. A, a series of five deletions were constructed that spanned the human Orc1 coding sequence. Two of them are diagrammed along with the bromo-adjacent homology (BAH) required for Orc1 association with chromatin (8), the NLS (40), the AAA+ motif required for ORC(1–5) binding to DNA (11, 13), and the winged-helix motif. The ΔAAA+ deletion mutant was missing residues 482–744, and the ΔW-H deletion mutant was missing residues 783–861. The −NLS mutation replaced 261RIKRK265 to 261GLEGV265. B, FH-Orc1 was immunoprecipitated (IP) from each of the indicated cell lines with anti-FLAG antibodies; the precipitate was fractionated by SDS-PAGE and then subjected to Western immunoblotting using anti-FLAG, anti-Orc3 and anti-Orc2 antibodies. C, HeLa-RR cells constitutively expressing the indicated FH-Orc1 protein were stained with anti-FLAG antibodies to detect FH-Orc1 and Hoechst to stain DNA.
Transient overexpression of Orc1 deletion mutants in human cells has also revealed that the C terminus of Orc1 containing the AAA+ domain (residues 501–861) is necessary and sufficient for assembly of ORC(1–5) (11). Because this region does not include the NLS, Orc1 may be capable of nuclear localization independently of the other ORC subunits. To test this hypothesis, HeLa cells that constitutively expressed various FH-Orc1 mutants were tested for their ability to localize Orc1 to the nucleus and for their ability to form ORC complexes. Deletion of the C-terminal winged-helix motif (amino acids 783–861) prevented association of Orc1 with either Orc2 or Orc3 (Fig. 9B), but it did not prevent Orc1 localization to the nucleus (Fig. 8C). Therefore, Orc1 can accumulate in the nucleus without associating with other ORC subunits. Moreover, none of the remaining regions of Orc1 were required for ORC assembly, including residues 482–744 that contain the AAA+ motif. Thus, we conclude that the winged-helix motif is the only region of Orc1 required for its association with other ORC subunits. Taken together with the data of Siddiqui and Stillman (11), the winged-helix domain is necessary, but not sufficient, to bind ORC(2–5); it must be extended by additional amino acid residues that are provided either by the N-terminal region or the AAA+ region of the Orc1 protein.
DISCUSSION
The results presented here demonstrate three novel characteristics of the human ORC. First, human Orc6 localizes to the nucleus independently of other ORC subunits through a karyopherin-α, phosphorylation-modulated NLS that is essential for ORC function but not for ORC assembly. Similarly, the Orc1 NLS and ORC assembly domains also function independently, thereby allowing Orc1 to localize to the nucleus independently of the other ORC subunits. Finally, stable ORC assemblies that include Orc6 can be purified from human cells. The heretofore elusive nature of ORC(1–6) assemblies results both from weak interactions between Orc6 and the other ORC subunits and a low ratio of endogenous Orc6 to ORC(2–5). Moreover, Kpna may interfere with association of Orc6 and other ORC subunits outside of the nucleus.
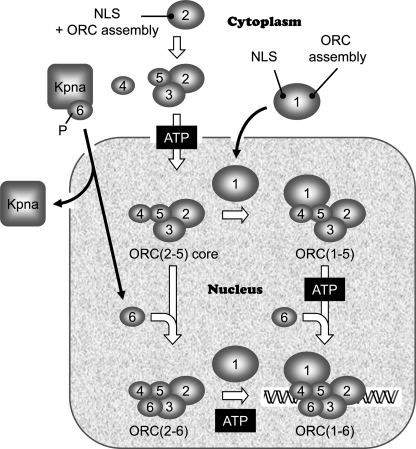
These results, together with those reported previously from this and other laboratories support a dynamic model for ORC assembly in human cells in which Orc6, Orc1, and ORC(2–5) localize independently to the nucleus where either they can assemble into various combinations of ORC subunits or they can function independently of one another (Fig. 10). Such a model allows ORC subunits to serve in a variety of roles during cell proliferation and differentiation.
FIGURE 10.
Dynamic model for ORC assembly in human cells. A stable ORC(2–5) core complex forms in the presence of ATP and is transported into the nucleus by a mechanism that is dependent on the Orc2 NLS and ORC assembly domain and perhaps the Orc3 NLS as well. Orc1 enters the nucleus by a mechanism that is dependent on the Orc1 NLS but not on the Orc1 ORC assembly domain. Orc1 then binds to ORC(2–5) to form a complex that in the presence of ATP can bind to DNA in vitro and to the chromatin fraction in vivo. Karyopherin-α6/1 (Kpna) binds to Orc6 in the cytoplasm and then transports it into the nucleus where it is assembled into ORC(2–6) and Orc(1–5). Orc6 phosphorylation facilitates Orc6 nuclear localization.
Nuclear Localization of Orc6
Human Orc6 is localized to the nucleus through association of its single NLS with Kpna6 and/or Kpna1, two of seven Kpna paralogs whose function is to transport other proteins to the nuclear pore complex where Kpna is released by Kpnab and the cargo protein, Orc6 in this case, is transported into the nucleus. Kpna6/1 was not associated with any of the other ORC subunits, suggesting that Orc6 is transported into the nucleus independently of the other ORC subunits. The fact that inactivation of the Orc6 NLS prevents both nuclear localization of Orc6 and cell proliferation demonstrates that nuclear localization of Orc6 is essential to Orc6 function. In the absence of Orc6 function, cells accumulated both in G1 and in G2 or M phase, consistent with previous reports that siRNA suppression of either human or Drosophila Orc6 expression retards both DNA replication and cytokinesis (27, 28).
The fact that the Orc6 NLS is essential for nuclear localization and for Orc6 function in vivo, but not for association of Orc6 with other ORC subunits in cell extracts, reveals that assembly of ORC(1–6) must occur within the nucleus. If Orc6 was transported into the nucleus by associating with other ORC subunits that had an NLS, then the Orc6 NLS would be dispensable. Consistent with this conclusion is the fact that Kpna dissociates from its cargo only after transporting it into the nucleus (34, 35) and the observation that Kpna co-sedimented with affinity-purified FH-Orc6 during glycerol gradient sedimentation but not with ORC(2–6). Nuclear localization was facilitated by phosphorylation of Orc6 at its single CDK consensus phosphorylation site in transient expression assays, but a comparable effect was not observed in cells that expressed Orc6 constitutively. The fact that this phosphorylation site is adjacent to the NLS suggests that it may regulate Orc6 activity by regulating Orc6 nuclear localization, a hypothesis that was supported by transient expression of Orc6 mutants. Orc6 phosphorylation was not required for Orc6 association with other ORC subunits.
The bonds between Orc6 and ORC(2–5) were significantly weaker than the bonds between ORC subunits 2, 3, 4, and 5. ORC(2–6) was stable enough to survive immunoprecipitation and then glycerol gradient sedimentation, but very little of it survived a second round of affinity purification. In addition, the level of Orc6 relative to other ORC subunits limited ORC(2–6) assembly. The cellular level of endogenous Orc2 was 5 times that of endogenous Orc6, thereby limiting the ratio of ORC(2–5) to ORC(2–6) to about 4:1. Because cells that constitutively expressed recombinant Orc6 contained about 30 times the level of endogenous Orc6, the amount of ORC(2–6) was proportionally increased to a detectable level.
Nuclear Localization of Orc1
Previous studies have shown that the ORC1 subunit is required for ORC to initiate DNA binding, pre-replication complex assembly, and DNA replication in the cells of yeast, insects, frogs, and humans (1, 13, 14). Although human ORC(2–5) accumulates in the nucleus, only ORC(1–5) is associated with the chromatin fraction (Ref. 9 and Fig. 3), consistent with earlier studies showing that Orc1 is required both for binding ORC(2–5) to DNA in vitro and for initiation of ORC-dependent DNA replication in Xenopus egg extract (13, 14). In vitro, Orc1 first associates with ORC(2–5), and then ORC(1–5) binds to DNA in a reaction that requires binding of ATP to Orc1 (10, 11, 13).
The results presented here show that, in vivo, Orc1 localizes to the nucleus independently of the other ORC subunits. However, in contrast to Orc6, Kpna was not detected in association with Orc1 by either of the two Kpna antibodies used in this study. These antibodies were prepared against either Kpna1 or Kpna6. Other ORC subunits may bind a different Kpna paralog (at least seven exist in human cells). Kpna serves as an adaptor that links cargos and importin-β. Six different NLS classes have been identified that bind specifically to distinct domains of Kpna proteins (37). Alternatively, the NLSs identified in Orc1, Orc2, and Orc3 may associate directly with one of the 19 human karyopherin-β/importin-β molecules that bind cargo to the nuclear pore complex (34).
Orc1 contains a single NLS that is required for nuclear localization. Inactivation of this NLS resulted in both nuclear and cytoplasmic distribution of Orc1 (Fig. 7), but it did not affect assembly of ORC(1–5) (Fig. 7). Therefore, this putative NLS may actually be a nuclear retention signal, and exclusive Orc1 nuclear localization may result from association with other ORC subunits. However, this was not the case. Orc1 contains a single ORC binding domain that was required for assembly of ORC(1–5) but not for nuclear localization of Orc1. Therefore, Orc1 is translocated to the nucleus where it subsequently associates with other ORC subunits. In early G1, Orc1 is distributed throughout the cell nucleus, but as cells progress through late G1 and S-phase, Orc1 preferentially associates with heterochromatin, a phenomenon associated with amino acids 151–269 (40).
Nuclear Localization of ORC(2–5)
The demonstration that either Orc1 or Orc2 protein tethered to a plasmid DNA can trigger initiation of plasmid DNA replication, whereas either Orc5 or Orc6 protein could not (41), suggests that Orc1 and Orc2 can initiate assembly of prereplication complexes, whereas Orc5 and Orc6 cannot. Mammalian ORC(2–5) can assemble spontaneously in vitro from individual ORC subunits under a variety of conditions (7–14). Assembly begins with formation of an ORC(2, 3, 5) complex to which Orc4 binds in a manner that requires binding of ATP to both the Orc4 and Orc5 subunits (11, 12) (Fig. 8). The available evidence suggests that ORC(2–5) is assembled in the cytoplasm and then rapidly transported into the nucleus where it interacts either with Orc1 or with Orc6 (Fig. 8). Associations between Orc2, Orc3, Orc4, and Orc5 proteins have been demonstrated in the cytoplasm of living cells, in the cytosol of cell extracts, in baculovirus expressed proteins, and in yeast two-hybrid screens (7, 9, 10, 15). Orc2 plays a central role in maintaining ORC(2–5) within the nucleus, independent of its role in binding ORC to DNA. Orc2 contains a single “ORC assembly domain” that is required to form a stable complex with all other ORC subunits, and two NLSs, one that lies outside the ORC assembly domain and one that lies within (9). Both NLSs are required to localize Orc2 exclusively to the nucleus; wild-type Orc2 does so as an ORC(2–5) complex, but Orc2 mutants that do not associate with other ORC subunits but retain both NLSs also localize to the nucleus. Orc2 NLS mutants that localized exclusively to the cytoplasm failed to assemble ORC(2–5), suggesting either that ORC(2–5) assembly is restricted to the nucleus or that NLS-B, which is part of the ORC assembly domain, is required for ORC(2–5) assembly as well as for nuclear localization. The second option appears more likely. Selective inactivation of NLS-A (located outside the ORC assembly domain) resulted in distribution of Orc2 throughout both nucleus and cytoplasm, but it did not interfere with ORC(2–5) assembly, suggesting that assembly can occur in both cellular compartments. In support of this conclusion, analysis of murine Orc2 and Orc3 interactions in living cells detected Orc2-Orc3 complexes of transiently expressed Orc2 and Orc3 proteins in the cytoplasm as well as in the nucleus, where prereplication complexes are assembled (18). However, Orc3 lacking the N-terminal 28 amino acids that contain a putative NLS localized to the cytoplasm where it remained in contact with Orc2, suggesting that ORC(2–5) core complexes require both the Orc2 and Orc3 NLSs to translocate to the nucleus. An NLS has not been reported for either Orc4 or Orc5, but these could be transported into the nucleus through association with Orc2 and Orc3. In either case, nuclear localization does not depend on association with chromatin, because both Orc1 and Orc2 mutants that cannot form ORCs still localize to the nucleus as long as their NLSs are intact. Nuclear localization of ORC(2–5) also does not appear to involve Kpna1 or Kpna6, as the antibodies used in this study detected these proteins associated only with free Orc6 protein.
This work was supported, in whole or in part, by National Institutes of Health intramural research programs (NICHD).
- ORC
- origin recognition complex
- CDK
- cyclin-dependent kinase
- FH
- FLAG-hemagglutinin epitopes
- Kpna
- karyopherin-α
- CAPSO
- 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid
- NLS
- nuclear localization signal
- Hs-
- human.
REFERENCES
- 1. Sivaprasad U., Dutta A., Bell S. P. (2006) in DNA Replication and Human Disease (DePamphilis M. L. ed.) pp. 63–88, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 2. DePamphilis M. L., Bell S. D. (2010) Genome Duplication (Concepts, Mechanisms, Evolution, and Disease) Garland Science, New York [Google Scholar]
- 3. Bell S. P. (2002) Genes Dev. 16, 659–672 [DOI] [PubMed] [Google Scholar]
- 4. Duncker B. P., Chesnokov I. N., McConkey B. J. (2009) Genome Biol. 10, 214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chesnokov I. N. (2007) Int. Rev. Cytol. 256, 69–109 [DOI] [PubMed] [Google Scholar]
- 6. Gillespie P. J., Li A., Blow J. J. (2001) BMC Biochem. 2, 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dhar S. K., Delmolino L., Dutta A. (2001) J. Biol. Chem. 276, 29067–29071 [DOI] [PubMed] [Google Scholar]
- 8. Noguchi K., Vassilev A., Ghosh S., Yates J. L., DePamphilis M. L. (2006) EMBO J. 25, 5372–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Radichev I., Kwon S. W., Zhao Y., DePamphilis M. L., Vassilev A. (2006) J. Biol. Chem. 281, 23264–23273 [DOI] [PubMed] [Google Scholar]
- 10. Vashee S., Simancek P., Challberg M. D., Kelly T. J. (2001) J. Biol. Chem. 276, 26666–26673 [DOI] [PubMed] [Google Scholar]
- 11. Siddiqui K., Stillman B. (2007) J. Biol. Chem. 282, 32370–32383 [DOI] [PubMed] [Google Scholar]
- 12. Ranjan A., Gossen M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4864–4869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Giordano-Coltart J., Ying C. Y., Gautier J., Hurwitz J. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vashee S., Cvetic C., Lu W., Simancek P., Kelly T. J., Walter J. C. (2003) Genes Dev. 17, 1894–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kneissl M., Pütter V., Szalay A. A., Grummt F. (2003) J. Mol. Biol. 327, 111–128 [DOI] [PubMed] [Google Scholar]
- 16. DePamphilis M. L., Blow J. J., Ghosh S., Saha T., Noguchi K., Vassilev A. (2006) Curr. Opin. Cell Biol. 18, 231–239 [DOI] [PubMed] [Google Scholar]
- 17. Diaz-Trivino S., del Mar Castellano M., de la Paz Sanchez M., Ramirez-Parra E., Desvoyes B., Gutierrez C. (2005) Nucleic Acids Res. 33, 5404–5414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Brand N., Faul T., Grummt F. (2007) Mol. Genet. Genomics 278, 623–632 [DOI] [PubMed] [Google Scholar]
- 19. Balasov M., Huijbregts R. P., Chesnokov I. (2007) Mol. Cell. Biol. 27, 3143–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee D. G., Bell S. P. (1997) Mol. Cell. Biol. 17, 7159–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kong D., DePamphilis M. L. (2001) Mol. Cell. Biol. 21, 8095–8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Semple J. W., Da-Silva L. F., Jervis E. J., Ah-Kee J., Al-Attar H., Kummer L., Heikkila J. J., Pasero P., Duncker B. P. (2006) EMBO J. 25, 5150–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen S., de Vries M. A., Bell S. P. (2007) Genes Dev. 21, 2897–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Asano T., Makise M., Takehara M., Mizushima T. (2007) FEMS Yeast Res. 7, 1256–1262 [DOI] [PubMed] [Google Scholar]
- 25. Chesnokov I., Remus D., Botchan M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 11997–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chesnokov I. N., Chesnokova O. N., Botchan M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9150–9155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huijbregts R. P., Svitin A., Stinnett M. W., Renfrow M. B., Chesnokov I. (2009) Mol. Biol. Cell 20, 270–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Prasanth S. G., Prasanth K. V., Stillman B. (2002) Science 297, 1026–1031 [DOI] [PubMed] [Google Scholar]
- 29. Sasaki T., Gilbert D. M. (2007) Curr. Opin. Cell Biol. 19, 337–343 [DOI] [PubMed] [Google Scholar]
- 30. Stuermer A., Hoehn K., Faul T., Auth T., Brand N., Kneissl M., Pütter V., Grummt F. (2007) Eur. J. Cell Biol. 86, 37–50 [DOI] [PubMed] [Google Scholar]
- 31. Hemerly A. S., Prasanth S. G., Siddiqui K., Stillman B. (2009) Science 323, 789–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhou G., Li H., Gong Y., Zhao Y., Cheng J., Lee P., Zhao Y. (2005) Proteomics 5, 3814–3821 [DOI] [PubMed] [Google Scholar]
- 33. Hübner S., Xiao C. Y., Jans D. A. (1997) J. Biol. Chem. 272, 17191–17195 [DOI] [PubMed] [Google Scholar]
- 34. Chook Y. M., Suel K. E. (2010) Biochim. Biophys. Acta, in press [Google Scholar]
- 35. Conti E., Müller C. W., Stewart M. (2006) Curr. Opin. Struct. Biol. 16, 237–244 [DOI] [PubMed] [Google Scholar]
- 36. Plant K. E., Everett D. M., Gordon Gibson G., Lyon J., Plant N. J. (2006) Pharmacogenet. Genomics 16, 647–658 [DOI] [PubMed] [Google Scholar]
- 37. Kosugi S., Hasebe M., Matsumura N., Takashima H., Miyamoto-Sato E., Tomita M., Yanagawa H. (2009) J. Biol. Chem. 284, 478–485 [DOI] [PubMed] [Google Scholar]
- 38. Elion E. A. (2007) Curr. Protoc. Protein Sci., Chapter 19, Unit 19.4 [DOI] [PubMed] [Google Scholar]
- 39. Conti E., Kuriyan J. (2000) Structure 8, 329–338 [DOI] [PubMed] [Google Scholar]
- 40. Lidonnici M. R., Rossi R., Paixão S., Mendoza-Maldonado R., Paolinelli R., Arcangeli C., Giacca M., Biamonti G., Montecucco A. (2004) J. Cell Sci. 117, 5221–5231 [DOI] [PubMed] [Google Scholar]
- 41. Takeda D. Y., Shibata Y., Parvin J. D., Dutta A. (2005) Genes Dev. 19, 2827–2836 [DOI] [PMC free article] [PubMed] [Google Scholar]