Abstract
Niemann-Pick type C (NP-C) disease is a fatal lysosomal lipid storage disorder for which no effective therapy exists. A genome-wide, conditional synthetic lethality screen was performed using the yeast model of NP-C disease during anaerobiosis, an auxotrophic condition that requires yeast to utilize exogenous sterol. We identified 12 pathways and 13 genes as modifiers of the absence of the yeast NPC1 ortholog (NCR1) and quantified the impact of loss of these genes on sterol metabolism in ncr1Δ strains grown under viable aerobic conditions. Deletion of components of the yeast NuA4 histone acetyltransferase complex in ncr1Δ strains conferred anaerobic inviability and accumulation of multiple sterol intermediates. Thus, we hypothesize an imbalance in histone acetylation in human NP-C disease. Accordingly, we show that the majority of the 11 histone deacetylase (HDAC) genes are transcriptionally up-regulated in three genetically distinct fibroblast lines derived from patients with NP-C disease. A clinically approved HDAC inhibitor (suberoylanilide hydroxamic acid) reverses the dysregulation of the majority of the HDAC genes. Consequently, three key cellular diagnostic criteria of NP-C disease are dramatically ameliorated as follows: lysosomal accumulation of both cholesterol and sphingolipids and defective esterification of LDL-derived cholesterol. These data suggest HDAC inhibition as a candidate therapy for NP-C disease. We conclude that pathways that exacerbate lethality in a model organism can be reversed in human cells as a novel therapeutic strategy. This “exacerbate-reverse” approach can potentially be utilized in any model organism for any disease.
Keywords: Cholesterol, Genetics, Histone Acetylase, Histone Deacetylase, Intracellular Trafficking, Lipid Transport, Lysosomal Storage Disease, Neurodegeneration, Sphingolipid, Trafficking
Introduction
The mechanisms by which endocytosed lipids such as cholesterol and sphingolipids leave the lysosomal compartment of a eukaryotic cell are obscure, despite the severe consequences of impaired transport in the pathology of neurodegenerative diseases (1). Two strikingly conserved genes encode proteins that when defective result in NP-C7 disease, a fatal pediatric neurodegenerative disease caused by a lysosomal accumulation of cholesterol and sphingolipids. The clinical hallmark of NP-C disease, for individuals that survive past the neonatal stage, is progressive dementia with markedly reduced executive function and global cognitive impairment, typically culminating in loss of life before adolescence. NP-C disease is conferred by mutations in either the NPC1 gene (95% of cases) or NPC2 gene (5% of cases). NPC1 encodes an ∼13-pass lysosomal transmembrane domain protein, and NPC2 encodes a soluble low molecular weight lumenal protein in the lysosome. Both proteins possess lipid-binding domains and have enigmatic functions as putative lipid transporters or chaperones (2, 3). NPC2 likely “hands off” cholesterol to NPC1 (4), but the interactions of this pathway before and after this event are unknown. As a result, treatments are currently experimental and limited to the suspected targets, i.e. modulating the metabolism of cholesterol (5) or sphingolipids (6). Approved therapies for NP-C disease, particularly in the United States, are thus limited to palliative treatment of the symptoms, and there is a striking clinical need for a novel intervention in what is an invariably fatal disease.
There is no doubting the conservation of NP-C disease across evolution. The mammalian and yeast NPC orthologs are interchangeable to the extent that the yeast orthologs of NPC1 and NPC2 complement lesions in the corresponding genes in cultured mammalian cells (7, 8). There is 35% amino acid sequence identity between the human and yeast NPC1 orthologs, and both proteins localize to the limiting membranes of the lysosomal (vacuolar)/endosomal systems. Moreover, 43% of missense mutations and 15% of missense polymorphisms in NPC1 in NP-C patients are at evolutionarily conserved residues (9). Together, these data suggest an evolutionarily conserved function is disturbed in NPC patients. Model organisms thus have the potential to identify conserved modifiers of the disease. Investigation of the function of NP-C disease orthologs is active in many model systems, including murine, feline, nematode, insect, and yeast (reviewed in Ref. 10). Given the NP-C disease phenotypes of lysosomal lipidosis and neurodegeneration, the ideal modifier in terms of a treatment strategy would be one with the capacity to impact multiple aspects of lipid transport and be amenable to compounds that cross the blood-brain barrier.
Here, we report the results of an unbiased genome-wide screen to identify genetic modifiers of NP-C disease. We exploited the yeast model of NP-C disease (7), a strain in which NCR1 (a functional and structural ortholog of human NPC1) is deleted ((ncr1Δ)). Our aim was to identify genes and pathways that exacerbate lethality of the yeast model of NP-C disease and then pharmacologically manipulate the orthologous pathways in the opposite direction as a potential therapeutic strategy in human cells. We present this “exacerbate-reverse” strategy to find new targets for disease intervention. Using this approach, we identify histone deacetylase inhibition as a potential therapy for NP-C disease and in particular identify suberoylanilide hydroxamic acid (SAHA) as a Food and Drug Administration-approved, brain-penetrative, lead candidate.
EXPERIMENTAL PROCEDURES
Anaerobic Synthetic Genetic Arrays
All yeast strains used in this study were derived from BY4741. The query strain for all synthetic genetic array analyses was Y3656 (MATα can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 lys2Δ0 (11)) with the addition of ncr1::NATR, which was introduced by PCR-mediated gene disruption (12). Synthetic genetic array analysis was performed as described previously (11, 13). Growth of each double mutant was quantitatively compared in the presence and absence of oxygen using ScreenMill (14). Anaerobiosis was achieved with the addition of 20 μg/ml ergosterol and 0.5% Tween 80 in anaerobic jars (BBL) with Anaero Packs (Mitsubishi). To validate interactions, we isolated haploids of double mutants and compared growth of double mutants with the parental single mutants and BY4741 in the presence and absence of oxygen.
Gene Annotations
Biological processes of interacting yeast genes were derived from the on-line Saccharomyces Genome Database. Orthologous human proteins and complexes were identified using Princeton Protein Orthology Database (43).
Sterol Profiles
Liquid cultures (100 ml) of the indicated yeast strains were grown to an A600 of 1.0 in YPD, pelleted, and frozen at −80 °C. Sterols were separated and quantified by GC-MS as described previously (15).
Cell Culture
All fibroblasts were cultured in DMEM supplemented with 10% FBS, 2 mm l-glutamine, and 50 μg/ml gentamicin in a humidified incubator at 37 °C with 5% CO2. Human control fibroblasts (GM06114) were obtained from Coriell Cell Repositories. The NPC-26, NPC-2, and NPC-29 mutant human fibroblasts were obtained from the National Institutes of Health; these fibroblasts were derived from unrelated patients with genetically distinct compound heterozygous missense mutations in NPC1. The NPC-J4 fibroblast line was obtained from a skin biopsy of a Japanese patient homozygous for the NPC1 H510P allele (16).
Drugs
SAHA (Cayman), MC1568 (Selleck), MGCD0103 (Selleck), and trichostatin A (Sigma) were applied to fibroblasts for 18 h. Concentrations used were previously described to achieve global (SAHA, trichostatin A) or class-specific (MC1568, MGCD0103) inhibition of HDAC genes.
Cholesterol and Sphingolipid Trafficking
Control and NPC1-deficient human fibroblasts were assessed for trafficking of cholesterol and sphingolipids. Cholesterol and the glycosphingolipid globotriosylceramide (GL-3) accumulation were monitored using filipin (Polysciences) or verotoxin, respectively, as described previously (17, 18). The trafficking of lactosylceramide was monitored using BODIPY-LacCer; cells in a glass bottom dish (Iwaki, Japan) were washed with Hanks' buffered saline solution, incubated with 5 μm BODIPY-LacCer with BSA complex (Molecular Probes) for 30 min at 4 °C, and incubated with DMEM 10% FBS at 37 °C for 30 min as described in the manufacturer's protocol. For detection of ganglioside GM1, cells in glass bottom dish were washed with DMEM, 25 mm Hepes, pH 7.4, and 0.01% BSA, incubated with 20 nm Alexa Fluor 555-conjugated cholera toxin subunit B (Molecular Probes) for 1 h at 37 °C, and fixed with 4% paraformaldehyde (7). Images were obtained sequentially using confocal laser microscopy, and quantification analysis was performed using ImageJ64 or MetaVue (17).
Esterification of LDL-derived Cholesterol
Human fibroblasts were assessed for esterification of LDL-derived cholesterol in 4 h as described previously (19).
Gene Expression
Quantitative real time PCR was performed using a MyiQ machine (Bio-Rad) and SYBR Green chemistry (Bio-Rad) to measure gene expression. The sequences of the primers used in these determinations are listed in supplemental Table S1. The mRNA levels were expressed relative to GAPDH and were calculated by the ΔΔCT method (20).
RESULTS
Genome-wide Screen for Modifiers of NP-C Disease in Yeast
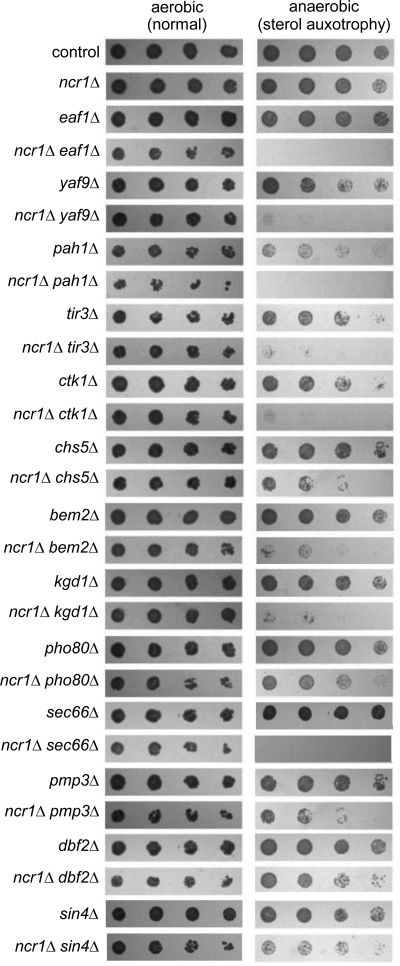
To identify genetic targets with NP-C disease-modifying potential, we performed a genome-wide screen for synthetic lethality in the yeast model of NP-C disease. Using synthetic genetic array methodology (11), we generated ∼5,500 haploid double mutants of ncr1Δ with deletions in ∼4,800 nonessential genes or knockdown alleles (21) of ∼700 essential genes (corresponding in total to ∼92% of the yeast genome). Under standard aerobic conditions, these double mutants conferred identical growth parameters (12) to their parental strains, despite unvalidated reports to the contrary (44). The accepted diagnostic criteria of NP-C disease are the lysosomal accumulation and defective esterification of LDL-derived exogenous cholesterol. We therefore optimized our screen to detect genetic interactions during anaerobiosis. Under these conditions, yeast cells depend on utilization of exogenous sterols due to the requirement of de novo sterol biosynthesis for molecular oxygen. The NP-C pathway in yeast was clearly impacted by anaerobiosis; we identified 13 genes in 12 pathways required for normal growth of NCR1-deficient cells (Fig. 1; Table 1). All 13 mutations were in nonessential genes that define novel genetic interactions and include the first set of validated genetic interactions for the yeast model of NP-C disease. We propose these represent either alternate routes of lipid transport or perhaps salvage strategies that compensate for accumulation of toxic metabolites that might be potential targets for disease treatment.
FIGURE 1.
Deletion of 13 genes confers significant growth defects to the ncr1Δ yeast model of NP-C disease as a consequence of sterol auxotrophy. Haploid double deletions were derived from a cross of ncr1Δ and the indicated haploid single gene deletions. 5-Fold dilutions (left to right) of saturated, aerobically grown cultures of the single mutants and their derived double mutant were grown at 30 °C aerobically on SCD and anaerobically on SCD + 20 μg/ml ergosterol + 0.5% Tween 80 for 3 days.
TABLE 1.
Functional annotation of genetic interactions with NCR1 during anaerobiosis
Anaerobic genetic interactions were defined as a minimum of 50% growth defect in double deletion strains compared with parental single deletions, only in anaerobiosis. Biological processes of interacting yeast genes were derived from the Princeton Protein Orthology Database (43). Orthologous human proteins and complexes were identified using the Princeton Protein Orthology Database and are solely based on protein sequence conservation.
| Gene | Biological process | Orthologous human protein-complex |
|---|---|---|
| EAF1 | Histone acetyltransferase | p400 |
| YAF9 | Histone acetyltransferase | GAS41 |
| PAH1 | Phosphatidic acid hydrolase | Lipins |
| TIR3 | Unknown | None |
| CTK1 | Phosphorylation of RNA polymerase II | CDK13 |
| CHS5 | Golgi to plasma membrane transport | Neurofilament H |
| BEM2 | Cytoskeleton organization | ARHGAP24 |
| KGD1 | 2-Oxoglutarate metabolism in TCA cycle | OGDH |
| PHO80 | Cyclin-dependent protein kinase | CDK5 complex |
| SEC66 | Protein targeting and import into the endoplasmic reticulum | SEC63 complex |
| PMP3 | Regulation of membrane potential | None |
| DBF2 | Stress-response kinase | None |
| SIN4 | Transcription from RNA polymerase II promoter | None |
The interaction screen was clearly successful in that it identified several pathways already associated with lipid metabolism and/or NP-C disease. For example, PHO80 encodes a regulatory component of the yeast cyclin-dependent kinase PHO85, of which the mammalian ortholog (CDK5) is dysregulated in the murine model of NP-C disease (22). Similarly, the CHS5 gene encodes a component of a Golgi to plasma membrane transport pathway that is also defective in the transport of cholesterol in NP-C disease (23). Among the remaining 11 genes, PAH1 encodes a key enzyme (phosphatidic acid phosphohydrolase) in glycerophospholipid metabolism (24), and BEM2 has been previously implicated in sterol uptake (25). Nine of the 13 genes are conserved in humans and thus may be important in modifying NPC1 disease pathogenesis. Notably, deletion of either EAF1 or YAF9, both of which are components of the NuA4 histone acetyltransferase complex, confer anaerobic inviability to ncr1Δ strains (Fig. 1). Compellingly, this was the only instance of multiple interactions in our dataset that are components of the same complex. Interestingly, mutations in EAF1 or YAF9 have enlarged vacuoles (26), the equivalent of the mammalian lysosome in yeast. EAF1 encoded activity has also been shown to regulate Golgi-vacuole vesicle-mediated transport (26).
Sterol Metabolism in the Yeast NP-C Model Is Impacted by Modifier Gene Deletions
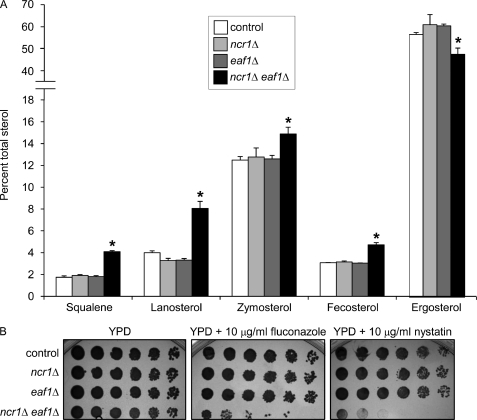
The anaerobic inviability of the double mutants indicates a genetic interaction, putatively dependent on exogenous sterol utilization. Inviability in the screen may be a consequence of alterations in fatty acid metabolism due to unsaturated fatty acid auxotrophy also associated with anaerobiosis. We therefore examined aerobic growth of the double mutants in the presence of fatty acids and sterols, a state that permits uptake and metabolism of the fatty acids but not sterol. The growth of all double mutants was normal under this condition (data not shown), a result that further implicates sterols as the toxic metabolites in the screen. To assess the impact of the 13 NCR1 modifiers on sterol levels in ncr1Δ strains, we used GC-MS to quantify endogenous levels of sterols in the 13 double deletions and a control strain under aerobic conditions in which the double deletions are viable. Compared with the control, three double deletions (ncr1Δ eaf1Δ, ncr1Δ sin4Δ, and ncr1Δ pah1Δ) displayed a 300% increase in at least one sterol intermediate (Table 2). Only ncr1Δ eaf1Δ strains exhibited an increase in more than one sterol. We further quantified the sterol accumulation in ncr1Δ eaf1Δ cells, this time relative to the parental strains. ncr1Δ eaf1Δ strains exhibited statistically significant increases in the intracellular accumulation of squalene, lanosterol, zymosterol, and fecosterol and a significant decrease in ergosterol relative to the single mutant strains (Fig. 2A).
TABLE 2.
Aerobically viable sterol profiles of the 13 anaerobically sensitive interactions distinguish three double mutants with >300% increases in intracellular accumulation of sterols
Cells of the control strain and all double mutants indicated in Fig. 1 were grown in triplicate under standard aerobic conditions in YPD at 30 °C to 100 OD units. Sterols were extracted as described and measured by GC-MS. The data are expressed here for each double mutant as a percentage relative to levels in the control strain.
| Percent sterol relative to control |
|||||||
|---|---|---|---|---|---|---|---|
| Squalene | Lanosterol | 4,4-Dimethylzymosterol | Zymosterol | Fecosterol | Episterol + Ergosta-5–7-dien-3β-ol | Ergosterol | |
| Control | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 |
| ncr1Δ eaf1Δ | 411.95 | 343.50 | 102.83 | 104.04 | 142.41 | 101.39 | 76.83 |
| ncr1Δ yaf9Δ | 83.12 | 85.16 | 27.88 | 81.04 | 15.99 | 114.84 | 108.87 |
| ncr1Δ pah1Δ | 216.30 | 84.96 | 335.85 | 85.86 | 160.54 | 65.92 | 102.06 |
| ncr1Δ tir3Δ | 168.92 | 117.95 | 98.02 | 92.59 | 97.48 | 98.48 | 100.00 |
| ncr1Δ ctk1Δ | 160.38 | 87.85 | 119.07 | 88.91 | 130.31 | 70.06 | 109.04 |
| ncr1Δ chs5Δ | 136.78 | 73.54 | 98.26 | 71.29 | 84.61 | 132.47 | 97.89 |
| ncr1Δ bem2Δ | 111.99 | 87.67 | 73.74 | 102.77 | 91.45 | 123.44 | 93.97 |
| ncr1Δ kgd1Δ | 116.76 | 165.44 | 203.08 | 107.01 | 160.80 | 95.66 | 89.45 |
| ncr1Δ pho80Δ | 74.31 | 111.19 | 76.25 | 114.59 | 116.45 | 125.14 | 88.83 |
| ncr1Δ sec66Δ | 215.20 | 129.25 | 107.31 | 100.29 | 118.95 | 97.22 | 95.89 |
| ncr1Δ pmp3Δ | 203.23 | 119.36 | 94.26 | 91.59 | 94.24 | 102.12 | 98.70 |
| ncr1Δ dbf2Δ | 159.74 | 62.26 | 125.85 | 110.48 | 126.92 | 114.15 | 92.42 |
| ncr1Δ sin4Δ | 529.54 | 183.34 | 117.19 | 141.56 | 207.83 | 86.23 | 76.98 |
FIGURE 2.
Sterol metabolism is disturbed in yeast ncr1Δ eaf1Δ cells. A, analysis of ncr1Δ eaf1Δ strains identifies a bottleneck in aerobic sterol synthesis with increased intracellular accumulation of ergosterol precursors and decreased ergosterol. Cells were grown in triplicate under aerobic conditions in YPD at 30 °C to 100 OD units. Sterol biosynthetic intermediates were measured by GC-MS and are expressed as a percentage of total sterols. *, p < 0.05, two-tailed Student's t test comparison of ncr1Δ eaf1Δ cells with control, ncr1Δ, or eaf1Δ strains. B, sensitivity of ncr1Δ eaf1Δ to fluconazole and nystatin. 5-Fold dilutions (left to right) of saturated, aerobically grown cultures were grown aerobically at 30 °C for 2 days in the presence of the indicated drug.
We further characterized ncr1Δ eaf1Δ cells by examining growth in the presence of two drugs that target sterol metabolism. The aerobic growth of ncr1Δ eaf1Δ strains in the presence of fluconazole or nystatin was markedly impaired relative to control or single deletion parental strains (Fig. 2B). Fluconazole is an inhibitor of lanosterol demethylation. The growth defect due to fluconazole therefore suggests that ncr1Δ eaf1Δ cells, which already display increased lanosterol and decreased ergosterol, cannot tolerate further sterol imbalance. By contrast, nystatin is a polyene antibiotic that binds ergosterol in membranes. The growth defect in response to nystatin, despite the lower levels of total ergosterol in ncr1Δ eaf1Δ cells relative to the single mutants or control, suggests that ncr1Δ eaf1Δ membranes may be more permeable to nystatin. Alternatively, ergosterol in ncr1Δ eaf1Δ strains may be mis-transported to a nystatin-sensitive compartment (e.g. the vacuole (27)) and thus manifest as elevated sensitivity.
Global Up-regulation of Histone Deacetylase Genes in NP-C-deficient Human Fibroblasts
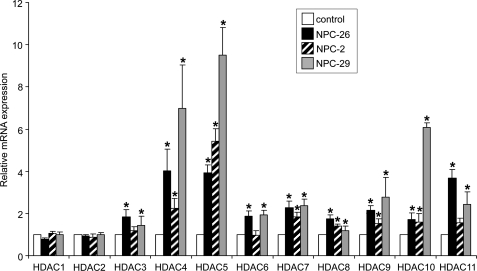
The anaerobic inviability and sterol accumulation of ncr1Δ eaf1Δ strains suggest the surprising interpretation that the activity of histone acetylases is key to cholesterol homeostasis in NP-C disease. Moreover, the histone acetyltransferase encoded by EAF1, and the complex in which it resides, is conserved in humans. We therefore hypothesized that an imbalance in histone acetylation contributes to human NPC1 disease pathophysiology. The relative activities of HDACs are maintained in a tightly controlled cellular equilibrium in part by transcriptional regulation of the deacetylases (28). Therefore, we used quantitative real time PCR to measure expression of the 11 HDAC genes (HDAC1–11) in fibroblasts derived from three unrelated NP-C patients with distinct NPC1 mutant alleles (cell lines NPC-26, NPC-2, and NPC-29). Significant up-regulation of 9 of the 11 genes in NPC-26 and NPC-29 and 7 of the 11 genes in NPC-2 relative to control fibroblasts was observed (Fig. 3). There were no significant changes in expression of HDAC1 and HDAC2 in any of the cell lines. The highest levels of up-regulation in the NP-C fibroblasts were exhibited by HDAC4 and HDAC5 (average increases of 4.4- and 6.3-fold, respectively, compared with control fibroblasts). p21, p53, and the CLU genes are transcriptional targets of the HDACs and were misregulated in untreated NP-C cells (Fig. 4). This was consistent with the predicted histone acetylation imbalance in the ncr1Δ eaf1Δ yeast strain particularly given the regulation of p21 by p400, the human ortholog of yeast EAF1 (29).
FIGURE 3.
Histone deacetylase genes are up-regulated in human NP-C fibroblasts. RNA from the indicated mutants and control fibroblast lines were assessed for HDAC gene expression by quantitative real time PCR using the primers described in supplemental Table S1. The majority of the 11 HDAC genes are up-regulated in fibroblasts derived from three patients with NP-C disease (NPC-26, NPC-2, and NPC-29). *, p < 0.05, two-tailed Student's t test for each NP-C fibroblast relative to the control fibroblast. Mis-regulation of HDAC11 has been reported elsewhere (42).
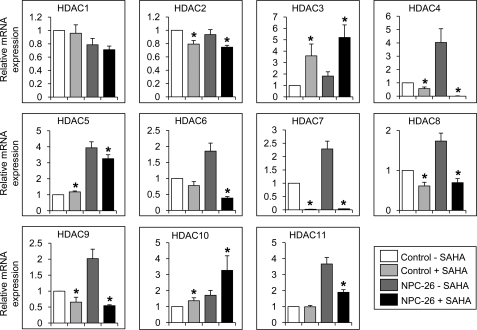
FIGURE 4.
Targets of HDAC genes are dysregulated in NP-C fibroblasts and regulated by SAHA. NPC1 mutant and control fibroblast lines were grown in the presence and absence of SAHA and assessed for expression of genes known to be responsive to HDAC activity. A statistical comparison was conducted separately for untreated control versus untreated NP-C, untreated control versus treated control, and untreated NP-C versus treated NP-C cells using a two-tailed Student's t test (* indicates a significant difference based on p < 0.05).
HDAC Inhibition in Human Fibroblasts Ameliorates Multiple Biochemical Hallmarks of NP-C Disease
Having demonstrated global HDAC dysregulation in NP-C fibroblasts, we sought to determine whether these cells could be normalized by HDAC inhibition. To inhibit HDAC, we used SAHA (Vorinostat, Zolinza®) because it is a potent global inhibitor of histone deacetylation (28). Furthermore, SAHA has been shown to cross the blood-brain barrier (30) and is Food and Drug Administration-approved to treat cutaneous T-cell lymphoma, a critical incentive given the short life span of NP-C patients who cannot wait for the development and approval of a new drug. A 24-h treatment of 5 μm SAHA in NP-C fibroblasts had the expected outcome for several known transcriptionally regulated targets of this drug (Fig. 4) (28). Many HDAC genes are transcriptionally regulated by HDAC enzymatic inhibitors such as SAHA (28). Strikingly, after a 24-h treatment of 5 μm SAHA, the expression of seven of the nine up-regulated HDAC genes in NPC-26 fibroblasts were significantly restored toward control levels (Fig. 5).
FIGURE 5.
Treatment of NPC-26 fibroblasts with SAHA restores expression of HDAC genes. NPC1 mutant and control fibroblast lines were grown in the presence and absence of SAHA and assessed for expression of the HDAC genes. *, p < 0.05, two-tailed Student's t test compared treated and untreated cells separately for control and NP-C fibroblasts.
We reasoned that inhibiting HDAC might reverse the cellular and biochemical phenotypes of NP-C disease. We characterized the effect of 2.5, 5.0, and 10.0 μm SAHA on the trafficking of LDL-derived cholesterol by evaluating filipin fluorescence in human fibroblasts. We determined that a dose of 5 μm SAHA significantly ameliorated filipin accumulation in NPC-26 cells. After 24 h of treatment with 5 μm SAHA, the lysosomal accumulation of cholesterol in NPC-26 or NPC-J4 was significantly reduced by 50–60% compared with untreated NP-C mutant cells (Fig. 6A). Significant reductions in filipin fluorescence were also observed for NPC-2 and NPC-29 (data not shown). We then examined the effect of 5 μm SAHA in NPC-26 cells on the esterification of LDL-derived cholesterol, a defect that is also a biochemical hallmark of NP-C disease (19). Following a 24-h treatment with 5 μm SAHA, esterification of LDL-cholesterol (added at 20 h) was significantly increased by ∼200% (Fig. 6B) to a value that approximated esterification in untreated controls, which were also elevated by SAHA treatment.
FIGURE 6.
Global histone deacetylase inhibition corrects cholesterol homeostasis in NP-C fibroblasts. Mutant fibroblasts (NPC-26 and NPC-J4) were incubated for 18 h in the presence of 5 μm SAHA, stained with filipin, and assessed for esterification of LDL-cholesterol. A, reduction in lysosomal accumulation of unesterified cholesterol as measured by filipin fluorescence. Quantification of filipin fluorescence was obtained and is expressed as arbitrary units. B, restoration of deficient esterification of LDL-derived cholesterol as measured by percent cholesteryl [3H]oleate formation relative to total lipids. Cells were grown for 4 days in lipoprotein-deficient serum, followed by a 24-h treatment with or without 5 μm SAHA, the last 4 h of which included LDL plus [3H]oleate. Incorporation was quantified by scintillation and normalized to total lipid. *, p < 0.05, treated versus untreated cells by two-tailed Student's t test.
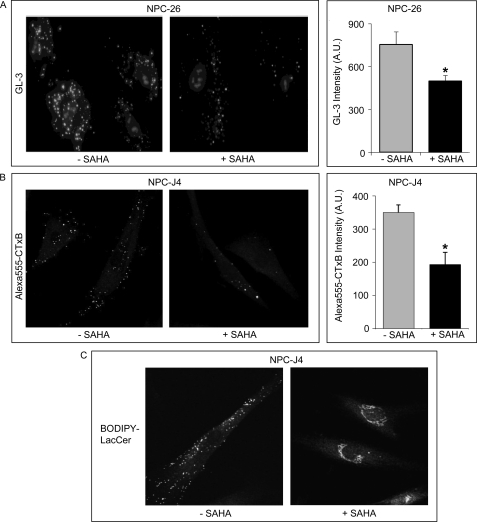
The NP-C lipidosis was also typified by aberrant lysosomal/endosomal accumulation of sphingolipids such as monosialotetrahexosylganglioside (GM1 (31)) and trihexosylceramide8 (GL-3) or defective transport of fluorescent sphingolipid analogs such as 4,4-difluoro-4-bora-3a,4a-diaza-s-indacene lactosylceramide (BODIPY-LacCer (32)). We evaluated remediation of cholesterol and sphingolipid transport in NPC1 mutant fibroblasts (NPC-26 and NPC-J4) cultured with inhibitors of the HDACs. Treatment with 5 μm SAHA significantly reduced the accumulation of GL-3 by ∼30% compared with untreated NP-C cells (Fig. 7A). Similarly, 5 μm SAHA completely cleared GM1 from the lysosomal compartment in the NPC-J4 NPC1 mutant line as assessed by Alexa Fluor 555-cholera toxin subunit B fluorescence (Fig. 7B). Finally, we assessed the transport of a fluorescent analog of lactosylceramide (BODIPY-LacCer) in NPC-J4 cells treated with SAHA. BODIPY-LacCer accumulates in the Golgi of normal cells but is retained in the lysosomal/endosomal compartment of NPC1 mutants (32), a localization that was reversed following treatment with SAHA (Fig. 7C). Similarly, treating the NPC-J4 mutant fibroblast line with another global HDAC inhibitor, 300 nm trichostatin A (TSA), produced a striking restoration of cholesterol and sphingolipid trafficking (supplemental Fig. S1).
FIGURE 7.
Global histone deacetylase inhibition corrects lysosomal accumulation of sphingolipids in NP-C fibroblasts. Mutant fibroblasts (NPC-26 and NPC-J4) were incubated for 18 h in the presence of 5 μm SAHA and assessed for sphingolipid accumulation. A, reduction in lysosomal accumulation of globotriosylceramide (GL-3) as measured with verotoxin. NPC-26 fibroblasts were incubated for 18 h in the presence of 5 μm SAHA and stained with verotoxin. B, NPC-J4 fibroblasts in the presence and absence of SAHA were stained with Alexa Fluor 555-cholera toxin subunit B to visualize GM1. C, transport of BODIPY-LacCer in NPC-J4 cells was assessed by fluorescent microscopy following a 30-min loading at 4 °C and a 30-min chase. Quantitation of fluorescence was obtained and is expressed as arbitrary units. Statistically significant differences are based on a two-tailed Student's t test (* indicates a significant difference based on p < 0.05).
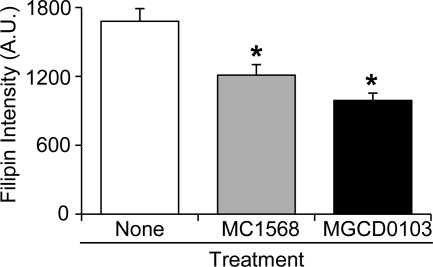
To elucidate the mechanism by which SAHA- or TSA-mediated HDAC inhibition ameliorates NP-C disease phenotypes, we next determined whether treatment with isoform-specific HDAC inhibitors can rescue the lysosomal accumulation of cholesterol in fibroblasts. Using MC1568 (33), an inhibitor of only HDAC class IIa genes (HDAC4, -5, -7, and -9), and MGCD0103 (34), an inhibitor of class I and class IV HDAC genes (HDAC1–3 and -11, respectively), we assessed sterol accumulation in the NPC-26 mutant fibroblast line. Like SAHA, the HDAC class-specific inhibitors MC1568 and MGCD0103 (5 μm) also reduced filipin accumulation in NPC-26 compared with untreated cells after 24 h of treatment (Fig. 8). MC1568 and MGCD0103 reduced filipin accumulation by 28 and 41% compared with untreated cells, values that are significant but markedly below the 60% impact of global HDAC inhibitors such as SAHA and TSA.
FIGURE 8.
Class-specific histone deacetylase inhibition ameliorates cholesterol accumulation. Mutant fibroblasts (NPC-26) were incubated for 18 h in the presence of the HDAC class-specific inhibitors MC1568 and MGCD0103 (5 μm) and assessed for cholesterol accumulation by filipin fluorescence. Quantification of filipin fluorescence is expressed as arbitrary units. *, p < 0.05, treated versus untreated cells by two-tailed Student's t test.
DISCUSSION
The identification of genetic or physical interactions underlying NP-C disease remains elusive. For example, both proteins defective in this disease are clearly associated with binding and transport of the same molecule, cholesterol, but have yet to be shown to interact at any level. NPC1 likely encodes a transporter that directly or indirectly affects the status of multiple lipids. We hypothesized that NPC1 and its orthologs would display numerous genetic interactions. We approached this problem using the yeast model of NP-C disease and identified 13 interacting loci that potentially identify modulators of the syndrome. Our premise is that an unbiased identification of genetic modifiers provides a means to identify previously unsuspected pharmacological targets to treat NP-C disease. The focus of this study emanates from two model systems separated by over 2 billion years of evolution. The genetic interaction of yeast NCR1 with EAF1, encoding a histone acetylase, conferred altered sterol metabolism and was manifested in human NP-C fibroblasts as a striking HDAC imbalance; the majority of HDAC genes are up-regulated in NP-C fibroblasts derived from patients. Pharmacological intervention in this pathway in several human NP-C lines by inhibiting HDAC corrected multiple intracellular lipid transport defects associated with NP-C disease. Consequently, we propose that SAHA, an Food and Drug Administration-approved agent, provides a novel point of entry into rapidly treating this devastating disorder. Moreover, we consider this as proof-of-concept that unbiased identification of genetic modifiers is a means to identify pharmacological targets to treat NP-C disease.
How does the activity of transcriptional regulators such as histone acetylases/deacetylases impact sterol homeostasis? The impact of HDAC inhibition on lipid accumulation in NP-C-deficient fibroblasts was not due to enhanced expression of NPC1, the chaperone HSP90, or RAB9 a genetic modifier that has been shown to ameliorate lipid accumulation in NP-C fibroblasts upon overexpression (35). Interestingly, SAHA treatment did induce expression of NPC2 in control and NPC1 mutant fibroblasts relative to untreated fibroblasts (data not shown), indicating NPC2 is a transcriptional target of SAHA. Increased expression of NPC2 in NPC1 mutant cells is established but does not however alter disease phenotypes (36). The amelioration of NPC disease phenotypes in the absence of increased expression of the NPC1 gene is in agreement with the use of valproate (a relatively nonspecific inhibitor of HDAC, glycogen synthase kinase 3, and ion channel transport) to rescue cholesterol accumulation in neural stem cells derived from an NPC1-null mouse (37). The histone-independent effects of SAHA, valproate, and other HDAC inhibitors are well established (28), raising the possibility that salvage of lipid transport may be unrelated to transcriptional regulation. Thus, the mechanism by which HDAC inhibitors correct lipid homeostasis in the human NP-C fibroblasts remains to be determined.
Our demonstration of greater rescue of cholesterol regulation with the global HDAC inhibitor than with isoform-specific HDAC inhibitors (MC1568 and MGCD0103) suggests this pathway is additively regulated by several HDAC genes, a prediction consistent with the global up-regulation of multiple HDAC genes observed here in human NP-C-deficient cells. HDAC4 was up-regulated in human NPC1 mutant cells and markedly responsive to SAHA suggesting HDAC4 may be a contributor to lipid homeostasis in NP-C disease. Interestingly, HDAC4 activity is critical to neuronal survival (38) achieving its impact via both histone and non-histone targets of HDAC genes. The major neurological pathology of NP-C disease is the loss of Purkinje cells in the cerebellum. HDAC6 and -11 are the most highly expressed HDAC genes in Purkinje cells (39), and our findings would suggest an HDAC11-specific inhibitor is a better target for NP-C disease.
The global inhibitor SAHA is Food and Drug Administration-approved and crosses the blood-brain barrier, critical parameters to NP-C patients who can ill afford to wait for development of a new drug. Despite this promise, we should be skeptical until drugs such as SAHA have been tested in the NPC1 mutant animal models to assess efficacy with respect to neurodegeneration. Testing the effectiveness of SAHA with cyclodextrin or imatinib might also be illuminating. Cyclodextrin treatment extends the longevity of npc1−/− mice yet inefficiently crosses the blood-brain barrier. By contrast, a SAHA-cyclodextrin complex is efficient at crossing the blood-brain barrier and ameliorates neurodegeneration in the murine model of Huntington disease (30). A similar synergy could also be investigated with imatinib, an anti-cancer drug that has been shown to improve neurological symptoms and increase survival of NPC1-deficient mice without impacting lipid accumulation (40). Intriguingly, SAHA and imatinib are synergistic in treatment of cancer (28).
Here, we describe a novel concept in the use of model systems to study and treat human disease. In the exacerbate-reverse approach utilizing a screen for conditional synthetic lethality in yeast, we identify a Food and Drug Administration-approved drug that improved the diagnostic criteria of NP-C disease, a fatal pediatric neurodegenerative disease for which there is currently no effective approved treatment. Our approach of identifying pathways that exacerbate lethality in a model organism and then pharmacologically manipulating the orthologous pathways in the opposite direction as a therapy in human cells could be utilized in any model organism. Proof of this concept is provided by our findings and a study published during the revision of this manuscript (41) that HDAC inhibition corrects defective subcellular lipid transport, the phenomena that precedes neurodegeneration in NP-C disease.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK54320 (to S. L. S.). This work was also supported by the Ara Parseghian Medical Research Foundation and the Dana Angels Research Trust.
This article was selected as a Paper of the Week.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Table S1.
Y. A. Ioannou and F. W. Chen, unpublished data.
- NP-C
- Niemann-Pick type C
- SAHA
- suberoylanilide hydroxamic acid
- HDAC
- histone deacetylase
- TSA
- trichostatin A.
REFERENCES
- 1. Madra M., Sturley S. L. (2010) Clin. Lipidol. 5, 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ohgami N., Ko D. C., Thomas M., Scott M. P., Chang C. C., Chang T. Y. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12473–12478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ko D. C., Binkley J., Sidow A., Scott M. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2518–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kwon H. J., Abi-Mosleh L., Wang M. L., Deisenhofer J., Goldstein J. L., Brown M. S., Infante R. E. (2009) Cell 137, 1213–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu B., Turley S. D., Burns D. K., Miller A. M., Repa J. J., Dietschy J. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 2377–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patterson M. C., Vecchio D., Prady H., Abel L., Wraith J. E. (2007) Lancet Neurol. 6, 765–772 [DOI] [PubMed] [Google Scholar]
- 7. Malathi K., Higaki K., Tinkelenberg A. H., Balderes D. A., Almanzar-Paramio D., Wilcox L. J., Erdeniz N., Redican F., Padamsee M., Liu Y., Khan S., Alcantara F., Carstea E. D., Morris J. A., Sturley S. L. (2004) J. Cell Biol. 164, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Berger A. C., Vanderford T. H., Gernert K. M., Nichols J. W., Faundez V., Corbett A. H. (2005) Eukaryot. Cell 4, 1851–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park W. D., O'Brien J. F., Lundquist P. A., Kraft D. L., Vockley C. W., Karnes P. S., Patterson M. C., Snow K. (2003) Hum. Mutat. 22, 313–325 [DOI] [PubMed] [Google Scholar]
- 10. Munkacsi A. B., Porto A. F., Sturley S. L. (2007) Future Lipidol. 2, 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tong A. H., Evangelista M., Parsons A. B., Xu H., Bader G. D., Pagé N., Robinson M., Raghibizadeh S., Hogue C. W., Bussey H., Andrews B., Tyers M., Boone C. (2001) Science 294, 2364–2368 [DOI] [PubMed] [Google Scholar]
- 12. Almanzar-Paramio D. (2006) NCR1, the Yeast Ortholog of the Niemann-Pick C1 Disease Gene NPC1. Ph.D. thesis, Columbia University, New York, NY [Google Scholar]
- 13. Breslow D. K., Cameron D. M., Collins S. R., Schuldiner M., Stewart-Ornstein J., Newman H. W., Braun S., Madhani H. D., Krogan N. J., Weissman J. S. (2008) Nat. Methods 5, 711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dittmar J. C., Reid R. J., Rothstein R. (2010) BMC Bioinformatics 11, 353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shakoury-Elizeh M., Protchenko O., Berger A., Cox J., Gable K., Dunn T. M., Prinz W. A., Bard M., Philpott C. C. (2010) J. Biol. Chem. 285, 14823–14833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamamoto T., Nanba E., Ninomiya H., Higaki K., Taniguchi M., Zhang H., Akaboshi S., Watanabe Y., Takeshima T., Inui K., Okada S., Tanaka A., Sakuragawa N., Millat G., Vanier M. T., Morris J. A., Pentchev P. G., Ohno K. (1999) Hum. Genet. 105, 10–16 [DOI] [PubMed] [Google Scholar]
- 17. Chen F. W., Li C., Ioannou Y. A. (2010) PLoS ONE 5, e15054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeidner K. M., Desnick R. J., Ioannou Y. A. (1999) Anal. Biochem. 267, 104–113 [DOI] [PubMed] [Google Scholar]
- 19. Pentchev P. G., Comly M. E., Kruth H. S., Vanier M. T., Wenger D. A., Patel S., Brady R. O. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 8247–8251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 21. Schuldiner M., Collins S. R., Weissman J. S., Krogan N. J. (2006) Methods 40, 344–352 [DOI] [PubMed] [Google Scholar]
- 22. Bu B., Li J., Davies P., Vincent I. (2002) J. Neurosci. 22, 6515–6525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Garver W. S., Krishnan K., Gallagos J. R., Michikawa M., Francis G. A., Heidenreich R. A. (2002) J. Lipid Res. 43, 579–589 [PubMed] [Google Scholar]
- 24. Han G. S., Wu W. I., Carman G. M. (2006) J. Biol. Chem. 281, 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sullivan D. P., Georgiev A., Menon A. K. (2009) Eukaryot. Cell 8, 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mitchell L., Lambert J. P., Gerdes M., Al-Madhoun A. S., Skerjanc I. S., Figeys D., Baetz K. (2008) Mol. Cell. Biol. 28, 2244–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhuiyan M. S., Ito Y., Nakamura A., Tanaka N., Fujita K., Fukui H., Takegawa K. (1999) Biosci. Biotechnol. Biochem. 63, 1075–1082 [DOI] [PubMed] [Google Scholar]
- 28. Marks P. A., Dokmanovic M. (2005) Expert Opin. Investig. Drugs 14, 1497–1511 [DOI] [PubMed] [Google Scholar]
- 29. Park J. H., Sun X. J., Roeder R. G. (2010) Mol. Cell. Biol. 30, 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hockly E., Richon V. M., Woodman B., Smith D. L., Zhou X., Rosa E., Sathasivam K., Ghazi-Noori S., Mahal A., Lowden P. A., Steffan J. S., Marsh J. L., Thompson L. M., Lewis C. M., Marks P. A., Bates G. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2041–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taniguchi M., Shinoda Y., Ninomiya H., Vanier M. T., Ohno K. (2001) Brain Dev. 23, 414–421 [DOI] [PubMed] [Google Scholar]
- 32. Choudhury A., Sharma D. K., Marks D. L., Pagano R. E. (2004) Mol. Biol. Cell 15, 4500–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Duong V., Bret C., Altucci L., Mai A., Duraffourd C., Loubersac J., Harmand P. O., Bonnet S., Valente S., Maudelonde T., Cavailles V., Boulle N. (2008) Mol. Cancer Res. 6, 1908–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fournel M., Bonfils C., Hou Y., Yan P. T., Trachy-Bourget M. C., Kalita A., Liu J., Lu A. H., Zhou N. Z., Robert M. F., Gillespie J., Wang J. J., Ste-Croix H., Rahil J., Lefebvre S., Moradei O., Delorme D., Macleod A. R., Besterman J. M., Li Z. (2008) Mol. Cancer Ther. 7, 759–768 [DOI] [PubMed] [Google Scholar]
- 35. Walter M., Davies J. P., Ioannou Y. A. (2003) J. Lipid Res. 44, 243–253 [DOI] [PubMed] [Google Scholar]
- 36. Storch J., Xu Z. (2009) Biochim. Biophys. Acta 1791, 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim S. J., Lee B. H., Lee Y. S., Kang K. S. (2007) Biochem. Biophys. Res. Commun. 360, 593–599 [DOI] [PubMed] [Google Scholar]
- 38. Majdzadeh N., Wang L., Morrison B. E., Bassel-Duby R., Olson E. N., D'Mello S. R. (2008) Dev. Neurobiol. 68, 1076–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Southwood C. M., Peppi M., Dryden S., Tainsky M. A., Gow A. (2007) Neurochem. Res. 32, 187–195 [DOI] [PubMed] [Google Scholar]
- 40. Alvarez A. R., Klein A., Castro J., Cancino G. I., Amigo J., Mosqueira M., Vargas L. M., Yévenes L. F., Bronfman F. C., Zanlungo S. (2008) FASEB J. 22, 3617–3627 [DOI] [PubMed] [Google Scholar]
- 41. Pipalia N. H., Cosner C. C., Huang A., Chatterjee A., Bourbon P., Farley N., Helquist P., Wiest O., Maxfield F. R. (2011) Proc. Natl. Acad. Sci. U.S.A., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reddy J. V., Ganley I. G., Pfeffer S. R. (2006) PLoS ONE 1, e1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heinicke S., Livstone M. S., Lu C., Oughtred R., Kang F., Angiuoli S. V., White O., Botstein D., Dolinski K. (2007) PloS ONE 2, e766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E. D., Sevier C. S., Ding H., Koh J. L., Toufighi K., Mostafavi S., Prinz J., St Onge R. P., VanderSluis B., Makhnevych T., Vizeacoumar F. J., Alizadeh S., Bahr S., Brost R. L., Chen Y., Cokol M., Deshpande R., Li Z., Lin Z. Y., Liang W., Marback M., Paw J., San Luis B. J., Shuteriqi E., Tong A. H., van Dyk N., Wallace I. M., Whitney J. A., Weirauch M. T., Zhong G., Zhu H., Houry W. A., Brudno M., Ragibizadeh S., Papp B., Pál C., Roth F. P., Giaever G., Nislow C., Troyanskaya O. G., Bussey H., Bader G. D., Gingras A. C., Morris Q. D., Kim P. M., Kaiser C. A., Myers C. L., Andrews B. J., Boone C. (2010) Science 327, 425–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.