Abstract
Elevated production of the matrix glycosaminoglycan hyaluronan is strongly implicated in epithelial tumor progression. Inhibition of synthesis of the hyaluronan precursor UDP-glucuronic acid (UDP-GlcUA) therefore presents an emerging target for cancer therapy. Human UDP-glucose 6-dehydrogenase (hUGDH) catalyzes, in two NAD+-dependent steps without release of intermediate aldehyde, the biosynthetic oxidation of UDP-glucose (UDP-Glc) to UDP-GlcUA. Here, we present a structural characterization of the hUGDH reaction coordinate using crystal structures of the apoenzyme and ternary complexes of the enzyme bound with UDP-Glc/NADH and UDP-GlcUA/NAD+. The quaternary structure of hUGDH is a disc-shaped trimer of homodimers whose subunits consist of two discrete α/β domains with the active site located in the interdomain cleft. Ternary complex formation is accompanied by rigid-body and restrained movement of the N-terminal NAD+ binding domain, sequestering substrate and coenzyme in their reactive positions through interdomain closure. By alternating between conformations in and out of the active site during domain motion, Tyr14, Glu161, and Glu165 participate in control of coenzyme binding and release during 2-fold oxidation. The proposed mechanism of hUGDH involves formation and breakdown of thiohemiacetal and thioester intermediates whereby Cys276 functions as the catalytic nucleophile. Stopped-flow kinetic data capture the essential deprotonation of Cys276 in the course of the first oxidation step, allowing the thiolate side chain to act as a trap of the incipient aldehyde. Because thiohemiacetal intermediate accumulates at steady state under physiological reaction conditions, hUGDH inhibition might best explore ligand binding to the NAD+ binding domain.
Keywords: Dehydrogenase, Enzyme Mechanisms, Enzyme Structure, Glycosaminoglycan, Hyaluronate, NAD, Tumor Marker, Conformational Changes and Catalysis, Covalent Intermediate, UDP-glucuronic Acid
Introduction
UDP-glucuronic acid (UDP-GlcUA)2 fulfills a range of vitally important functions in human physiology. It is a precursor for synthesis of extracellular matrix glycosaminoglycans (heparin, hyaluronan, and chondroitin sulfate), which in turn play significant roles in diverse cellular processes like signaling, wound healing, inflammation, morphogenesis, and matrix organization as well as in the pathobiology of cancer (1–6). UDP-GlcUA is the substrate for enzymatic glucuronidation, a key step in numerous detoxification pathways carried out in liver and intestine. Biosynthetic routes toward various UDP-sugars involve UDP-GlcUA as the central intermediate. UDP-GlcUA is derived from UDP-glucose (UDP-Glc) whereby UDP-glucose 6-dehydrogenase (UGDH; EC 1.1.1.22) catalyzes, in two successive NAD+-dependent steps without release of the intermediate aldehyde, the oxidation of the substrate C6 alcohol into the corresponding carboxylic acid (7–9). Elevated production of hyaluronan strongly promotes epithelial cancer progression, and it was shown that limitation of hyaluronan synthesis at the level of UDP-GlcUA has significant potential in slowing tumor growth (10–14). Moreover, UGDH was recently proposed as a novel biomarker for prostate cancer (15). Therefore, restriction of UDP-GlcUA availability in cancer cells through inhibition of human UGDH (hUGDH) presents a clear target for therapy. Design of useful antagonists for hUGDH relies on known relationships of structure and function for this enzyme, which in the absence of high resolution structural information on hUGDH are lacking in their basis. UGDH from the bacterium Streptococcus pyogenes (SpUGDH) is the only member of the large and divergent UGDH protein family (16) for which a crystal structure has so far been reported (17). However, considering the low level of sequence identity (23%) between hUGDH and SpUGDH, it would be a pointless exercise trying to infer relevant properties of the human enzyme from structure-function relationship data for the bacterial ortholog (8, 17–21). This study reports on a comprehensive structural characterization of hUGDH.
Four-electron enzymatic oxidation of UDP-Glc is complex and involves the consecutive activities of alcohol dehydrogenase and aldehyde dehydrogenase, both recruited from a single UGDH catalytic center. Combined evidence from previous mechanistic studies of bovine liver UGDH (7, 22–24) and SpUGDH (8, 20, 21) supports enzymatic reaction in three catalytic steps via thiohemiacetal and thioester intermediates, as shown in Scheme 1. The proposed mechanism necessitates that the different chemical steps of the enzymatic reaction are precisely timed with intermediate physical steps of coenzyme binding and release. A central question of UGDH catalytic function therefore is how the enzyme achieves exact orchestration of these multiple steps. We present crystal structures of hUGDH that provide useful insight. It will be shown that protein conformational changes occurring at different time scales and level of structure have key roles in promoting the overall enzymatic reaction and are directly conducive to catalysis. Results of stopped-flow kinetic experiments enabled us to analyze oxidation of substrate alcohol and aldehyde as kinetically separated reaction steps. The data provide new evidence on the role of Cys276 as catalytic nucleophile of the reaction and lead to a comprehensive proposal for the enzymatic mechanism. Moreover, using the kinetic data, we were able to locate the rate-limiting step in the reaction of hUGDH, and this provides important information to guide the development of inhibitors for the enzyme.
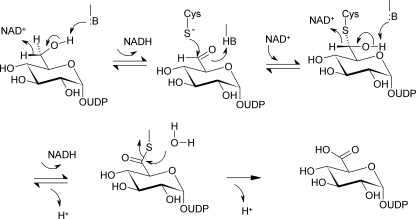
SCHEME 1.
Proposed three-step enzymatic transformation of UDP-Glc into UDP-GlcUA catalyzed by UGDH.
EXPERIMENTAL PROCEDURES
Materials
Unless stated otherwise, all materials were of highest purity available from Sigma. NAD+ was obtained from Roth (Karlsruhe, Germany) at a purity of >98%.
Expression and Purification of Recombinant Human UGDH
Recombinant hUGDH and site-directed mutants thereof were produced as C-terminally truncated forms (residues 1–467) of the native protein, which is 494 amino acids in length. Expression was performed in Escherichia coli BL21(DE3)-R3 using a pBEN-derived plasmid vector that encoded the target protein fused to an N-terminal extension, which comprised a solubility enhancement tag, a streptavidin tag, and a tobacco etch virus protease cleavage site. Enzymes were isolated from crude cell extracts using a three-step procedure consisting of affinity chromatography, gel filtration (Superdex 200 16/60 HiLoad, GE Healthcare), and anion exchange chromatography (HiTrap-Q HP, GE Healthcare). The N-terminal extension was removed prior to gel filtration using tobacco etch virus protease. Site-directed mutagenesis was performed using QuikChange site-directed mutagenesis kit (Stratagene) according to standard protocols (see supplemental material).
Crystallization
Crystals of wild-type hUGDH were grown at 4 °C in 150-nl sitting drops, equilibrated against mother liquor containing 16–20% PEG3350, 10% ethylene glycol, 0.1 m Bistris propane (pH 6.5), and 0.08–0.2 mm NaBr. The protein solution (20 mg/ml) contained 5 mm NADH, 10 mm UDP-Glc or 5 mm NAD+, 1 mm UDP-GlcUA. Crystals of apo-T131A were obtained at 20 °C using 0.1 m MES buffer (pH 6.0) that contained 20 mg/ml protein and 15% PEG “smears” (a mixture of 10 PEG polymers with molecular masses ranging from 400 to 10,000 Da).
Data Collection and Refinement
Diffraction data were collected at 100 K at Swiss Light Source station X10SA or Diamond beamline I03. They were processed with MOSFLM (25, 26) and subsequently scaled using the program SCALA (25). Structures were solved by molecular replacement using the Phaser (27) program and the structure of Caenorhabditis elegans UGDH (PDB code 2O3J) as the search model. The structures were manually rebuilt in COOT (28), and restrained refinement with appropriate TLS groups was performed using REFMAC5 (29). Data collection and refinement statistics are summarized in Table 1.
TABLE 1.
Crystallographic data collection and refinement statistics
| Complex | hUGDH-NADH-UDP-Glc | hUGDH-NAD+-UDP-GlcA | Apo-hUGDH, T131A |
|---|---|---|---|
| PDB accession code | 2q3e | 2qg4 | 3itk |
| Synchrotron beamline | SLS, X10SA | SLS, X10SA | Diamond I03 |
| Wavelength | 0.95362 Å | 1.03315 Å | 0.9763 Å |
| Space group | P21 | R3 | P212121 |
| Unit cell dimensions | a = 116.0, b = 184.1, c = 170.9 Å | a = b = 193.9, c = 352.2 Å | a = 89.1, b = 106.6, c = 349.1 Å |
| α = γ = 90.0° | α = β = 90.0° | α = β = γ = 90.0° | |
| β = 109.2° | γ = 120.0° | ||
| Resolution rangea | 50.0–2.00 Å(2.11–2.00) | 60.00–2.10 Å(2.21–2.10) | 52.90–2.40 Å(2.53–2.40) |
| No. of unique reflectionsa | 452,865 (65,636) | 288,088 (42,048) | 130,411 (18,766) |
| Completenessa | 99.6% (99.1%) | 100.0% (100.0%) | 99.8% (99.4%) |
| I/σIa | 9.7 (2.0) | 7.6 (1.8) | 7.9 (2.1) |
| Rmergea | 0.089% (0.636%) | 0.139% (0.711%) | 0.131% (0.550%) |
| Redundancya | 3.9 (3.9) | 3.9(3.8) | 4.0 (3.3) |
| Refinement | |||
| No. of atoms P/L/Ob | 35,787/800/2755 | 28,546/580/3023 | 21,656/0/1321 |
| Rfact/Rfree | 0.173/0.205% | 0.178/0.225% | 0.199/0.241% |
| r.m.s. bond lengthc | 0.010 Å | 0.010 Å | 0.016 Å |
| r.m.s. bond anglec | 1.234° | 1.238° | 1.463° |
| Bmean P/L/Ob | 33.0/25.7/35.0 Å2 | 17.0/15.1/23.9 Å2 | 15.4/-/20.6 Å2 |
a Values in parentheses show the statistics for the highest resolution shells.
b P/L/O represents protein/ligand/other (water, ion, and solvent).
c r.m.s. indicates root mean square.
Kinetic Studies
Initial rates of oxidation of UDP-Glc were recorded from the NADH produced in the reaction (25 °C), by absorbance at 340 nm (ϵNADH = 6220 m−1 cm−1). Potassium phosphate buffer (50 mm, pH 7.5) was used. The concentration of wild-type or mutated enzyme was chosen to obtain rates between about 0.001 and 0.1 ΔA/min. Reaction times were between 10 (wild-type enzyme) and 500 min (mutants). It was confirmed that rates were obtained under conditions in which the enzyme used had undergone at least 2–3 turnovers. For C276S, only transient kinetic data could be collected. Kinetic parameters (Vmax and Km) were calculated from initial rate data obtained at varied concentrations of UDP-Glc or NAD+ using a constant saturating concentration of the respective other substrate (10 mm NAD+, 1 mm UDP-Glc). Turnover numbers (kcat) were calculated by dividing ½Vmax (2 NADH produced/1 UDP-Glc converted) with the molar concentration of enzyme active sites (E) that was determined from the protein concentration obtained from absorbance (280 nm) measurements and applying a molar extinction coefficient of 48360 m−1 cm−1.
Stopped-flow kinetic experiments were carried out at 25 °C using reported procedures (30) and applying absorbance detection at 340 nm. Traces were recorded in triplicate and averaged. The limiting concentration of enzyme active sites was in the range 10–15 μm, and a solution of hUGDH saturated with UDP-Glc (1 mm) was mixed with UDP-Glc (1 mm) and NAD+ solution (0.2–20 mm). Phosphate buffer (50 mm (pH 7.5)) was used except for studies measuring formation of both NADH and proton that employed 0.5 mm Tris buffer (pH 7.5) containing 30 μm phenol red as pH indicator and 150 mm NaCl for the adjustment of ionic strength to that of 50 mm phosphate. Wild-type enzyme was doubly gel-filtered to this Tris buffer immediately prior to reaction. Proton release was measured by absorbance decrease at 556 nm and calibrated using titration with HCl as well as from an enzymatic reference having known relationship between formation of NADH and proton during the reaction (31).
Stopped-flow time courses of formation of NADH or proton were fitted, with nonlinear least squares regression analysis, using Equation 1,
where [Y] is the concentration of the measured compound; [Yburst] is the amplitude of the transient (“burst”) phase; kobs is a transient rate constant (s−1); Vss is the steady-state rate, and [Y0] corrects for absorbance at zero time. The relationship ½Vss(NADH)/E gives the apparent steady-state rate constant under the conditions used. It equals kcat when all substrates are present in a saturating concentration.
Isothermal Titration Calorimetry (ITC)
A VP-ITC titration calorimeter from Microcal was used. Protein samples were dialyzed against 10 mm Hepes (pH 7.5) containing 150 mm NaCl. Solutions containing 1–2 mg/ml protein were used. Each experiment consisted of a first 2-μl injection of ligand solution (UDP-Glc. 1 mm; NAD+, 0.5 mm) followed by 29 injections of 8 μl. Binding heats were corrected for dilution heats. Data were normalized and evaluated using ORIGIN with a single binding site model.
Electrospray-TOF-MS Analysis
An Agilent LC/MSD TOF electrospray ionization time-of-flight mass spectrometer coupled to an 1100 series HPLC from Agilent was used. Protein samples at 1 mg/ml were diluted 1:50 (v/v) in H2O containing 0.1% formic acid. Sample volumes of 10 μl were loaded onto a Zorbax 300SB-C3 column (4.6 × 500 mm, 5-μm particles) at a flow rate of 0.5 ml/min. A gradient was developed from 5 to 95% (B:A) over 6 min, where solvent A was H2O (0.1% formic acid) and solvent B was MeOH (0.1% formic acid). The mass spectrometer was operated in positive ion mode using a standard electrospray ionization source. Data analysis and spectral deconvolution were performed using the Protein/MagTrans analysis software package (Agilent).
Besides mass spectrometric characterization of enzyme preparations as isolated, the MS analysis was used to measure covalent intermediate formation in the reaction catalyzed by an hUGDH variant in which Cys276 was substituted by Ser (C276S). Incubation was done at 25 °C in 50 mm phosphate buffer (pH 7.5) using C276S (20 μm subunits), 15 mm NAD+, and 1 mm UDP-Glc. Samples were taken at certain times for up to 24 h and analyzed. Formation of NADH in the reaction was measured spectrophotometrically.
HPLC Product Analysis
Reactions were performed at 25 °C in 50 mm phosphate buffer (pH 7.5) using 15 mm NAD+ and 1 mm UDP-Glc. The enzyme concentration was varied as required to observe significant conversion in about 12 h that was monitored spectrophotometrically. Addition of acetonitrile in a 1:1 volume ratio to buffer was used to stop the reactions. Precipitated protein was removed by centrifugation (10 min; 13,000 rpm). The sample was analyzed using an Agilent 1200 HPLC system equipped with a 5-μm ZORBAX SAX analytical HPLC column (4.6 × 250 mm; Agilent). A linear gradient between buffer A (5 mm potassium phosphate (pH 3.2)) and 60% buffer B (600 mm potassium phosphate (pH 3.2)) over 20 min was used. This was followed by 100% B and 100% A, for each 20 min to re-equilibrate the column. The flow rate was 1.5 ml/min.
Differential Scanning Fluorometry
Thermal stability shift assays were performed to examine protein stabilization upon substrate binding. Experiments were carried out using an Agilent Mx3005p RT-PCR machine according to a protocol reported elsewhere (32). The protein concentration was 1 μm in 10 mm Hepes (pH 7.5) containing 150 mm NaCl. UDP-Glc, UDP-GlcUA, NAD+, and/or NADH were added at a concentration of 1 mm. SYPRO Orange (Invitrogen) was present as fluorescence probe in a 1:1000 dilution. Excitation and emission filters for SYPRO Orange were at 465 and 590 nm, respectively. Reactions were carried out in 96-well format using 20 μl of total reaction volume each. Temperature was raised at 1 °C/min from 25 to 96 °C, and fluorescence intensity was measured every 1–3 °C. The Boltzmann sigmoid function was used for data fitting.
Other Analytical Methods
Dynamic light scattering and gel filtration analyses are described in the supplemental material.
RESULTS AND DISCUSSION
Overall Structure of the Functional hUGDH Hexamer
Crystal structures of a truncated form of hUGDH (residues 1–467) were determined (Table 1) because crystallization of the full-length enzyme (residues 1–494) had failed. However, the C-terminal deletion did not affect the enzyme activity, as demonstrated in initial rate assays using purified hUGDH preparations (Table 2). Fig. 1A depicts the tertiary fold of the enzyme protomer with UDP-Glc and NADH bound (PDB code 2Q3E). Structural superimposition of hUGDH and SpUGDH subunits reveals conservation of the overall fold (root mean square deviation of 1.5 Å for 351 equivalent Cα atoms; Fig. 1B). However, clear differences among the two enzymes are observed at the C-terminal domain wherein the shorter bacterial UGDH (residue 1–402), a relatively flexible tail, packs on top of the substrate binding pocket (Fig. 1B). Fig. 1C shows assembly of the hUGDH subunits into a dimer, which serves as basic oligomerization unit in formation of the functional enzyme hexamer. The overall structure of the hexamer is depicted in Fig. 1D. It was confirmed by using in-solution methods (analytical gel filtration and dynamic light scattering) that wild-type hUGDH and all enzyme variants to be described later fold into functional hexamers.
TABLE 2.
Kinetic parameters for hUGDH and mutants thereof determined at 25 °C and pH 7.5
Initial rates were obtained in triplicate measurements. Averaged data (relative S.D. ≤10%) were used for determination of kinetic parameters.
| Enzyme | kcat,app | -Fold decrease | KmUDP-Glc | KmNAD+ |
|---|---|---|---|---|
| s−1 | μm | mm | ||
| Wild type | 0.85 ± 0.1 | 35 ± 5 | 0.7 ± 0.1 | |
| Wild type, full length | 0.75 ± 0.1 | 1 | 35 ± 5 | 1.0 ± 0.2 |
| C276A | 8.7 ± 1.5 × 10−5 | 1.8 × 104 | 80 ± 12 | 1.3 ± 0.2 |
| C276Sa | 1.9 ± 0.3 × 10−4 | NA | NA | NA |
| T131A | 0.10 ± 0.02 | 7 | 100 ± 15 | 0.8 ± 0.1 |
a Transient kinetic data collected prior to formation of a stable ester intermediate.
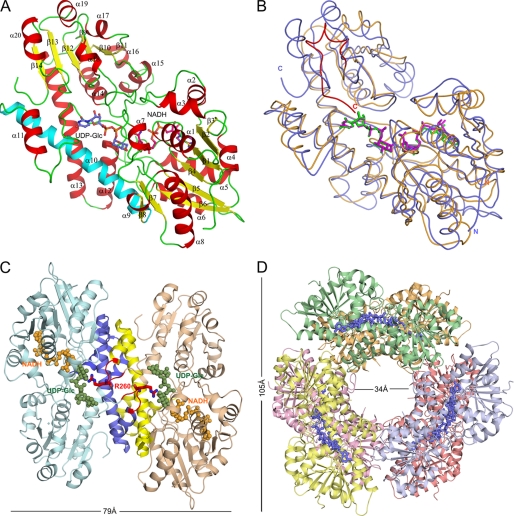
FIGURE 1.
Overall structure of hUGDH bound with UDP-Glc and NADH. A, protein subunit with α-helices and β-strands numbered in red and yellow, respectively. The central interdomain α10 helix is shown in cyan. B, structural superimposition of the subunits of hUGDH (PDB code 2Q3E) (blue) and SpUGDH (PDB code 1DLJ) (orange). The flexible C-terminal region of SpUGDH is shown in red. C, assembly of hUGDH subunits into dimers. Regions forming dimer contacts are shown in blue and yellow. Arg260, which extends into the adjacent substrate binding pocket, is shown in red. D, formation of the functional hUGDH hexamer.
The subunit of hUGDH consists of two domains, each of which adopts a similar α/β fold whereby a six-stranded parallel β-sheet core is flanked by α-helices on both sides (Fig. 1A). The domain interface is located in the central region of the subunit and consists mainly of interdomain helix α10 (residues 213–244), which bridges the last β-strand (β8) of the N-terminal domain and the first helix of a three-helical bundle (α11-α13) of the C-terminal domain.
hUGDH forms a disc-shaped double-layer hexameric structure assembled from a trimer of dimers. Each dimer pair is related by a 3-fold symmetry axis running through a central channel, perpendicular to the view in Fig. 1D. The dimer interface is located adjacent to the UDP-Glc binding pocket and is created through “head-to-tail” arrangement of the individual subunits approaching each other in a “back-to-back” orientation (Fig. 1C). Dimer contacts mainly in the form of hydrophobic interactions and hydrogen bond networks are provided by the interdomain helix as well as by the helical bundle α11–α13. Despite global similarity, dimerization in hUGDH and SpUGDH is markedly different due to sequence variation (supplemental Fig. S1). For example, the hydrogen bond network in the central region of the dimer interface appears to be substantially stronger in hUGDH, where Ser233, Tyr299, and Gln229 substitute for the corresponding residues (Tyr217, Leu279, and Leu213) in SpUGDH, respectively.
Intersubunit contacts between adjacent dimer pairs forming the hUGDH hexamer are distinct from the ones involved in dimerization (supplemental Fig. S1). They exploit a completely different oligomeric interface, which has ∼48% less contact area than the dimer interface (∼2673 Å2). The loop connecting β4 and α5 (residues 88–105) in the N-terminal domain of subunit A is located close to the central channel and extends to the C-terminal edge of subunit C where it forms intersubunit contacts with the loop between α13 and β9 (residues 322–330). Dimer-dimer interactions include hydrogen bonds of Lys94(A) with Glu360(C) and Asn324(C), Tyr96(A) with Tyr327(C), Met98(A) with Glu360(C), and Asp105(A) with Thr325(C). Further stabilization of the hexamer is derived from a salt bridge between Glu110(A) and Lys329(C) as well as from a hydrogen bond network involving Arg114(A), Lys434(C), and Asn147(A). Site-directed mutagenesis data were previously interpreted to support a critical role of Ala222, Ser233, and Lys279 in hUGDH hexameric assembly (9, 33, 34). However, because of their positions in the structure remote from the relevant interface, stabilization of the enzyme hexamer by these residues must be indirect. The importance of Lys279 is plausibly linked to structural stabilization of the β4-α5 loop and to formation of the NAD+ binding pocket. Stabilization by Ala222 and Ser233 would seem to facilitate stability of the hUGDH dimer, prior to formation of the “trimer of dimers.” Structural and sequence comparisons reveal that interactions of residues leading to higher order oligomerization in hUGDH, which are mainly conserved in enzymes from higher phylogenetic lineages, are completely absent in dimeric enzymes from bacteria (supplemental Fig. S1) (16).
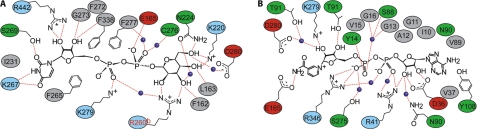
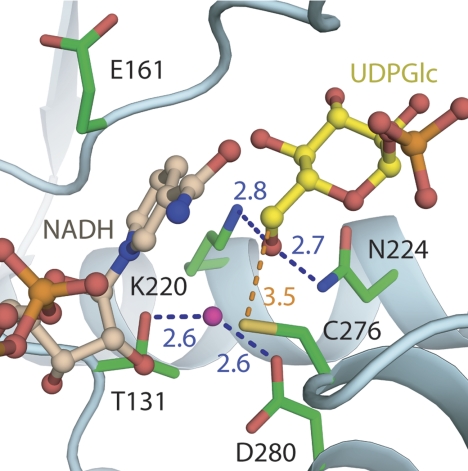
The structures of ternary complexes of hUGDH bound with UDP-Glc/NADH and UDP-GlcUA/NAD+ (PDB code 2QG4) reveal that NAD+ (Fig. 2A) is accommodated by the N-terminal domain in a cleft between α1 and α4 (Fig. 2B). Asp36 is the determinant of coenzyme specificity of hUGDH, which utilizes NAD+ but not NADP+. The binding pocket for UDP-Glc is built from amino acids of both domains whereby anchoring of substrate is achieved primarily by residues from the C-terminal domain (UMP) and assistance from some N-terminal domain residues interacting with the α-glucose 1-phosphate portion of the substrate. The active site of hUGDH is situated in a cleft at the domain interface and contains six highly conserved residues (Fig. 3): Thr131, Glu161, Lys220, Asn224, Cys276, and Asp280. Cys276 is the clear candidate catalytic nucleophile of the reaction. A water molecule forming hydrogen bonds (∼2.6 Å) to Asp280 and Thr131 is conserved in all ternary complex hUGDH structures described (Table 1). Its position in the active site is compatible with different functions in catalysis, as will be discussed later. However, the water also forms a hydrogen bond with the C2 ribose hydroxyl of NAD+ (2.9 Å), arguably facilitating the association of the reactive groups of substrate and coenzyme in the active site. Contribution of the loop connecting helices α11 and α12 (residues 258–275) to subunit-subunit interactions (Fig. 1C) positions Arg260 in the UDP-Glc binding pocket of the adjacent subunit. The guanidinium group of Arg260 forms a bidentate hydrogen bond with glucopyranosyl hydroxy groups at C2 and C3 of the substrate (Fig. 2B). This indicates a possible communication between the two active sites during binding of UDP-Glc and strongly supports a role for dimerization in creating a functional hUGDH enzyme.
FIGURE 2.
hUGDH binding pockets for coenzyme (A) and UDP-Glc (B). Residue coloring is as follows: red, acidic; blue, basic; gray, nonpolar; green, uncharged polar. Water molecules are shown as blue spheres, and hydrogen bonds are shown as dashed red lines.
FIGURE 3.
Close-up view of the active site of wild-type hUGDH bound with UDP-Glc and coenzyme. Distances are in Å. The distance between the water bonded to Asp280 and the sulfur of Cys276 is 3.2 Å.
Protein Conformational Changes That Accompany Binding of Substrate and Coenzyme
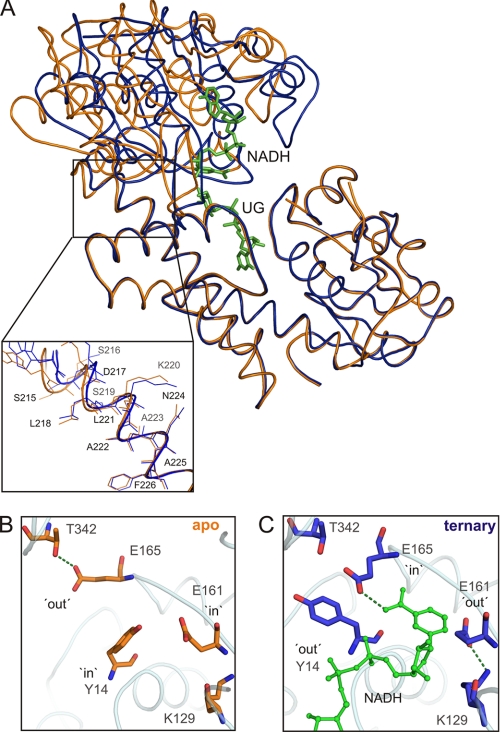
Crystallization of hUGDH in apo-form failed, but a relevant structure for the T131A mutant was obtained (PDB code 3ITK). T131A displays activity within 1 order of magnitude of that of wild-type enzyme (Table 2). Structural overlay of apo-T131A and wild-type hUGDH bound with UDP-Glc and NADH (Fig. 4A) indicates that differences among the two protein structures are best emphasized on angular variation between the N- and C-terminal domain. With the assumption that the C-terminal domain remains in place (Fig. 4A), the Rossmann-fold domain exhibits an ∼13° rotation toward the immobile region of the subunit when UDP-Glc and NADH are anchored to the enzyme. This “domain-closure” conformational change of hUGDH was further analyzed using DYNDOM (35). It is shown to result from a combination of rigid body and restrained movements of the N-terminal domain and involves Leu218–Leu227 from the α10 helix as hinge region (Fig. 4A, inset). Its immediate consequence for the catalytic mechanism of hUGDH is that substrate and coenzyme are sequestered in their reactive positions, and the active site is closed up. However, there is the additional implication that exchange of NADH by NAD+ in the course of the two-step oxidation requires intermittent domain opening. Superimposition of the two structures in Fig. 4A provides strong support in favor of the notion that NAD(H) exchange steps in the catalytic cycle of hUGDH are coupled to “opening” and “closing” motions of the N-terminal domain.
FIGURE 4.
Analysis of the domain closure conformational change of hUGDH induced by ligand binding. A, global structural rearrangements. Structural superimposition of apo-T131A and wild-type hUGDH bound with UDP-Glc and NADH. The inset shows the “hinge” region involved in the motion of the N-terminal domain. B, C, conformational switches of residues in immediate vicinity of the active site in response to closing of the N-terminal domain in the ternary complex as compared with apo-T131A.
The structural trajectory from the “open” to the “closed” conformation of hUGDH is also traceable as a change of residue occupancy within the coenzyme-binding site whereby Tyr14 and Glu161 alternate with Glu165 between conformations inside and outside of the binding pocket (Fig. 4B). Interactions with Thr342 and Lys129 stabilize Glu165 and Glu161 in their respective “out” conformations. Bonding of Glu165 with the carboxamide of NAD+ brings the si face of the nicotinamide ring into a suitable position for pro-S stereospecific hydride transfer to its C4 atom. Therefore, this implies that global structural rearrangements of hUGDH in response to substrate/coenzyme binding (Fig. 4A) are precisely relayed into the active site where they will be directly conducive to stereospecific catalysis (Fig. 4B). The “in” conformation of Glu161 would generate a steric conflict with the nicotinamide ring of NAD+, suggesting that out → in movement of Glu161 could serve as trigger for release of NADH at the steps following the formation of thiohemiacetal and thioester intermediates. Tyr14, Glu161, and Glu165 are conserved across UGDH primary structures (supplemental Fig. S1), but their roles were previously not known.
Biochemical Characterization of Substrate/Coenzyme Binding to hUGDH
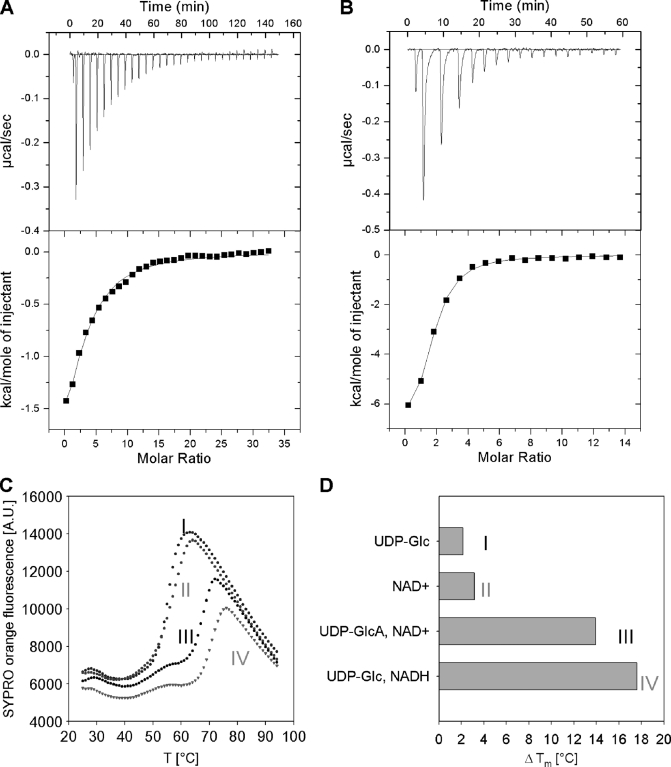
Using ITC we showed that UDP-Glc binds to wild-type apoenzyme with a dissociation constant (Kd) of 17 ± 2 μm and a large favorable free energy of −6.2 kcal/mol (ΔH = −3.3 ± 0.5 kcal/mol; TΔS = 2.9 kcal/mol) (Fig. 5A). The observed Kd is in close agreement with the kinetically determined Michaelis constant for UDP-Glc (Table 2). Addition of NAD+ to apoenzyme failed to produce a heat signal under otherwise identical conditions, suggesting that NAD+ does not bind to hUGDH unless the enzyme-substrate complex has formed. Ordered binding of substrate, UDP-Glc before NAD+, was kinetically determined for liver UGDH (22) and SpUGDH (18) and is consistent with our observations for the human enzyme. We employed a C276A mutant of hUGDH that showed negligible activity toward substrate conversion in the time span of the ITC experiment, to determine a Kd of 1.6 ± 0.3 μm for binding of NAD+ to the complex of enzyme and UDP-Glc (ΔG = −7.5 kcal/mol; ΔH = −8.6 ± 0.5 kcal/mol; TΔS = −1.1 kcal/mol) (Fig. 5B). Comparison of this Kd with the Km of 1.3 mm (Table 2) reveals that the affinity of C276A for binding NAD+ in the first round of the 2-fold oxidation (∼1/Kd) was about 440-fold larger than the kinetically determined apparent affinity for NAD+ (∼1/Km).
FIGURE 5.
Ligand binding to hUGDH measured by ITC (A and B) and differential scanning fluorometry (C and D). ITC data for binding of UDP-Glc (1 mm) to wild-type hUGDH (33 μm) (A) and binding of NAD+ (0.5 mm) to a complex of a C276A mutant (22 μm) and UDP-Glc (1 mm) (B). C, differential scanning fluorometry with hUGDH (1 μm) in the presence of 1 mm each UDP-Glc (I), NAD+ (II), UDP-GlcUA and NAD+ (III), as well as UDP-Glc and NADH (IV). SYPRO Orange fluorescence is used as reporter of enzyme denaturation. D, difference (ΔTm) in apparent melting temperature (Tm) for enzyme incubated in the presence and absence of the added compounds. The Tm of hUGDH in buffer is 50 °C. The Tm was calculated by fitting a Boltzmann sigmoid to the data. Measurements were done in triplicate, and standard errors on all parameters were ≤15%.
Data from differential scanning fluorometry provided further insight into hUGDH conformational changes induced by ligand binding. Substrate (or product) and coenzyme acted in synergy toward enhancing the apparent melting temperature (Tm) of apoenzyme (Tm ≈50 °C), the observed increase in Tm being as large as 18 ± 1 and 14 ± 1 °C when either UDP-Glc/NADH or UDP-GlcUA/NAD+ was added to the enzyme solution (Fig. 5, C and D). Considering that the apo-hexamer has ∼12% more exposed surface area than the average surface area of the also hexameric ternary complex (103,170 versus 91880 Å2), it is plausible that the crystallographically observed conformational rearrangement of hUGDH upon ternary complex formation is due to binding of NAD+ to enzyme/UDP-Glc, and the resulting compaction of the active site confers this extra stability to the protein structure.
Structural Comparison of Substrate and Product Complexes
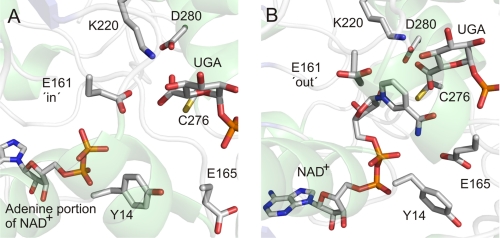
We obtained a crystal structure of a nonproductive ternary complex in which wild-type enzyme was bound with UDP-GlcUA and NAD+ (PDB code 2QG4; Table 1). The overall structure of this complex is highly similar to the above described structure of enzyme bound with UDP-Glc and NADH. All subunits of the hUGDH hexamer have their two domains in the closed conformation. Interestingly, half of the subunits contain Glu161 adopting the in conformation, resulting in a displacement of the nicotinamide ring of coenzyme from the active site (Fig. 6A). The nicotinamide-riboside portion of NAD+ is disordered in these subunits, and only the ADP moiety is seen bound to the enzyme. The remainder of the subunits feature the alternative out conformation for Glu161 and have an ordered nicotinamide moiety on NAD+ accommodated in their active sites (Fig. 6B). Conformational flexibility of Glu161 distinguishes the product complex structure from the structure of the substrate complex where Glu161 was found exclusively in the out conformation.
FIGURE 6.
Close-up views of two active-site conformations seen in the product complex structure of hUGDH (PDB code 2Q3E). See text for further discussion of the role of conformational flexibility of Glu161.
Kinetic Dissection of Catalytic Steps during 2-Fold Oxidation of UDP-Glc
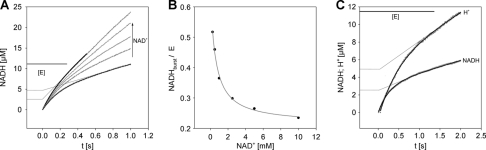
Stopped-flow experiments were performed with the aim of analyzing oxidation of substrate alcohol and aldehyde as kinetically separated reaction steps. Time courses of UDP-Glc conversion by wild-type hUGDH (Fig. 7A) showed a weak transient “burst” of NADH formation that was followed by the linear steady-state phase. From fits of the data with Equation 1, we determined the concentration of NADH produced in the burst phase ([Yburst]) and found that [Yburst] decreased as the concentration of NAD+ was raised (Fig. 7B). This result implies that there is a slow step in the enzymatic reaction that occurs after the formation of NADH, and the contribution of this step to overall rate limitation decreases at high NAD+ concentration. The pre-steady-state rate constant of NADH formation (kobs) increased only 2.5-fold, to a limiting value of about 10 s−1, in response to an increase in NAD+ concentration from 0.1 to 10 mm, although there was a 10-fold change in the steady-state rate constant (Vss/E) within the same NAD+ range. Scheme 1 shows that oxidation of thiohemiacetal and hydrolysis of thioester succeed the formation of NADH in the enzymatic mechanism so that both must be considered as candidate rate-limiting steps. However, thioester hydrolysis lacks NAD+ dependence, and its participation in rate limitation would therefore be completely inconsistent with observations in Fig. 7B. Oxidation of thiohemiacetal to thioester, by contrast, depends on NAD+ and proceeds only after the NADH formed in the first oxidation step has been replaced by new NAD+. The overall process of conversion of thiohemiacetal to thioester is thus strongly suggested to be chiefly rate-determining in the reaction of hUGDH at the steady state. At physiological reaction conditions where the level of UDP-Glc (155 μm) (36) is saturating and that of NAD+ (∼0.36 mm) (37, 38) is limiting (see Km values in Table 2), reaction of hUGDH would therefore be governed by coenzyme binding in this second oxidation step.
FIGURE 7.
Transient kinetic analysis of UDP-Glc oxidation by wild-type hUGDH. A, stopped-flow progress curves for reactions at varied NAD+ concentration in the range 0.25–10 mm. The concentration of UDP-Glc was 1 mm. Gray lines show data and black lines are fits with Equation 1. The pre-steady-state burst is indicated by extrapolation. E is the concentration of hUGDH subunits. B, dependence of the pre-steady-state burst on the concentration of NAD+ used. C, formation of NADH and release of proton during oxidation of UDP-Glc (1.0 mm) by NAD+ (0.1 mm). Black lines are fits of data with Equation 1.
The enzymatic reaction of hUGDH, UDP-Glc + 2 NAD+ + H2O → UDP-GlcUA + 2 NADH + 2 H+, implies, that two protons are released as result of substrate oxidation. At neutral pH, a third proton will appear in solution because of ionization of UDP-GlcUA. A molar ratio of 2:3 for NADH and proton formed at the steady state was confirmed. Because the relative timing of the transfer of hydride and proton in the kinetic transient potentially contains important mechanistic information, we performed the stopped-flow experiments depicted in Fig. 7C and found that in the burst phase the change in total proton concentration was twice (1.9 ± 0.1) the molar equivalent of NADH formed. In other words, two protons were released prior to the rate-limiting step.
Kinetic Consequences of Mutation of Cys276
Considering the insight from prior studies of SpUGDH (8, 18) and UGDH from bovine liver (7), the most probable function of the conserved Cys276 in the active site of hUGDH was that of the catalytic nucleophile of the enzymatic reaction, participating in formation of thiohemiacetal and thioester intermediates. Tanner and co-workers (8) mutated Cys260 of SpUGDH into a Ser and demonstrated accumulation of a covalent ester adduct on Ser260 upon incubation of the mutant in the presence of UDP-Glc and NAD+. Cys276 of hUGDH is homologous to Cys260 of SpUGDH both in sequence and three-dimensional protein structure. Simpson and co-workers (9) prepared a C276S mutant of hUGDH and showed that it performed only a single round of oxidation of UDP-Glc. However, other significant characteristics of the reaction of C276S have not been resolved, including the identity of the product formed and the possible intermediacy of a hemiacetal enzyme on Ser276.
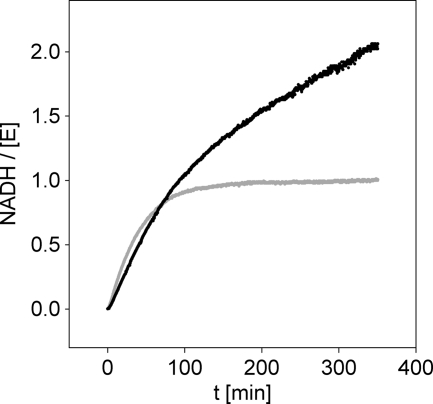
We therefore substituted Cys276 by Ala and Ser and performed a kinetic characterization of the purified mutants. C276S was hardly active under steady-state assay conditions (≥10,000-fold lower activity than wild-type enzyme). The time course of NADH formation by C276S displayed an initial release of product in amounts corresponding to precisely 1 m eq of the enzyme used, followed by the very slow steady-state phase of reaction (Fig. 8). Electrospray ionization-MS analysis of a sample taken from the reaction mixture at steady state (after 200 min of incubation) revealed accumulation of a macromolecular species displaying the mass increase (experimental Δmass, +562 ± 2 g/mol; calculated Δmass, +564 g/mol for hemiacetal; +562 g/mol for ester) relative to apoenzyme that would be expected if a covalent hemiacetal/ester adduct of C276S was formed (supplemental Fig. S2). Deconvolution of the MS data showed two peaks of comparable size, one corresponding to native (i.e. unmodified) C276S and another corresponding to the enzyme intermediate (supplemental Fig. S2). Transient release of 1 NADH per equivalent C276S would thus be best consistent with a long lived acyl enzyme intermediate produced in two successive steps of oxidation. The kinetic behavior of C276S would thus be comparable with that of the analogous SpUGDH mutant (8).
FIGURE 8.
Time course analysis for reactions of Cys276 mutants of hUGDH. The gray line represents C276S, and the black line C276A. The concentrations of UDP-Glc and NAD+ were 1 and 15 mm, respectively. The molar enzyme concentration was 100 μm.
Contrary to C276S, the C276A mutant converted UDP-Glc to UDP-GlcUA in the absence of a transient burst of formation of NADH (Fig. 8). Using HPLC analysis of samples taken at certain times, we showed within error limit (±20%) that for each pair of NADH produced, one UDP-Glc was converted into UDP-GlcUA. This result suggests that complete removal of the catalytic nucleophile from the enzyme, as in C276A but not in C276S, does not specifically disrupt the second step of 2-fold oxidation of UDP-Glc as one might expect for a reaction in which formation and covalent trapping of the intermediate aldehyde are kinetically uncoupled one from another.
Proposed Catalytic Mechanism of hUGDH and Its Implication for Enzyme Inhibition
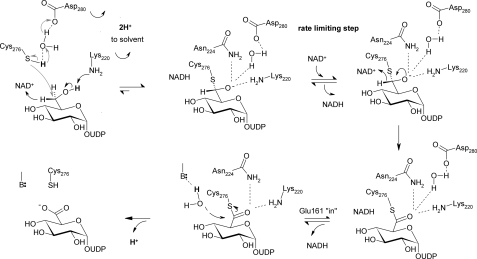
Based on evidence from crystal structures, site-directed mutagenesis, and kinetic data, we suggest a catalytic mechanism for hUGDH that is summarized in Scheme 2. In this mechanism, the different steps of chemical catalysis are linked to associated protein conformational changes in and out of the active site that are required to promote the reaction. It is recognized that domain closing/opening conformational changes are crucial for coenzyme binding and release in each step of 2-fold oxidation. The active-site conformational dynamics accompanying the large scale domain motion facilitates in precise positioning (Glu165 in and Glu161 out) or displacement (Glu165 out and Glu161 in) of the nicotinamide moiety of NAD+ and NADH, respectively. They are therefore directly conducive to stereospecific enzymatic catalysis.
SCHEME 2.
Proposed catalytic mechanism of hUGDH. See text for further explanations. Conversion of alcohol to (anionic) thiohemiacetal is thought to proceed through hydride transfer oxidation (black arrow) kinetically coupled to nucleophilic attack from deprotonated Cys276 (gray arrows). The indicated roles of Lys220 and water-bonded to Asp280 are tentative. Glu161 is the favored candidate catalytic base facilitating attack of water during thioester hydrolysis.
A central feature of the mechanistic proposal is that nucleophilic attack from (anionic) Cys276 takes place in a strongly coupled fashion with hydride abstraction from alcohol. Kinetic data showing that two protons are released to bulk solvent in the course of the first oxidation step (Fig. 7) are consistent with coupling of chemical reaction and “activation” of Cys276 through deprotonation. Hydride transfer from substrate probably involves a coupled motion from the second hydrogen of the C6 primary alcohol. With steric conflict to orbital overlap removed by this motion, attack of the thiolate side chain of Cys276 on carbon could start immediately, resulting in trapping of the incipient aldehyde. The proposed action of Cys276 is consistent with the absence of intermediate aldehyde release in the course of reaction of the wild-type enzyme. It is likewise fully consistent with the notion of Tanner and co-workers (8, 21) studying a C260A mutant of SpUGDH that the catalytic Cys could have a role in making the first oxidation step thermodynamically more favorable. Also note that the proposed coupling between hydride transfer oxidation and nucleophilic attack does not rule out that under certain assay conditions chemical (e.g. reductive) trapping of aldehyde can be successful, as was shown in early studies of UGDH, employing a chemically modified form of the bovine liver enzyme that had its active-site cysteine derivatized by cyanide (39). The thiohemiacetal formed in the reaction of hUGDH is suggested to be an oxyanion that could recruit suitable stabilization by strong hydrogen bonding interactions with Lys220, Asn224, and water coordinated by Asp280 and Thr131.
Catalytic assistance to generation of the thiohemiacetal intermediate will be provided by the side chains of Lys220 and active-site water bonded to Asp280 (Fig. 3). Of the two residues, water appears to be ideally positioned to pull off the proton from the thiol side chain of Cys276. A role of Lys220 as a catalytic base for the alcohol dehydrogenase step of the hUGDH reaction would be consistent with the proposed analogous function of the homologous Lys in the active site of 6-phosphogluconate dehydrogenase, one of the closest structural neighbors of UGDH enzymes (40, 41). Following proton abstraction from substrate, deprotonation of Lys220 to solvent would regenerate the uncharged ϵ-amino side chain, which would then be ideally primed for function, together with the carboxamide side chain of Asn224 and water bonded to Asp280, as oxyanion stabilizer in the following steps of the catalytic cycle. A role for Lys220 in “stabilizing anionic transition states” was also proposed by Simpson and co-workers (42).
Reloading the active site with NAD+ for the second oxidation to occur requires a full round of domain opening and closing. In the presence of physiological concentrations of NAD+, this will be the slowest step of the overall reaction. At steady state under these conditions, a substantial fraction of the enzyme will be present as a thiohemiacetal intermediate. Search for selective inhibitors of hUGDH would thus best explore ligand binding to the N-terminal coenzyme binding domain of the covalently modified enzyme.
NAD+-dependent conversion of thiohemiacetal to thioester that is presumably followed by release of NADH from the active site, hydrolysis of thioester, and UDP-GlcUA product dissociation complete the reaction of hUGDH. Now, irrespective of whether NADH is actually dissociated from hUGDH at the time of the hydrolysis step (Fig. 6), we believe, in agreement with interpretation of structural evidence for SpUGDH (17), that the presence of Glu161 in the active site is important for breakdown of the thioester. The preferred mechanism of two possible mechanisms therefore is that Glu161 provides catalytic assistance to the attack of substrate water (see Fig. 6A, showing Glu161 in the in conformation). An alternative possibility of hUGDH is that the ever present active-site water functions as the nucleophile, and Asp280 would be well positioned for providing base catalytic assistance to its reaction. There is a chain of water molecules connecting active-site water to bulk solvent, providing a plausible route through which the catalytic site could recruit a fresh water molecule post-hydrolysis. Glu161 is too far away from nucleophilic water to fulfill a catalytic role in this mechanism, but its presence could generate a favorable electrostatic environment for hydrolysis to proceed. To distinguish between the possible catalytic scenarios for thioester hydrolysis by hUGDH, it is also relevant to consider mechanistic proposals for aldehyde dehydrogenase enzymes that display an active-site organization highly similar to that of hUGDH, including the well positioned active-site water (42). A “mobile” Glu that resembles Glu161 of hUGDH was implicated in base catalysis to thioester hydrolysis in aldehyde dehydrogenases (43–45). In the proposed mechanism of hUGDH, UDP-GlcUA is the last product to dissociate from the enzyme, completing the catalytic cycle.
In summary, this paper presents a comprehensive elucidation of structure-function relationships of hUGDH and describes the first structural analysis of the reaction coordinate for any member of this class of enzymes. Considering hUGDH as a potential drug target for cancer therapy, the results may be important not only because they provide the requisite molecular basis for inhibitor development but also because they identify the coenzyme-binding site in thiohemiacetal intermediate to be potentially useful for reversible inhibition of the enzyme in vivo. These findings together with literature precedent (46–49) showing selective inhibition of different dehydrogenases by compounds directed toward their coenzyme binding sites might be the useful points of departure for designing hUGDH antagonists.
Supplementary Material
Acknowledgments
We thank Dr. Kunde Guo (Structural Genomics Consortium) for contributing to protein crystallization (PDB codes 2Q3E and 2QG4) and for cloning of the hUGDH mutants. Dr. Mario Klimacek (Graz University of Technology), Dr. Panagis Filippakopoulos (Structural Genomics Consortium), Dr. Ivan Alfano (Structural Genomics Consortium), and Prof. Stefan Knapp (Structural Genomics Consortium) are gratefully acknowledged for helpful discussions and suggestions. We thank Dr. Frank von Delft (Structural Genomics Consortium) for data collection at the Swiss Light Source.
This work was supported by the Austrian Science Fund DK Molecular Enzymology W901-B05 and the Structural Genomics Consortium registered charity (number 1097737) funded by the Wellcome Trust, GlaxoSmithKline, Genome Canada, the Canadian Institutes of Health Research, the Ontario Innovation Trust, the Ontario Research and Development Challenge Fund, the Canadian Foundation for Innovation, Vinnova, the Swedish Strategic Research Foundation, the Knut and Alice Wallenberg Foundation, and the Karolinska Institutet. This work was additionally supported by the NIHR Oxford Biomedical Research Unit.

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods,” Figs. S1 and S2, and additional references.
- UDP-GlcUA
- UDP-glucuronic acid
- PDB
- Protein Data Bank
- h
- human
- UGDH
- UDP-glucose 6-dehydrogenase
- Bistris propane
- 1,3-bis[tris(hydroxymethyl)methylamino]propane
- ITC
- isothermal titration calorimetry
- UDP-Glc
- UDP-glucose.
REFERENCES
- 1. Bharadwaj A. G., Rector K., Simpson M. A. (2007) J. Biol. Chem. 282, 20561–20572 [DOI] [PubMed] [Google Scholar]
- 2. Lin S. L., Chang D., Ying S. Y. (2007) Carcinogenesis 28, 310–320 [DOI] [PubMed] [Google Scholar]
- 3. Slomiany M. G., Toole B. P. (2009) in Hyaluronan in Cancer Biology (Stern R. ed) pp. 19–35, Academic Press, San Diego [Google Scholar]
- 4. Tammi R. H., Kultti A., Kosma V. M., Pirinen R., Auvinen P., Tammi M. I. (2008) Semin. Cancer Biol. 18, 288–295 [DOI] [PubMed] [Google Scholar]
- 5. Toole B. P., Slomiany M. G. (2008) Semin. Cancer Biol. 18, 244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Bertozzi C. R., Hart G. W., Etzler M. E. (2009) Essentials of Glycobiology, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 7. Feingold D. S., Franzen J. S. (1981) Trends Biochem. Sci. 6, 103–105 [Google Scholar]
- 8. Ge X., Campbell R. E., van de Rijn I., Tanner M. E. (1998) J. Am. Chem. Soc. 120, 6613–6614 [Google Scholar]
- 9. Sommer B. J., Barycki J. J., Simpson M. A. (2004) J. Biol. Chem. 279, 23590–23596 [DOI] [PubMed] [Google Scholar]
- 10. Auvinen P., Tammi R., Parkkinen J., Tammi M., Agren U., Johansson R., Hirvikoski P., Eskelinen M., Kosma V. M. (2000) Am. J. Pathol. 156, 529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huh J. W., Choi M. M., Yang S. J., Yoon S. Y., Choi S. Y., Cho S. W. (2005) Biotechnol. Lett. 27, 1229–1232 [DOI] [PubMed] [Google Scholar]
- 12. Simpson M. A., Wilson C. M., McCarthy J. B. (2002) Am. J. Pathol. 161, 849–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang T. P., Pan Y. R., Fu C. Y., Chang H. Y. (2010) Exp. Cell Res. 316, 2893–2902 [DOI] [PubMed] [Google Scholar]
- 14. Wei Q., Galbenus R., Raza A., Cerny R. L., Simpson M. A. (2009) Cancer Res. 69, 2332–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang D., Casale G. P., Tian J., Lele S. M., Pisarev V. M., Simpson M. A., Hemstreet G. P., 3rd (2010) Int. J. Cancer 126, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Egger S., Chaikuad A., Kavanagh K. L., Oppermann U., Nidetzky B. (2010) Biochem. Soc. Trans. 38, 1378–1385 [DOI] [PubMed] [Google Scholar]
- 17. Campbell R. E., Mosimann S. C., van De Rijn I., Tanner M. E., Strynadka N. C. (2000) Biochemistry 39, 7012–7023 [PubMed] [Google Scholar]
- 18. Campbell R. E., Sala R. F., van de Rijn I., Tanner M. E. (1997) J. Biol. Chem. 272, 3416–3422 [DOI] [PubMed] [Google Scholar]
- 19. Campbell R. E., Tanner M. E. (1997) Angew. Chem. Int. Ed. Engl. 36, 3 [Google Scholar]
- 20. Campbell R. E., Tanner M. E. (1999) J. Org. Chem. 64, 9487–9492 [Google Scholar]
- 21. Ge X., Penney L. C., van de Rijn I., Tanner M. E. (2004) Eur. J. Biochem. 271, 14–22 [DOI] [PubMed] [Google Scholar]
- 22. Ordman A. B., Kirkwood S. (1977) Biochim. Biophys. Acta 481, 25–32 [DOI] [PubMed] [Google Scholar]
- 23. Schiller J. G., Bowser A. M., Feingold D. S. (1972) Carbohydr. Res. 21, 249–253 [DOI] [PubMed] [Google Scholar]
- 24. Strominger J. L., Kalckar H. M., Axelrod J., Maxwell E. S. (1954) J. Am. Chem. Soc. 76, 6411–6412 [Google Scholar]
- 25. Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 15299374 [Google Scholar]
- 26. Leslie A. G. (1999) Acta Crystallogr. D Biol. Crystallogr. 55, 1696–1702 [DOI] [PubMed] [Google Scholar]
- 27. McCoy A. J. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 29. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30. Klimacek M., Nidetzky B. (2010) Biochem. J. 425, 455–463 [DOI] [PubMed] [Google Scholar]
- 31. Pival S. L., Klimacek M., Nidetzky B. (2009) Biochem. J. 421, 43–49 [DOI] [PubMed] [Google Scholar]
- 32. Niesen F. H., Berglund H., Vedadi M. (2007) Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 33. Huh J. W., Yang S. J., Hwang E. Y., Choi M. M., Lee H. J., Kim E. A., Choi S. Y., Choi J., Hong H. N., Cho S. W. (2007) J. Biochem. Mol. Biol. 40, 690–696 [DOI] [PubMed] [Google Scholar]
- 34. Lee H. S., Son Y. J., Chong S. H., Bae J. Y., Leem C. H., Jang Y. J., Choe H. (2009) Arch. Biochem. Biophys. 486, 35–43 [DOI] [PubMed] [Google Scholar]
- 35. Hayward S., Kitao A., Berendsen H. J. (1997) Proteins 27, 425–437 [DOI] [PubMed] [Google Scholar]
- 36. Traut T. W. (1994) Mol. Cell. Biochem. 140, 1–22 [DOI] [PubMed] [Google Scholar]
- 37. Yamada K., Hara N., Shibata T., Osago H., Tsuchiya M. (2006) Anal. Biochem. 352, 282–285 [DOI] [PubMed] [Google Scholar]
- 38. Yang H., Yang T., Baur J. A., Perez E., Matsui T., Carmona J. J., Lamming D. W., Souza-Pinto N. C., Bohr V. A., Rosenzweig A., de Cabo R., Sauve A. A., Sinclair D. A. (2007) Cell 130, 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ordman A. B., Kirkwood S. (1977) J. Biol. Chem. 252, 1320–1326 [PubMed] [Google Scholar]
- 40. Adams M. J., Ellis G. H., Gover S., Naylor C. E., Phillips C. (1994) Structure 2, 651–668 [DOI] [PubMed] [Google Scholar]
- 41. Zhang L., Chooback L., Cook P. F. (1999) Biochemistry 38, 11231–11238 [DOI] [PubMed] [Google Scholar]
- 42. Easley K. E., Sommer B. J., Boanca G., Barycki J. J., Simpson M. A. (2007) Biochemistry 46, 369–378 [DOI] [PubMed] [Google Scholar]
- 43. Steinmetz C. G., Xie P., Weiner H., Hurley T. D. (1997) Structure 5, 701–711 [DOI] [PubMed] [Google Scholar]
- 44. Graham C. E., Brocklehurst K., Pickersgill R. W., Warren M. J. (2006) Biochem. J. 394, 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marchal S., Rahuel-Clermont S., Branlant G. (2000) Biochemistry 39, 3327–3335 [DOI] [PubMed] [Google Scholar]
- 46. Day J. M., Tutill H. J., Purohit A., Reed M. J. (2008) Endocr. Relat. Cancer 15, 665–692 [DOI] [PubMed] [Google Scholar]
- 47. Ge X., Wakim B., Sem D. S. (2008) J. Med. Chem. 51, 4571–4580 [DOI] [PubMed] [Google Scholar]
- 48. Singh R., Mozzarelli A. (2009) Methods Mol. Biol. 575, 93–122 [DOI] [PubMed] [Google Scholar]
- 49. Vander Jagt D. L., Deck L. M., Royer R. E. (2000) Curr. Med. Chem. 7, 479–498 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.