Abstract
The molecular basis of ligand binding and activation of family B G protein-coupled receptors is not yet clear due to the lack of insight into the structure of intact receptors. Although NMR and crystal structures of amino-terminal domains of several family members support consistency in general structural motifs that include a peptide-binding cleft, there are variations in the details of docking of the carboxyl terminus of peptide ligands within this cleft, and there is no information about siting of the amino terminus of these peptides. There are also no empirical data to orient the receptor amino terminus relative to the core helical bundle domain. Here, we prepared a series of five new probes, incorporating photolabile moieties into positions 2, 15, 20, 24, and 25 of full agonist secretin analogues. Each bound specifically to the receptor and covalently labeled single distinct receptor residues. Peptide mapping of labeled wild-type and mutant receptors identified that the position 15, 20, and 25 probes labeled residues within the distal amino terminus of the receptor, whereas the position 24 probe labeled the amino terminus adjacent to TM1. Of note, the position 2 probe labeled a residue within the first extracellular loop of the receptor, a region not previously labeled, providing an important new constraint for docking the amino-terminal region of secretin to its receptor core. These additional experimentally derived constraints help to refine our understanding of the structure of the secretin-intact receptor complex and provide new insights into understanding the molecular mechanism for activation of family B G protein-coupled receptors.
Keywords: G Protein-coupled Receptors (GPCR), Hormone Receptors, Ligand-binding Protein, Photoaffinity Labeling, Receptors, Ligand Binding, Molecular Modeling, Secretin, Secretin Receptor
Introduction
The secretin receptor is a prototypic member of family B G protein-coupled receptors (GPCRs)3 that includes multiple potentially important drug targets (1). Understanding the molecular basis of ligand binding and activation of these receptors will facilitate the rational design of receptor-active drugs. However, because of limited information regarding the structures of the intact receptors, the molecular basis of ligand binding and activation of these receptors is not yet clear.
Since the resolution of the three-dimensional structure of the β2-adrenergic receptor (2), a growing number of such structures have been solved for members of the family A GPCRs (3–8). Although family B GPCRs share the heptahelical topology and G protein coupling with family A GPCRs, they do not have the typical signature sequences of that family and have their own typical motifs. These include a relatively long, disulfide-bonded amino-terminal domain, which functions as the predominant site of ligand docking, with determinants for interaction with the peptide carboxyl terminus. To date, NMR and crystal structures have been described for the isolated ligand-bound amino-terminal domains of several family members including receptors for corticotropin-releasing factor (9–11), glucose-dependent insulinotropic polypeptide (GIP) (12), glucagon-like peptide 1 (GLP1) (13, 14), pituitary adenylate cyclase-activating peptide (PACAP) (15), parathyroid hormone (PTH1) (16), and calcitonin gene-related peptide (17, 18). Although these suggest a shared general mode of ligand binding, there are inconsistencies in the specific docking of the ligands within these structures (19), suggesting some variation in binding mechanisms among family members. In addition, these structures do not provide any insights into the mode of docking of the flexible amino-terminal regions of the ligands that are critical for their biological activity.
Currently, no three-dimensional structures are available for the intact receptors in family B GPCRs, and even the orientation of amino-terminal and core helical bundle domains is not yet clear. The most useful insights into the ligand binding to the intact receptor come from photoaffinity labeling studies of the secretin receptor. Using this approach, we previously demonstrated that secretin residues 6, 12, 13, 16, 18, 21, 22, 23, and 26 are spatially approximated with the receptor amino terminus (20, 21). Photolabile residues in positions 1 and 5 labeled the top of the sixth transmembrane segment and third extracellular loop of the receptor, respectively (22, 23). These studies support the two-domain tethering mechanism that has been proposed for activation of family B GPCRs (23–25).
In this work, we developed, characterized, and applied five additional secretin probes, incorporating a photolabile residue into positions spread throughout the secretin peptide including residues 2, 15, 20, 24, and 25. These were used to gain additional insights into spatial approximations that might help to refine our insights into structure and mode of docking. These probes were all full agonists that bound to the secretin receptor specifically and that covalently labeled distinct single residues within the receptor. The position 15, 20, and 25 probes continued to label receptor residues within the distal amino terminus, whereas the position 24 probe labeled a residue within the carboxyl-terminal region of the amino-terminal domain adjacent to the first transmembrane segment (TM1). Of particular interest, the amino-terminal position 2 probe labeled a residue within the first extracellular loop (ECL1), a region that has not previously been labeled. These five pairs of experimentally derived constraints were used to refine our working molecular model of the ligand-bound secretin receptor that had previously been proposed based on 11 pairs of spatial approximation constraints (21). These findings provide new insights into the relative orientation of the two major receptor domains and how ligand binding might contribute to receptor activation.
EXPERIMENTAL PROCEDURES
Materials
Rat secretin, [Tyr10]rat secretin and endoglycosidase F were prepared in our laboratory (26, 27). Endoproteinase Lys-C (Lys-C) was from Roche Applied Science. Cyanogen bromide (CNBr), solid-phase oxidant, N-chlorobenzenesulfonamide (iodobead), and m-maleimidobenzoyl-N-hydroxysulfosuccinimide ester were purchased from Pierce. 3-Isobutyl-1-methylxanthine and N-(2-aminoethyl-1)-3-aminopropyl glass beads were from Sigma. Fetal Clone II was from Hyclone Laboratories (Logan, UT), and tissue culture medium was from Invitrogen. All other reagents were of analytical grade.
Peptides
The photolabile secretin probes were designed to incorporate a photolabile p-benzoyl-l-phenylalanine (Bpa) to replace Asp15, Gln20, Gln24, and Gly25 of the natural peptide ligand. These represented the following: [Tyr10,Bpa15]rat secretin(1–27) (Bpa15 probe), [Tyr10,Bpa20]rat secretin(1–27) (Bpa20 probe), [Tyr10,Bpa24]rat secretin(1–27) (Bpa24 probe), and [Tyr10,Bpa25]rat secretin(1–27) (Bpa25 probe). Each probe incorporated a tyrosine residue in position 10 to replace a leucine as a site for radioiodination that has previously been well tolerated (29, 30). All these peptides were synthesized using manual solid-phase synthesis techniques with Pal resin (Advanced ChemTech, Louisville, KY) and Fmoc-protected amino acids as described previously (31). After completion of the synthesis, each peptide was cleaved from the resin using a solution of 6.25% (w/v) phenol, 2% (v/v) triisopropylsilane, 4% (v/v) thioanisole, 4% (v/v) distilled water, and 83% (v/v) trifluoroacetic acid and purified by reversed-phase HPLC as we described previously (31).
The photolabile secretin probe [N3-Phe2,Tyr10]rat secretin(1–27) (N3-Phe2 probe) was designed to replace Ser2 with an N3-Phe. To minimize the negative impact on binding affinity and biological activity of making modifications within the structurally critical amino-terminal region of the peptide, N3-Phe was used instead of a Bpa to take advantage of its smaller size. For this, an Fmoc-(p-Boc-amino)phenylalanine was incorporated into position 2 to replace serine during synthesis. The Fmoc protection of the α-amino group was initially removed by treatment with piperidine. Then, the peptide was cleaved from the resin, and all other side chain-protecting groups were removed using the trifluoroacetic acid solution described above. The p-amino group of the (p-amino)phenylalanine was subsequently converted to an azido group (N3-) using the procedure described previously (32), and the peptide was purified to homogeneity by reversed-phase HPLC (31). All the procedures were performed under reduced light to minimize photoactivation of the azido group.
All the above probes and the radioligand [Tyr10]rat secretin were radioiodinated oxidatively using 1 mCi of Na125I (PerkinElmer Life Sciences) and exposure to the solid-phase oxidant iodobead for 15 s and were purified using reversed-phase HPLC to yield specific radioactivities of 2,000 Ci/mmol (27).
Receptor Resources
CHO cell lines stably expressing the wild-type (CHO-SecR) and HA-tagged rat secretin receptors that had been previously established (CHO-HA37-SecR and CHO-HA79-SecR) (27, 33) were used as receptor sources for the current study. These cells were cultured in Ham's F-12 medium supplemented with 5% Fetal Clone II in a humidified atmosphere containing 5% CO2 at 37 °C, and they were passaged approximately twice per week.
It was necessary to develop two new secretin receptor mutants to eliminate sites of CNBr cleavage for the current work. These were secretin receptor constructs M123L and M197L in which residues Met123 and Met197 were each replaced by a leucine. They were generated using the QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA), and the products were verified by direct DNA sequencing. They were expressed transiently in COS-1 cells (American Type Culture Collection, Manassas, VA) grown in Dulbecco's modified Eagle's medium supplied with 5% Fetal Clone II after transfection using a modification of the diethylaminoethyl-dextran method (34).
Receptor-expressing CHO and COS-1 cells were harvested mechanically after being grown to confluence. Plasma membranes were prepared from these cells using discontinuous sucrose gradient centrifugation (26) and suspended in Krebs-Ringer-HEPES (KRH) medium (25 mm HEPES, pH 7.4, 104 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm KH2PO4, 1.2 mm MgSO4) containing 0.01% soybean trypsin inhibitor and 1 mm phenylmethylsulfonyl fluoride (PMSF) before being stored at −80 °C.
Receptor Binding Assay
A radioligand competition binding assay (26) with membranes prepared from CHO or COS-1 cells expressing wild-type or mutant secretin receptors was used to determine the ability of each of the photolabile secretin probes to bind to the receptor. In brief, membranes (∼5 μg) were incubated for 1 h at room temperature with a constant amount of radioligand, [125I-Tyr10]rat secretin-27 (5 pm; ∼20,000 cpm), in the presence of increasing concentrations (ranging from 0 to 1 μm) of secretin or the photolabile secretin probes in KRH medium containing 0.01% soybean trypsin inhibitor, 1 mm PMSF, and 0.2% bovine serum albumin. Bound and free radioligand were separated using a Skatron cell harvester with glass fiber filter mats that had been soaked in 0.3% Polybrene, and bound radioactivity was quantified in a γ-spectrometer. Nonspecific binding was determined in the presence of 1 μm secretin and represented less than 15% of total radioligand binding. The same assay was also utilized to characterize the binding activity of the M123L and M197L secretin receptor mutants expressed in COS-1 cells. Binding curves were analyzed and plotted using the non-linear regression analysis program in the Prism software suite v3.02 (GraphPad Software, San Diego, CA). Binding kinetics were determined by analysis with the LIGAND program of Munson and Rodbard (35). Data are reported as the means ± S.E. of duplicate determinations from a minimum of three independent experiments.
Biological Activity Assay
To determine the biological activity of each of the photolabile secretin probes, their abilities to stimulate cAMP responses in receptor-bearing CHO-SecR cells were examined. In brief, ∼8,000 CHO-SecR cells/well were grown in 96-well plates for 2 days. After cells were washed with PBS, they were stimulated for 30 min at 37 °C with increasing concentrations of secretin or each of the photolabile secretin probes (ranging from 0 to 1 μm) in KRH medium containing 0.01% soybean trypsin inhibitor, 0.2% bovine serum albumin, 0.1% bacitracin, and 1 mm 3-isobutyl-1-methylxanthine. After incubation, supernatants were removed by aspiration, and cells were lysed with 6% ice-cold perchloric acid. The cell lysates were adjusted to pH 6 with 30% KHCO3 and introduced into an assay of cAMP levels using a LANCETM cAMP-384 kit from PerkinElmer Life Sciences. The assay was performed according to the manufacturer's instructions and repeated in at least three independent experiments. This assay was also used for characterizing the M123L and M197L secretin receptor mutants transiently expressed in COS-1 cells.
Photoaffinity Labeling
Membranes (∼50 μg) from CHO-SecR cells or COS-1 cells expressing secretin receptor mutants were incubated in the dark for 1 h at room temperature with each of the radioiodinated photolabile secretin probes (∼0.1 nm) in KRH medium containing 0.01% soybean trypsin inhibitor and 1 mm PMSF in the presence of increasing amounts of competing secretin (ranging from 0 to 1 μm). The reactions were then exposed to photolysis for 30 min at 4 °C using a Rayonet photochemical reactor (Southern New England Ultraviolet Co., Bradford, CT) equipped with 3500-Å lamps. After being washed with ice-cold KRH medium, membranes were solubilized in SDS sample buffer and separated on 10% SDS-polyacrylamide gels. Bands of interest were visualized by autoradiography, and densitometric analysis was performed by the NIH ImageJ software. The apparent molecular weights of the radioactive bands were determined by interpolation on a plot of the mobility of the appropriate ProSieve protein markers (Cambrex, Rockland, ME) versus the log values of their apparent masses.
Peptide Mapping
This required larger scale preparation of affinity-labeled receptors. Approximately 200 μg of cell membranes were incubated with each of the radioiodinated photolabile secretin probes (∼0.5 nm) in the absence of competing secretin. After electrophoresis, radioactive bands of interest were excised from gels, eluted in water, lyophilized, and ethanol-precipitated as described previously (33). For selected experiments, receptor samples were deglycosylated by treatment with endoglycosidase F (26). Cleavage of the labeled wild-type or mutant secretin receptors with CNBr and Lys-C was performed using protocols that we described previously (33), and the products of cleavage were analyzed on 10% Bis-Tris NuPAGE gels (Invitrogen) using MES running buffer system under reducing conditions. The radiolabeled bands were visualized by autoradiography, and their apparent molecular weights were determined by interpolation on a plot of the mobilities of the appropriate Multimark multicolored standards (Invitrogen) versus the log values of their apparent masses.
Radiochemical Sequencing
To identify the specific receptor residues labeled with each of the photolabile secretin probes, manual Edman degradation radiochemical sequencing was performed (36). It should be noted that for the position 2 probe, after photoaffinity labeling but prior to CNBr cleavage, the labeled membranes were treated by acetic anhydride (23). This was performed to block the amino terminus of this probe by acetylation and to prevent it from being cleaved during sequencing. Radioactively pure receptor fragments labeled with the position 2 (Ile198–Met205), 15 (Leu17–Met51), 20 (Ala1–Met51), 24 (Pro97–Lys119), or 25 (Leu17–Met51) probe were covalently coupled through the Cys203, Cys24/Cys44, Cys11/Cys24/Cys44, Cys101, and Cys24/Cys44 residues, respectively, to m-maleimidobenzoyl-N-succinimide-activated N-(2-aminoethyl-1)-3-aminopropyl glass beads. Immobilized fragments were subjected to manual Edman degradation sequencing as described previously (36). Radioactivity eluted in each cycle was quantified using a γ-spectrometer.
Molecular Modeling
All molecular modeling was conducted using a stochastic global energy optimization procedure in Internal Coordinate Mechanics (ICM) (37) with the ICM-Pro package version 3.7 (MolSoft LLC, San Diego, CA). This procedure consisted of three iterative steps: (a) random conformational change of a dihedral angle according to the biased probability Monte Carlo method (38), (b) local minimization of all free dihedral angles, and (c) acceptance or rejection of the new conformation based on the Metropolis criterion at the simulation temperature (usually 600 K) (39). The initial model of the amino-terminal domain of the secretin receptor was generated using the x-ray structure of the amino terminus of the GIP receptor complexed with the GIP peptide (Protein Data Bank code 2QKH (12)) as template. As the structure for this template terminates near the third disulfide bond (Cys67-Cys101), we extended an α helical structure to the top of TM1 based on the secondary structure prediction algorithms in ICM that suggest the propensity for α helix in this region of the family B GPCRs. A pentasaccharide, Man3(GlcNAc)2, was attached to secretin receptor residues Asn50, Asn78, Asn84, and Asn106 to mimic their glycosylated state. The initial conformation of the secretin peptide was generated using the NMR structure of receptor-bound PACAP(1–21)-NH2 as template and aligning this with GIP in the GIP-GIP receptor complex to determine its initial docking pose.
It was noted that this initial pose did not satisfy all experimentally determined photoaffinity labeling constraints. Therefore, the whole complex was globally optimized in the presence of the following spatial approximation constraints coming from photoaffinity labeling experiments (peptide residue to its labeled receptor residue): Phe6 to Val4, Arg12 to Val6, Leu13 to Val103, Asp15 to Glu19, Ser16 to Leu99, Arg18 to Arg14, Gln20 to His2, Arg21 to Arg15, Leu22 to Leu17, Leu23 to Arg21, Gln24 to Pro97, Gly25 to His23, and Leu26 to Leu36. Additionally, four FRET distance constraints (40) were also incorporated. The backbone dihedral angles around the first (Cys24-Cys53) and last (Cys67-Cys101) disulfide bonds of the amino-terminal domain were relaxed to allow for rotational freedom, resulting in a better fit of the experimental constraint data. Twenty-five of the lowest energy complexes were retained. The TM bundle was constructed using the x-ray structure of CXCR4 (Protein Data Bank code 3ODU) as template (8). To accommodate distance restraints between secretin Ser2 and Phe199 in the receptor, TM3 was rotated clockwise relative to the template to orient Phe199 toward the core of the transmembrane domain while maintaining the integrity of the disulfide bond between Cys193 of TM3 and Cys263 of ECL2. The peptide ligand-amino-terminal receptor domain complexes were then docked onto 200 diverse helical bundle domain models from 20 different TM bundles, each completed with 10 different loop conformations, in the presence of three additional photoaffinity labeling constraints involving the receptor core (peptide residue to its labeled receptor residue), His1 to Phe338, Ser2 to Phe199, and Thr5 to Phe349, and 12 FRET distance constraints between the peptide and the transmembrane domain as described previously (22). All the resultant models were clustered, ranked by their ICM energetics and their health as established by PROCHECK and WHAT_CHECK evaluations (41, 42). The three best models from 200 independent runs were selected.
RESULTS
Photolabile Probe Characterization
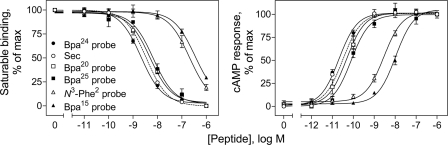
Each of the photolabile secretin probes was synthesized and purified to homogeneity, and its identity was verified by mass spectrometry. Probes were functionally characterized to determine their abilities to bind to the wild-type secretin receptor and to stimulate intracellular cAMP accumulation in secretin receptor-bearing CHO-SecR cells. Fig. 1 shows that position 20, 24, and 25 probes bound to the receptor specifically and saturably with affinities (Ki values for position 20, 24, and 25 probes, 5.0 ± 0.7, 2.2 ± 0.3, and 7.1 ± 1.5 nm, respectively) similar to that of natural secretin (Ki = 3.5 ± 0.2 nm), whereas the affinities of the position 2 and 15 probes were much lower (Ki values for position 2 and 15 probes, 231 ± 71 and 370 ± 71 nm, respectively). Fig. 1 also shows that all probes were full agonists, stimulating intracellular cAMP responses in CHO-SecR cells in a concentration-dependent manner. Although the potencies for position 20, 24, and 25 probes (EC50 values for position 20, 24, and 25 probes, 48 ± 10, 20 ± 5, and 79 ± 19 pm, respectively) were similar to that of secretin (EC50 = 32 ± 4 pm), potencies for position 2 and 15 probes were much lower (EC50 values for position 2 and 15 probes, 2.9 ± 0.8 and 10.5 ± 2.3 nm, respectively).
FIGURE 1.
Functional characterization of photolabile secretin analogues. Left, binding curves of increasing concentrations of secretin and each of the photolabile secretin probes to compete for binding of the secretin-like radioligand [125I-Tyr10]rat secretin-27 to secretin receptor-bearing CHO-SecR cells. The values illustrated represent saturable binding as percentages of maximal binding observed in the absence of the competing secretin and are expressed as the means ± S.E. of duplicate values from a minimum of three independent experiments. Right, curves of intracellular cAMP responses in CHO-SecR cells stimulated by increasing concentrations of secretin and each of the noted probes. Data points represent the means ± S.E. of three independent experiments performed in duplicate, normalized relative to the maximal response to secretin. Basal (5.7 ± 1.3 pmol/million cells) and maximal (189 ± 38 pmol/million cells) cAMP levels by each of the peptides were similar. Sec, secretin.
Photoaffinity Labeling of Secretin Receptor
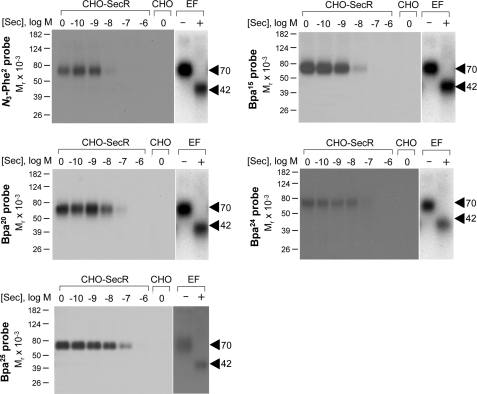
Each of the position 2, 15, 20, 24, and 25 probes was used in photoaffinity labeling experiments with CHO-SecR cell membranes in the absence and presence of competing secretin to explore their abilities to covalently label the secretin receptor (Fig. 2). Fig. 3 shows that each probe labeled this receptor specifically and saturably with labeling by each probe inhibited by secretin in a concentration-dependent manner (IC50 values for position 2, 15, 20, 24, and 25 probes, 4.5 ± 1.2, 4.9 ± 1.3, 9.9 ± 2.1, 51 ± 12, and 101 ± 20 nm, respectively). The labeled secretin receptor migrated at approximately Mr = 70,000 and shifted to approximately Mr = 42,000 after deglycosylation with endoglycosidase F. No receptor bands were detected in affinity-labeled non-receptor-bearing CHO cell membranes. The receptor bands migrated identically to those labeled with other secretin probes that we reported previously (20–22).
FIGURE 2.
Photoaffinity labeling of secretin receptor. Shown are representative autoradiographs of 10% SDS-polyacrylamide gels used to separate the products of affinity labeling membranes from CHO-SecR cells with each of the noted photolabile probes in the presence of increasing concentrations of competing unlabeled secretin (Sec; from 0 to 1 μm). As controls, labeling of the non-receptor-bearing CHO cell membranes by each probe in the absence of competitor is also shown. Each of the probes labeled the secretin receptor specifically and saturably with the labeling being competed by secretin in a concentration-dependent manner. The receptor bands labeled by each probe migrated at approximately Mr = 70,000 and shifted to approximately Mr = 42,000 after deglycosylation with endoglycosidase F (EF). No radioactive band was observed in the affinity-labeled non-receptor-bearing CHO cell membranes. Data are representative of at least three independent experiments.
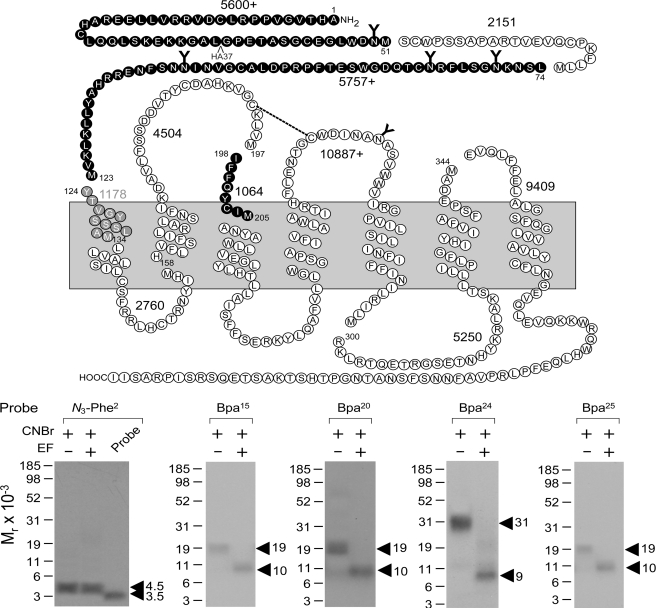
FIGURE 3.
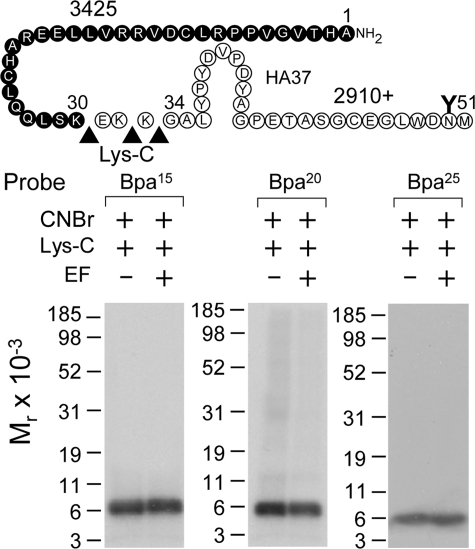
CNBr cleavage of labeled secretin receptor. Top, diagram illustrating the theoretical fragments of the secretin receptor resulting from CNBr cleavage. Bottom, results of CNBr cleavage of the secretin receptor labeled by each of the noted probes. Cleavage of the secretin receptor labeled by the position 2 probe yielded a non-glycosylated fragment migrating at approximately Mr = 4,500, representing either the fragment Tyr124–Met134 (beginning of TM1; gray circles) or Ile198–Met205 (beginning of TM3; black circles). Cleavage of the secretin receptor labeled by the position 15, 20, and 25 probes each yielded a fragment migrating at approximately Mr = 19,000 that shifted to approximately Mr = 10,000 after deglycosylation by endoglycosidase F (EF), likely representing the first CNBr fragment between Ala1 and Met51 (20). Cleavage of the secretin receptor labeled by the position 24 probe yielded a fragment migrating at approximately Mr = 31,000 that shifted to approximately Mr = 9,000 after deglycosylation, likely representing the third CNBr fragment between Leu74 and Met123 (21, 43). Data are representative of at least three independent experiments.
Site Identification
The secretin receptor contains nine methionine residues distributed throughout the molecule whereby CNBr cleavage should yield 10 fragments with molecular masses ranging from 1 to 11 kDa of which three contain sites of glycosylation. CNBr cleavage has been proven very useful in identification of regions and even sites of labeling for secretin probes (20–22). Here again, it was used to provide the first indication of the region of labeling with each probe. As shown in Fig. 3, CNBr cleavage of the secretin receptor labeled with the position 2 probe yielded a single radioactive band migrating at approximately Mr = 4,500 that did not further shift after deglycosylation by endoglycosidase F. Given its non-glycosylated nature and the mass of the attached N3-Phe2 probe (3,180 Da), there were two possible candidate fragments that match these data, Tyr124–Met134 including TM1 and Ile198–Met205 spanning the ECL1 and TM3.
Fig. 3 also shows that CNBr cleavage of the secretin receptor labeled with the position 15, 20, and 25 probes each yielded a fragment migrating at approximately Mr = 19,000 that shifted to approximately Mr = 10,000 after deglycosylation by endoglycosidase F. This most likely represented the first CNBr fragment between Ala1 and Met51 as we previously demonstrated for other secretin probes (20), and this was confirmed by immunoprecipitation of HA-tagged fragments from CNBr cleavage of a well characterized receptor mutant (HA37-SecR) incorporating an HA epitope within this fragment (data not shown).
Fig. 3 further shows that CNBr cleavage of the secretin receptor labeled by the position 24 probe yielded a fragment migrating at approximately Mr = 31,000 that shifted to approximately Mr = 9,000 after deglycosylation. This most likely represented the third CNBr fragment between Leu74 and Met123 that had previously been labeled with the Bpa13 and Bpa16 secretin probes (21, 43), and this was confirmed by immunoprecipitation of HA-tagged fragments from CNBr cleavage of a well characterized receptor mutant (HA79-SecR) incorporating an HA epitope within this fragment (data not shown).
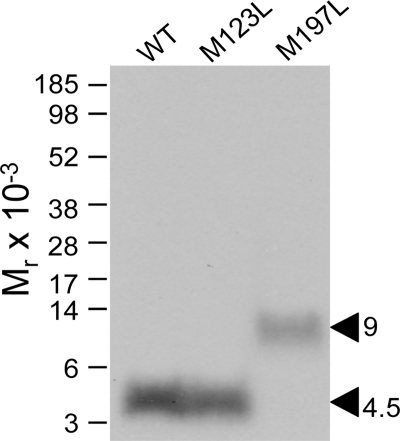
Two secretin receptor mutants, M123L and M197L, were used to identify which of the two above mentioned candidate fragments contained the site of labeling by the position 2 probe. Both receptor mutants bound secretin (Ki values: wild-type, 1.9 ± 0.5 nm; M123L, 3.5 ± 0.9 nm; M197L, 3.1 ± 0.5 nm) and signaled similarly to the wild-type receptor (EC50 values: wild-type, 21 ± 8 pm; M123L, 32 ± 11 pm; M197L, 50 ± 14 pm). They were also specifically and saturably labeled with the position 2 probe (data not shown). CNBr cleavage of the labeled M123L receptor mutant yielded a labeled fragment that migrated at approximately Mr = 4,500 on a 10% NuPAGE gel, similar to that originating from the wild-type receptor. In contrast, CNBr cleavage of the labeled M197L receptor mutant yielded a much larger fragment migrating at approximately Mr = 9,000, representing the His158–Met205 fragment spanning TM2 to TM3 of the receptor (Fig. 4). This identified the receptor fragment Ile198–Met205 spanning ECL1 and TM3 as the region of labeling by the position 2 probe.
FIGURE 4.
CNBr cleavage of mutant secretin receptors labeled by position 2 probe. CNBr cleavage of the labeled M197L mutant receptor resulted in a radioactive band migrating at approximately Mr = 9,000, clearly distinct from the migration pattern of the CNBr cleavage products from the wild-type (WT) and M123L mutant receptors (Mr = 4,500). This indicates that the Ile198–Met205 fragment spanning ECL1 and TM3 contained the site of labeling by the position 2 probe (see diagram in Fig. 3).
Endoproteinase Lys-C, which specifically cleaves at the carboxyl terminus of lysine residues, was used to further refine the receptor region labeled by the position 15, 20, and 25 probes. As shown in Fig. 5, Lys-C cleavage of the first CNBr fragment (Ala1–Met51) labeled by each of these probes yielded a fragment migrating at approximately Mr = 6,000 that did not shift after deglycosylation. This identified the non-glycosylated Ala1–Lys30 fragment at the distal amino terminus of the secretin receptor as the region of labeling by each of the position 15, 20, and 25 probes.
FIGURE 5.
Lys-C cleavage of secretin receptor labeled by each of position 15, 20, and 25 probes. Top, diagram of the sites of Lys-C cleavage of the first CNBr fragment (Ala1–Met51) of the secretin receptor along with the masses of the protein cores of the resultant fragments. Bottom, Lys-C cleavage of the CNBr fragment of the secretin receptor labeled by each probe resulted in a non-glycosylated band migrating at approximately Mr = 6,000, representing the labeling of the Ala1–Lys30 fragment at the distal amino terminus of the receptor. EF, endoglycosidase F.
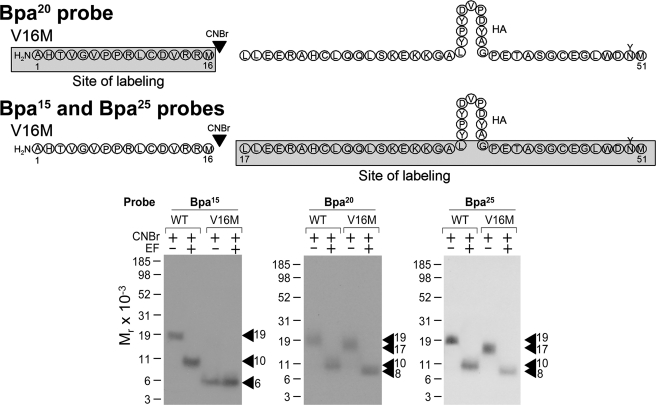
A previously characterized V16M mutant secretin receptor (44) was used to further refine the region of labeling by the position 15, 20, and 25 probes. As shown in Fig. 6, CNBr cleavage of the V16M mutant receptor labeled by the position 20 probe yielded a non-glycosylated band migrating at approximately Mr = 6,000, representing the labeling of the amino-terminal Ala1–Met16 fragment. This was further localized to the Ala1–Pro8 segment by cleaving the previously characterized P8M mutant receptor (45) labeled by this probe (data not shown). CNBr cleavage of the V16M mutant receptor labeled by the position 15 and 25 probes each yielded a fragment migrating at approximately Mr = 17,000 that shifted to approximately Mr = 8,000 after deglycosylation, representing the labeling of the glycosylated carboxyl-terminal Leu17–Met51 fragment. Taking into account the above identification by Lys-C cleavage, the sites of labeling by each of the position 15 and 25 probes were localized between Leu17 and Lys30 of the secretin receptor.
FIGURE 6.
CNBr cleavage of V16M secretin receptor labeled by position 15, 20, and 25 probes. Top, diagrams of the sites of CNBr cleavage of the Ala1–Met51 region of the V16M mutant secretin receptor labeled by each probe. Bottom, CNBr cleavage of the V16M mutant receptor labeled by the position 20 probe yielded a non-glycosylated band migrating at approximately Mr = 6,000, representing the labeling of the amino-terminal Ala1–Met16 fragment. CNBr cleavage of the V16M mutant receptor labeled by position 15 and 25 probes each yielded a fragment migrating at approximately Mr = 17,000 that shifted to approximately Mr = 8,000 after deglycosylation, representing the labeling of the carboxyl-terminal Leu17–Met51 fragment. EF, endoglycosidase F.
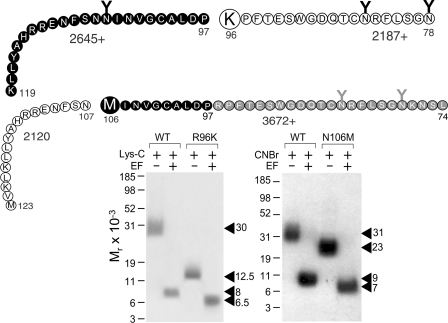
Two previously characterized secretin receptor mutants, R96K and N106M (21), were used to further refine the region of labeling by the position 24 probe. As shown in Fig. 7, Lys-C cleavage of the labeled wild-type secretin receptor yielded a band migrating at approximately Mr = 30,000 and shifting to approximately Mr = 8,000 after deglycosylation, consistent with labeling the Asn78–Lys119 fragment. Lys-C cleavage of the labeled R96K receptor resulted in a band migrating at approximately Mr = 12,500 and shifting to approximately Mr = 6,500 after deglycosylation, consistent with labeling the carboxyl-terminal Pro97–Lys119 segment (21, 43). Fig. 7 also shows that CNBr cleavage of the labeled N106M receptor yielded a band migrating at approximately Mr = 23,000 and shifting to approximately Mr = 7,000, distinct from the pattern of cleavage of the wild-type secretin receptor (Mr = 31,000 shifting to Mr = 9,000 after deglycosylation), indicating that the Leu74–Met106 fragment was the region of labeling (21). Taken together, the site of labeling with the position 24 probe was localized to the region between Pro97 and Asn106 of the secretin receptor.
FIGURE 7.
Cleavage of R96K and N106M secretin receptor mutants labeled by position 24 probe. Top, diagrams of the sites of Lys-C and CNBr cleavage of the R96K and N106M mutant secretin receptor constructs, respectively, along with the masses of the protein cores of the resultant fragments. Bottom, Lys-C cleavage of the wild-type secretin receptor (WT) and the R96K mutant labeled by the position 24 probe resulted in bands migrating at approximately Mr = 30,000 and Mr = 12,500 that further shifted to approximately Mr = 8,000 and Mr = 6,500 after deglycosylation, respectively, indicating that the site of labeling was within the Pro97–Met123 fragment (21, 43). CNBr cleavage of the labeled N106M receptor yielded a band migrating at approximately Mr = 23,000 and shifting to approximately Mr = 7,000, distinct from the pattern of cleavage of the wild-type secretin receptor (approximate Mr = 31,000 shifting to approximate Mr = 9,000 after deglycosylation), indicating that the fragment Leu74–Asn106 contained the site of labeling (21). Taken together, the site of labeling with the position 24 probe was within the region between Pro97 and Asn106 of the secretin receptor. EF, endoglycosidase F.
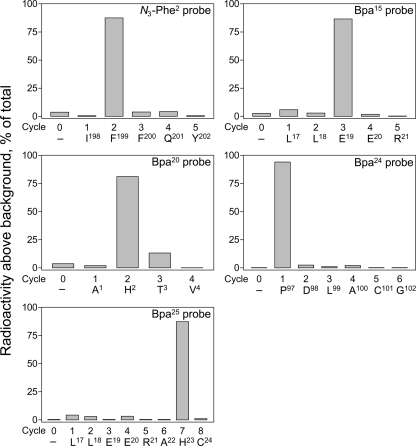
The specific receptor residues labeled with each of the position 2, 15, 20, 24, and 25 probes were identified by manual Edman degradation radiochemical sequencing of relevant receptor fragments that had been purified to radioactive homogeneity. As shown in Fig. 8, radiochemical sequencing of receptor fragments Ile198–Met205, Leu17–Met51, Ala1–Met51, Pro97–Lys119, and Leu17–Met51 labeled by position 2, 15, 20, 24, and 25 probes, respectively, yielded peaks in eluted radioactivity in cycles 2, 3, 2, 1, and 7. This corresponds to covalent attachment to receptor residues Leu199, Glu19, His2, Pro97, and His23 by position 2, 15, 20, 24, and 25 probes, respectively.
FIGURE 8.
Identification of labeled receptor residues by radiochemical sequencing. Shown are radioactive elution profiles of Edman degradation sequencing of the purified receptor fragments labeled by each of the position 2 (Ile198–Met205), 15 (Leu17–Met51), 20 (Ala1–Met51), 24 (Pro97–Lys119), and 25 (Leu17–Met51) probes. There were peaks in eluted radioactivity in cycles 2, 3, 2, 1, and 7 that correspond with covalent attachment to receptor residues Leu199, Glu19, His2, Pro97, and His23 by position 2, 15, 20, 24, and 25 probes, respectively.
Molecular Model of Intact Secretin Receptor
The x-ray crystal structure of the amino terminus of the GIP receptor complexed with the GIP peptide provided a starting point for modeling the amino-terminal domain of the secretin receptor. As noted in our previous work (21), a direct superposition of the secretin receptor-secretin complex to the GIP receptor-GIP complex did not satisfy the experimental constraints imposed by data from photoaffinity labeling experiments with a series of full agonist peptide ligands. Therefore, the secretin peptide was docked and globally optimized with the secretin receptor in the presence of these experimental spatial approximation constraints. The recently released x-ray crystal structure of CXCR4 (8) provided a different template for core helical bundle modeling from the β2-adrenergic receptor template we had used previously (2). Each of the 25 conformations of amino-terminal secretin receptor domain-secretin peptide complexes was docked to 200 transmembrane domain models in the presence of all experimental constraints followed by all-atom refinements, generating the final models. Three of the best models were selected as being representative.
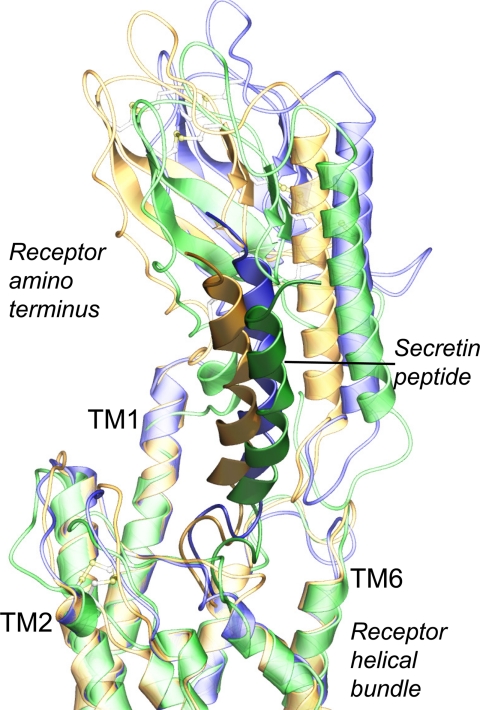
Table 1 shows the distances between positions of cross-linked residues in these three models, each of which satisfies all current experimentally derived constraints. Distances were measured from the Cβ of the natural amino acid in the position of the Bpa in the peptide ligand (except when it was a glycine) to the noted atom within the labeled receptor residue. Given the size of the photolabile Bpa moiety, an approximate 8–10-Å distance is typical. Fig. 9 shows the spatial approximation between the photolabile residues within the secretin probe and the receptor residues labeled in one of the best molecular models of the secretin-bound secretin receptor. Fig. 10 shows how similar all three of the best molecular models were to each other. In this, the backbones of the transmembrane segments were superimposed, and the amino-terminal domains and docked peptides were shown to vary only minimally.
TABLE 1.
Interatomic distances (Å) between cross-linked residues in three best models
| Secretin peptide residuea | Secretin receptor residue, proximal atom | Distance in model 1 | Distance in model 2 | Distance in model 3 |
|---|---|---|---|---|
| His1 | Phe338, Cζ | 8.1 | 8.1 | 8.2 |
| Ser2 | Phe199, Cζ | 7.3 | 7.3 | 7.6 |
| Thr5 | Phe349, Cζ | 8.9 | 9.1 | 8.7 |
| Phe6 | Val4, Cγ1 | 7.2 | 9.1 | 7.2 |
| Arg12 | Val6, Cγ1 | 7.8 | 7.7 | 7.8 |
| Leu13 | Val103, Cγ1 | 8.1 | 8.4 | 8.1 |
| Asp15 | Glu19, Cδ | 11.1 | 12.1 | 11.1 |
| Ser16 | Leu99, Cδ1 | 8.1 | 7.9 | 8.1 |
| Arg18 | Arg14, Cζ | 8.2 | 7.9 | 8.2 |
| Gln20 | His2, Cβ | 9.2 | 8.7 | 9.2 |
| Arg21 | Arg15, Cζ | 9.0 | 8.3 | 9.0 |
| Leu22 | Leu17, Cδ1 | 8.2 | 8.6 | 8.2 |
| Leu23 | Arg21, Cζ | 5.7 | 4.9 | 5.7 |
| Gln24 | Pro97, Cβ | 9.7 | 10.0 | 9.7 |
| Gly25 | His23, Cϵ1 | 12.8 | 10.1 | 12.8 |
| Leu26 | Leu36, Cδ1 | 7.9 | 8.8 | 7.9 |
a Measured from the Cβ of the corresponding peptide residue, except for Gly25, which was measured from Cα, to the proximal atom of noted receptor residue.
FIGURE 9.
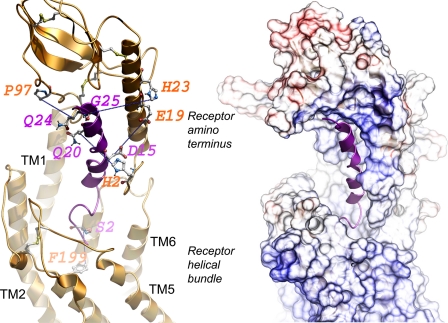
Molecular model of ligand-bound secretin receptor. Shown is a lateral view of the best model of the secretin receptor-secretin peptide complex with the amino-terminal domain of the receptor above and the transmembrane helical bundle domain below. The secretin peptide ligand is illustrated in magenta with its carboxyl terminus within the peptide-binding cleft at the top and its amino terminus extending into the helical bundle at the bottom. The left panel highlights the five pairs of residues contributing to the experimental spatial approximation constraints (residues linked with blue dotted lines). The sites of incorporation of Bpa into the peptide ligand are identified in magenta, and the sites of covalent labeling of the receptor are identified in orange. The right panel illustrates the surfaces of the peptide-binding cleft within the receptor amino terminus and extending to the helical bundle.
FIGURE 10.
Three best molecular models of ligand-bound secretin receptor. Shown is the high degree of structural similarity among the three best molecular models, superimposed by the backbone of their transmembrane domains. Model 1 is colored gold, model 2 is green, and model 3 is blue (in each model, the receptor is in lighter shade, whereas the peptide is darker).
DISCUSSION
Understanding the molecular basis of the docking of a ligand to a receptor requires insights into the structures of each of these molecules. The docking of the natural peptide secretin ligand to its family B GPCR is challenged by the flexibility of the ligand and by lack of insight into the structure of the intact receptor. Although considerable progress has been achieved with the solution of high resolution NMR and crystal structures for the amino-terminal domains of several members of this receptor family (9–18), much less is understood about the core helical bundle domains of these receptors, and virtually nothing is known about the relative orientation of these two major domains.
In the current work, we developed, characterized, and applied five new photolabile probes to establish spatial approximations between residues spread throughout the secretin pharmacophore and residues within the secretin receptor. This brings the total of such spatial approximation constraints to 16 of the 27 residues comprising natural secretin. We previously described the application of 11 such probes; nine of them were found to covalently label the receptor amino-terminal domain (20, 21, 33, 44–47), and only probes with sites of covalent attachment in positions 5 and 1 (or an amino-terminal extension before the 1-position) were found to label the receptor core helical bundle domain (22, 23). Application of those spatial approximation constraints was insufficient to refine a general working model of the intact secretin receptor (21). In the current report, the new spatial approximation constraints have provided substantial refinement of our understanding of the structure of the intact secretin receptor and provided more meaningful insights into the docking of the amino-terminal region of secretin, which is known to be critical for its biological activity.
It is important to draw attention to the N3-Phe2 probe used in this work. The amino-terminal region of secretin is critically important for the biological activity of this hormone (23) as it is for other natural ligands of family B GPCRs (48–50). When modifying secretin to allow incorporation of a photolabile moiety for covalent labeling of the receptor, it is challenging to accommodate a residue with optimal photochemical characteristics without disrupting binding and activity. Although benzophenone moieties, like the Bpa used in most of the secretin analogue probes we have developed, have ideal characteristics, typically having high efficiency of incorporation and low levels of background labeling, this represents a bulky hydrophobic group that is not readily accommodated. Indeed, in developing amino-terminal secretin probes, Bpa in position 1 was poorly accommodated (23), necessitating situating it in the −1 and −2 positions for receptor labeling (23). Use of N3-Phe2 in the position 2 probe provided a functional peptide labeled within this key region of the peptide and the opportunity to determine proximity with the receptor.
In this work, four new probes having a photolabile residue in positions 15, 20, 24, and 25 labeled the amino terminus of the secretin receptor, the same receptor region most commonly labeled in previous studies as well. This reflects the major determinants of natural ligand binding within the peptide-binding cleft of the receptor amino terminus. In contrast, the probe with its photolabile residue in position 2 labeled receptor residue Phe199 within the ECL1-TM3 region of this receptor. This region is distinct from the TM6-ECL3 region labeled by the amino-terminal position 1 and 5 probes, representing the only other probes that did not label the receptor amino terminus prior to the current study (22, 23). Spatial approximation of this secretin probe with the ECL1-TM3 region of the receptor is consistent with mutagenesis data for family B GPCRs (28, 34). For example, residues Lys173 and Asp174 within the ECL1 region of the secretin receptor have been proposed to interact with Asp3 of secretin based on mutagenesis data (28), and these regions are closely approximated in the current model. Also consistent with the model is a correlative photoaffinity labeling study performed with the structurally related GLP1 receptor in which receptor residue Tyr205 within ECL1 was labeled by an amino-terminal peptide probe (51). The ECL1-TM3 region has also been identified as the region of labeling of the PTH receptor and corticotropin-releasing factor 1 receptor by midregion PTH (52, 53) and urocortin (54) probes, respectively. Of note, although the binding affinities and potencies of the probes substituted with Bpa at positions 20, 24, and 25 were equivalent to these parameters for the natural secretin peptide, the position 15-substituted probe had significantly reduced affinity and potency. Although the Bpa15 probe labeled receptor residue Glu19, examination of the model reveals close approximation between secretin residue Asp15 and secretin receptor residue Arg15 where they might form an important salt bridge with loss of ligand binding affinity with disruption of that interaction. In fact, we previously mutated receptor residue Arg15 to an alanine and found that the ability of natural secretin to bind to and stimulate cAMP at that mutant were reduced more than 12-fold (47).
The new molecular model being proposed represents a substantial improvement over our previous working model. The CXCR4 receptor structure (8), as a family A GPCR that binds a natural peptide ligand, seems to provide a more suitable structural template than the β2-adrenergic receptor structure (2) that was previously utilized. Comparison of the structures of these two receptors reveals the following key differences that contributed to the improvement in our current secretin receptor model. (a) TM1 in the CXCR4 receptor is bent toward the rest of the transmembrane helical bundle, bringing the receptor amino-terminal domain closer to the center of the top of the helical bundle. (b) TM6 and TM7 in the CXCR4 receptor adopt a more open conformation than they do in the β2-adrenergic receptor and therefore more readily accept the insertion of the peptide amino terminus into the transmembrane helical bundle. Indeed, using this template in the current model resulted in new adjacency and possible contact between the docked secretin peptide and receptor residue Trp264; this residue is analogous to Trp297 in the GLP1 receptor that has been shown to represent an important determinant for peptide binding (55). Furthermore, in our previous working model (21), the peptide amino terminus primarily occupied the region at the top of TM1, TM2, and TM7, whereas using the CXCR4 receptor as template allows the peptide to come into direct contact with TM6 and TM7. Both of these regions are known to be adjacent to the docked secretin peptide as shown in the photoaffinity labeling of receptor residues Phe338 and Phe349 (22, 23). This new model also suggests that docked secretin will be adjacent to ECL2 and the top of TM5, a prediction that should be further explored in future studies.
Additionally, the new photoaffinity labeling data between peptide residue 2 and Phe199 of the receptor provides an important additional constraint for positioning the peptide amino terminus within the receptor core. Phe199 is present below the conserved Cys193 that is known to be disulfide-bonded to Cys263 in ECL2. As a result, the amino terminus of the secretin peptide is more confidently placed between Phe338 and Phe349 of TM6-ECL3-TM7 and Phe199 of TM3-ECL1. In our previous study (21), there was considerable variation in the relative orientation of the peptide ligand amino terminus to the receptor core transmembrane domain even among the best scoring models. In contrast, the three best models in this study generated significantly less variation in the juxtapositions of the two domains as illustrated in Fig. 10. The positioning of Ser2 within this aromatic cage also provides a possible reason for the loss in affinity and potency of the N3-Phe2 analogue with even this photoactive substitution likely to sterically clash within the binding pocket.
As the quality of the molecular model of secretin-occupied intact secretin receptor is improved, the insights into molecular mechanisms can be refined. The current model helps to provide support for the “two-domain tethering” concept that has been proposed for family B GPCRs (23–25). It also supports the insights that came from the previous fluorescence studies utilizing secretin analogues with fluorescent Alexa at the amino terminus, carboxyl terminus, and midregions of the peptide (56). In those studies, the amino terminus of secretin was most exposed to the hydrophilic milieu and most easily quenched using potassium iodide with the rest of the peptide protected, such as by its insertion into the peptide-binding cleft. With activation, this cleft opened up to provide more mobility and a shorter lifetime of the midregion fluorophores. It is reassuring to see all the existing data including photoaffinity labeling constraints with 16 probes, 16 FRET distance constraints (40), fluorescence microdomain insights (56), and all extant structure-activity data fully accommodated with a working molecular model. Although this model does not yet provide the resolution necessary for drug development, it clearly provides new detail to yield testable questions for the future.
Acknowledgments
We thank Mary Lou Augustine and Alicja M. Skalecka Ball for excellent technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant DK46577 (to L. J. M.). This work was also supported by Australian Research Council Grant DP0985210 (to P. M. S.).
- GPCR
- G protein-coupled receptor
- Bpa
- p-benzoyl-l-phenylalanine
- CHO-SecR
- rat secretin receptor-bearing CHO
- ECL
- extracellular loop
- GIP
- glucose-dependent insulinotropic polypeptide
- KRH
- Krebs-Ringer-HEPES
- Lys-C
- endoproteinase Lys-C
- PACAP
- pituitary adenylate cyclase-activating peptide
- PTH
- parathyroid hormone
- TM
- transmembrane domain
- GLP1
- glucagon-like peptide 1
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- Boc
- t-butoxycarbonyl
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- ICM
- Internal Coordinate Mechanics.
REFERENCES
- 1. Ulrich C. D., 2nd, Holtmann M., Miller L. J. (1998) Gastroenterology 114, 382–397 [DOI] [PubMed] [Google Scholar]
- 2. Cherezov V., Rosenbaum D. M., Hanson M. A., Rasmussen S. G., Thian F. S., Kobilka T. S., Choi H. J., Kuhn P., Weis W. I., Kobilka B. K., Stevens R. C. (2007) Science 318, 1258–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jaakola V. P., Griffith M. T., Hanson M. A., Cherezov V., Chien E. Y., Lane J. R., Ijzerman A. P., Stevens R. C. (2008) Science 322, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., Hofmann K. P., Ernst O. P. (2008) Nature 455, 497–502 [DOI] [PubMed] [Google Scholar]
- 5. Warne T., Serrano-Vega M. J., Baker J. G., Moukhametzianov R., Edwards P. C., Henderson R., Leslie A. G., Tate C. G., Schertler G. F. (2008) Nature 454, 486–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rosenbaum D. M., Zhang C., Lyons J. A., Holl R., Aragao D., Arlow D. H., Rasmussen S. G., Choi H. J., Devree B. T., Sunahara R. K., Chae P. S., Gellman S. H., Dror R. O., Shaw D. E., Weis W. I., Caffrey M., Gmeiner P., Kobilka B. K. (2011) Nature 469, 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chien E. Y., Liu W., Zhao Q., Katritch V., Han G. W., Hanson M. A., Shi L., Newman A. H., Javitch J. A., Cherezov V., Stevens R. C. (2010) Science 330, 1091–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu B., Chien E. Y., Mol C. D., Fenalti G., Liu W., Katritch V., Abagyan R., Brooun A., Wells P., Bi F. C., Hamel D. J., Kuhn P., Handel T. M., Cherezov V., Stevens R. C. (2010) Science 330, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grace C. R., Perrin M. H., DiGruccio M. R., Miller C. L., Rivier J. E., Vale W. W., Riek R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12836–12841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grace C. R., Perrin M. H., Gulyas J., Digruccio M. R., Cantle J. P., Rivier J. E., Vale W. W., Riek R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4858–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pioszak A. A., Parker N. R., Suino-Powell K., Xu H. E. (2008) J. Biol. Chem. 283, 32900–32912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parthier C., Kleinschmidt M., Neumann P., Rudolph R., Manhart S., Schlenzig D., Fanghänel J., Rahfeld J. U., Demuth H. U., Stubbs M. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 13942–13947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Runge S., Thøgersen H., Madsen K., Lau J., Rudolph R. (2008) J. Biol. Chem. 283, 11340–11347 [DOI] [PubMed] [Google Scholar]
- 14. Underwood C. R., Garibay P., Knudsen L. B., Hastrup S., Peters G. H., Rudolph R., Reedtz-Runge S. (2010) J. Biol. Chem. 285, 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun C., Song D., Davis-Taber R. A., Barrett L. W., Scott V. E., Richardson P. L., Pereda-Lopez A., Uchic M. E., Solomon L. R., Lake M. R., Walter K. A., Hajduk P. J., Olejniczak E. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7875–7880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pioszak A. A., Xu H. E. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5034–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. ter Haar E., Koth C. M., Abdul-Manan N., Swenson L., Coll J. T., Lippke J. A., Lepre C. A., Garcia-Guzman M., Moore J. M. (2010) Structure 18, 1083–1093 [DOI] [PubMed] [Google Scholar]
- 18. Koth C. M., Abdul-Manan N., Lepre C. A., Connolly P. J., Yoo S., Mohanty A. K., Lippke J. A., Zwahlen J., Coll J. T., Doran J. D., Garcia-Guzman M., Moore J. M. (2010) Biochemistry 49, 1862–1872 [DOI] [PubMed] [Google Scholar]
- 19. Parthier C., Reedtz-Runge S., Rudolph R., Stubbs M. T. (2009) Trends Biochem. Sci. 34, 303–310 [DOI] [PubMed] [Google Scholar]
- 20. Dong M., Lam P. C., Gao F., Hosohata K., Pinon D. I., Sexton P. M., Abagyan R., Miller L. J. (2007) Mol. Pharmacol. 72, 280–290 [DOI] [PubMed] [Google Scholar]
- 21. Dong M., Lam P. C., Pinon D. I., Orry A., Sexton P. M., Abagyan R., Miller L. J. (2010) J. Biol. Chem. 285, 9919–9931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dong M., Lam P. C., Pinon D. I., Sexton P. M., Abagyan R., Miller L. J. (2008) Mol. Pharmacol. 74, 413–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dong M., Li Z., Pinon D. I., Lybrand T. P., Miller L. J. (2004) J. Biol. Chem. 279, 2894–2903 [DOI] [PubMed] [Google Scholar]
- 24. Bisello A., Adams A. E., Mierke D. F., Pellegrini M., Rosenblatt M., Suva L. J., Chorev M. (1998) J. Biol. Chem. 273, 22498–22505 [DOI] [PubMed] [Google Scholar]
- 25. Dong M., Pinon D. I., Cox R. F., Miller L. J. (2004) J. Biol. Chem. 279, 31177–31182 [DOI] [PubMed] [Google Scholar]
- 26. Hadac E. M., Ghanekar D. V., Holicky E. L., Pinon D. I., Dougherty R. W., Miller L. J. (1996) Pancreas 13, 130–139 [DOI] [PubMed] [Google Scholar]
- 27. Ulrich C. D., 2nd, Pinon D. I., Hadac E. M., Holicky E. L., Chang-Miller A., Gates L. K., Miller L. J. (1993) Gastroenterology 105, 1534–1543 [DOI] [PubMed] [Google Scholar]
- 28. Di Paolo E., De Neef P., Moguilevsky N., Petry H., Bollen A., Waelbroeck M., Robberecht P. (1998) FEBS Lett. 424, 207–210 [DOI] [PubMed] [Google Scholar]
- 29. Gardner J. D., Conlon T. P., Beyerman H. C., Van Zon A. (1977) Gastroenterology 73, 52–56 [PubMed] [Google Scholar]
- 30. Kofod H. (1991) Int. J. Pept. Protein Res. 37, 185–190 [DOI] [PubMed] [Google Scholar]
- 31. Powers S. P., Pinon D. I., Miller L. J. (1988) Int. J. Pept. Protein Res. 31, 429–434 [DOI] [PubMed] [Google Scholar]
- 32. Kurose T., Pashmforoush M., Yoshimasa Y., Carroll R., Schwartz G. P., Burke G. T., Katsoyannis P. G., Steiner D. F. (1994) J. Biol. Chem. 269, 29190–29197 [PubMed] [Google Scholar]
- 33. Dong M., Wang Y., Pinon D. I., Hadac E. M., Miller L. J. (1999) J. Biol. Chem. 274, 903–909 [DOI] [PubMed] [Google Scholar]
- 34. Holtmann M. H., Ganguli S., Hadac E. M., Dolu V., Miller L. J. (1996) J. Biol. Chem. 271, 14944–14949 [DOI] [PubMed] [Google Scholar]
- 35. Munson P. J., Rodbard D. (1980) Anal. Biochem. 107, 220–239 [DOI] [PubMed] [Google Scholar]
- 36. Ji Z., Hadac E. M., Henne R. M., Patel S. A., Lybrand T. P., Miller L. J. (1997) J. Biol. Chem. 272, 24393–24401 [DOI] [PubMed] [Google Scholar]
- 37. Abagyan R., Totrov M., Kuznetsov D. (1994) J. Comput. Chem. 15, 488–506 [Google Scholar]
- 38. Abagyan R., Totrov M. (1994) J. Mol. Biol. 235, 983–1002 [DOI] [PubMed] [Google Scholar]
- 39. Metropolis N., Rosenbluth A. W., Rosenbluth M. N., Teller A. H., Teller E. (1953) J. Chem. Phys. 21, 1087–1092 [Google Scholar]
- 40. Harikumar K. G., Lam P. C., Dong M., Sexton P. M., Abagyan R., Miller L. J. (2007) J. Biol. Chem. 282, 32834–32843 [DOI] [PubMed] [Google Scholar]
- 41. Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Nature 381, 272–272 [DOI] [PubMed] [Google Scholar]
- 42. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 43. Zang M., Dong M., Pinon D. I., Ding X. Q., Hadac E. M., Li Z., Lybrand T. P., Miller L. J. (2003) Mol. Pharmacol. 63, 993–1001 [DOI] [PubMed] [Google Scholar]
- 44. Dong M., Wang Y., Hadac E. M., Pinon D. I., Holicky E., Miller L. J. (1999) J. Biol. Chem. 274, 19161–19167 [DOI] [PubMed] [Google Scholar]
- 45. Dong M., Li Z., Zang M., Pinon D. I., Lybrand T. P., Miller L. J. (2003) J. Biol. Chem. 278, 48300–48312 [DOI] [PubMed] [Google Scholar]
- 46. Dong M., Asmann Y. W., Zang M., Pinon D. I., Miller L. J. (2000) J. Biol. Chem. 275, 26032–26039 [DOI] [PubMed] [Google Scholar]
- 47. Dong M., Zang M., Pinon D. I., Li Z., Lybrand T. P., Miller L. J. (2002) Mol. Endocrinol. 16, 2490–2501 [DOI] [PubMed] [Google Scholar]
- 48. Adelhorst K., Hedegaard B. B., Knudsen L. B., Kirk O. (1994) J. Biol. Chem. 269, 6275–6278 [PubMed] [Google Scholar]
- 49. Igarashi H., Ito T., Pradhan T. K., Mantey S. A., Hou W., Coy D. H., Jensen R. T. (2002) J. Pharmacol. Exp. Ther. 303, 445–460 [DOI] [PubMed] [Google Scholar]
- 50. Nicole P., Lins L., Rouyer-Fessard C., Drouot C., Fulcrand P., Thomas A., Couvineau A., Martinez J., Brasseur R., Laburthe M. (2000) J. Biol. Chem. 275, 24003–24012 [DOI] [PubMed] [Google Scholar]
- 51. Chen Q., Pinon D. I., Miller L. J., Dong M. (2010) J. Biol. Chem. 285, 24508–24518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gensure R. C., Shimizu N., Tsang J., Gardella T. J. (2003) Mol. Endocrinol. 17, 2647–2658 [DOI] [PubMed] [Google Scholar]
- 53. Greenberg Z., Bisello A., Mierke D. F., Rosenblatt M., Chorev M. (2000) Biochemistry 39, 8142–8152 [DOI] [PubMed] [Google Scholar]
- 54. Kraetke O., Holeran B., Berger H., Escher E., Bienert M., Beyermann M. (2005) Biochemistry 44, 15569–15577 [DOI] [PubMed] [Google Scholar]
- 55. Miller L. J., Chen Q., Lam P. C., Pinon D. I., Sexton P. M., Abagyan R., Dong M. (2011) J. Biol. Chem. 286, 15895–15907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Harikumar K. G., Hosohata K., Pinon D. I., Miller L. J. (2006) J. Biol. Chem. 281, 2543–2550 [DOI] [PubMed] [Google Scholar]