Abstract
Peptidoglycan (PG) N-acetyl muramic acid (MurNAc) O-acetylation is widely spread in Gram-positive bacteria and is generally associated with resistance against lysozyme and endogenous autolysins. We report here the presence of O-acetylation on N-acetylglucosamine (GlcNAc) in Lactobacillus plantarum PG. This modification of glycan strands was never described in bacteria. Fine structural characterization of acetylated muropeptides released from L. plantarum PG demonstrated that both MurNAc and GlcNAc are O-acetylated in this species. These two PG post-modifications rely on two dedicated O-acetyltransferase encoding genes, named oatA and oatB, respectively. By analyzing the resistance to cell wall hydrolysis of mutant strains, we showed that GlcNAc O-acetylation inhibits N-acetylglucosaminidase Acm2, the major L. plantarum autolysin. In this bacterial species, inactivation of oatA, encoding MurNAc O-acetyltransferase, resulted in marked sensitivity to lysozyme. Moreover, MurNAc over-O-acetylation was shown to activate autolysis through the putative N-acetylmuramoyl-l-alanine amidase LytH enzyme. Our data indicate that in L. plantarum, two different O-acetyltransferases play original and antagonistic roles in the modulation of the activity of endogenous autolysins.
Keywords: Bacteria, Bacterial Metabolism, Carbohydrate, Cell Wall, Lactic Acid, Autolysis, Lactobacillus plantarum, Lysozyme, Peptidoglycan O-Acetylation, Glucosaminidase
Introduction
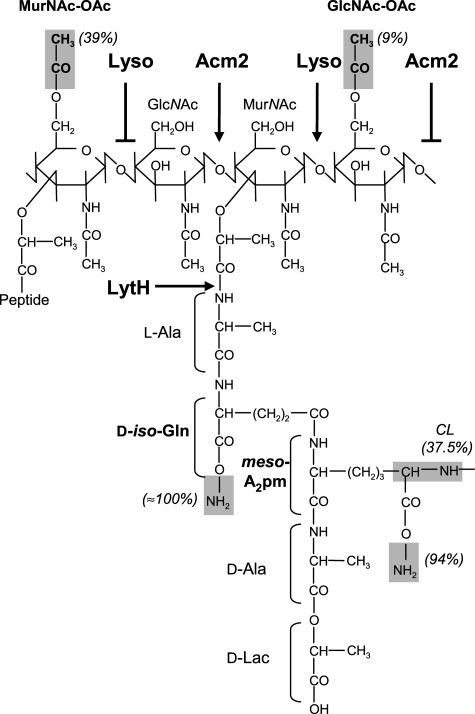
Peptidoglycan (PG),6 the major constituent of the cell envelope of Gram-positive bacteria, is a polymer of the disaccharide N-acetylmuramic acid-(β-1,4)-N-acetylglucosamine (MurNAc-GlcNAc) associated with a peptidic stem linked to MurNAc (1, 2). The composition of the peptidic stem varies from one bacterium to another and, in Lactobacillus plantarum, is composed of l-alanine, d-glutamic acid, meso-diaminopimelic acid, d-alanine, and a d-lactate as last moiety (see Fig. 1) (3–6). The different glycan strands are cross-linked between the fourth amino acid of the donor stem and the third amino acid of the acceptor peptide forming a three-dimensional network around the cell called the sacculus. The main functions of this sacculus are to ensure cell integrity against the internal osmotic pressure and to provide the shape of the bacteria (1, 2, 7).
FIGURE 1.
Structure of PG and modulation of PGH activity in L. plantarum. Two disaccharides are represented. The different identified modifications (O-acetylation, amidation, and cross-linking) are highlighted in gray, and their occurrences (%) are indicated in parentheses. The O-acetylation of GlcNAc on C6-OH is predicted by analogy to MurNAc O-acetylation. Modulation of PGH activities by O-acetylation are indicated by an arrow (cleavage) or a broken arrow (no cleavage). Lyso, lysozyme (N-acetylmuramidase); Acm2, N-acetylglucosaminidase; LytH, N-acetylmuramoyl-l-alanine amidase; meso-A2 pm, meso-diaminopimelic acid; CL, cross-linking index.
Because of its rigidity, PG has to be remodeled by the PG hydrolases (PGH) to allow cell growth and cell division. PGH are divided into different classes according to their cleavage site: the carboxy- and endopeptidases that cleave the peptidic stem, the N-acetylmuramoyl-l-alanine amidases that cleave between the first amino acid and MurNAc, and finally the N-acetylglucosaminidases and N-acetylmuramidases that cut inside the glycan chain between GlcNAc and MurNAc or MurNAc and GlcNAc, respectively (7, 8).
Extracellular synthesis of PG begins with the addition of MurNAc-GlcNAc-pentadepsipeptide from lipid II to an existing glycan strand and the cross-linking of the peptidic stem, both catalyzed by the high molecular weight penicillin binding proteins (2, 9). In addition, the MurNAc-GlcNAc disaccharide can be modified by the addition of an acetyl group on the C6-OH position of MurNAc or by removing the acetyl group N-linked to MurNAc or GlcNAc (10–20). These modifications confer resistance against lysis induced by lysozyme, an N-acetylmuramidase found in human tears, saliva, or gastrointestinal tract (10, 13, 14, 18, 19). O-Acetylation inhibits lysozyme activity through steric hindrance (10) but also lytic transglycosylases that need a free –OH group at position 6 of MurNAc to form the anhydro ring concomitantly to the cleavage of GlcNAc-MurNAc linkage (20, 21).
MurNAc O-acetylation is described in Gram-negative bacteria as well as in Gram-positive bacteria but involves a different set of dedicated proteins (16, 20). Moynihan and Clarke (16) have recently demonstrated that two different proteins are involved in this process in Gram-negative bacteria: 1) an integral membrane protein called PatA is dedicated to the transport of the acetyl-donor and 2) a periplasmic membrane-anchored protein called PatB catalyzes the acetylation reaction. In contrast, in Gram-positive bacteria, MurNAc O-acetylation can be catalyzed by a unique membrane-bound protein named OatA for O-acetyltransferase. OatA is composed of two domains, 11 transmembrane helices form the first domain at the N terminus and an extracellularly exposed globular domain, which contain the catalytic site at the C terminus of the protein (10). Importantly, a recent study showed the presence of both types of O-acetyltransferases involved in PG O-acetylation in Bacillus anthracis (22).
Despite an extensive study of the role of MurNAc O-acetylation as a pathogenic determinant, little is known about its basic function in cell wall assembly and degradation (10, 12–14). MurNAc O-acetylation is described as a means to increase PG resistance. In Streptococcus pneumoniae and Enteroccocus faecalis, it provides resistance against general autolysis (13, 23). Emirian et al. (23) reported a modest impact of O-acetylation on the activities of the PGH AtlA and AtlB in E. faecalis. Crisostomo et al. (13) described MurNAc O-acetylation as conferring resistance to β-lactams in S. pneumoniae, probably through a more robust PG.
Here, we identify the presence of O-acetylation on GlcNAc residues of PG and describe the first bacterium containing two types of O-acetylation. These two modifications play opposing roles in the control of autolysis within L. plantarum. Strikingly, MurNAc O-acetylation increases L. plantarum autolysis via LytH, a putative l-alanine-muramyl amidase, whereas GlcNAc O-acetylation provides resistance against N-acetylglucosaminidase Acm2-mediated autolysis.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in the present study are listed in supplemental Table S1. Plasmids were constructed in Escherichia coli MC1061. E. coli and Bacillus subtilis were grown in LB medium with shaking at 37 °C. L. plantarum was grown in MRS broth (Difco Laboratories, Inc., Detroit, MI) at 30 °C. When required, erythromycin (250 μg/ml for E. coli, 5 μg/ml for L. plantarum) or chloramphenicol (10 μg/ml for E. coli and L. plantarum) was added to the medium. Solid agar plates were prepared by adding 2% (w/v) agar to the medium. For lysozyme plate assays, chicken egg white lysozyme powder (Sigma-Aldrich) was directly added at a final concentration of 2 mg/ml. Nisin A (Sigma-Aldrich) was routinely used at a concentration of 20 ng/ml for the induction of genes under the control of the nisA expression signals. Cephalothin, ampicillin, and methicillin were purchased from Sigma-Aldrich.
DNA Techniques and Electrotransformation
General molecular biology techniques were performed according to the instructions given by Sambrook et al. (24). Electrotransformation of E. coli was performed as described by Dower et al. (25). Electrocompetent L. plantarum cells were prepared as described previously (26). PCRs were performed with the Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland) in a GeneAmp PCR system 2400 (Applied Biosystems, Foster City, CA). The primers used in this study were purchased from Eurogentec (Seraing, Belgium) and are listed in supplemental Table S2.
Construction of Deletion Mutants
Construction of the L. plantarum gene deletion mutants for oatA (lp_0856) and oatB (lp_0925) was performed as described previously (27). A double crossover gene replacement strategy was used to replace the target gene(s) by a chloramphenicol resistance cassette (lox66-P32cat-lox71; (27)). Briefly, the upstream and downstream flanking regions of the target genes were amplified by PCR using L. plantarum WCFS1 chromosomal DNA. Subsequently, amplicons were respectively cloned in the SwaI (oatA and oatB) and SmaI (oatA) or Ecl136II (oatB) restriction sites of the suicide vector pNZ5319 (27). The mutagenesis plasmids were transformed in L. plantarum WCFS1 and colonies displaying a chloramphenicol-resistant and erythromycin-sensitive phenotype represent candidate double crossover gene replacements. The anticipated cat replacement genotype was confirmed by PCR using primers flanking the sites of recombination (supplemental Table S2). Subsequently, the lox66-P32cat-lox71 cassette was excised by temporal expression of the cre recombinase using the unstable cre expression plasmid pNZ5348 as described previously (27). Erythromycin-resistant and chloramphenicol-sensitive colonies were checked by PCR for Cre-mediated recombination and correct excision of the cassette by using primers flanking the recombination locus (supplemental Table S2). This strategy was applied for the construction of oatA::lox72 (OatA−, EB002) and oatB::lox72 (OatB−, EB003) deletion mutants. To obtain the double oatA::lox72 oatB::P32cat (OatAB−, EB004) the oatA::lox72 mutant was used for a next round of mutagenesis targeting for the deletion of oatB following the same procedures as described above.
Cloning and Overexpression of oatAWT and oatAD510A/S511A
The oatA gene was amplified by PCR using the primer pair 5′oatAPstI-3′OatASpeI, generating a 1990-bp DNA fragment. The PstI/SpeI-restricted fragment was ligated to a PstI/SpeI-restricted pNZ8048 (28) and transformed in E. coli, leading to the pGIEB003 expression plasmid. To generate a catalytic mutant enzyme of OatA (OatA(D510A/S511A)), point mutations were introduced by PCR using the primer pair OatAS511A and OatAD510A, containing the S511A and D510A mutations respectively, with pGIEB003 as a template. The PCR product was phosphorylated and then ligated, leading to the pGIEB011 expression plasmid. The integrity of oatA and oatAD510A/S511A were verified by DNA sequencing.
Purification and Structural Analysis of PG
PG from L. plantarum strains was prepared as described previously (29) with some modifications. DNase (50 μg/ml) and RNase (50 μg/ml) treatment were applied before hydrofluoric acid treatment. PG was digested with mutanolysin from Streptomyces globisporus (Sigma-Aldrich), and the resulting muropeptides were analyzed by RP-HPLC and MALDI-TOF mass spectrometry as reported previously (29). Alkaline treatments of muropeptide solutions were performed by increasing pH to pH 13 with 5 m NaOH for 2 h at 37 °C. For lysozyme digestions, PG was incubated with chicken egg white lysozyme (Sigma-Aldrich) at a final concentration of 2 mg/ml in 50 mm Tris-HCl buffer, pH 7.0, at 37 °C with gentle agitation during 16 h. For MSn structural analysis, muropeptides were desalted on a Betasil C18 column (4.6 × 250 mm, Thermo Electron Corp.) with an acetonitrile/formic acid buffer system and dried with a speed vacuum. Samples were solubilized in 2% acetonitrile, 0.1% formic acid in Milli-Q water (1 μl for 1 mAU detected at 214 nm in the previous HPLC system). Each purified muropeptide was injected and analyzed at a flow rate of 0.2 μl/min on the mass spectrometers (LTQ-ETD or LTQ-Orbitrap, Thermo Fisher Scientific) located on the PAPPSO platform (INRA, UMR1319 Micalis, France).
Triton X-100-induced Autolysis in Buffer Solution
L. plantarum strains were grown in MRS medium to midexponential phase (A600 = 0.8). Cells were harvested by centrifugation at 5000 × g for 10 min at 4 °C, washed once with 50 mm potassium phosphate buffer, pH 7.0, and resuspended at an A600 of 1.0 in 50 mm potassium phosphate buffer, pH 7.0, supplemented with 0.05% Triton X-100 (30). Cell suspensions were then transferred into 96-well sterile microplates with a transparent bottom (Greiner, Alphen a/d Rjin, the Netherlands) and incubated at 30 °C. Autolysis was monitored by measuring the A600 of the cell suspensions every 20 min with a Varioskan Flash multimode reader (Thermo Fisher Scientific). The extent of autolysis was expressed as the percentage decrease in A600.
Assay of Acm2 Activity Using L. plantarum Dead Cells
L. plantarum-autoclaved cells were used as a substrate for measuring Acm2 activity and were prepared as follows. L. plantarum strains were grown in MRS medium to midexponential phase (A600 = 0.8). Cells were harvested by centrifugation at 5000 × g for 10 min at 4 °C, washed twice with deionized H2O, resuspended at A600 = 200 in deionized H2O, autoclaved at 120 °C during 20 min, and stored at 4 °C. Autoclaved cells were diluted in 50 mm Tris-HCl, pH 7.0, to A600 = 0.8. Purified His6-tagged Acm2 (2.5 μg/ml final concentration) was added in a final volume of 300 μl, and the turbidity of the cell suspension was monitored by measuring the A600 every 10 min with a Varioskan Flash multimode reader (Thermo Fisher Scientific).
SDS-PAGE and Zymogram
SDS-PAGE was performed with 8% (w/v) polyacrylamide separating gels. Zymogram was performed as described previously (31). The polyacrylamide gels contained L. plantarum autoclaved cells resuspended at A600 of 4.0 as enzyme substrates. Lysed cells were used as a sample. Briefly, overnight culture was washed in 50 mm potassium phosphate buffer, pH 7.0, and then cells were lysed with glass beads using a Precellys cell disrupter (Bertin Technologies) at 5000 rpm for 5 × 30 s. Cell lysates were boiled in denaturing sample buffer and centrifuged for 1 min at 20,000 × g prior loading. After sample migration in the gels, the gels were washed for 30 min in deionized H2O at room temperature and then incubated in 50 mm Tris-HCl, pH 6.0, 1 mm DTT containing 0.1% (v/v) Triton X-100 overnight at room temperature. The gels were subsequently washed for 30 min in deionized H2O and then stained with 0.1% methylene blue in 0.01% (w/v) KOH for 2 h at room temperature and destained in deionized H2O.
Bioinformatic Analysis
Homology searches and phylogenic analysis were performed using the BLASTP program. Phylogenic tree was drawn with TreeGraph 2 software (32). Sequence alignments were carried out with the PRALINE algorithm (33). Secondary structures were predicted using HMMTOP (version 2.1) (34), whereas the tertiary structures were predicted with LOMETS metaserver (35) and drawn using PyMOL software.
RESULTS
Peptidoglycan of L. plantarum Contains Two Types of O-Acetylation
The PG structure of L. plantarum NZ7100 (WCFS1 derivative) was determined by analysis of muropeptides obtained after mutanolysin digestion by RP-HPLC and MALDI-TOF mass spectrometry. The measured m/z values for the different muropeptides confirmed the previously proposed structure with a direct meso-diaminopimelic acid cross-bridge (Fig. 1, supplemental Fig. S1, and supplemental Table S3) (3–6). The PG of L. plantarum NZ7100 displays a cross-linking index of 37.5%. Muropeptides with a tripeptide chain as acceptor chain are the most abundant (46%), revealing carboxypeptidase activity. Concerning PG structural modifications, amidation was found on d-iso-glutamate and meso-diaminopimelic acid (100 and 94%, respectively). Around 44% of monomers harbor at least one acetylation.
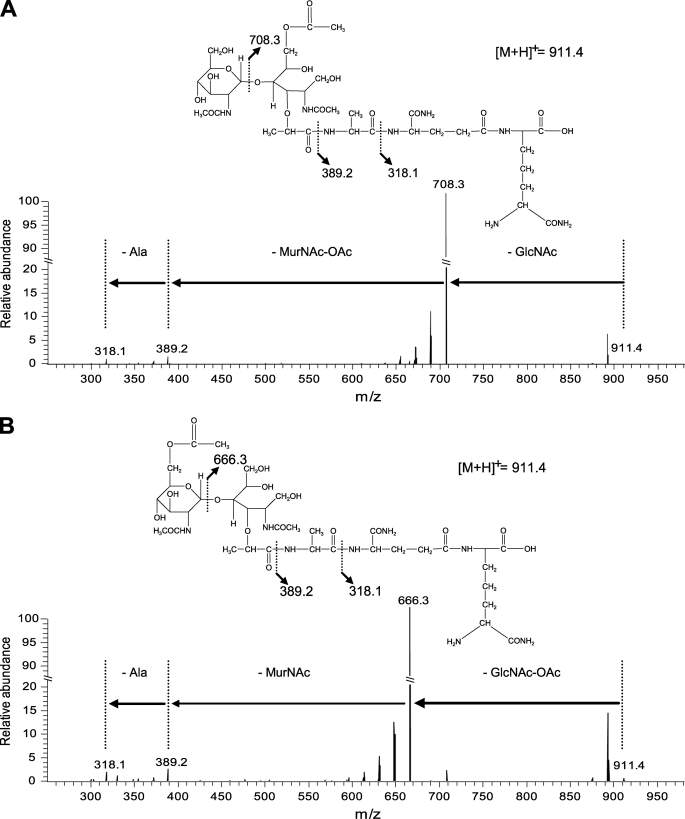
Intriguingly, we identified acetylated muropeptides presenting a nearly identical mass (m/z of 933.45 for sodiated molecular ions) but with a different elution time on the HPLC profile (Fig. 2A and supplemental Table S3). Tandem mass spectrometry (MS/MS) performed on these muropeptides showed a different localization of the O-acetyl group in the molecules. As shown for peak 12 (Fig. 2A and supplemental Fig. S1), corresponding to the more abundant acetylated disaccharide tripeptide, the fragmentation pattern showed the presence of one GlcNAc (mass loss of 203.08 Da) and one O-acetylated MurNAc (mass loss of 319.13 Da) (Fig. 3A). In contrast, for peak 14 (Fig. 2A and supplemental Fig. S1), corresponding to a second acetylated disaccharide tripeptide, fragments resulting from the loss of one O-acetylated GlcNAc (245.10 Da) and one MurNAc (277.11 Da) were identified (Fig. 3B). All of the predicted O-acetylated muropeptides were sensitive to alkaline pH (NaOH treatment) and lost their acetyl group due to their chemical properties (data not shown). Globally, O-acetylation of MurNAc and GlcNAc was found in ∼39 and ∼9% of monomers, respectively (supplemental Table S3). Disaccharide tripeptide with a double O-acetylation (peak 23, supplemental Fig. S1) as well as dimers with three O-acetylations could also be detected, showing that adjacent O-acetylations of MurNAc and GlcNAc are not mutually exclusive (supplemental Fig. S1 and Table S3). To our knowledge, L. plantarum is the first identified bacterium whose PG contains two types of O-acetylation on glycan strands and in particular with O-acetylation of GlcNAc that has never been reported previously.
FIGURE 2.
RP-HPLC separation of muropeptides from L. plantarum PG. WT (A), oatA mutant (B), oatB mutant (C), and oatA oatB mutant (D). Peak 12 (supplemental Table S3), the dissacharide tripeptide with O-acetylated MurNAc, is indicated by an asterisk, and peak 14 (supplemental Table S3), the dissacharide tripeptide with O-acetylated GlcNAc, is indicated by an arrow.
FIGURE 3.
Fragmentation of O-acetylated dissacharide tripeptides by MS-MS. A, localization of the O-acetyl group on MurNAc. Peak 12 (supplemental Table S3) yielded fragments resulting from a mass loss of 203.08 Da (GlcNAc residue) and 319.13 Da (MurNAc-OAc residue). B, localization of the O-acetyl group on GlcNAc. Peak 14 (supplemental Table S3) yielded fragments resulting from a mass loss of 245.10 Da (GlcNAc-OAc residue) and 277.11 Da (MurNAc residue). Fragmentation was performed on the [M + H]+ ion at m/z 911.4. The indicated m/z values correspond to ions obtained by cleavage of peptide bonds as represented on the chemical structures. The indicated masses correspond to [M + H]+.
O-Acetylation of MurNAc and GlcNAc Results from Activity of Two Dedicated O-Acetyltransferases
Identification of the O-acetyltransferase content of L. plantarum WCFS1 was performed using the BLASTP algorithm with the staphylococcal OatA protein (SAV2567) as the query. Two candidates encoded by lp_0856 and lp_0925 were found with amino acid similarity of 51 and 43%, respectively. The closer homologue was named OatA (lp_0856, 660 amino acids), and the most distal candidate was named OatB (lp_0925, 615 amino acids). These two O-acetyltransferases only displayed 21% identity to each other.
Phylogenetic analysis showed that these two O-acetyltransferases belong to two different protein clusters (supplemental Fig. S2). OatA is closer to O-acetyltransferases present in lactobacilli and lactococci, whereas OatB seems to be related to streptococcal O-acetyltransferases. In this analysis, we also identified two other bacterial species (Lactobacillus sakei subsp. sakei 23K and Weissella paramesenteroides ATCC33313) that harbor two potential O-acetyltransferases belonging to the OatA and OatB subgroups as found in L. plantarum.
Protein sequence comparison revealed some common features between OatA and OatB. Both proteins display a similar organization with an N-terminal domain composed of 11 predicted transmembrane segments (HMMtop prediction, version 2.1), potentially involved in acyl donor transport, and a surface-exposed C-terminal region corresponding to the predicted acetyltransferase catalytic domain (Fig. 4A). Both domains exhibit sequence identity with PatA (7 and 9% for OatA and OatB, respectively) and PatB (17 and 20%), which were identified, respectively, as the acyl donor transporter and O-acetyltransferase in Neisseria gonorrhoeae (supplemental Fig. S3) (16). Both acetyltransferase domains of OatA and OatB were assigned to the family of SGNH/GDSL hydrolases (LOMETS prediction) as reported for PatB with a conserved Ser-Asp-His predicted catalytic triad (Fig. 4B and supplemental Fig. S4) (16).
FIGURE 4.
Comparison between OatA and OatB. A, schematic representation of the topology of OatA and OatB. The position and the orientation of transmembrane segments are drawn based on HMMTOP prediction. The numbers denote sizes (in amino acids) of the different loops. The putative C-terminal acetyltransferase domain (AT) is predicted as surface-exposed. Regions displaying identity and similarity (% in parentheses) with N. gonorrhoeae PatA and PatB are presented in blue and red, respectively. B, partial sequence alignment of the AT domain of OatA and OatB. Conservation based on the PRALINE algorithm is represented by a color code where dark blue and red represent the less and most conserved residues, respectively. Similarity scores are indicated below the alignment. Values in parentheses denote the residue number of OatA from L. plantarum used as reference, and black asterisks indicate the putative catalytic residues. Abbreviations (locus tag) are as follows: OatA_Lpl, L. plantarum (lp_0856); OatA_Wpa, W. paramesenteroides (HMPREF0877_0552); OatA_Lla, L. lactis (llmg_2391); OatA_Sau, Staphylococcus aureus (SAV2567); OatA_Lsa, L. sakei (LSA_1044); OatA_Efa, E. faecalis (EF_0783); OatA_Lmo, Listeria monocytogenes (lmo1291); OatB_Lpl, L. plantarum (lp_0925); OatB_Lsa, L. sakei (LSA_0646); OatB_Wpa, W. paramesenteroides (HMPREF0877_1514); PatB_Ngo, N. gonorrhoeae (NGO0533).
To confirm the role of these two putative O-acetyltransferases, oatA and oatB were independently deleted and a double oatA oatB mutant was also constructed. PG analysis of the oatA mutant showed a complete disappearance of O-acetylated MurNAc containing muropeptides and the persistence of a similar proportion of O-acetylated GlcNAc containing muropeptides (Fig. 2B). Conversely, the deletion of oatB led to a complete absence of O-acetylated GlcNAc containing muropeptides with the maintenance of O-acetylated MurNAc containing muropeptides (Fig. 2C). The PG of the double mutant was totally depleted of both O-acetylated MurNAc and GlcNAc containing muropeptides (Fig. 2D). A more detailed analysis of the muropeptide content of the three mutants did not reveal any other major PG structure modification (data not shown). These results show that O-acetylation of MurNAc and GlcNAc in L. plantarum are separately performed by two dedicated O-acetyltransferases.
OatA Alone Confers Resistance to Lysozyme
The lysozyme resistance conferred by O-acetylation of MurNAc is well documented (10, 13, 14, 18), but nothing is known about O-acetylation of GlcNAc and its impact on lysozyme resistance. To assess their contribution to this function in vivo, plate assays with a fixed concentration of lysozyme (2 mg ml−1) were performed with the various mutant strains (Fig. 5A, upper panel). The oatA mutant showed a higher sensitivity to lysozyme compared with the wild-type strain, whereas the deletion of oatB has no effect in both wild-type and OatA-deficient backgrounds. Complementation of the oatA mutant with oatAWT on a multicopy plasmid restores lysozyme resistance (Fig. 5A, lower panel), whereas a catalytic mutant of OatA (OatA(D510A/S511A)) was not able to complement the lysozyme resistance phenotype (data not shown).
FIGURE 5.
Involvement of MurNAc O-acetylation in lysozyme resistance. A, comparison of lysozyme resistance of O-acetyltransferase mutants by serial dilutions (100 to 10−5) on MRS medium supplemented or not with lysozyme (2 mg/ml). For complementation experiments (bottom panels), chloramphenicol (10 μg/ml) and nisin (20 ng/ml) were added to MRS medium. OatA−, oatA mutant; OatB−, oatB mutant; OatAB−, oatA oatB mutant; OatA−/ctl, oatA mutant carrying the empty plasmid pNZ8048 (control); OatA−/OatA+, oatA mutant complemented with plasmid pGIEB003 (OatAWT), WT/ctl, WT carrying the empty plasmid pNZ8048 (control). B, RP-HPLC separation of muropeptides resulting from PG digestion by lysozyme. I, WT; II, oatA mutant; III, oatB mutant. Asterisks indicate GlcNAc-OAc containing muropeptides (peaks 14 and 33, supplemental Table S3).
To confirm these in vivo observations, PG from wild-type, oatA, and oatAB mutants was digested by lysozymes, and the resulting products were analyzed by RP-HPLC (Fig. 5B). Wild-type PG was poorly digested by lysozyme, whereas PG depleted of O-acetylated MurNAc from the oatA mutant was well digested, confirming the established role of MurNAc O-acetylation in lysozyme resistance (Fig. 5B, I and II). Notably, the lysozyme was able to release O-acetylated GlcNAc containing muropeptides from the PG of the oatA mutant, which were absent in the digestion profile of the PG from the oatAB mutant (Fig. 5B, II and III). These data clearly show that the lysozyme (N-acetylmuramidase) is able to cleave the β-1–4-linkage next to an O-acetylated GlcNAc (Fig. 1) and that this O-acetylation is not involved in lysozyme resistance.
O-Acetylation of GlcNAc Inhibits Activity of Acm2 N-Acetylglucosaminidase
Control of autolysis by PG modifications is well established (13, 15, 17, 23, 36). O-acetylation of MurNAc in some species has been reported to confer resistance to endogenous PGH (13, 17, 23). As L. plantarum PG contains two types of O-acetylation, the control of the PGH pool in this species could be unique compared with previous reports. To evaluate the impact of O-acetylation on PGH activity, Triton X-100-induced autolysis tests were performed with the different O-acetyltransferase mutants (Fig. 6A). Notably, the mutant strain depleted of O-acetylated GlcNAc (OatB−) was more sensitive to autolysis compared with the wild-type, whereas the mutant strain without O-acetylated MurNAc (OatA−) was more resistant. This opposite behavior was confirmed by the autolytic profile of the double mutant strain (OatAB−), which is similar to the wild-type, revealing a compensatory effect.
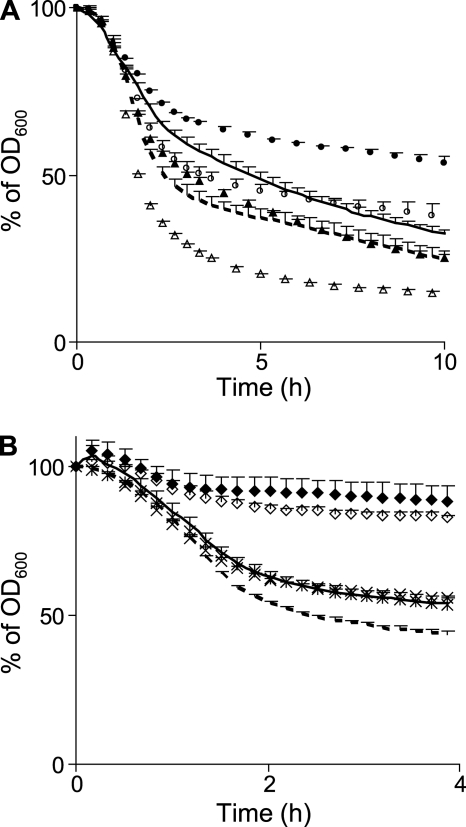
FIGURE 6.
Effect of GlcNAc O-acetylation on autolysis and Acm2 activity. A, autolysis of L. plantarum and its mutant derivatives in presence of Triton X-100 (0.05%). Wild-type is represented by a line, oatA mutant by circles, oatB mutant by triangles, and oatA oatB mutant by squares. Mean values ± S.D. of one representative experiment of three independent experiments (n = 5 for each). B, zymogram with cell extracts of B. subtilis (Bsu), L. plantarum wild-type (Lpl or Lpl WT), and acm2 mutant (Lpl Acm2−) against dead cells of WT or derivative strains either lacking OatA (OatA−) or OatB (OatB−). C, Acm2 activity against autoclaved cells of L. plantarum. Mean values ± S.D. of one representative experiment of two independent experiments (n = 3 for each). Symbols are as described in A.
To confirm these data, the PGH activity content of L. plantarum NZ7100 was evaluated by zymogram analysis using wild-type, OatA−, and OatB− autoclaved cells as substrates (Fig. 6B). B. subtilis 168 cell extracts were used as control because the PG of this species is very similar to L. plantarum (meso-diaminopimelic acid direct linkage) and only contains O-acetylated MurNAc (22). The same amount of crude cell extracts of the two species was tested on the different substrates. The PGH activity profile of B. subtilis was globally similar on the different substrates. In contrast, the PGH activity was strongly increased for L. plantarum using OatB− dead cells compared with wild-type and OatA− substrates, confirming that O-acetylated GlcNAc inhibits one or more PGH in this species. We have recently shown that the N-acetylglucosaminidase Acm2 is the major autolysin of L. plantarum WCFS1.7 To evaluate its contribution to the autolysin profile observed on zymogram with the OatB− substrate, the same amount of crude cell extracts from the wild-type and an acm2-deleted mutant were compared (Fig. 6B). All of the autolytic activity bands detected on the OatB− substrate are attributed to Acm2 and its various processed forms. To definitively prove that Acm2 activity is modulated by O-acetylation of GlcNAc, purified His6-tagged Acm2 was incubated with wild-type, OatA−, and OatB− dead cells. As shown in Fig. 6C, Acm2 activity was similar on wild-type and OatA− substrates but strongly increased with the OatB− substrate deprived of O-acetylated GlcNAc, whereas the activity of mutanolysin, an N-acetylmuramidase insensitive to O-acetylation used as control, was not affected (supplemental Fig. S5). Altogether, these data show an inhibitory effect of O-acetylated GlcNAc on autolysis of L. plantarum, which is mediated by an inhibition of the N-acetylglucosaminidase Acm2 (Fig. 1).
O-Acetylation of MurNAc Activates LytH N-Acetylmuramoyl-l-alanine Amidase
Another strategy to evaluate the contribution of PG O-acetylation on PGH activity is to increase the O-acetylation level by O-acetyltransferase overproduction. Overproduction of OatAWT and OatA(D510A/S511A) were achieved by the use of the nisin-inducible controlled expression system. Unfortunately, we were unable to clone oatB under the control of the nisin-inducible controlled expression system in various hosts except a truncated version deprived of the acetyltransferase domain, suggesting a toxic effect of GlcNAc O-acetylation. Concerning overproduction of OatAWT, the growth rate slightly decreased in a nisin dose-dependent manner both in wild-type and OatA− backgrounds, whereas overproduction of the catalytic site mutant OatA(D510A/S511A) had no impact on growth. Together, these data confirm the role of O-acetylation rather than protein overproduction in this slower growth (supplemental Fig. S6, A and B). This slight effect on growth is in sharp contrast with the major growth defect due to OatA overproduction previously reported in Lactococcus lactis (18). Concerning the impact on MurNAc O-acetylation, we showed that overproduction of OatAWT leads to an increase of ∼6% of O-acetylated MurNAc containing muropeptides (supplemental Table S4) compared with the control strain containing the empty plasmid.
To evaluate the contribution of over-O-acetylation on PGH activity, Triton X-100-induced autolysis tests were performed with wild-type and the O-acetyltransferase mutants (OatA− and OatB−) overproducing OatAWT. An increased autolysis was observed in each overproducing strain (Fig. 7A). In contrast, overproduction of OatAD510A/S511A did not increase autolysis confirming a direct role of over-O-acetylation rather than an indirect effect due to overproduction (data not shown). To identify which autolysin is activated by an increase of O-acetylated MurNAc, OatAWT overproduction was achieved in a collection of eight different PGH mutants of L. plantarum (four predicted N-acetylmuramidases/N-acetylglucosaminidases, Acm1, Acm2, Lys, and Lp_3093; one N-acetylmuramoyl-l-alanine amidase, LytH; two endopeptidases, Lp_2162 and Lp_1242; and the lytic transglycosylase Lp_0302) (37). Triton X-100-induced autolysis tests revealed that over-O-acetylation was able to increase autolysis of all strains (illustrated for the acm2 mutant in Fig. 7B), except for the lytH mutant where the increased autolysis phenotype was completely abolished (Fig. 7B).
FIGURE 7.
Effect of MurNAc over-O-acetylation on autolysis. A, autolysis curves (Triton X-100-induced) of oatAWT overexpressing strains L. plantarum WT (dashed line), oatA mutant (open circles), and oatB mutant (open triangles) compared with control strains (empty vector) of WT (line), oatA mutant (filled circles), and oatB mutant (filled triangles). Mean values ± S.D. (n = 3). B, autolysis curves of oatAWT overexpressing strains: WT (dashed line), acm2 mutant (open diamonds), and lytH mutant (asterisks) compared to control strains of WT (line), acm2 mutant (filled diamonds), and lytH mutant (crosses). WT (line) and lytH mutant (asterisks and crosses) overlap. Mean values ± S.D. of one representative experiment of two independent experiments (n = 6 for each).
DISCUSSION
L. plantarum is the first bacterium described to harbor two different types of O-acetylation in its cell wall PG. MurNAc-specific O-acetyltransferases are widespread among Gram-positive bacteria as well as in a range of Gram-negative bacteria (12, 20), whereas GlcNAc-specific O-acetyltransferases (OatB), described for the first time in this study, seem to be restricted to a smaller number of bacterial species. In Gram-positive bacteria, OatA proteins are derived from at least three different ancestors (supplemental Fig. S2), one of which led to two different types of O-acetyltransferases: the MurNAc-specific O-acetyltransferases mainly present in streptococci and the GlcNAc-specific O-acetyltransferases (OatB subgroup, supplemental Fig. S2). As the phylogeny of GlcNAc O-acetyltransferases does not reflect the phylogeny of the bacteria included in this protein cluster, we hypothesized that the oatB gene was acquired by horizontal gene transfer. The GC content of the oatB gene of L. plantarum (49%) is higher than that of the whole genome and of the oatA gene (both 44%) (37), which would be in favor of such an event. The origin of OatB remains unclear because the GlcNAc O-acetyltransferase could derive from a MurNAc O-acetyltransferase ancestor or the opposite.
Sequence comparison between OatA and OatB proteins exhibited a similar global organization with two predicted domains. The N-terminal transmembrane domain, composed of 11 transmembrane segments, could be involved in the transport of acetyl-CoA, as previously proposed to be the acetyl donor (10). Alignment of this domain between OatA and OatB members showed a higher similarity in five transmembrane segments (TM1, -2, -4, -6, and -9), including a range of well conserved aromatic residues (Phe, Tyr, Trp) and the conserved region Fxx(HR)RxxR (91–98 amino acids in OatALp) in the intracellular loop L2 (23 amino acids) (supplemental Fig. S3A). However, there is no clear clue to assign any of these conservations regarding to substrate specificity. The globular C-terminal domain, predicted to be surface exposed and assigned to the family of SGNH/GDSL hydrolases, displays a similar conservation level between OatA, OatB, and PatB (32–33% of similarity), the latter being reported to display a MurNAc-specific O-acetyltransferase activity (16). The two most conserved regions GDSV (509–512 amino acids in OatALp) and DxxH (634–637 amino acids in OatALp) are supposed to contain the predicted Ser-Asp-His catalytic triad (underlined amino acids) (supplemental Fig. S3B), for which we showed the importance of Asp-510 and Ser-511 as their concomitant substitution by alanine residues leads to OatA inactivation. The use of various modeling tools shows that the residues of the catalytic triad are indeed close to each other in OatA, OatB, and PatB (supplemental Fig. S4) but does not allow to define residues or structural elements involved in the recognition of MurNAc or GlcNAc as substrates.
MurNAc O-acetylation is seen as a mean for the bacteria to increase their PG resistance against lysozyme, autolysins, β-lactams, or as a step leading to the entry in a dormant state (10, 12–14, 17, 18, 23). Indeed, L. plantarum MurNAc O-acetylation also confers resistance to lysozyme and β-lactams (supplemental Fig. S7). However, in contrast to previous reports, L. plantarum MurNAc O-acetylation does not enhance the PG resistance against its own PGH, but instead induces autolysis through the LytH enzyme, a putative N-acetylmuramoyl-l-alanine amidase. This was observed by the over-O-acetylation of MurNAc and corroborates the observation of a slight decrease in autolysis observed with the oatA mutant compared with the wild-type. Future biochemical characterization of this amidase will be needed to ascertain the effect of the O-acetyl group of MurNAc on its activity.
L. plantarum is the first species identified containing GlcNAc O-acetylation in its PG. Notably, we show that the major autolysin, N-acetylglucosaminidase Acm2, displays a higher autolytic activity on PG deprived of GlcNAc O-acetylation. The underlying mechanism could very well be a lack of access to cleavage of the β-1–4 bond between GlcNAc and MurNAc due to the presence of the acetyl group on GlcNAc as demonstrated for acetylated MurNAc regarding to lysozyme activity (Fig. 1) (10). Remarkably, GlcNAc O-acetylation in L. plantarum plays a similar role as MurNAc O-acetylation in other bacteria regarding inhibition of autolysins. This independent evolution toward a similar functional role of both types of O-acetylation highlights their importance for the control, probably in a localization-dependent manner, of the activity of autolysins without directly altering their intrinsic activity. Hypothetically, the regulation of autolysis by O-acetylated GlcNAc in L. plantarum was horizontally acquired; therefore, we propose a divergent evolutionary path for this species. Initially, the role of MurNAc O-acetylation was probably similar to other bacteria; however, the acquisition and maintenance of the second O-acetylation has led to an adaptation of synthesis/maturation PG machineries. This resulted in the co-evolution of GlcNAc O-acetylation and its dedicated PGH (Acm2). This is a novel example of the plasticity of PG synthesis/degradation machineries similar to what we have reported before regarding to d-lactate incorporation in the PG of this species (4–6).
Therefore, L. plantarum represents a unique model for studying the functional role of PG O-acetylation because it contains both MurNAc and GlcNAc O-acetylation, catalyzed by two dedicated proteins with a similar global organization, and both members of the Oat family. Furthermore, we show that these two modifications could independently modulate PG degradation, either by triggering the N-acetylmuramoyl-l-alanine amidase LytH for MurNAc O-acetylation or by inhibiting the N-acetylglucosaminidase Acm2 through GlcNAc O-acetylation. This affords L. plantarum the capacity to control PG degradation and/or remodel by modulating the abundance and localization of either O-acetylation on glycan strands. Future biochemical studies on the relationship between these modifications and their associated PGH will provide new insight into the control of cell wall assembly and degradation.
Supplementary Material
Acknowledgment
We thank Saulius Kulakauskas for critical reading of the manuscript.
This work was supported by a Jeune Equipe grant from the Institut National de la Recherche Agronomique (INRA) (to M.-P. C.-C.) and the National Fund for Scientific Research, the Université catholique de Louvain (Fonds Spéciaux de Recherche), and the Research Department of the Communauté Française de Belgique (Concerted Research Action) (all to P. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S4, Figs. S1–S7, and additional references.
T. Rolain, E. Bernard, P. Courtin, M. P. Chapot-Chartier, and P. Hols, manuscript in preparation.
- PG
- peptidoglycan
- MurNAc
- N-acetylmuramic acid
- GlcNAc
- N-acetylglucosamine
- PGH
- peptidoglycan hydrolase(s)
- Oat
- O-acetyltransferase
- RP-HPLC
- reverse-phase high performance liquid chromatography
- MRS
- de Man, Rogosa, and Sharpe.
REFERENCES
- 1. Delcour J., Ferain T., Deghorain M., Palumbo E., Hols P. (1999) Antonie Van Leeuwenhoek 76, 159–184 [PubMed] [Google Scholar]
- 2. Vollmer W., Blanot D., de Pedro M. A. (2008) FEMS Microbiol. Rev. 32, 149–167 [DOI] [PubMed] [Google Scholar]
- 3. Schleifer K. H., Kandler O. (1972) Bacteriol. Rev. 36, 407–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deghorain M., Goffin P., Fontaine L., Mainardi J. L., Daniel R., Errington J., Hallet B., Hols P. (2007) J. Bacteriol. 189, 4332–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ferain T., Hobbs J. N., Jr., Richardson J., Bernard N., Garmyn D., Hols P., Allen N. E., Delcour J. (1996) J. Bacteriol. 178, 5431–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goffin P., Deghorain M., Mainardi J. L., Tytgat I., Champomier-Vergès M. C., Kleerebezem M., Hols P. (2005) J. Bacteriol. 187, 6750–6761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vollmer W., Joris B., Charlier P., Foster S. (2008) FEMS Microbiol. Rev. 32, 259–286 [DOI] [PubMed] [Google Scholar]
- 8. Chapot-Chartier M. P. (2010) in Prokaryotic Cell Wall Compounds (König H., Claus H., Varma A. eds) pp. 383–406, Springer Verlag Berlin, Heidelberg, Germany [Google Scholar]
- 9. Sauvage E., Kerff F., Terrak M., Ayala J. A., Charlier P. (2008) FEMS Microbiol. Rev. 32, 234–258 [DOI] [PubMed] [Google Scholar]
- 10. Bera A., Herbert S., Jakob A., Vollmer W., Götz F. (2005) Mol. Microbiol. 55, 778–787 [DOI] [PubMed] [Google Scholar]
- 11. Boneca I. G., Dussurget O., Cabanes D., Nahori M. A., Sousa S., Lecuit M., Psylinakis E., Bouriotis V., Hugot J. P., Giovannini M., Coyle A., Bertin J., Namane A., Rousselle J. C., Cayet N., Prévost M. C., Balloy V., Chignard M., Philpott D. J., Cossart P., Girardin S. E. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clarke A. J., Dupont C. (1992) Can. J. Microbiol. 38, 85–91 [DOI] [PubMed] [Google Scholar]
- 13. Crisóstomo M. I., Vollmer W., Kharat A. S., Inhülsen S., Gehre F., Buckenmaier S., Tomasz A. (2006) Mol. Microbiol. 61, 1497–1509 [DOI] [PubMed] [Google Scholar]
- 14. Hébert L., Courtin P., Torelli R., Sanguinetti M., Chapot-Chartier M. P., Auffray Y., Benachour A. (2007) Infect. Immun. 75, 5390–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meyrand M., Boughammoura A., Courtin P., Mézange C., Guillot A., Chapot-Chartier M. P. (2007) Microbiology 153, 3275–3285 [DOI] [PubMed] [Google Scholar]
- 16. Moynihan P. J., Clarke A. J. (2010) J. Biol. Chem. 285, 13264–13273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pfeffer J. M., Strating H., Weadge J. T., Clarke A. J. (2006) J. Bacteriol. 188, 902–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Veiga P., Bulbarela-Sampieri C., Furlan S., Maisons A., Chapot-Chartier M. P., Erkelenz M., Mervelet P., Noirot P., Frees D., Kuipers O. P., Kok J., Gruss A., Buist G., Kulakauskas S. (2007) J. Biol. Chem. 282, 19342–19354 [DOI] [PubMed] [Google Scholar]
- 19. Vollmer W., Tomasz A. (2000) J. Biol. Chem. 275, 20496–20501 [DOI] [PubMed] [Google Scholar]
- 20. Vollmer W. (2008) FEMS Microbiol. Rev. 32, 287–306 [DOI] [PubMed] [Google Scholar]
- 21. Blackburn N. T., Clarke A. J. (2002) Biochemistry 41, 1001–1013 [DOI] [PubMed] [Google Scholar]
- 22. Laaberki M. H., Pfeffer J., Clarke A. J., Dworkin J. (2011) J. Biol. Chem. 286, 5278–5288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emirian A., Fromentin S., Eckert C., Chau F., Dubost L., Delepierre M., Gutmann L., Arthur M., Mesnage S. (2009) FEBS Lett. 583, 3033–3038 [DOI] [PubMed] [Google Scholar]
- 24. Sambrook J., Russel D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 25. Dower W. J., Miller J. F., Ragsdale C. W. (1988) Nucleic Acids Res. 16, 6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aukrust T. W., Brurberg M. B., Nes I. F. (1995) Methods Mol. Biol. 47, 201–208 [DOI] [PubMed] [Google Scholar]
- 27. Lambert J. M., Bongers R. S., Kleerebezem M. (2007) Appl. Environ. Microbiol. 73, 1126–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuipers O. P., de Ruyter P. G., Kleerebezem M., de Vos W. M. (1998) J. Biotechnol. 64, 15–21 [Google Scholar]
- 29. Courtin P., Miranda G., Guillot A., Wessner F., Mézange C., Domakova E., Kulakauskas S., Chapot-Chartier M. P. (2006) J. Bacteriol. 188, 5293–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cornett J. B., Shockman G. D. (1978) J. Bacteriol. 135, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huard C., Miranda G., Wessner F., Bolotin A., Hansen J., Foster S. J., Chapot-Chartier M. P. (2003) Microbiology 149, 695–705 [DOI] [PubMed] [Google Scholar]
- 32. Stöver B. C., Müller K. F. (2010) BMC. Bioinformatics 11, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Simossis V. A., Heringa J. (2005) Nucleic Acids Res. 33, W289–W294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tusnády G. E., Simon I. (2001) Bioinformatics. 17, 849–850 [DOI] [PubMed] [Google Scholar]
- 35. Wu S., Zhang Y. (2007) Nucleic Acids Res. 35, 3375–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Veiga P., Erkelenz M., Bernard E., Courtin P., Kulakauskas S., Chapot-Chartier M. P. (2009) J. Bacteriol. 191, 3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kleerebezem M., Boekhorst J., van Kranenburg R., Molenaar D., Kuipers O. P., Leer R., Tarchini R., Peters S. A., Sandbrink H. M., Fiers M. W., Stiekema W., Lankhorst R. M., Bron P. A., Hoffer S. M., Groot M. N., Kerkhoven R., de Vries M., Ursing B., de Vos W. M., Siezen R. J. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1990–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.