Abstract
Resistance to antibiotics is a problem not only in terms of healthcare but also biodefense. Engineering of resistance into a human pathogen could create an untreatable biothreat pathogen. One such pathogen is Yersinia pestis, the causative agent of plague. Previously, we have used a bioinformatic approach to identify proteins that may be suitable targets for antimicrobial therapy and in particular for the treatment of plague. The serine protease inhibitor ecotin was identified as one such target. We have carried out mutational analyses in the closely related Yersinia pseudotuberculosis, validating that the ecotin gene is a virulence-associated gene in this bacterium. Y. pestis ecotin inhibits chymotrypsin. Here, we present the structure of ecotin in complex with chymotrypsin to 2.74 Å resolution. The structure features a biologically relevant tetramer whereby an ecotin dimer binds to two chymotrypsin molecules, similar to what was observed in related serine protease inhibitor structures. However, the vast majority of the interactions in the present structure are distinctive, indicating that the broad specificity of the inhibitor for these proteases is based largely on its capacity to recognize features unique to each of them. These findings will have implications for the development of small ecotin inhibitors for therapeutic use.
Keywords: Crystallography, Enzyme Structure, Protease, Protease Inhibitor, Protein-Protein Interactions, Molecular Recognition
Introduction
Antibiotic resistance is an ever-increasing problem in the fight against microbial disease. It is a major public health issue, where naturally resistant strains are emerging against all classes of antibiotics. In addition, genetic engineering has raised the specter that resistance could be engineered into an organism to create an untreatable biothreat pathogen. Current bactericidal antibiotics inhibit only a few targets: DNA, RNA, cell wall, or protein synthesis (1), and as such, there is a need to identify new targets against which antimicrobial compounds could be developed.
Three species of Yersinia are known to be pathogenic for man; Yersinia enterocolitica and Yersinia pseudotuberculosis are gastrointestinal pathogens, whereas Yersinia pestis is the etiological agent of bubonic and pneumonic plague. Y. pestis is considered to be a clonal derivative of Y. pseudotuberculosis (2) but is awarded species designation due to the severity of the disease it causes. The infectivity of Y. pestis by the aerosol route, the acute disease, and high associated mortality rate meant that plague attracted attention as a potential bioweapon in the offensive biological weapons programs of the last century and is still an agent of concern in biodefense. Antibiotic-resistant strains are rare, but a multiple antibiotic-resistant strain was isolated in Madagascar (3), and efficient transfer of resistance genes to Y. pestis in the midgut of the flea has been demonstrated (4). Therefore, there is a need for novel effective therapeutics for use in treatment of plague.
It is notable that the close relatives Y. pestis and Y. pseudotuberculosis cause remarkably different diseases yet are very closely genetically related (99% nucleotide identity for most shared genes), and data based on multiple locus sequence typing analysis suggest that Y. pestis diverged from Y. pseudotuberculosis only 1500–20,000 years ago (2). The evolution from a mild intestinal pathogen into a systemic pathogen, which has repeatedly devastated the human race, has occurred in an eye blink of evolutionary time. A major step along this pathway appears to be the acquisition of two plasmids by Y. pestis. Pathogenic Y. pseudotuberculosis retains a single plasmid (pCD1), whereas Y. pestis also possesses plasmids pMT1 and pPCP1. It is therefore widely accepted that Y. pestis was once a simple enteropathogen, and gene acquisition and loss allowed it to change its environmental niches and its lifestyle. The complete genome sequence of Y. pestis CO92 (biovar orientalis) was determined and revealed a highly fluid genome, with large inversions occurring between insertion sequence (IS) elements (5). A large number of pseudogenes are also present in the Y. pestis genome, disrupted by IS elements or by other mutations. These pseudogenes are thought to have been required for an enteropathogenic lifestyle but are now redundant. However, the close relationship between Y. pestis and Y. pseudotuberculosis means that often the proteins in one organism retain the same role in the other. This allows us to gain insight into Yersinia virulence using the less pathogenic enteropathogen, which can then be followed up by focused efforts on the highly virulent plague bacillus.
When considering novel antimicrobial development, ideally, a target will be common to a range of pathogens. We have previously reported a bioinformatic approach to identify genes more commonly found in pathogens than in non-pathogens (6). One of the targets identified as potentially being of interest was ecotin. Ecotin is a serine protease inhibitor that inhibits a range of serine proteases, including trypsin, chymotrypsin, and elastase (7). In this study, we validated eco as an attenuating locus. We elucidated the crystal structure of ecotin in complex with activated bovine chymotrypsin and described the molecular basis for the serine protease inhibition by ecotin. These molecular details provide a useful platform on which to base further research into the development of novel therapeutics for plague.
EXPERIMENTAL PROCEDURES
Materials
Unless stated otherwise, chemicals were purchased from Sigma, and enzymes were purchased from Promega.
Preparation of Y. pseudotuberculosis Mutants
Construction of Y. pseudotuberculosis mutants was carried out as reported previously (8). Briefly, primers were designed for the target gene to be disrupted that included 20 bp complementary to the 5′- or 3′-sequence of the kanamycin gene of plasmid pK2 and pUC4K, followed by 50 bp of upstream or downstream sequence flanking the gene to be disrupted. The following primers were used: 5′-AATCGAGTTTTAGAGACGTATTGCTGATAAAACTTAGAGAAAAAAAGATG-3′ (forward) and 5′-ATGTTAATATTTAGCCCAGGCGCGGAATAGCGCCTGGTTAATGAGTGCTA-3′ (reverse). PCR products were generated using plasmid pK2 as a template, and excess template was digested with DpnI. PCR products were purified using Millipore Microcon Ultracel YM-100 membranes and were then transformed into Y. pseudotuberculosis YPIII pAJD434 (9) by electroporation. Following overnight incubation at 28 °C in LB medium supplemented with 0.8% arabinose, transformants were selected on LB agar supplemented with kanamycin (50 μg/ml) and trimethoprim (100 μg/ml) for 48 h at 28 °C. Transformants were screened by PCR using target gene-specific and kanamycin gene-specific primers: 5′-GATCCTACCCTGTTGTTGTC-3′ (forward) and 5′-TTAGCCCAGGCGCGGAATAG-3′ (reverse).
Mutant strains were cured of the pAJD434 plasmid by growth at 37 °C in LB medium supplemented with kanamycin (50 μg/ml). Cured mutant strains were screened for the virulence plasmid pYV by PCR for two genes located on this plasmid: virF and yscC. The retention of the Yersinia virulence plasmid (pYV) was also confirmed by culture on Congo red-magnesium oxalate plates, where plasmid retention results in small red colonies, and plasmid loss results in large pink colonies (10).
Competitive Index Studies
For in vivo competitive index studies, mutant and wild-type strains were grown separately to exponential phase in 20 ml of LB broth with shaking. Broth cultures were then centrifuged (10 min, 4000 × g), and the pellet was resuspended in 10 ml of sterile PBS and centrifuged again (10 min, 4000 × g). The bacteria were washed and resuspended in 10 ml of PBS, and the A600 was adjusted to 0.55–0.60 with sterile PBS. Wild-type and mutant bacterial suspensions were then mixed in a 1:1 ratio and serially diluted with sterile PBS to give an inoculation concentration of ∼1 × 103 colony-forming units/ml. Groups of six mice were dosed with 0.1 ml of this solution by the intravenous route. Retrospective viable counts were determined by plating out dilutions (in triplicate) on LB agar and LB agar supplemented with kanamycin to determine the input ratio. After 5 days, spleens were recovered and passed through 70 μm sieves (BD Biosciences) to produce a cell suspension in 3 ml of PBS. Cell suspensions were serially diluted in sterile PBS and plated onto LB agar and LB agar supplemented with kanamycin to determine the output ratio. The competitive index is defined as the output ratio (mutant/wild-type) divided by the input ratio (mutant/wild-type) (11, 12).
Cloning of the Ecotin Gene
The eco gene minus the signal peptide was PCR-amplified from Y. pseudotuberculosis IP32953 DNA using primers 5′-ACGCCTACGCCTCTCAATCA-3′ (forward) and 5′-CTATTTGACCTGCGCACTTCTGA-3′ (reverse) and cloned into the expression vector pCRT7/NT-TOPO (Invitrogen). The plasmid was transformed by heat shock into Escherichia coli strain TOP10F′. The resultant constructs were sequenced, and those in which ecotin was appropriately orientated for translation were chosen and transformed into E. coli BL21*(DE3) cells for protein expression studies.
Expression and Purification of Ecotin
E. coli BL21*(DE3) cells harboring recombinant pCRT7/NT-TOPO plasmids were cultured in LB broth supplemented with 1% glucose and ampicillin (50 μg/ml). Cultures were grown with shaking (170 rpm) at 37 °C until an A600 of 0.4 was reached. Protein expression was induced with 1 mm isopropyl β-d-thiogalactopyranoside with incubation for a further 4 h, followed by harvesting by centrifugation (10 min, 1700 × g). Cell pellets were resuspended in PBS and sonicated in an ice water bath. The suspension was clarified by centrifugation at 27,000 × g for 30 min. Supernatants were loaded onto a HisTrapTM column (GE Healthcare) equilibrated with 40 mm Tris (pH 7.5) and 750 mm NaCl. The column was washed with the equilibration buffer supplemented with 10 and 20 mm imidazole prior to elution with elution buffer (40 mm Tris (pH 7.5), 750 mm NaCl, and 250 mm imidazole). Elution fractions containing ecotin as determined by SDS-PAGE analysis were pooled and buffer-exchanged into 40 mm Tris (pH 7.5).
Chymotrypsin Inhibition Assay
Inhibition of chymotrypsin activity was determined by hydrolysis of N-benzoyl-l-tyrosine ethyl ester based on the method of Wirnt (13). Varying concentrations of ecotin were mixed with 1.4 nm chymotrypsin in a 1:1 (v/v) ratio and incubated for 5 min at 25 °C prior to addition to a 1 ml reaction volume containing 465 nm N-benzoyl-l-tyrosine ethyl ester in 80 mm Tris and 20 mm CaCl2 (pH 7.8) prewarmed to the same temperature. The A256 was monitored for 3 min using an Ultraspec 4000 spectrophotometer (GE Healthcare).
Crystallization of the Ecotin-Chymotrypsin Complex
Ecotin at 3 mg/ml and chymotrypsin (Sigma) at 6 mg/ml in 40 mm Tris (pH 7.5) were mixed in a 1:1 volume ratio before incubation at 16 °C for 2 h prior to crystallization trials. Initial screens to identify conditions in which the ecotin-chymotrypsin complex crystallized were carried out using pre-prepared 96-well screens (Molecular Dimensions) on an Art Robbins Phoenix nanodispenser. 300-nl drops were set up using the sitting drop method. Crystals were obtained under various conditions, and optimization was carried out in 24 well plates using the hanging drop method. Improved crystals of the ecotin-chymotrypsin complex grew over ∼8 weeks in a 2 μl drop, which contained a 1:1 volume ratio of protein solution to mother liquor. The mother liquor contained 100 mm BisTris2 (pH 5.5), 200 mm ammonium sulfate, and 20% (w/v) PEG 3350.
Structure Determination and Refinement
X-ray data were collected from a single cryocooled crystal with 25% PEG 3350 at the Diamond Light Source (Didcot, Oxon, United Kingdom) on an ADSC Q315 CCD detector on station I03 (λ = 0.97 Å). Diffraction images were collected at an oscillation angle of 1.0°. Data were processed in the monoclinic space group P21 using the HKL2000 package (14). Initial phases were obtained through molecular replacement using the program PHASER (15). The search models used for molecular replacement were the 1.68 Å structure of bovine chymotrypsin (Protein Data Bank code 4CHA) and a model of ecotin generated based on its sequence alignment with E. coli ecotin (code 1ECZ). Model building was performed with Coot (16), and refinement was performed with REFMAC5 (17). A set of reflections was set aside for Rfree calculation (18).
RESULTS
Competitive Index Study
The eco gene was previously identified as being present predominantly in pathogenic bacteria while rarely found in non-pathogens (6). To validate eco as an attenuating locus when mutated in Yersinia, the gene encoding ecotin was disrupted in Y. pseudotuberculosis to create a mutant. Mice were then infected with mutant and wild-type Y. pseudotuberculosis (see “Experimental Procedures”), and the competitive index was calculated. A competitive index value of 0.2 or less indicates that the locus is attenuating. The competitive index of the ecotin-defective mutant was 0.03 ± 0.02 (mean ± S.D.), indicating that this is a highly attenuating locus, thus identifying ecotin as a virulence-associated protein.
Ecotin-Chymotrypsin Inhibition Assay
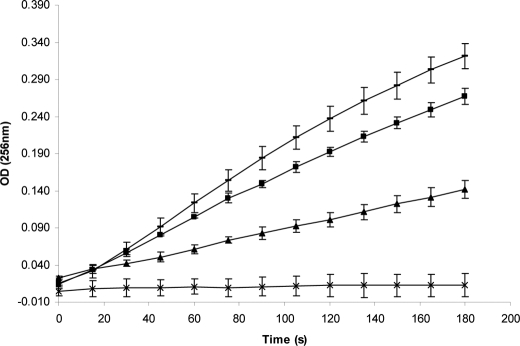
An assay was carried out to determine the effect of Yersinia ecotin on chymotrypsin. Chymotrypsin activity was tested subsequent to incubation with varying concentrations of ecotin. The result, shown in Fig. 1, indicates that chymotrypsin activity was impaired upon incubation with ecotin.
FIGURE 1.
Chymotrypsin inhibition assay data. Chymotrypsin activity was assayed subsequent to incubation with varying concentrations of ecotin. 1.4 nm chymotrypsin was incubated with no ecotin (lines), 0.5 nm ecotin (■), 2.5 nm ecotin (▴), or 3.5 nm ecotin (×) prior to assaying. The A256 of the N-benzoyl-l-tyrosine ethyl ester hydrolysis reaction mixture was measured at various time intervals over the 180-s time course. The reaction rate is related to change in A256 and is indicative of chymotrypsin activity.
Quality of the Ecotin-Chymotrypsin Structure
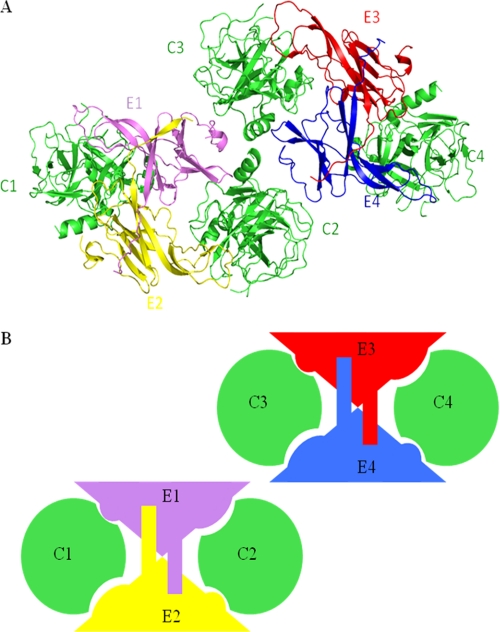
The crystallographic asymmetric unit contains four copies each of ecotin and chymotrypsin molecules (Fig. 2), with a solvent content of 44%. Molecular replacement with the aid of the structure of bovine chymotrypsin (Protein Data Bank code 4CHA) as a search model was used to place four molecules of chymotrypsin in the asymmetric unit. The model of Yersinia ecotin based on its sequence alignment with E. coli ecotin (Protein Data Bank code 1ECZ) was then used to find two molecules of ecotin in the asymmetric unit. Positioning of the third molecule of ecotin was carried out by rotation of a symmetry-related molecule. The remaining difference density was indicative of the presence of a fourth molecule of ecotin. However, the orientation was unclear. Model building was carried out, initially using polyalanine chains, until the density was improved such that the fourth molecule of ecotin could be positioned through rotation of a symmetry-related molecule.
FIGURE 2.
Structure of the ecotin-chymotrypsin complex. Chymotrypsin molecules are colored green and are designated C1–C4. Ecotin molecules are colored pink, yellow, red, and blue and are designated E1–E4. A, there are two tetramers in the asymmetric unit, orientated as shown. B, shown is a schematic of molecules in the asymmetric unit. This figure was prepared using PyMOL.
The final refined structure has an Rcryst of 24.4% and an Rfree of 31.4% (Table 1). The final model consists of 33 water molecules and four sulfate ions in the asymmetric unit. The Ramachandran plot generated by PROCHECK (19) shows that 88.5% of the residues are in allowed regions, 11.4% are in additionally allowed regions, and 0.1% (1 residue) is in a generously allowed region.
TABLE 1.
Crystallographic data for the ecotin-chymotrypsin complex
Numbers in parentheses are for the upper resolution shell (2.84–2.74 Å), where appropriate.
| Cell dimensions | a = 97.31, b = 48.28, c = 174.64 Å; α = 90°, β = 103.96°, γ = 90° |
| Space group (monoclinic) (monoclinic) | P21 |
| Resolution range (Å) | 50.0–2.74 |
| Completeness (%) | 97.5 (96.9) |
| No. of reflections | 273,325 |
| No. of unique reflections | 42,247 |
| Redundancy (%) | 3.2 (3.2) |
| I/σI | 13.3 (5.5) |
| Rmerge (%)a | 7.6 (21.3) |
| Rcryst (%)b | 24.4 |
| Rfree (%)c | 31.4 |
| Wilson B-factor (Å2) | 54.6 |
| Average B-factors (Å2) | |
| Chymotrypsin C1 main chain/side chain | 30.2/30.0 |
| Chymotrypsin C2 main chain/side chain | 34.0/34.1 |
| Chymotrypsin C3 main chain/side chain | 34.6/34.8 |
| Chymotrypsin C4 main chain/side chain | 33.2/33.2 |
| Ecotin E1 main chain/side chain | 38.8/39.3 |
| Ecotin E2 main chain/side chain | 42.5/42.5 |
| Ecotin E3 main chain/side chain | 35.8/36.2 |
| Ecotin E4 main chain/side chain | 44.0/44.5 |
| r.m.s.d. from ideal values | |
| Bonds (Å) | 0.008 |
| Angles | 1.084° |
a Rmerge = ΣhklΣi|Ii(hkl − 〈I(hkl)〉|/ΣhklΣiIi(hkl), where 〈I〉 is the averaged intensity of the i observations of reflection hkl.
b Rcryst = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
c Rfree is equal to Rcryst for a random set of reflections (5.1%) not used in refinement (18).
Activated bovine chymotrypsin is composed of three disulfide-linked polypeptide chains of 11, 130, and 97 residues, and density can be observed for all three chains (Cys-1–Ser-11, Ile-16–Thr-147, and Ala-149–Asn-245). The functional Yersinia ecotin protein is composed of a single polypeptide chain (Asp-22–Lys-169). The electron density observed at the N terminus differs between each copy of ecotin, commencing at Gln-29, Asn-28, Gln-30, and Pro-32 for chains E1, E2, E3, and E4, respectively. Clear density was observed through to the C-terminal Lys-169 residue in all four copies of ecotin.
Overall Topology of the Ecotin-Chymotrypsin Structure
Chymotrypsin is a globular monomeric protein composed of 12 β-strands, two α-helices, and a number of loop regions. In the present complex structure, chymotrypsin molecules superimpose on each other with a root mean square deviation (r.m.s.d.) of Cα atoms ranging from 0.35 to 0.47 Å. The r.m.s.d. between the chymotrypsin molecules in the present structure and those of the previously published structure (Protein Data Bank code 4CHA) ranges from 0.49 to 0.72 Å.
Ecotin is an elongated protein with a β-barrel fold whereby the β-strands are connected by flexible loops. The four ecotin molecules in the asymmetric unit can be superimposed with an r.m.s.d. of Cα atoms ranging from 0.38 to 0.70 Å. The r.m.s.d. between the ecotin molecules in the present structure and those of the previously published structure of native ecotin (Protein Data Bank code 1ECY) ranges from 1.48 to 1.51 Å. The r.m.s.d. between the ecotin molecules in the present structure and those of the previously published structure of ecotin complex (code 1N8O) ranges from 0.82 to 0.92 Å. Ecotin takes the form of a homodimer that binds to two chymotrypsin molecules. The ecotin dimer is shaped like a “butterfly,” and this shape provides two clefts into which chymotrypsin can fit (Fig. 2). Binding of two molecules of chymotrypsin to the ecotin dimer is facilitated through two distinct binding sites for chymotrypsin on monomeric ecotin. A primary interaction occurs between ecotin and the active site of chymotrypsin, and a secondary interaction between the partner ecotin and a different part of chymotrypsin acts to stabilize binding. This results in a biologically relevant tetramer, which in the structure forms half of the asymmetric unit (Fig. 2). The shape complementarity between the dimer and each chymotrypsin monomer is good, with an average shape correlation statistic of 0.73 (20). The details of the interfaces found within the asymmetric unit due to molecular recognition of the complex are provided in Table 2. The details of buried surface areas and solvation free energy gain for the interfaces discussed below were calculated using the program PISA (21).
TABLE 2.
Details of interfaces within the asymmetric unit
Buried surface areas and solvation free energy gains were calculated using PISA (21). Hydrogen bonding interactions were identified with the program HBPLUS (33). Contact distances were calculated using the program CONTACT (17), and the maximum allowed contact distances for van der Waals contacts are as follows: C–C, 4.1 Å; C–N, 3.8 Å; C–O, 3.7 Å; O–O, 3.3 Å; O–N, 3.4 Å; N–N, 3.4 Å; C–S, 4.1 Å; O–S, 3.7 Å; and N–S, 3.8 Å.
| Interface | Total buried surface area | Solvation free energy gain upon complex formation | No. of hydrogen bonds | No.of van der Waals contacts |
|---|---|---|---|---|
| Å2 | kcal/mol | |||
| E1/E2 dimer | 3142 | −12.6 | 20 | 174 |
| E3/E4 dimer | 2982 | −12.1 | 18 | 161 |
| Average | 3062 | −12.4 | ||
| Primary interface | ||||
| E1/C1 | 1924 | −14.3 | 11 | 102 |
| E2/C2 | 1945 | −13.0 | 9 | 126 |
| E3/C3 | 1923 | −16.7 | 11 | 109 |
| E4/C4 | 1917 | −16.2 | 10 | 112 |
| Average | 1927 | −15.1 | ||
| Secondary interface | ||||
| E1/C2 | 1051 | −3.3 | 4 | 64 |
| E2/C1 | 1064 | −2.2 | 4 | 56 |
| E3/C4 | 1109 | −0.7 | 7 | 69 |
| E4/C3 | 1077 | −0.9 | 6 | 65 |
| Average | 1076 | −1.8 | ||
Ecotin Homodimer
The C-terminal arm of ecotin (Glu-158–Lys-169) exchanges with its dimeric partner to create a two-stranded antiparallel β-ribbon. 10 potential hydrogen bonds exist between each exchanged arm and residues of the partner ecotin molecule, giving a total of 20 potential hydrogen bonds between monomers of the E1/E2 dimer. These hydrogen bonds are predominantly between main chain atoms of the exchanged arm and Val-152–Lys-169 of the partner ecotin. 15% of the solvent-accessible area of each ecotin monomer is buried at the dimer interface, giving a total buried surface area of 3142 Å2.
Primary Ecotin-Chymotrypsin Binding Interface (E1/C1)
Two extended loops of ecotin bind at the chymotrypsin active site: an outer loop and a supporting inner loop. 10% of the solvent-accessible area of ecotin is buried at the interface with chymotrypsin, and the total buried surface area is 1924 Å2, with a solvation free energy gain of −14.3 kcal/mol (21). There are some 25 ecotin residues at the interface. The interfacing residues of ecotin are derived predominantly from the two loop regions of ecotin: Gln-105–Glu-115 on the outer loop and Asp-75–Asn-84 on the inner loop. Additional contacts are made by Thr-125 and Asn-127. The extended loops are stabilized by a disulfide bridge between Cys-76 on the inner loop and Cys-113 on the outer loop. In addition, there are 13 potential hydrogen bonds between each loop, predominantly between main chain atoms.
There are 35 chymotrypsin residues at the interface with ecotin. These residues are derived predominantly from loop regions at and adjacent to the active site, which is composed of a catalytic triad of His-57, Ser-195, and Asp-102. There are 11 potential hydrogen bonds between ecotin and chymotrypsin at and adjacent to the chymotrypsin active site. Met-110 of ecotin is known as the P1 residue, based on the standard nomenclature of binding sites as described by Schechter and Berger (22). Met-110 resides on the main binding loop of ecotin and is involved in three hydrogen bonds; the main chain oxygen atom of ecotin Met-110 faces the oxyanion hole of chymotrypsin and is hydrogen-bonded to the main chain nitrogen atoms of Ser-195 and Gly-193. There is also a hydrogen bond between the main chain nitrogen atom of Met-110 and the Oγ of Ser-195, and the Met-110 side chain fits into the substrate-binding pocket of chymotrypsin (Fig. 3). In addition to hydrogen bonding, a number of van der Waals interactions occur (Table 2). Residues at the interface display chemical character as follows: 45% hydrophobic, 40% polar uncharged, and 15% charged.
FIGURE 3.
Ecotin binding loop at the chymotrypsin interface. A, residues of the ecotin Gln-105–Pro-114 binding loop are represented as sticks and are colored pink. Oxygen, nitrogen, and sulfur atoms are colored red, blue, and yellow, respectively. The ecotin Cys-113 side chain has been omitted for clarity. A surface representation of chymotrypsin is shown, and residues that make direct contacts with ecotin are colored green. The side chain of Met-110 can be seen extending into the chymotrypsin substrate-binding pocket. B, hydrogen bonding of ecotin Met-110 and Ala-112 is represented by dashed lines, and contacting chymotrypsin residues are shown as green sticks. This figure was prepared using PyMOL.
Secondary Ecotin-Chymotrypsin Binding Interface (E1/C2)
The second binding site is located at the other end of the elongated ecotin molecule. There are 14 ecotin residues at this interface (Thr-89–Asp-96 and Arg-135–Leu-140). This second binding site serves to stabilize the first binding site, with ∼5% of the accessible surface area of ecotin being buried at this site to give a total buried surface area of 1051 Å2. The solvation free energy gain upon formation of the interface ranges from −3.3 to −0.7 kcal/mol. with an average of −1.8 kcal/mol. This low number reflects the fact that this binding site is subsidiary. Despite the subsidiary nature of this binding site, there are at least four hydrogen bonds between ecotin and chymotrypsin at this site, along with a number of van der Waals interactions (Table 2). The large buried surface area and the presence of a number of van der Waals contacts and potential hydrogen bonds identify this as biologically relevant interface as opposed to a crystal contact interface. The most extensive interactions involve the hydrophobic ecotin residues Trp-93 and Phe-95 from the Thr-89–Asp-96 loop and Leu-140, Arg-135, and Arg-139 from the Arg-135–Leu-140 loop (Fig. 4). There are 18 interfacing chymotrypsin residues at the secondary binding site. These residues are derived predominantly from the 90s loop and the Thr-232–Gln-240 section of the chymotrypsin C-terminal α-helix. Residues at the interface are 45% hydrophobic, 33% polar uncharged, and 21% charged.
FIGURE 4.
Interactions at the secondary ecotin-chymotrypsin interface (E3/C4). Ecotin is colored pink, and chymotrypsin is colored green. Interfacing residues from each protein are shown as sticks, and oxygen and nitrogen atoms are colored red and blue, respectively. Atoms that are within hydrogen bonding distance are connected by black dashed lines, and van der Waals interactions are indicated by orange dashed lines. A, hydrogen bonding and van der Waals interactions between residues on the Thr-89–Asp-96 loop and residues of the chymotrypsin C-terminal α-helix are shown. The potential hydrogen bond between Lys-93 and Gly-94 shown is not observed in the other three copies of the interface. B, the potential hydrogen bonds made by Arg-135 side chain atoms are not observed in two of the four copies of the interface. There is a potential hydrogen bond between Arg-139 and chymotrypsin Ser-92 instead. The position of the sulfate ion (Sul) is shown, and its hydrogen bonding interactions with surrounding residues are indicated. The side chains of ecotin Arg-139 and chymotrypsin Tyr-94 have been omitted for clarity. This figure was prepared using PyMOL.
Variation between Interfaces
Local variation between the four copies of the interfaces in terms of which atoms are involved in hydrogen bonding and van der Waals contacts occurs as a result of alternative arginine side chain conformations. For example, at the E1/C1 primary interface, there is hydrogen bond between the side chain of ecotin Arg-78 and the side chain of chymotrypsin Tyr-94. At the E2/C2 primary interface, there is a potential hydrogen bond between ecotin Arg-78 and chymotrypsin Ser-96. At the E3/C3 and E4/C4 interfaces, potential hydrogen bonds are not observed for this arginine side chain. At the E3/C4 and E4/C3 secondary interfaces, potential hydrogen bonds are observed between side chain atoms of Arg-135 and main chain atoms of Lys-93 and Tyr-94, interactions that are not observed in the E1/C2 and E2/C1 interfaces. Despite this local variation, key recognition interactions, such as those involving residues of the chymotrypsin catalytic triad, are conserved.
Ligands
Four sulfate molecules (from the crystallization medium) are bound in the complex structure. One sulfate molecule resides at the periphery of each secondary binding site. The area is solvent-exposed, and the sulfate molecules are close to the side chains of chymotrypsin Asn-95 and Lys-93 and ecotin Arg-135. All four sulfate molecules are within hydrogen bonding distance of the side chain of chymotrypsin Asn-95. Additional hydrogen bonding characteristics vary between the four sulfate molecules due to the variation in the side chain conformation of adjacent residues: one sulfate molecule is within hydrogen bonding distance of the nearby chymotrypsin Lys-93 side chain, and two sulfate molecules are within hydrogen bonding distance of the nearby ecotin Arg-135 side chain. Sulfate-mediated hydrogen bonding between ecotin and chymotrypsin provides additional stabilization at these interfaces (Fig. 4B).
DISCUSSION
The need for new antibiotics has become pressing in light of the emergence of antibiotic-resistant strains of human pathogens. Development of such drugs is important in terms of not only healthcare but also biodefense. Y. pestis, the causative agent of plague, is an agent of concern in biodefense. One potential new target for the development of novel antimicrobials is the serine protease inhibitor ecotin. The gene encoding ecotin was found to be more common in the genomes of pathogens compared with non-pathogens, therefore identifying ecotin as a potential target (6). We have confirmed that eco is an attenuating locus in Y. pseudotuberculosis, identifying it as a potential target in this bacterium and the closely related bacterium Y. pestis. It has previously been postulated that ecotin plays a role in pathogenicity in other pathogens; the presence of ecotin in the E. coli periplasm led to the suggestion that ecotin acts to protect the bacterium from serine proteases in the intestines of the mammalian host (7). As Y. pseudotuberculosis is also an enteric pathogen, it is possible that it plays a similar role in this organism. However, we observed attenuation after intravenous challenge, thus by-passing the intestines, and the gene is functional in Y. pestis, implying further functions in pathogenesis. Yersinia ecotin inhibits neutrophil elastase, and so there is a potential role for ecotin in protection against intracellular host immunity (23). Yersinia species are susceptible to killing by neutrophils, and ablation of neutrophils results in increased colonization (24); therefore, it is possible that the main function of ecotin in vivo is to protect Yersinia from neutrophil killing. It has been observed that E. coli ecotin enhances recovery following neutrophil elastase treatment (23).
Yersinia ecotin inhibits neutrophil elastase, trypsin, chymotrypsin, factor Xa, and thrombin (23). This pan-specificity is shared by E. coli ecotin (25, 26) and is thought to be partly facilitated by the presence of a methionine at the P1 position. Serine protease inhibitors traditionally have a target-specific residue at the P1 position, whereas a methionine is considered to be a good “all-rounder.” It has been suggested that the contribution of the secondary binding site to the free energy of the E. coli ecotin-trypsin complex (27) allows for the less than favorable methionine at the P1 site (28).
We have observed inhibition of chymotrypsin by Yersinia ecotin, and we have provided a structural basis for this observed inhibition. Ecotin inhibits chymotrypsin in a “substrate-like” fashion (32), and the Ki for Yersinia ecotin inhibition of chymotrypsin is <0.002 nm (23). The 100% sequence identity of ecotin across a range of species of Yersinia makes this structural result relevant to a range of Yersinia species, including Y. pestis, although inhibition of chymotrypsin may be less important for Y. pestis than for Y. pseudotuberculosis. We have shown that the Yersinia ecotin-chymotrypsin complex features a biologically relevant tetramer, in line with what is observed in a number of related structures. A number of structures of E. coli ecotin in complex with a target protease have identified the biologically significant unit as a tetramer that is formed when a dimer of ecotin binds two target proteases. In the structure of the E. coli ecotin-rat ionic trypsin complex, there is one tetramer per asymmetric unit (27). Wang et al. (29) created an E. coli ecotin mutant and solved the structure of the mutant in complex with thrombin, and the asymmetric unit in this structure is composed of half of the tetramer. The crystal packing in the Yersinia ecotin-chymotrypsin complex shown here has displayed the unique arrangement of the asymmetric unit, which contains two tetramers related by non-crystallographic symmetry. A key element of the biologically relevant tetramer is the ecotin dimer. Dimerization of ecotin provides a large surface area (17,802 Å2 for the Yersinia ecotin-chymotrypsin complex) that features two clefts into which two molecules of chymotrypsin can fit. This large surface area has meant that E. coli ecotin has attracted attention as a potential scaffold for engineering novel protease inhibitors (30).
The primary interface between ecotin and chymotrypsin buries a large surface area. The buried surface area of 1927 Å2 is comparable with that of the related structures of E. coli ecotin-protease complexes: the primary interface of the ecotin-trypsin complex buries 2000 Å2 (27), that of the ecotin mutant-thrombin complex buries 2290 Å2 (29), and the ecotin-crab collagenase complex buries 1950 Å2 at this interface (31). The ecotin binding loop on which the P1 site methionine resides is stabilized through binding to an inner loop. There is a disulfide bridge between the two loops anchored by a number of hydrogen bonds. The architecture of this binding loop is characteristic of that of the substrate-like family of serine protease inhibitors and their protease ligands (32). The hydrogen bonding characteristics of the P1 methionine residue are also characteristic of this type of inhibitor.
Some specific comparisons can be made between the Yersinia ecotin-chymotrypsin primary interface and the related E. coli ecotin-protease interfaces. The Ki value for E. coli ecotin is also <0.002 nm (23); hence, any structural differences between the E. coli and Y. pestis ecotins should not affect the ability to inhibit chymotrypsin. The Yersinia and E. coli ecotins share 70% identity and 79% similarity across the C-terminal 139 residues of the functional protein (Fig. 5). Key residues are conserved between E. coli and Yersinia ecotin residues at the primary interface, including the methionine residue at the P1 position. Hydrogen bonding features of the P1 residue in E. coli ecotin at the interface with chymotrypsin (Protein Data Bank code 1N8O) are also found in this structure and the related E. coli ecotin-rat anion trypsin structure (27). Among the residues of ecotin at binding site 1, Ser-108–Pro-114 (P3–P4′) are conserved. Adjacent to this section is a glutamic acid residue (Glu-115) not present in E. coli ecotin, the side chain of which extends toward chymotrypsin in the Yersinia structure and makes contacts. The P6 serine of E. coli ecotin is substituted for a glutamine (Gln-105) in Yersinia ecotin, which contributes more to the solvation free energy gain upon complex formation than the equivalent serine and makes a potential hydrogen bond with chymotrypsin. Arg-78 on the inner loop of Yersinia ecotin shows potential hydrogen bonds in two of the four interfaces; it is replaced with a leucine residue in E. coli ecotin, which is not involved in hydrogen bonding. Asn-84 of Yersinia ecotin lies at the periphery of the residues involved in binding at binding site 1, extending this section in comparison with E. coli ecotin, where the substituted lysine makes no contacts with chymotrypsin.
FIGURE 5.
Alignment of amino acid sequences of Yersinia (B1JSA0) and E. coli (P23827) ecotins. Alignment was carried out using the ClustalW sequence alignment program (34).
Ecotin is unique among serine protease inhibitors in that it makes an additional discrete interaction with the protease. In the absence of structural data for the secondary ecotin-chymotrypsin interaction, McGrath et al. (27) modeled 10 putative secondary site interactions for the E. coli ecotin-chymotrypsin complex, nine of which featured ecotin residues that are conserved between E. coli and Yersinia ecotins. However, our structural data (based on the structure of the Yersinia ecotin-chymotrypsin complex) show that only three of those hydrogen bonds are predicted correctly in the modeled structure.
Various features of the ecotin-chymotrypsin structure reflect the inherent flexibility of ecotin. It can be seen that the solvation free energy gain upon complex formation for the primary and secondary interfaces varies throughout the asymmetric unit. There is a complementation process whereby the sum of the energy gain for the interfaces within each tetramer in the asymmetric unit is approximately the same; however, the distribution of energies across the interfaces within the tetramers varies: compare the solvation free energy gain upon complexation at the E1/C1 interface of −14.3 kcal/mol with that of the E1/C2 interface of −3.3 kcal/mol, with the equivalent primary and secondary interface values of −16.7 kcal/mol (E3/C3) and −0.7 kcal/mol (E3/C4) in the E3/E4/C3/C4 tetramer. It can be postulated that alterations in binding occur as a result of crystal packing and that the flexibility of ecotin allows the molecule to compensate for this by altering binding elsewhere, resulting in a stable tetrameric complex arrangement. Flexibility is a trait shared with E. coli ecotin, which has been characterized as an inherently flexible molecule (28).
CONCLUSIONS
Ecotin has been identified as a potential protein target for antimicrobial development, and we have validated the eco gene as an attenuating locus in Y. pseudotuberculosis, identifying that ecotin is a virulence factor for this bacterium. Ecotin contributes to Yersinia pathogenicity through its inhibition of host serine proteases, and the ecotin-chymotrypsin structure described represents an important host-pathogen interaction. The molecular details shown here will provide an adequate platform on which to base further research; for example, an inhibitor of the ecotin-protease interaction may reduce the pathogenicity of Y. pestis such that it has potential for development as a treatment for plague.
Acknowledgments
We thank the scientists at station IO3 of the Diamond Light Source and Nethaji Thiyagarajan for support during x-ray data collection. We also thank Helen E. Jones, Carwyn Davies, and Melanie L. Duffield for technical support.
This work was supported by the Ministry of Defence, Wellcome Trust (U.K.) Equipment Grant 088464 and by a Royal Society (U.K.) industry fellowship (to K. R. A.).
The atomic coordinates and structure factors (code 2Y6T) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Kohanski M. A., Dwyer D. J., Collins J. J. (2010) Nat. Rev. Microbiol. 8, 423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Achtman M., Zurth K., Morelli G., Torrea G., Guiyoule A., Carniel E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14043–14048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guiyoule A., Gerbaud G., Buchrieser C., Galimand M., Rahalison L., Chanteau S., Courvalin P., Carniel E. (2001) Emerg. Infect. Dis. 7, 43–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hinnebusch B. J., Rosso M. L., Schwan T. G., Carniel E. (2002) Mol. Microbiol. 46, 349–354 [DOI] [PubMed] [Google Scholar]
- 5. Parkhill J., Wren B. W., Thomson N. R., Titball R. W., Holden M. T., Prentice M. B., Sebaihia M., James K. D., Churcher C., Mungall K. L., Baker S., Basham D., Bentley S. D., Brooks K., Cerdeño-Tárraga A. M., Chillingworth T., Cronin A., Davies R. M., Davis P., Dougan G., Feltwell T., Hamlin N., Holroyd S., Jagels K., Karlyshev A. V., Leather S., Moule S., Oyston P. C., Quail M., Rutherford K., Simmonds M., Skelton J., Stevens K., Whitehead S., Barrell B. G. (2001) Nature 413, 523–527 [DOI] [PubMed] [Google Scholar]
- 6. Stubben C. J., Duffield M. L., Cooper I. A., Ford D. C., Gans J. D., Karlyshev A. V., Lingard B., Oyston P. C., de Rochefort A., Song J., Wren B. W., Titball R. W., Wolinsky M. (2009) BMC Genomics 10, 501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chung C. H., Ives H. E., Almeda S., Goldberg A. L. (1983) J. Biol. Chem. 258, 11032–11038 [PubMed] [Google Scholar]
- 8. Derbise A., Lesic B., Dacheux D., Ghigo J. M., Carniel E. (2003) FEMS Immunol. Med. Microbiol. 38, 113–116 [DOI] [PubMed] [Google Scholar]
- 9. Maxson M. E., Darwin A. J. (2004) J. Bacteriol. 186, 4199–4208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Riley G., Toma S. (1989) J. Clin. Microbiol. 27, 213–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Freter R., Allweiss B., O'Brien P. C., Halstead S. A., Macsai M. S. (1981) Infect. Immun. 34, 241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Taylor R. K., Miller V. L., Furlong D. B., Mekalanos J. J. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 2833–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wirnt R. (1974) Methods of Enzymatic Analysis, 2nd Ed., Academic Press Inc., New York [Google Scholar]
- 14. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–325 [DOI] [PubMed] [Google Scholar]
- 15. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. CCP4 (1994) Acta Crystallogr. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 18. Brünger A. T. (1992) Nature 355, 472–475 [DOI] [PubMed] [Google Scholar]
- 19. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 20. Lawrence M. C., Colman P. M. (1993) J. Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
- 21. Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 22. Schechter I., Berger A. (1967) Biochem. Biophys. Res. Commun. 27, 157–162 [DOI] [PubMed] [Google Scholar]
- 23. Eggers C. T., Murray I. A., Delmar V. A., Day A. G., Craik C. S. (2004) Biochem. J. 379, 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Laws T. R., Davey M. S., Titball R. W., Lukaszewski R. (2010) Microbes Infect. 12, 331–335 [DOI] [PubMed] [Google Scholar]
- 25. Seymour J. L., Lindquist R. N., Dennis M. S., Moffat B., Yansura D., Reilly D., Wessinger M. E., Lazarus R. A. (1994) Biochemistry 33, 3949–3958 [DOI] [PubMed] [Google Scholar]
- 26. Ulmer J. S., Lindquist R. N., Dennis M. S., Lazarus R. A. (1995) FEBS Lett. 365, 159–163 [DOI] [PubMed] [Google Scholar]
- 27. McGrath M. E., Erpel T., Bystroff C., Fletterick R. J. (1994) EMBO J. 13, 1502–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gillmor S. A., Takeuchi T., Yang S. Q., Craik C. S., Fletterick R. J. (2000) J. Mol. Biol. 299, 993–1003 [DOI] [PubMed] [Google Scholar]
- 29. Wang S. X., Esmon C. T., Fletterick R. J. (2001) Biochemistry 40, 10038–10046 [DOI] [PubMed] [Google Scholar]
- 30. Stoop A. A., Craik C. S. (2003) Nat. Biotechnol. 21, 1063–1068 [DOI] [PubMed] [Google Scholar]
- 31. Perona J. J., Tsu C. A., Craik C. S., Fletterick R. J. (1997) Biochemistry 36, 5381–5392 [DOI] [PubMed] [Google Scholar]
- 32. Bode W., Huber R. (1992) Eur. J. Biochem. 204, 433–451 [DOI] [PubMed] [Google Scholar]
- 33. McDonald I. K., Thornton J. M. (1994) J. Mol. Biol. 238, 777–793 [DOI] [PubMed] [Google Scholar]
- 34. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]