Abstract
Lysophosphatidic acid (LPA) is a bioactive phospholipid that affects various biological functions, such as cell proliferation, migration, and survival, through LPA receptors. Among them, the motility of cancer cells is an especially important activity for invasion and metastasis. Recently, AMP-activated protein kinase (AMPK), an energy-sensing kinase, was shown to regulate cell migration. However, the specific role of AMPK in cancer cell migration is unknown. The present study investigated whether LPA could induce AMPK activation and whether this process was associated with cell migration in ovarian cancer cells. We found that LPA led to a striking increase in AMPK phosphorylation in pathways involving the phospholipase C-β3 (PLC-β3) and calcium/calmodulin-dependent protein kinase kinase β (CaMKKβ) in SKOV3 ovarian cancer cells. siRNA-mediated knockdown of AMPKα1, PLC-β3, or (CaMKKβ) impaired the stimulatory effects of LPA on cell migration. Furthermore, we found that knockdown of AMPKα1 abrogated LPA-induced activation of the small GTPase RhoA and ezrin/radixin/moesin proteins regulating membrane dynamics as membrane-cytoskeleton linkers. In ovarian cancer xenograft models, knockdown of AMPK significantly decreased peritoneal dissemination and lung metastasis. Taken together, our results suggest that activation of AMPK by LPA induces cell migration through the signaling pathway to cytoskeletal dynamics and increases tumor metastasis in ovarian cancer.
Keywords: AMP-activated Kinase (AMPK), Cell Migration, Chemotaxis, Cytoskeleton, Phospholipase C, Lysophosphatidic Acid (LPA), Ovarian Cancer
Introduction
Lysophosphatidic acid (LPA)2 has been shown to participate in diverse biological actions, including changes in cell shape, motility, and proliferation, in a variety of cell types (1). Previous studies have shown that LPA has a role in early signaling events, such as Ca2+ mobilization, changes in cAMP accumulation, and the activation of several protein kinases (1–4). Among these pathological processes, the role of LPA in ovarian cancer has been most extensively studied. LPA contributes to the development, progression, and metastasis of ovarian cancer, and its concentration is increased up to 80 μm (in comparison to the basal 1–5 μm concentration) in both plasma and ascites of ovarian cancer patients (5). In vitro studies have shown that production of LPA levels was constitutively increased in ovarian cancer cells but not in normal ovarian epithelial cells (6, 7). Moreover, in a study of the expression of LPA receptor mRNA and protein levels in ovarian cancer tissues, LPA2 and LPA3 were aberrantly up-regulated, but LPA1 was not changed (8, 9). Overexpression of LPA2 and LPA3 are closely associated with tumor progression in ovarian cancer cells (10–13). As evidence of intracellular signaling in cancer cell migration, LPA induces activation of Ras-MEKK1 (14), Rac1 (15), Ca2+-dependent Pyk2 (16), and the Rho/ROCK pathway (17), which indicates that dynamic cytoskeletal rearrangement in LPA-mediated cell migration is regulated through the coordination of complex contexts (such as small GTPases, focal adhesion, and Ca2+-dependent signaling). However, the exact regulatory factors of these molecular mechanisms underlying LPA-induced cell migration have not been fully elucidated.
AMP-activated protein kinase (AMPK) is a highly conserved sensor of cellular energy status in eukaryotes and is widely known as a regulator of cell metabolism (18). It consists of a heterotrimeric complex of a catalytic α subunit and regulatory β/γ subunits (19, 20). AMPK is activated in response to an increase in the ratio of AMP-to-ATP within the cell, and it is phosphorylated at Thr-172 within the activation domain of the α subunit by upstream kinases, LKB1 (21–23) and calmodulin-dependent protein kinase kinase β (CaMKKβ) (24–26). Recent studies introduced AMPK as an important regulatory factor in cell migration (27–31). Activation of AMPK facilitates microtubule dynamics (27) and tube formation (28) through the increasing phosphorylation of cytoplasmic linker protein-170 and triggering the endothelial nitric oxide synthase pathway. Specifically, in cancer cells, AMPK increases cell migration through the transcriptional up-regulation of integrins (29, 30) and down-regulation of microRNA-451 levels (31). Therefore, it is possible that AMPK promotes LPA-induced cell migration by regulating dynamic cytoskeletal rearrangement in cancer cells.
In this study, we investigated the role of AMPK in LPA-induced cell migration in ovarian cancer cells. We found that LPA activates AMPK through Ca2+-dependent signaling, including PLC-β3 and CaMKKβ. The activation of AMPK is essential for LPA-induced cell migration by modulating the activation of ezrin/radixin/moesin (ERM) proteins, which are involved in actin filament/plasma membrane interactions, through the Rho pathway. Therefore, these findings provided new insight into the molecular mechanism of AMPK activation in cell migration and indicated that AMPK may be a potential therapeutic target in ovarian cancer.
EXPERIMENTAL PROCEDURES
Materials
Lysophosphatidic acid (1-oleoyl-2-hydroxy-sn-glycerol-3-phosphate) was purchased from Avanti Lipid (Alabaster, AL). Anti-phospho-acetyl-CoA carboxylase (ACC) (Ser-79), anti-phospho-AMPK (Thr-172), anti-phospho-ERM, anti-AMPKα1, anti-ACC, and anti-ERM antibodies were purchased from Cell Signaling Technology (New England Biolabs, Beverly, MA). Anti-PLC-β3, anti-Rac1, and anti-RhoA antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and a β-actin antibody and rhodamine-conjugated phalloidin were purchased from Sigma-Aldrich (St. Louis, MO). Horseradish peroxidase-conjugated secondary antibodies were obtained from the Kirkegaard and Perry Laboratory (Gaithersburg, MD). Compound C, U73122, STO-609, pertussis toxin, PD98059, and Y27632 were obtained from Calbiochem.
Cell Culture
Human ovarian cancer SKOV3 and OVCAR3 cell lines were purchased from the American Type Culture Collection. Human ovarian surface epithelial cell line HIO-80 (immortalized by SV40 large T) was obtained from Dr. Andrew K. Godwin (Fox Chase Cancer Center, Philadelphia, PA). SKOV3 cells were cultured in RPMI 1640 medium with 10% FBS, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. OVCAR3 cells were cultured in DMEM containing 10% FBS and antibiotics. HIO-80 cells were cultured in a 1:1 mixture of medium 199 (Earle's salts, l-glutamine, 2.2 g/liter sodium bicarbonate, 25 mm HEPES) and MCDB-105 (l-glutamine, 25 mm HEPES) with 4% FBS, 0.2 units/ml pork insulin, and antibiotics. All cells were grown at 37 °C in a humidified 5% CO2 incubator.
Immunoblot Analysis
Cells were serum-starved overnight prior to treatment with the indicated agents. Whole cell lysates were prepared in lysis buffer, as described previously (32). Lysates were then centrifuged at 14,000 × g for 10 min at 4 °C. Supernatants were electrophoresed on SDS-PAGE (8%) gels and transferred to nitrocellulose membranes. Membranes were incubated overnight at 4 °C with primary antibodies and then washed three times in Tris-buffered saline/0.1% Tween 20 prior to 1 h incubation with horseradish peroxidase-conjugated secondary antibodies at room temperature. Proteins were then detected via ECL reagents (Amersham Biosciences).
Small Interfering RNA Transfection
siRNA duplexes directed against LPA2 (nucleotides 867–885), PLC-β3 (nucleotides 483–501), AMPKα1, and CaMKKβ were synthesized or purchased from Dharmacon, Inc. (Lafayette, CO). The presynthesized control siRNA duplexes (luciferase GL3 duplex) were also purchased and used as control oligonucleotides. The cells were transfected with 20 nm siRNA using Lipofectamine (Invitrogen) in serum-free conditions. After 4 h of transfection, the cells were washed and supplemented with fresh medium containing 10% FBS. The cells were incubated for 48 h prior to use.
Transwell Migration Analysis
Chemotactic directional migration was evaluated using a modified Boyden chamber (Neuroprobe, Inc., Gaithersburg, MD). Porous filters (8 μm) were coated by passive adsorption of type I collagen (Sigma) by incubation with 20 μg/ml collagen in 0.1 m acetic acid overnight at 4 °C. Cells (2 × 104) were plated in the upper chamber in medium containing 1 μm LPA with or without agents (pertussis toxin, U73122, STO-609, compound C, PD98059, or Y27632) as indicated and allowed to migrate for 4 h. Non-migrating cells were removed from the upper chamber with a cotton swab, whereas migrating cells adherent to the underside of the filter were fixed with 4% paraformaldehyde, permeated with 0.2% Triton X-100, and stained with Hoechst 33342 solution. The migrated cells were photographed and quantified by fluorescent microscopy at a magnification of 100× by counting the stained cells from three randomly chosen high power fields. The mean values obtained from vehicle-treated cells were expressed as 1.0, and others were relative values. Each value is expressed as the mean ± S.D. of three independent experiments performed in triplicate.
Wound Healing Analysis
Control or AMPKα1 siRNA-transfected SKOV3 or OVCAR3 cells (2 × 105) were seeded on a type I collagen-coated 35-mm dish and incubated for 24 h until a confluent monolayer was formed. Cells were incubated overnight in RPMI 1640 supplemented with 0.1% (w/v) BSA. The cell monolayer was then scratched with a pipette tip. Subsequently, cells were washed twice with PBS, and the migration of cells into the wound area was induced by treatment of 1 μm LPA. For kinetic analysis, two individual fields of the wound area on a 35-mm dish were photographed at 12 h post-wounding.
Thymidine Incorporation Assay
The assay was performed as described previously (33) with some modifications. Control or LPA2 siRNA-transfected SKOV3 cells were seeded into six-well plates at 2 × 105 cells/well for 24 h. Upon serum deprivation for 24 h to reach quiescence, the cells were treated with LPA. After 18 h incubation, cells were pulse-labeled with [3H]thymidine (0.8 μCi/ml) for 6 h. The pulse-labeled cells were followed by three quick washes with PBS to remove the remaining unincorporated labels and then fixed with ice-cold 10% trichloroacetic acid for 15 min at 4 °C. The fixed cells were then lysed with 0.4 N NaOH with 1% SDS, and the radioactivity was counted by a scintillation counter.
Rho GTPase Activation Assay
The activation of RhoA or Rac1 was measured using the GST-Rho binding domain of Rhotekin or GST-Rac binding domain of Pak bound to glutathione-Sepharose beads. Cells were lysed in lysis buffer (50 mm Tris (pH 7.5), 10 mm MgCl2, 0.5 m NaC1, and 1% Triton X-100), and the lysates were incubated with GST-Rho binding domain of Rhotekin or GST-Rac binding domain of Pak beads at 4 °C for 40 min. The beads were collected by centrifugation and washed three times with washing buffer (25 mm Tris·HCl (pH 7.6), 1 mm DTT, 30 mm MgCl2, 40 mm NaCl, and 1% (v/v) Nonidet P-40). Proteins were eluted from beads by boiling in 2× sample buffer for 5 min. The precipitates were analyzed with antibodies specific for RhoA or Rac1. To equalize the cellular proteins in each sample, total cell lysates were also run on gels and subjected to Western blot analysis.
RT-PCR and Real-time Quantitative RT-PCR
cDNA (1 μg) was reverse-transcribed from 3 μg of total cellular RNA prepared using TRIzol reagent (Invitrogen) according to the manufacturer's method, using oligo(dT) primers and murine leukemia virus reverse transcriptase (Promega, Madison, WI). Amplification parameters were as follows: 30 s at 94 °C for denaturation, 30 s at 58 °C for primer annealing, and 30 s at 72 °C for polymerization. PCRs were performed for 35 cycles. The same amount of cDNA was amplified for 25 cycles using the following specific GAPDH primers. Primer sequences were as follows: LPA1, 5′-ATGGCTGGGAGTTGATTGAG-3′ and 5′-GAAGTTGGTGTCCCGTGTCT-3′; LPA2, 5′-TTGTCTTCCTGCTCATGGTG-3′ and 5′-CTCGGCAAGAGTACACAGCA-3′; LPA3, 5′-CTCATGGCCTTCCTCATCAT-3′ and 5′-GCCATACATGTCCTCGTCCT-3′; GAPDH, 5′-CCATGACAACTTTGGCATTG-3′ and 5′-CCTGCTTCACCACCTTCTTG-3′; CaMKKβ, 5′-GCAGGTGTACCAGGAAATTG-3′ and 5′-CTTCCCAGAGAAGATCTTGCG-′3.
For real-time quantitative RT-PCR, total RNA (100 ng) was amplified with a One Step SYBR RT-PCR kit using a LightCycler 2.0 PCR system (Roche). PCR conditions consisted of a 10-min hot start at 95 °C, followed by 45 cycles of 15 s at 95 °C, 10 s at 60 °C, and 30 s at 72 °C. Expression levels of each mRNA were compared after normalization against the expression of GAPDH. This experiment was repeated three times.
Immunofluorescence Assay
Control or AMPKα1 knockdown cells were cultured overnight on 20 μg/ml collagen I-coated coverslips and then treated with 1 μm LPA for 5 min. The cells were washed twice with PBS and fixed with 4% paraformaldehyde for 10 min at room temperature. The coverslips were washed twice with PBS and blocked with BSA (5%) in tris-buffered saline with tween-20 and permeabilized with PBS containing 1% horse serum and 0.2% Triton X-100 for 30 min at room temperature. To visualize F-actin, the cells were incubated with rhodamine-conjugated phalloidin for 1 h at room temperature and washed four times with PBS. To visualize focal adhesions, permeabilized cells were incubated with p-ERM antibody (1/100) diluted in PBS containing 1% horse serum overnight at 4 °C. The cells were then washed four times with PBS and incubated with FITC-conjugated anti-rabbit secondary antibody (1/500) for 1 h at room temperature. After being washed four times with PBS, cells were then analyzed with a laser-scanning confocal microscope imaging system (Olympus, FV 1000).
Lentivirus Transduction and Stable Cell Line Generation
293T cells were transfected with plasmids encoding control or AMPKα1 shRNA (sequence: CCGGCCTGGAAGTCACACAATAGAACTCGAGTTCTATTGTGTGACTTCCAGGTTTTT) in pLKO vectors (Sigma-Aldrich), vesicular stomatitis virus G and Δ 8.9 for 16 h, after which the medium was replaced. Following 48-h culture, media containing lentiviruses were harvested and used for transduction of target cells for 24 h. Stable cell lines were selected in the presence of 3 μg/ml puromycin (Sigma-Aldrich) and then used for xenograft models.
Animals and Xenograft Models
All mice were bred and maintained in specific pathogen-free conditions at the animal facility of the Seoul National University College of Medicine. All animal experiments were performed with the approval of the institutional animal care and use committee of Seoul National University.
To assess tumor metastasis of the cancer cells, control knockdown SKOV3 (SKOV3shControl) or AMPKα1 knockdown SKOV3 cells (SKOV3shAMPKα1, 3 × 105) were injected intraperitoneally into 8-week-old NOD/scid/IL-2Rγ−/−(NOG) mice (The Jackson Laboratory, Bar Harbor, ME). Eight weeks after the injection, the mice were euthanized, and primary tumor masses in the peritoneum for ovarian cancer, liver, and lung were fixed in 4% paraformaldehyde for 24 h. Sections (4 μm) were stained with hematoxylin and eosin. The number of tumor masses in the liver and lung was counted under a dissecting microscope.
Statistical Analysis
Student's t test was used to compare data between two groups. Values are expressed as means ± S.D. of at least triplicate samples. p < 0.05 was considered statistically significant.
RESULTS
LPA Induces Activation of AMPK in Ovarian Cancer Cells
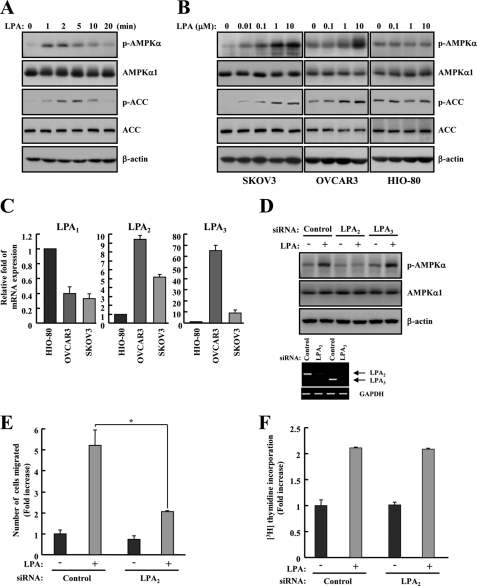
Several studies have recently reported that elevated activation of AMPK occurs in human cancer tissues (34), and the molecular mechanisms linking agonist-mediated AMPK regulation are important for cell motility (28–30). Thus, we sought to characterize the phosphorylation of AMPK by LPA known as the chemotatic factor (1) in SKOV3 ovarian cancer cell lines. The phosphorylation of AMPK was gradually increased and peaked at 2 min after LPA treatment. After 20 min it had returned to basal levels. Consistent with the increase in AMPK activation, the phosphorylation of its substrate, ACC, was also increased in SKOV3 cells (Fig. 1A). Next, we examined whether this phosphorylation of AMPK was cancer-specific. Three different cell lines (SKOV3, OVCAR3, and HIO-80) were prepared, and different doses of LPA (0–10 μm) were applied for 2 min. Both ovarian cancer cell lines, SKOV3 and OVCAR3, showed gradually increased phosphorylation of AMPK and ACC (Fig. 1B). However, HIO-80, a non-cancer ovarian surface epithelial cell line, was not affected by LPA (Fig. 1B). These data showed that LPA increases the phosphorylation of AMPK and its substrate, ACC, in both SKOV3 and OVCAR3 cells.
FIGURE 1.
LPA activates AMPK through LPA2 in ovarian cancer cells. A, SKOV3 cells were treated with 1 μm LPA under serum-free conditions, and cell lysates were prepared at different times as indicated. B, SKOV3, OVCAR3, and HIO-80 cells were treated with LPA for 2 min under serum-free conditions, and cell lysates were prepared at the indicated concentrations. The phosphorylation of AMPK and ACC was analyzed by immunoblot analysis. AMPKα1 and ACC were used as experimental controls, and β-actin was used as the loading control. These results represent one of three independent experiments. C, the expression levels of LPA1, LPA2, LPA3, and GAPDH were analyzed by quantitative real-time PCR. The obtained results were quantified and normalized relative to GAPDH levels. D, SKOV3 cells were transfected with control, LPA2, or LPA3 siRNA for 48 h. Cell lysates were prepared after 1 μm LPA treatment for 2 min under serum-free conditions and then subjected to immunoblot analysis using specific antibodies against p-AMPK, AMPKα1, and β-actin. The efficiency of siRNA interference against LPA2 or LPA3 was tested by RT-PCR. SKOV3 cells were transfected with control or LPA2 siRNA for 48 h. E, control or LPA2 siRNA-transfected cells were examined for the migration activity after the treatment with 1 μm LPA for 4 h. The number of migrated cells was determined by counting them in three randomly chosen high power fields. Each value is expressed as the mean ± S.D. of three independent experiments, each performed in triplicate. *, p < 0.05 compared with LPA alone. F, for the study of cell proliferation activity induced by LPA2, LPA2 siRNA-transfected cells were incubated in 10 μm LPA for 18 h and then pulse-labeled with [3H]thymidine, and the amount of [3H]thymidine incorporation was measured by scintillation counting.
LPA Induces Activation of AMPK through the LPA2 in Ovarian Cancer Cells
Different responses to LPA for normal and cancer cells might be caused by differential expression of LPA receptors (8, 9). Thus, we sought to confirm mRNA expression of LPA receptor subtypes using quantitative real-time PCR. LPA2 and LPA3, but not LPA1, are aberrantly up-regulated in ovarian cancer cells (Fig. 1C). To identify which LPA receptor subtypes is involved in phosphorylation of AMPK, we explored the activation of AMPK in LPA2 or LPA3 knockdown cells. The phosphorylation of AMPK in response to LPA was significantly attenuated by knockdown of LPA2 but not by knockdown of LPA3 (Fig. 1D). Furthermore, to verify the role of LPA2, we assessed the effects of LPA2 knockdown on LPA-induced cell migration and proliferation. Interestingly, the knockdown of LPA2 suppressed LPA-induced cell migration but not cell proliferation (Fig. 1, E and F). These results indicated that phosphorylation of AMPK by LPA is associated with LPA2 activation in ovarian cancer cell migration.
Activation of AMPK Is Required for LPA-induced Cell Migration
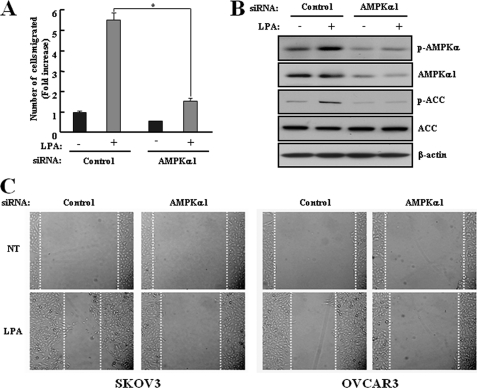
Considering the result in Fig. 1, activation of AMPK by LPA might be involved in changes of ovarian cancer cell motility. To define the specific relevance in LPA-induced cell migration, the chemotactic migration assay after transfection with control or AMPKα1 siRNA was accomplished in SKOV3 cells. AMPKα1 siRNA-transfected cells significantly attenuated LPA-induced cell migration compared with control siRNA-transfected cells (Fig. 2A) and suppressed LPA-induced phosphorylation of AMPK and ACC (B). In addition, a wound healing assay was used to assess the potential effects of constant doses of LPA on cell migration in vitro. When a confluent monolayer was scratched, the migration of AMPKα1 siRNA-transfected cells into the wounded area was significantly reduced in the presence of LPA compared with control siRNA-transfected cells. We also confirmed that the LPA-induced cell migration was reduced by knockdown of AMPKα1 in OVCAR3 cells, another ovarian cancer cell line, using the wound healing assay (Fig. 2C). These data demonstrated that AMPK acts as a key regulator in LPA-induced cell migration.
FIGURE 2.
LPA-activated AMPK induces cell migration in ovarian cancer cells. SKOV3 cells were transfected with control or AMPKα1 siRNA for 48 h. A, the cells were used for a transwell migration assay after treatment with 1 μm LPA for 4 h as described under “Experimental Procedures.” Each value is expressed as the mean ± S.D. of three independent experiments, each performed in triplicate. *, p < 0.05 compared with LPA alone. B, the cell lysates were prepared after 1 μm LPA treatment for 2 min under serum-free conditions and then subjected to immunoblot analysis using specific antibodies against p-AMPK, p-ACC, AMPKα1, ACC, and β-actin. These results represent one of three independent experiments. C, SKOV3 or OVCAR3 cells were transfected with control or AMPKα1 siRNA for 48 h. The cells were plated on a collagen type I-coated 35-mm dish and incubated until a confluent monolayer was formed. After being serum-starved overnight in RPMI 1640 or DMEM containing 0.1% BSA, a cell-free space was created by scraping through the monolayer. Migration was induced by treatment with 1 μm LPA for 12 h. Magnifications, 200×.
LPA Induces Activation of AMPK through Ca2+-dependent Signaling in Cell Migration
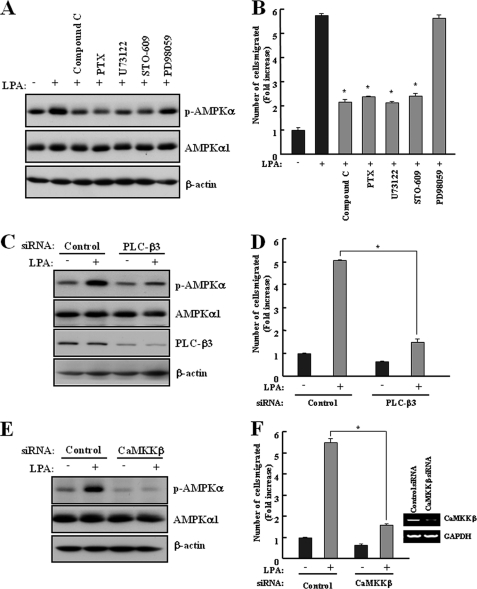
It has been reported that LPA increases the intracellular calcium concentration in ovarian cancer cells (35) and induces cell migration through Ca2+-dependent Pyk2 activation (16). Additionally, activation of AMPK is triggered by an increase in the intracellular calcium concentration without detectable changes in the AMP-to-ATP ratio (24). Thus, we wished to characterize a possible Ca2+-dependent signaling pathway responsible for the LPA-induced phosphorylation of AMPK. SKOV3 cells that were incubated with the Gi/o inhibitor pertussis toxin, a PLC inhibitor (U73122), or a specific CaMKK inhibitor (STO-609) were suppressed in the LPA-stimulated phosphorylation of AMPK. This effect was not observed with a selective MEK inhibitor (PD98059) (Fig. 3A). In addition, these inhibitors significantly reduced LPA-induced cell migration (Fig. 3B). Consistent with our results, previous report revealed that PD98059 did not affect LPA-induced cell migration in ovarian cancer cells (17). We have shown previously that a ternary complex consisting of the LPA2 receptor NHERF2 and PLC-β3 may play a key role in the LPA-mediated Ca2+ signaling pathway (36). Therefore, AMPK might be regulated by intracellular Ca2+ induced through PLC-β3 activation. To understand the role of PLC-β3 in the LPA-induced activation of AMPK, we explored the activation of AMPK and cell migration in PLC-β3 knockdown cells upon LPA treatment. Knockdown of PLC-β3 abolished the activation of AMPK and cell migration upon LPA treatment (Fig. 3, C and D). In addition, knockdown of CaMKKβ, an upstream AMPK kinase in Ca2+ signaling, severely impaired the LPA-induced activation of AMPK and cell migration (Fig. 3, E and F). These results suggested that Ca2+-dependent signaling, including PLC-β3 and CaMKKβ, is the upstream pathway of LPA-induced AMPK phosphorylation and increases LPA-induced cell migration through the activation of AMPK.
FIGURE 3.
LPA-activated AMPK induces cell migration through Ca2+-dependent signaling. A and B, SKOV3 cells were pretreated with 5 μm AMPK inhibitor (Compound C), 10 μm MEK inhibitor (PD98059), 5 μm PLC inhibitor (U73122), 10 μΜ CaMKK inhibitor (STO-609), or 0.1% DMSO for 30 min and then with 2 μg/ml pertussis toxin overnight before the addition of LPA. C and D, SKOV3 cells were transfected with control or PLC-β3 siRNA for 48 h. E and F, SKOV3 cells were transfected with control or CaMKKβ siRNA for 48 h. A, C, and E, cells were treated with 1 μm LPA for 2 min under serum-free conditions. Cell lysates were prepared and subjected to immunoblot analysis using specific antibodies against p-AMPK, AMPKα1, PLC-β3, and β-actin. These results represent one of three independent experiments. B, D, and F, the cells were used for a transwell migration assay after treatment with 1 μm LPA for 4 h, as described under “Experimental Procedures.” Each value is expressed as the mean ± S.D. of three independent experiments, each performed in triplicate. *, p < 0.05 compared with LPA alone. The efficiency of siRNA interference against CaMKKβ was tested by RT-PCR (F).
LPA Activates RhoA through AMPK Activation
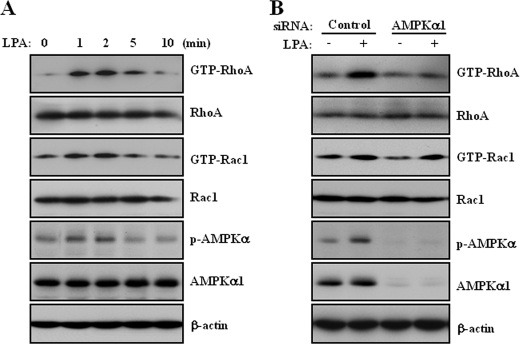
Cell migration begins with an initial protrusion or extension of the plasma membrane via cytoskeletal rearrangement. The cytoskeleton is regulated by activation of Rho-GTPases (37). LPA activates RhoA through Rho-specific guanine nucleotide exchange factors that promote RhoA-GTP accumulation (38). Therefore, it is likely that RhoA or Rac1 may serve as a downstream effector of AMPK to facilitate LPA-induced cell migration through actin rearrangement. To assess this possibility, we first examined the effect of LPA on RhoA and Rac1 activation in SKOV3 cells. Consistent with the activation of AMPK, LPA activated RhoA and Rac1 as early as 30 s and peaked at 2 min (Fig. 4A). Moreover, knockdown of AMPKα1 by siRNA transfection significantly attenuated LPA-induced RhoA activation, but not Rac1 activation (Fig. 4B). Therefore, these results demonstrated that AMPK plays an important role in LPA-induced RhoA activation for actin rearrangement.
FIGURE 4.
LPA induces activation of RhoA through AMPK. A, SKOV3 cells were stimulated for the indicated times with 10 μm LPA under serum-free conditions. B, SKOV3 cells were transfected with control and AMPKα1 siRNA for 48 h and then stimulated with 10 μm LPA for 2 min under serum-free conditions. Activated RhoA or Rac1 was isolated using GST-Rhotekin or GST-Pak bound to glutathione-Sepharose beads. RhoA or Rac1 bound to the beads was detected by immunoblot analysis using specific antibodies against RhoA or Rac1. Total cell lysates were prepared and subjected to immunoblot analysis using specific antibodies against p-AMPK, AMPKα1, RhoA, Rac1, and β-actin. AMPKα1, RhoA, and Rac1 were used as experimental controls, and β-actin was used as the loading control. These results represent one of three independent experiments.
AMPK Activates ERM Proteins through RhoA Activation in LPA-induced Cell Migration
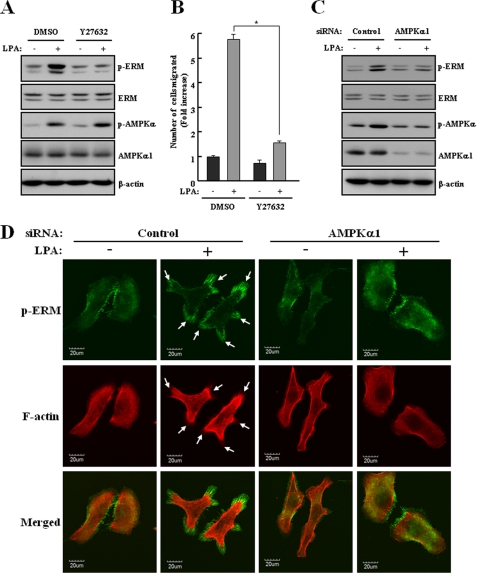
The ERM proteins, concentrated in actin-rich cell surface structures, cross-link the actin cytoskeleton with the plasma membrane. They are involved in the formation of microvilli, cell-cell adhesion, maintenance of cell shape, cell motility, and membrane trafficking (39, 40). In addition, they are directly phosphorylated by ROCK, a direct target of Rho (41). Therefore, we assessed the effects of LPA on ERM proteins and found that LPA increased the phosphorylation of ERM proteins in SKOV3 cells. To detect phosphorylation of ERM proteins in Fig. 5, A and C, an immunoblot analysis was performed, and two bands were detected at size of 75 kDa and 80 kDa. The upper band represents ezrin phosphorylated at Thr-567 and radixin phosphorylated at Thr-564, whereas the lower band represents moesin phosphorylated at Thr-558 as described previously (42). LPA-induced phosphorylation of ERM proteins and cell migration were clearly suppressed in SKOV3 cells pretreated with Y27632, a ROCK inhibitor (Fig. 5, A and B). Importantly, knockdown of AMPKα1 by siRNA transfection significantly reduced the LPA-induced phosphorylation of ERM proteins (Fig. 5C). To confirm this effect, we examined the subcellular localization of phosphorylated ERM proteins in SKOV3 cells by an immunofluorescence analysis. In agreement with Western blot analysis, treatment with LPA leads to a marked increase in the intensity of phosphorylation of ERM proteins. Moreover, phosphorylated ERM proteins were exactly colocalized with F-actin structures localized in these protrusions. However, knockdown of AMPKα1 significantly decreased the LPA-induced colocalization between phosphorylated ERM proteins and F-actin (Fig. 5D). These results indicated that AMPK triggers actin rearrangement for LPA-induced cell migration by activation of ERM proteins through ROCK.
FIGURE 5.
AMPK activates ERM proteins in LPA-induced cell migration. A and B, SKOV3 cells were pretreated with 10 μm Y27632, a ROCK inhibitor, for 30 min before the addition of LPA. C and D, SKOV3 cells were transfected with control or AMPKα1 siRNA for 48 h. A and C, the cells were stimulated with 1 μm LPA for 2 min under serum-free conditions. Cell lysates were prepared and subjected to immunoblot analysis using specific antibodies against p-ERM, p-AMPK, ERM, AMPKα1, and β-actin. These results represent one of three independent experiments. B, the cells were used for a transwell migration assay after treatment with 1 μm LPA for 4 h, as described under “Experimental Procedures.” Each value is expressed as the mean ± S.D. of three independent experiments, each performed in triplicate. *, p < 0.05 compared with LPA alone. D, transfected cells were plated on collagen type I-coated dishes and stimulated with 1 μm LPA for 5 min under serum-free conditions. Cells were labeled with p-ERM antibody (green) or rhodamine-labeled phalloidin (red). Cells were analyzed by confocal microscopy. The arrows indicate membrane protrusions.
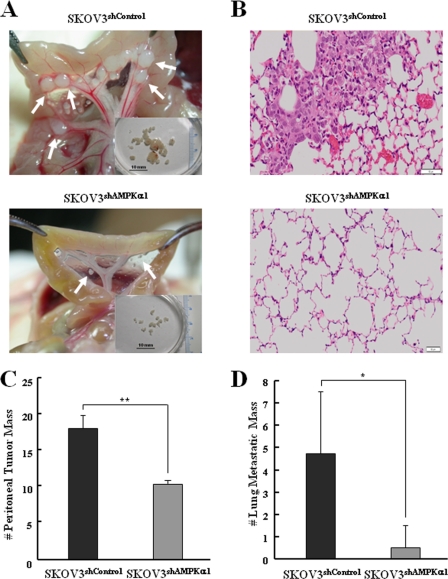
Knockdown of AMPK Decreases Peritoneal Dissemination and Lung Metastasis in Vivo
Ovarian cancer metastasizes primarily on the peritoneum and rarely to distant sites. Subsequent peritoneal implants were characterized by the adhesion and invasion of tumor cells into the peritoneum, leading to miliary dissemination (43). Therefore, to analyze the in vivo role of AMPK in peritoneal dissemination of ovarian cancer cells, xenograft models were accomplished. We used lentivirus-mediated shRNA to knock down the AMPKα1 in SKOV3 cells. Expression of AMPKα1 was significantly decreased to 80–90% in SKOV3 cells infected with AMPKα1 shRNA compared with control shRNA (data not shown). Female NOG mice were injected intraperitoneally with control (SKOV3shControl) or AMPKα1 knockdown SKOV3 cells (SKOV3shAMPKα1), and tumors were allowed to grow for 8 weeks. After 8 weeks, the tumor burden in the peritoneal surface was significantly higher in the SKOV3shControl group than in the SKOV3shAMPKα1 group (Fig. 6A). Moreover, the representative size of peritoneal tumor masses was 4–6 mm in the SKOV3shControl group and 2–3 mm in the SKOV3shAMPKα1 group (Fig. 6A). The number of peritoneal tumor masses was reduced up to 50% (mean ± S.D., p < 0.005, n = 6 each) in the SKOV3shAMPKα1 group when compared with the SKOV3shControl group (Fig. 6C). Furthermore, a significant decrease in lung metastases was observed in the SKOV3shAMPKα1 group (mean ± S.D., p < 0.05, n = 6 each) (Fig. 6, B and D). Taken together, these data indicated that knockdown of AMPK decreases peritoneal dissemination and lung metastasis in vivo.
FIGURE 6.
Knockdown of AMPK decreases peritoneal dissemination and lung metastasis in vivo. NOG mice (n = 6 per group) were injected with SKOV3shControl or SKOV3shAMPKα1 cells and euthanized 8 weeks after injection. A, representative views of the metastasis in the peritoneal cavity of mice. The arrows indicate peritoneal tumor masses. B, hematoxylin and eosin staining of lungs from NOG mice injected with SKOV3shControl or SKOV3shAMPKα1. Scale bar = 50 μm. C, the number of peritoneal tumor masses was counted. D, the number of lung metastatic masses (size; > 1 mm) was counted. Each value is expressed as the mean ± S.D. of two independent experiments. *, p < 0.05, **, p < 0.005, difference with SKOV3shControl.
DISCUSSION
The principal finding of this study was that activation of AMPK by LPA significantly contributes to cell migration through dynamic cytoskeletal rearrangements in ovarian cancer cells.
Several studies have suggested that cell migration is regulated by the activation of AMPK in various cells types (44–47). For example, activation of AMPK by adiponectin inhibits lipopolysaccharide-induced adventitial fibroblast migration (44), and insulin-like growth factor-1-induced vascular smooth muscle cell migration is also suppressed by AMPK activation (45). However, AMPK stimulates human umbilical vein endothelium cell migration (46) and induces transendothelial lymphocyte migration by endothelial nitric-oxide synthase activation (47). In addition, hypertonicity- and A769662-induced AMPK activation triggers actin cytoskeleton remodeling via phosphorylation of ROCK downstream targets in Madin-Darby Canine Kidney cells (48). Therefore, the role of AMPK in cell migration may be dependent on which extracellular factors are stimulated or occur in a cell type-specific manner. Because the role of AMPK in cancer cell migration is especially unclear, it is important to understand what factors trigger the activation of AMPK and how this activation is regulated in cancer cells. In this study, we found that the activation of AMPK by LPA was required for cell migration through the activation of ERM proteins in ovarian cancer cells. We thus considered the possibility that AMPK is a positive regulator of LPA-induced cell migration in ovarian cancer cells.
AMPK has been shown to be activated by the upstream kinase CaMKKβ (24, 25). The activation of AMPK by CaMKKβ is triggered by an increase in intracellular calcium concentration without detectable changes in the AMP-to-ATP ratio (24). CaMKKβ induces cell migration through receptor-mediated AMPK activation, such as vascular endothelial growth factor receptor 2 (28). Therefore, AMPK activation might be essential for receptor-mediated Ca2+ signaling in cell migration. LPA-modulated Ca2+ signaling has been shown previously to regulate cell migration (16). However, it is not fully understood how LPA regulates the Ca2+-mediated intracellular signaling pathway in cell migration. In this study, we found that the inhibition and knockdown of CaMKKβ significantly attenuated the LPA-induced phosphorylation of AMPK and cell migration (Fig. 3, E and F). In addition, treatment with 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR), a known AMPK activator, had no effect on ovarian cancer cell migration. Intracellular Ca2+ generation also did not have an effect upon AICAR treatment (data not shown). Taken together, these results suggest that the activation of AMPK might be important in coordinating other LPA-induced pathways, including Ca2+ signaling in LPA-induced cell migration.
Multiple intracellular pathways are well documented in LPA-induced migration of ovarian cancer cells. The Gi-Ras-MEKK1 signaling pathway mediates LPA-stimulated ovarian cancer cell migration by facilitating focal adhesion kinase redistribution to focal contacts (14). Also, LPA-induced Rac activation requires phosphoinositide 3-kinase activity through LPA1 and thereby promotes cell spreading, lamellipodium formation, and cell migration (15). In this study, we found that the knockdown of AMPKα1 significantly attenuated LPA-induced RhoA activation but not Rac activation (Fig. 4B). Because LPA-induced AMPK phosphorylation was associated with LPA2 activation, LPA-induced Rac activation through LPA1 might have no effect on ovarian cancer cell migration. In addition, the Gi/o inhibitor pertussis toxin significantly reduced LPA-induced phosphorylation of AMPK and cell migration (Fig. 3, A and B). We thus considered the possibility that AMPK activation by LPA induces ovarian cancer cell migration through the Gi-Ras-MEKK1 signaling pathway.
The migratory response to LPA was associated with LPA2-mediated action in ovarian cancer cells (Fig. 1E). It has been reported recently that overexpression of LPA2 in ovarian cancer cells was more efficient at increasing in vitro motility and invasion and in vivo tumor growth, particularly metastasis to distant organs including skeletal muscle, cervical lymph nodes, and the heart (49). In this study, we have shown that the expression levels of LPA2 and LPA3 aberrantly up-regulated in ovarian cancer cells compared with normal ovarian surface epithelial cells (Fig. 1C). Moreover, knockdown of LPA2 significantly attenuated LPA-induced AMPK phosphorylation but not knockdown of LPA3 (Fig. 1D). In addition, peritoneal dissemination and lung metastases were decreased by knockdown of AMPK in vivo using xenografts in NOG mice (Fig. 6). Collectively, our results indicated that the activation of AMPK by LPA might play a crucial role in LPA2-mediated tumor progression, particularly metastasis.
Cell migration begins with an initial protrusion or extension of the plasma membrane at the leading edge of the cell. The protrusion is driven by polymerization of a network of cytoskeletal actin filaments and is stabilized through the formation of an adhesive complex (50). To explain the underlying molecular mechanism of AMPK-mediated cytoskeleton rearrangements in LPA-induced cell migration, we analyzed the activation of small GTPases that induced particular surface protrusions generated by actin-remodeling reactions (51, 52). Interestingly, our data revealed that LPA-activated AMPK enhances RhoA activation (Fig. 4). Our novel identification of a RhoA-ROCK-ERM axis through activation of AMPK by LPA led us to hypothesize that RhoA might play a role in the AMPK-mediated actin cytoskeleton remodeling in SKOV3 cells. However, the regulatory mechanism of AMPK-activated small GTPases is unclear. It has been reported that phosphorylation is the regulatory mechanism of Rho GTPase activation and is independent of GDP-GTP cycling (53). cAMP- and cGMP-dependent kinases (PKA and PKG, respectively) have been shown to phosphorylate RhoA on Ser-188 (54, 55). Additionally, LPA induces threonine phosphorylation of the Rac1-specific nucleotide exchange factor Tiam1 through protein kinase C (56). It is therefore possible that AMPK increases the activation of small GTPases or regulatory proteins of small GTPases, such as guanine nucleotide exchange factor, GDP dissociation inhibitor, and GTPase activating protein, by phosphorylation.
In conclusion, AMPK is an important regulatory factor for ovarian cancer cell migration through LPA2-mediated cytoskeleton reorganization and tumor metastasis in vivo. Thus, AMPK may be a key therapeutic target for the control of ovarian cancer progression.
Acknowledgment
We thank Dr. Andrew K. Godwin for providing the human ovarian surface epithelial cell line HIO-80.
This study was supported by National Research Foundation of Korea Grant KRF-2007-341-C00027 funded by the Korean Government and by a grant of the Korean Ministry of Education, Science and Technology (The Regional Core Research Program/Anti-aging and Well-being Research Center).
- LPA
- lysophosphatidic acid
- ROCK
- Rho-associated kinase
- AMPK
- AMP-activated protein kinase
- CaMKKβ
- calmodulin-dependent protein kinase kinase β
- PLC
- phospholipase C
- ERM
- ezrin/radixin/moesin
- ACC
- acetyl-CoA carboxylase.
REFERENCES
- 1. Mills G. B., Moolenaar W. H. (2003) Nat. Rev. Cancer 3, 582–591 [DOI] [PubMed] [Google Scholar]
- 2. Stracke M. L., Clair T., Liotta L. A. (1997) Adv. Enzyme Regul. 37, 135–144 [DOI] [PubMed] [Google Scholar]
- 3. Umezu-Goto M., Kishi Y., Taira A., Hama K., Dohmae N., Takio K., Yamori T., Mills G. B., Inoue K., Aoki J., Arai H. (2002) J. Cell Biol. 158, 227–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tokumura A., Majima E., Kariya Y., Tominaga K., Kogure K., Yasuda K., Fukuzawa K. (2002) J. Biol. Chem. 277, 39436–39442 [DOI] [PubMed] [Google Scholar]
- 5. Fang X., Gaudette D., Furui T., Mao M., Estrella V., Eder A., Pustilnik T., Sasagawa T., Lapushin R., Yu S., Jaffe R. B., Wiener J. R., Erickson J. R., Mills G. B. (2000) Ann. N.Y. Acad. Sci. 905, 188–208 [DOI] [PubMed] [Google Scholar]
- 6. Shen Z., Belinson J., Morton R. E., Xu Y., Xu Y. (1998) Gynecol. Oncol. 71, 364–368 [DOI] [PubMed] [Google Scholar]
- 7. Eder A. M., Sasagawa T., Mao M., Aoki J., Mills G. B. (2000) Clin. Cancer Res. 6, 2482–2491 [PubMed] [Google Scholar]
- 8. Nakamoto T., Yasuda K., Yasuhara M., Yoshimura T., Kinoshita T., Nakajima T., Okada H., Ikuta A., Kanzaki H. (2005) J. Obstet. Gynaecol. Res. 31, 344–351 [DOI] [PubMed] [Google Scholar]
- 9. Wang P., Wu X., Chen W., Liu J., Wang X. (2007) Gynecol. Oncol. 104, 714–720 [DOI] [PubMed] [Google Scholar]
- 10. Lee Z., Swaby R. F., Liang Y., Yu S., Liu S., Lu K. H., Bast R. C., Jr., Mills G. B., Fang X. (2006) Cancer Res. 66, 2740–2748 [DOI] [PubMed] [Google Scholar]
- 11. Pustilnik T. B., Estrella V., Wiener J. R., Mao M., Eder A., Watt M. A., Bast R. C., Jr., Mills G. B. (1999) Clin. Cancer Res. 5, 3704–3710 [PubMed] [Google Scholar]
- 12. Estrella V. C., Eder A. M., Liu S., Pustilnik T. B., Tabassam F. H., Claret F. X., Gallick G. E., Mills G. B., Wiener J. R. (2007) Int. J. Oncol. 31, 441–449 [PubMed] [Google Scholar]
- 13. Sengupta S., Xiao Y. J., Xu Y. (2003) FASEB J. 17, 1570–1572 [DOI] [PubMed] [Google Scholar]
- 14. Bian D., Su S., Mahanivong C., Cheng R. K., Han Q., Pan Z. K., Sun P., Huang S. (2004) Cancer Res. 64, 4209–4217 [DOI] [PubMed] [Google Scholar]
- 15. Van Leeuwen F. N., Olivo C., Grivell S., Giepmans B. N., Collard J. G., Moolenaar W. H. (2003) J. Biol. Chem. 278, 400–406 [DOI] [PubMed] [Google Scholar]
- 16. Park S. Y., Schinkmann K. A., Avraham S. (2006) Cell. Signal. 18, 1063–1071 [DOI] [PubMed] [Google Scholar]
- 17. Sawada K., Morishige K., Tahara M., Ikebuchi Y., Kawagishi R., Tasaka K., Murata Y. (2002) Gynecol. Oncol. 87, 252–259 [DOI] [PubMed] [Google Scholar]
- 18. Hardie D. G. (2007) Nat. Rev. Mol. Cell Biol. 8, 774–785 [DOI] [PubMed] [Google Scholar]
- 19. Hardie D. G., Carling D. (1997) Eur. J. Biochem. 246, 259–273 [DOI] [PubMed] [Google Scholar]
- 20. Hardie D. G., Scott J. W., Pan D. A., Hudson E. R. (2003) FEBS Lett. 546, 113–120 [DOI] [PubMed] [Google Scholar]
- 21. Carling D. (2004) Trends Biochem. Sci. 29, 18–24 [DOI] [PubMed] [Google Scholar]
- 22. Hardie D. G. (2004) J. Cell Sci. 117, 5479–5487 [DOI] [PubMed] [Google Scholar]
- 23. Kahn B. B., Alquier T., Carling D., Hardie D. G. (2005) Cell Metab. 1, 15–25 [DOI] [PubMed] [Google Scholar]
- 24. Hawley S. A., Pan D. A., Mustard K. J., Ross L., Bain J., Edelman A. M., Frenguelli B. G., Hardie D. G. (2005) Cell Metab. 2, 9–19 [DOI] [PubMed] [Google Scholar]
- 25. Hurley R. L., Anderson K. A., Franzone J. M., Kemp B. E., Means A. R., Witters L. A. (2005) J. Biol. Chem. 280, 29060–29066 [DOI] [PubMed] [Google Scholar]
- 26. Woods A., Dickerson K., Heath R., Hong S. P., Momcilovic M., Johnstone S. R., Carlson M., Carling D. (2005) Cell Metab. 2, 21–33 [DOI] [PubMed] [Google Scholar]
- 27. Nakano A., Kato H., Watanabe T., Min K. D., Yamazaki S., Asano Y., Seguchi O., Higo S., Shintani Y., Asanuma H., Asakura M., Minamino T., Kaibuchi K., Mochizuki N., Kitakaze M., Takashima S. (2010) Nat. Cell Biol. 12, 583–590 [DOI] [PubMed] [Google Scholar]
- 28. Levine Y. C., Li G. K., Michel T. (2007) J. Biol. Chem. 282, 20351–20364 [DOI] [PubMed] [Google Scholar]
- 29. Chiu Y. C., Shieh D. C., Tong K. M., Chen C. P., Huang K. C., Chen P. C., Fong Y. C., Hsu H. C., Tang C. H. (2009) Carcinogenesis 30, 1651–1659 [DOI] [PubMed] [Google Scholar]
- 30. Tang C. H., Lu M. E. (2009) Prostate 69, 1781–1789 [DOI] [PubMed] [Google Scholar]
- 31. Godlewski J., Nowicki M. O., Bronisz A., Nuovo G., Palatini J., De Lay M., Van Brocklyn J., Ostrowski M. C., Chiocca E. A., Lawler S. E. (2010) Mol. Cell 37, 620–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lim S., Jang H. J., Kim J. K., Kim J. M., Park E. H., Yang J. H., Kim Y. H., Yea K., Ryu S. H., Suh P. G. (2010) Stem Cells Dev. [DOI] [PubMed] [Google Scholar]
- 33. Choi J. W., Lim S., Oh Y. S., Kim E. K., Kim S. H., Kim Y. H., Heo K., Kim J., Kim J. K., Yang Y. R., Ryu S. H., Suh P. G. (2010) Cell. Signal. 22, 1153–1161 [DOI] [PubMed] [Google Scholar]
- 34. Natsuizaka M., Ozasa M., Darmanin S., Miyamoto M., Kondo S., Kamada S., Shindoh M., Higashino F., Suhara W., Koide H., Aita K., Nakagawa K., Kondo T., Asaka M., Okada F., Kobayashi M. (2007) Exp. Cell Res. 313, 3337–3348 [DOI] [PubMed] [Google Scholar]
- 35. Xu Y., Fang X. J., Casey G., Mills G. B. (1995) Biochem. J. 309, 933–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oh Y. S., Jo N. W., Choi J. W., Kim H. S., Seo S. W., Kang K. O., Hwang J. I., Heo K., Kim S. H., Kim Y. H., Kim I. H., Kim J. H., Banno Y., Ryu S. H., Suh P. G. (2004) Mol. Cell Biol. 24, 5069–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Etienne-Manneville S., Hall A. (2002) Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 38. Schmidt A., Hall A. (2002) Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- 39. Crepaldi T., Gautreau A., Comoglio P. M., Louvard D., Arpin M. (1997) J. Cell Biol. 138, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Louvet-Vallée S. (2000) Biol. Cell 92, 305–316 [DOI] [PubMed] [Google Scholar]
- 41. Matsui T., Maeda M., Doi Y., Yonemura S., Amano M., Kaibuchi K., Tsukita S. (1998) J. Cell Biol. 140, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gautreau A., Louvard D., Arpin M. (2000) J. Cell Biol. 150, 193–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sawada K., Radjabi A. R., Shinomiya N., Kistner E., Kenny H., Becker A. R., Turkyilmaz M. A., Salgia R., Yamada S. D., Vande Woude G. F., Tretiakova M. S., Lengyel E. (2007) Cancer Res. 67, 1670–1679 [DOI] [PubMed] [Google Scholar]
- 44. Cai X. J., Chen L., Li L., Feng M., Li X., Zhang K., Rong Y. Y., Hu X. B., Zhang M. X., Zhang Y., Zhang M. (2010) Mol. Endocrinol. 24, 218–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Motobayashi Y., Izawa-Ishizawa Y., Ishizawa K., Orino S., Yamaguchi K., Kawazoe K., Hamano S., Tsuchiya K., Tomita S., Tamaki T. (2009) Hypertens. Res. 32, 188–193 [DOI] [PubMed] [Google Scholar]
- 46. Ouchi N., Kobayashi H., Kihara S., Kumada M., Sato K., Inoue T., Funahashi T., Walsh K. (2004) J. Biol. Chem. 279, 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Martinelli R., Gegg M., Longbottom R., Adamson P., Turowski P., Greenwood J. (2009) Mol. Biol. Cell 20, 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miranda L., Carpentier S., Platek A., Hussain N., Gueuning M. A., Vertommen D., Ozkan Y., Sid B., Hue L., Courtoy P. J., Rider M. H., Horman S. (2010) Biochem. Biophys. Res. Commun. 396, 656–661 [DOI] [PubMed] [Google Scholar]
- 49. Yu S., Murph M. M., Lu Y., Liu S., Hall H. S., Liu J., Stephens C., Fang X., Mills G. B. (2008) J. Natl. Cancer Inst. 100, 1630–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Horwitz A. R., Parsons J. T. (1999) Science 286, 1102–1103 [DOI] [PubMed] [Google Scholar]
- 51. Nobes C. D., Hall A. (1995) Biochem. Soc. Trans. 23, 456–459 [DOI] [PubMed] [Google Scholar]
- 52. Gauthier-Rouvière C., Vignal E., Mériane M., Roux P., Montcourier P., Fort P. (1998) Mol. Biol. Cell 9, 1379–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Forget M. A., Desrosiers R. R., Gingras D., Béliveau R. (2002) Biochem. J. 361, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lang P., Gesbert F., Delespine-Carmagnat M., Stancou R., Pouchelet M., Bertoglio J. (1996) EMBO J. 15, 510–519 [PMC free article] [PubMed] [Google Scholar]
- 55. Sawada N., Itoh H., Yamashita J., Doi K., Inoue M., Masatsugu K., Fukunaga Y., Sakaguchi S., Sone M., Yamahara K., Yurugi T., Nakao K. (2001) Biochem. Biophys. Res. Commun. 280, 798–805 [DOI] [PubMed] [Google Scholar]
- 56. Fleming I. N., Elliott C. M., Collard J. G., Exton J. H. (1997) J. Biol. Chem. 272, 33105–33110 [DOI] [PubMed] [Google Scholar]