Abstract
Adult hematopoietic progenitor cells (HPCs) are maintained by highly coordinated signals in the bone marrow. The molecular mechanisms linking intracellular signaling network of HPCs with their microenvironment remain poorly defined. The Rho family GTPase Rac1/Rac2 has previously been implicated in cell functions involved in HPC maintenance, including adhesion, migration, homing, and mobilization. In the present studies we have identified R-Ras, a member of the Ras family, as a key signal mediator required for Rac1/Rac2 activation. We found that whereas Rac1 activity is up-regulated upon stem cell factor, integrin, or CXCL12 stimulation, R-Ras activity is inversely up-regulated. Expression of a constitutively active R-Ras mutant resulted in down-regulation of Rac1-activity whereas deletion of R-Ras led to an increase in Rac1/Rac2 activity and signaling. R-Ras−/− HPCs displayed a constitutively assembled cortical actin structure and showed increased directional migration. Rac1/Rac2 inhibition reversed the migration phenotype of R-Ras−/− HPCs, similar to that by expressing an R-Ras active mutant. Furthermore, R-Ras−/− mice showed enhanced responsiveness to G-CSF for HPC mobilization and exhibited decreased bone marrow homing. Transplantation experiments indicate that the R-Ras deficiency-induced HPC mobilization is a HPC intrinsic property. These results indicate that R-Ras is a critical regulator of Rac signaling required for HPC migration, homing, and mobilization.
Keywords: Adhesion, Cell Migration, G Proteins, Hematopoiesis, Ras, Rho, Rac GTPase
Introduction
Adult hematopoietic stem and progenitor cells (HSPCs)2 are rare populations of cells residing in the bone marrow (BM) that undergo a complex but highly ordered differentiation and self-renewal program to support blood cell development (1–3). Recent genetic studies suggested that HSPCs need an active interaction with their BM microenvironment through a network of cytokines, developmental cues, chemokines, and adhesion molecules to be retained in the BM. The BM microenvironment supplies multiples of these extrinsic factors that are essential for the retention of HSPCs, whereas correspondingly, HSPCs express receptors for these extrinsic stimuli to transduce the signals leading to proper HSPC behaviors. One challenge in understanding how the HSPCs are maintained in such complex signaling networks is to identify critical intracellular components in HSPCs that link various stimuli to signaling effectors that ultimately determine their localization and cell fate. In this context, signaling events mediated by integrins, chemokines, and cytokines are essential in regulating HSPC residency in the BM, and alterations of the related pathways may be critical for HSPC mobilization regimens that are in use clinically (1–5).
Rac1/Rac2 are members of the Rho family of GTPases that function as intracellular signaling switches cycling between a GTP-bound active and a GDP-bound inactive state to transduce diverse signals in cells (6, 7). The Rac GTPases play a major role in the organization of the actin cytoskeleton, cell migration and adhesion and in the control of gene expression and the activation of proliferation and survival pathways. Specifically in HSPCs, a variety of cytokines, chemokines, growth factors, and integrins, including SCF/c-kit, CXCL12/CXCR4, and VCAM-1/fibronectin through β1-integrins, signal through Rac1/Rac2 (4, 5). Guanine nucleotide exchange factors, downstream of cytokine and chemokine receptors, are engaged to mediate Rac activation and initiate an array of cellular responses by endowing the active Rac1/Rac2 with the ability to engage specific effectors (8–10). Therefore, Rac GTPases are critical intracellular signaling molecules in HSPCs responsible for the regulation of migration, adhesion, homing, marrow retention, and peripheral mobilization (11–13). Although extensive efforts have been dedicated to understanding the architecture of the cytokine receptor/guanine nucleotide exchange factor signaling pathways, the complete picture on the mechanism on how Rac activities are regulated by various BM-derived stimuli is not clear.
R-Ras is a member of the Ras family of GTPases (14–17). The sequence of R-Ras is approximately 55% homologous to other members in the Ras family including H-, K-, and N-Ras. All Ras proteins share nearly all of their effector-binding properties and share the majority of their effectors. However, R-Ras and H-Ras have different functions on cell-extracellular matrix adhesion. For example, R-Ras enhances integrin-mediated cell adhesion, whereas H-Ras inhibits integrin activity (18, 19). The role of R-Ras in regulating cell adhesion and migration in different cell types is not well understood. In primary human coronary artery smooth muscle cells and in the breast epithelial cell line MCF10, R-Ras-GTP was reported to inhibit the PDGF-induced cell migration and disrupt epithelial cell motility (20, 21), whereas other reports indicated that R-Ras-GTP could promote cell migration in the cell lines including NIH3T3, 32D, MCF10A and T47D (22–25). Similarly, several reports suggested that R-Ras-GTP induces cell adhesion (14, 26–28), although Nakada et al. reported that R-Ras negatively regulated EphB2-mediated glioma cellular adhesion (29).
The functional relationship between R-Ras and Rac remains controversial. R-Ras has been reported as an upstream regulator of Rac1 (14, 24) but whether it may activate or suppress Rac1 activity appears to differ among cell systems (14, 23, 25). Recently, R-Ras was found to be exclusively expressed in endothelial lineage in mouse (21), raising the issue whether most findings of R-Ras studies in the literature bear physiologic relevance. In the current work, we found that R-Ras is expressed in hematopoietic progenitor cells. To determine the function of R-Ras in primary hematopoietic cells and to clarify the functional relationship between R-Ras and Rac GTPases in this cell population, we have examined an R-Ras knock-out mouse model and found that R-Ras deficiency induces hematopoietic progenitor cell (HPC) migration through an up-regulation of Rac1/Rac2 activity. Our in vitro and in vivo studies show that R-Ras serves as an upstream suppressor of Rac in response to multiple stimuli that control Rac activity and are essential for HPC mobilization and homing. These results indicate that R-Ras functionally cross-talks with Rac1/Rac2 in mediating signaling to actomyosin machinery in HPCs.
EXPERIMENTAL PROCEDURES
Animals and Cells
Knock-out (KO) R-Ras mice have been described previously (21, 30). WT and R-Ras-KO mice, backcrossed for >10 generations into C57BL/6 mice, were used in our experiments. B6.SJL-Ptprc Pepc/BoyJ mice were used for competitive repopulation experiments. BM cells were obtained by crunching murine femora and tibiae. Single BM or spleen cell suspensions were obtained by filtration through a 40-μm filter. The cells were laid on top of Histopaque-1083 (Sigma) and centrifuged at 2000 rpm for 10 min at room temperature to obtain low density BM (LDBM) cells. Murine embryonic fibroblasts and human vascular endothelial cells (HMVECs) were obtained and cultured as described elsewhere (31, 32).
Flow Cytometry Analysis
Isolated LDBM cells were washed in PBS and stained for 15 min at room temperature with a mixture of FITC-conjugated anti-mouse antibodies specific for the cell lineage antigens, including CD45R (B220; clone RA3-6B2), Gr-1 (Ly-6G and Ly-6C; clone RB6-8C5), CD4 (L3T4; clone RM4-5), CD8a (Ly-2; clone 53-6.7), CD3e (clone 145-2C11), CD11b (M1/70), and Ter119 (Ly-76) according to the manufacturer's recommendations (PharMingen, San Diego, CA). Negativity for the expression of these antigens constituted the lineage-negative (Lin−) cell population.
Fluorescence-activated Cell Sorting (FACS) of HPCs
For signaling experiments, Lin− cells were sorted by immunomagnetic selection by incubating labeled cells with anti-biotin beads (MACS, Auburn, CA) according to the manufacturer's instructions. Viable Lin−/c-Kit+, designated as HPCs, were isolated by a FACS under sterile conditions. FACS analysis and sorting were performed using a FACS Canto Flow Cytometer or a FACS Vantage SE DIVA flow cytometer (both from Becton Dickinson), respectively.
Colony-forming Unit (cfu) Assays
Isolated LDBM cells (5 × 104 cells) were cultured in 1 ml of methylcellulose medium (1% methylcellulose, 30% FBS, 2% penicillin and streptomycin, 1% BSA, and 10−4 m β-mercaptoethanol; Stem Cell Technologies, Vancouver, BC) containing 4 units/ml erythropoietin, 100 ng/ml recombinant rat stem cell factor, 100 ng/ml G-CSF, and 100 ng/ml IL-3 (all of them from Peprotech, Rocky Hill, NJ) for 10 days. Colonies containing more than 50 cells were counted under an inverted microscope.
Fluorescence Microscopy
To characterize F-actin assembly and the Rac activity, purified Lin−/c-Kit+ cells were serum-starved in Iscove modification of DMEM (IMDM; Mediatech, Herndon, VA) and stimulated with or without the chemokine CXCL12 (100 ng/ml, from Peprotech, Rocky Hill, NJ) for 15 s. The cells were fixed with 2% paraformaldehyde (pH 7.4; Sigma) and permeabilized with 0.1% Triton X-100 (Sigma) for F-actin or cold icetone (50 and 100%) for Rac-GTP. After blocking in 2% BSA, the cells were stained with TRITC-conjugated phalloidin (Molecular Probes) and DAPI for F-actin or Rac-GTP (Neweast Bioscience, Malwern, PA) antibody followed by Alexa Fluor® 568 goat anti-mouse secondary antibody (Molecular Probes) and mounted for fluorescence imaging analysis (Zeiss fluorescence microscope). Images shown of F-actin staining are representative of more than 100 cells examined for each genotype. The fluorescence intensity of Rac1-GTP was measured by using the ImageJ (1.44p) software. At least 50 cells were examined for each genotype.
R-Ras, H-Ras, Cdc42, RhoA, and Rac1/Rac2 Activity Assays
The relative levels of Rac1-GTP, Rac2-GTP Cdc42-GTP, R-Ras-GTP, and H-Ras-GTP in Lin− cells were examined by effector domains of GST-PAK1 or GST-cRaf1 pulldown protocols (33). Briefly, the isolated wild-type or mutant Lin− cells were starved in serum/cytokine-free IMDM overnight and challenged with rrSCF (100 ng/ml), CXCL12 (100 ng/ml), or IL-3 (20 ng/ml) for single or multiple time points of 2, 5, and 10 min prior to the effector domain pulldowns. Separately, the cells adhering to fibronectin fragment (CH296)-coated surfaces for 2 h were subjected to a similar GST-cRaf1 pulldown assay compared with the cells in suspension. The levels of active Rac1, Cdc42, Rac2, R-Ras, and H-Ras were detected by monoclonal anti-Rac1, anti-Cdc42 (both from BD Transduction), polyclonal anti-Rac2 (Novus Biologicals, Littleton, CO), polyclonal anti-R-Ras (Cell Signaling Technology, Boston, MA), and monoclonal anti-H-Ras (Santa Cruz Biotechnology, Santa Cruz, CA) antibodies, respectively.
Downstream Signaling Activation by Phospho-protein Immunoblotting
Isolated Lin− cells from both wild-type and mutant R-Ras mice were lysed and immunoblotted with rabbit polyclonal anti-R-Ras antibody (Cell Signaling Technology) to examine the expression of R-Ras in the WT and mutant R-Ras mice. To examine the activities of PAK1, MLC, AKT, and ERK, isolated Lin− cells were serum-starved for 24 h and then subjected with or without stimulation with 10% FBS, 100 ng/ml rrSCF, 100 ng/ml CXCL12, and 20 ng/ml IL-3 for 15 min or 100 ng/ml CXCL12 for 2, 5, 10 min, respectively, before cells were lysed and immunoblotted with anti-phospho-PAK1 (Ser199/Ser204), anti-phospho-MLC, anti-phospho-AKT, and anti-phospho-ERK (Cell Signaling Technology).
F-actin and Integrin Expression Assays
To determine the F-actin and integrin (α4β1 and α5β1) expression in HPCs, total BM cells were stained with rhodamine phalloidin for F-actin; α4β1-FITC or α5β1-PE for integrin as well as progenitor markers including c-Kit-APC, Sca-1-PE-Cy7, and biotinylated lineage-depletion mixture labeled with the secondary antibody Percp-Cy5-streptividin (all of the antibodies are from PharMingen). F-actin or integrin was quantified by flow cytometry as mean fluorescence intensity.
Cell Migration and Adhesion Assays
Isolated LDBM cells were subjected to adhesion and migration assays in vitro. For adhesion assays, the cells were incubated on plates coated with recombinant fibronectin fragment CH-296 (TaKaRa Biotechnology) containing the integrin α4β1 and α5β1 binding sites for 1 h in IMDM and were washed with PBS three times before the adherent cells were harvested by using the Cell Dissociation buffer (BD Biotechnology). For migration assays, the cells were plated in the upper well of a transwell chamber separated with a filter containing 5-μm pore size (Corning, Corning, NY) in IMDM medium with 2% BSA. After a 4-h incubation against a CXCL12 (or without CXCL12) gradient at the lower chamber, the cells that migrated through the filter were harvested. They were then assayed for colony-forming activity in methylcellulose medium containing rrSCF, IL-3, EPO, and G-CSF. The percentage of adherent or migrated HPCs was calculated and expressed as a ratio of formed colony numbers from the adherent or migrated cells and from the total input cells.
Mobilization of Peripheral Blood (PB) Hematopoietic Progenitors by Administration of G-CSF
We administered recombinant human G-CSF (Neupogen, Amgen) intraperitoneally every day for 6 days at a dose of 200 μg/kg per day. PB was collected by retroperitoneal puncture 2 h after administration of the last dose of drug (42).
Generation of R-Ras-deficient Hematopoietic and Nonhematopoietic Chimeric Mice
Full hematopoietic and nonhematopoietic chimeras were generated after transplantation of 3 × 106 bone marrow cells from either WT and R-Ras−/− mice into lethally irradiated (two doses of 7 and 4.75 gray, 3 h apart) CD45.1+ B6.SJL[Ptprca Pep3b/BoyJ] mice (hematopoietic chimeras) or after transplantation of CD45.1+ B6.SJ[LPtprca Pep3b/BoyJ] (WT) mice into lethally irradiated either WT and R-Ras−/− mice (reverse transplantation). Chimerism was analyzed in PB at 6 weeks after transplantation. PB of hematopoietic chimeric mice contained 98.8 ± 1.0% and 96.4 ± 4.3% CD45.1+ leukocytes for WT and R-Ras−/− groups, respectively. PB of reverse transplantation recipients of WT bone marrow contained 94.8 ± 1.4% and 94.2 ± 2.3% CD45.2+ leukocytes for WT and R-Ras−/− groups, respectively.
Homing Assay
Donor animals (R-Ras-KO or R-Ras WT) were sacrificed, and single-cell suspensions of the BM were prepared, counted, and used for transplantation. An aliquot was also plated in methylcellulose culture to quantify committed progenitor cells. Twenty million cells were injected through the tail vein into lethally irradiated mice. Sixteen hours later, recipients were sacrificed, and the BM and spleen were collected. Single-cell suspensions of BM were prepared and cultured in triplicate to assess donor colony-forming unit in culture (cfu-C) recovery in the recipient animals. To estimate the total BM homing, the BM content of one femur was assumed to represent 6.7% of total BM (34).
Statistical Analysis
Experimental data were analyzed and compared for statistically significant differences by a two-tailed Student's t test or two-way ANOVA. Data are presented as the mean ± S.D. or S.E.
RESULTS
R-Ras and Rac1 Activities Are Inversely Regulated by Cytokine, Chemokine, and Integrin
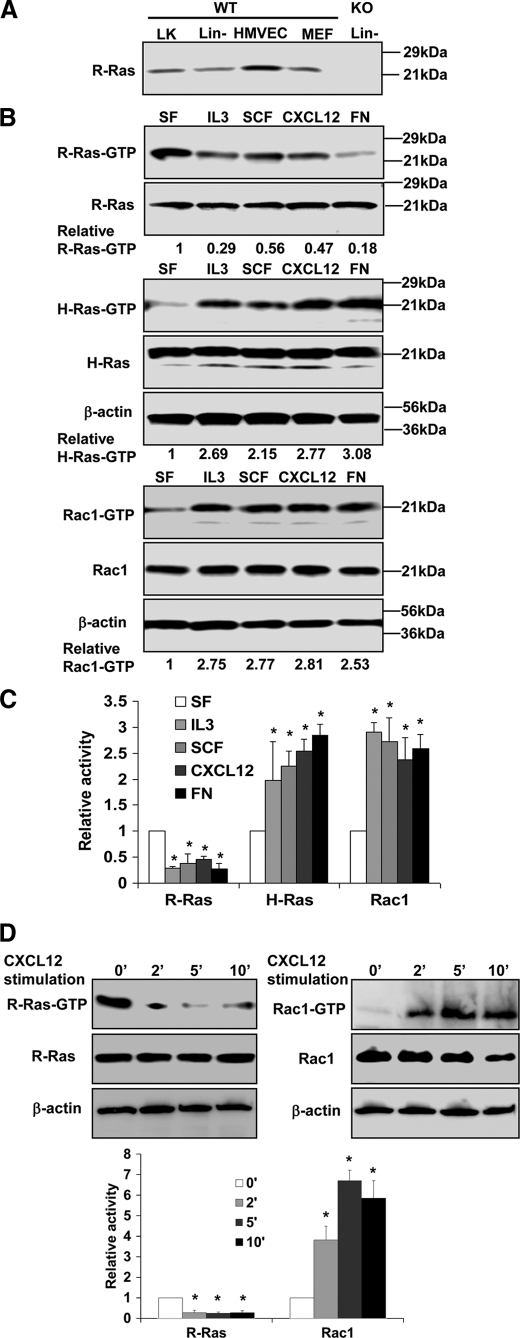
R-Ras was previously suggested to be exclusively expressed in the endothelial lineage in mice (16). We first carried out Western blotting to examine whether R-Ras is also expressed in hematopoietic progenitor cells. We used an R-Ras antibody to detect the endogenous expression of R-Ras in Lin−, Lin−/c-Kit+ BM hematopoietic cells, primary mouse embryonic fibroblast cells, and HMVECs, using the R-Ras−/− Lin− cells as controls. We found that R-Ras, in addition to HMVEC, is also expressed in Lin− and Lin−/c-Kit+ BM cells, populations enriched for HPCs (Fig. 1A).
FIGURE 1.
Analysis of R-Ras, H-Ras, and Rac1 activities in HPCs. A, expression of R-Ras in HPCs. Lin− and Lin−/c-Kit+ (LK) cells were harvested from WT or knock-out (KO) mouse BM. Expression of R-Ras of the indicated samples was examined by Western blotting. B, R-Ras, H-Ras, and Rac1 activities under various stimulatory conditions in Lin− cells. Lin− cells from WT mice under various stimulatory conditions were subjected to effector domain pulldown assays, and the activities of R-Ras, H-Ras, and Rac1 were examined. Blotting of the respective total cell lysates was carried out in parallel. Relative amounts of GTP-bound form of the GTPases were quantified by densitometry and normalized to the nonstimulated control. Data are representative of three independent experiments. C, relative activities quantified by densitometry and normalized to the nonstimulated control (mean ± S.D. from three independent experiments). D, R-Ras and Rac1 activities under CXCL12 stimulation at multiple time points (mean ± S.D. (error bars) from three independent experiments). *, p < 0.05.
Previous reports showed that in hematopoietic cells, Rac1 activity was regulated by multiple cytokine, chemokine, or integrin stimulations (4, 5). Because of the controversial relationship of R-Ras with Rac1 described in the literature, we next tested whether Rac1 activity is associated with R-Ras activity in hematopoietic cells. We measured R-Ras and Rac1 activation in Lin− BM cells after transient stimulation by the cytokine IL-3 or SCF, chemokine CXCL12, or fibronectin. R-Ras activity was significantly decreased (∼3–5-fold) under these stimulation conditions (Fig. 1B). In contrast, the Rac1-GTP level, as well as the H-Ras-GTP level, in the isolated Lin− BM cells were significantly elevated (∼ 2–3-fold) in response to similar stimulation by SCF, IL-3, CXCL12, or fibronectin (Fig. 1, B and C). The inverse effects of these different stimuli on R-Ras and Rac1 activities suggest a close association between Rac1 activation and R-Ras inhibition in the transduction of diverse signals in Lin− HPCs. To explore the dynamics of R-Ras and Rac1 activities in response to the agonists, we serum-starved and then stimulated the Lin− BM cells with 100 ng/ml CXCL12 for 2, 5, 10 min, respectively. Rac1 activities were increased, and R-Ras activities were inhibited at these time points after stimulation (Fig. 1D), further confirming the inverse relationship of R-Ras inhibition and Rac1 activation by these stimuli.
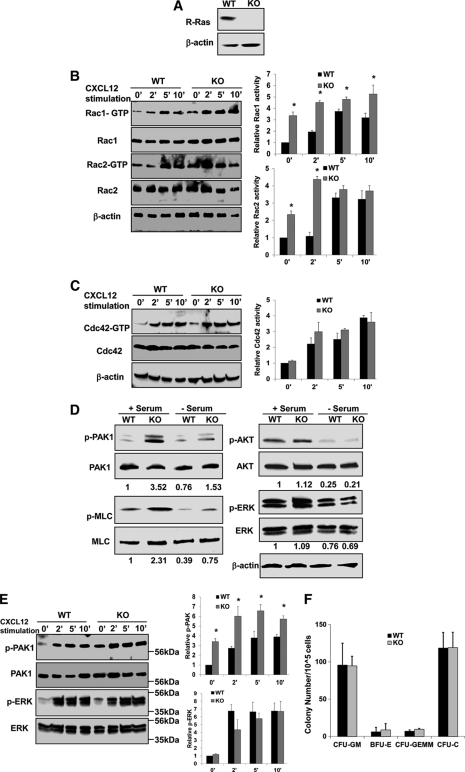
R-Ras Deficiency Causes Rac1/Rac2 Activation in HPCs
To determine whether the inverse relationship of R-Ras activity and Rac1 activity is linked, we utilized Lin− cells from R-Ras mutant (KO) mice and their litter control WT mice. Lin− cells from the mutant mice showed an absence of R-Ras protein expression (Fig. 2A). We examined the Rac1 activity under CXCL12 stimulation at multiple time points by the GST-Pak1 effector pulldown assay. As shown in Fig. 2B, the Rac1-GTP level in the isolated Lin− BM cells in the nonstimulatory condition was >3-fold higher in R-Ras mutant cells compared with WT control cells. Moreover, the Rac1-GTP levels at 2-, 5-, and 10-min stimulation points were all significantly higher in the R-Ras mutant cells than in the WT cells. In addition, R-Ras mutant cells contained a higher Rac2 activity than WT cells at the nonstimulatory and 2-min time point but not at later time points, suggesting that Rac2 activity is also inversely related with R-Ras activity but with a slightly different kinetics (Fig. 2B). R-Ras-deficient Lin− BM cells showed a higher basal level of Rac1 activation (Fig. 2B). These data indicate that R-Ras-deficient Lin− BM cells have reduced sensitivity to stimuli in Rac1 response and contain close to maximal activation of Rac1 at the basal level. Unlike Rac1, the activity of Cdc42 remained unchanged in the R-Ras mutant cells (Fig. 2C, from the same cell samples in Fig. 2B). Consistent with the elevated basal Rac1 status in R-Ras−/− cells, we found that phospho-PAK and phospho-MLC, downstream signal components of Rac1, were elevated in R-Ras-deficient Lin−/c-Kit+ cells under both serum-stimulatory and serum-free conditions, as well as under CXCL12 stimulation (Fig. 2, D and E). In contrast, we did not detect significant changes in the activities of AKT or ERK by phospho-antibody blotting (Fig. 2, D and E), suggesting that R-Ras deficiency impairs signals crucial in actomyosin machinery but does not affect signals controlling proliferation and/or survival. Indeed, there was no detectable difference in the cfu-C activity of R-Ras-KO BM compared with that of WT bone marrow (Fig. 2F), suggesting that proliferation and/or survival of R-Ras-KO HPCs is not compromised. Together, these results establish that R-Ras deficiency promotes Rac1/Rac2 activation in HPCs.
FIGURE 2.
R-Ras inversely regulates Rac1/Rac2 activity and signaling in HPCs. A, R-Ras signals from Lin− cells of WT or mutant R-Ras mice were detected by Western blotting using R-Ras antibody. B and C, Lin− cells were stimulated with CXCL12 for 0, 2, 5, and 10 min. Cells were then subjected to effector domain pulldown assays. The activities of Rac1 (B), Rac2 (B), and Cdc42 (C) were analyzed. Relative amounts of GTP-bound form of the GTPases were quantified by densitometry and normalized to the nonstimulated control. D, Western blotting of phospho-PAK1, phospho-MLC, phospho-AKT, phospho-ERK and relevant controls of the Lin− cells from WT or mutant mice is shown. Relative amounts phospho-PAK1, phospho-MLC, phospho-AKT and phospho-ERK were quantified by densitometry measurements and normalized to the nonstimulated control. Data are representative of three independent experiments. E, Western blotting of phospho-PAK1 and phospho-ERK and relevant controls of the Lin− cells from WT and mutant mice that were stimulated with CXCL12 of the indicated time points is shown. F, cfu-GM, BFU-E, cfu-GEMM, and cfu-C activities in BM were determined in the methylcellulose culture. cfu-GM, cfu that produces colonies of granulocytes and macrophages; cfu-GEMM, cfu that produces colonies of neutrophils, eosinophils, erythrocytes, macrophages, megakaryocytes, and mast cells in various combination; BFU-E, burst-forming unit-erythroid). Data are representative of three independent experiments.
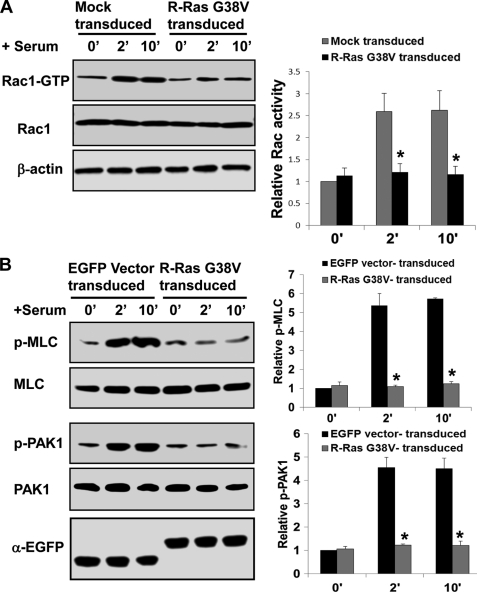
Constitutively Active R-Ras Mutant Suppresses Rac1 Activity
To investigate further whether R-Ras activation could inhibit Rac1 activity, we introduced a constitutively active R-Ras mutant (G38V) into the WT Lin− BM cells by retrovirus-mediated transduction. Expression of the R-Ras G38V rendered the Rac1 activity of the Lin− BM cells from WT mice insensitive to stimulation by serum and multiple cytokines at 2- and 10-min time points (Fig. 3A). In conjunction with the activity pulldown assay, Western blotting results showed that both p-PAK1 and p-MLC levels also became insensitive to serum/cytokine stimulation in the R-Ras G38V expression cells (Fig. 3B). These data indicate that R-Ras acts upstream of Rac1 to suppress its activity.
FIGURE 3.
R-Ras G38V inhibits cytokine-induced Rac1 activation and signaling in HPCs. Lin− cells from WT mice were transduced with retroviruses containing the MSCV-EGFP-R-Ras G38V constitutive active mutant vector, the empty MSCV vector (Mock), or the empty MSCV-EGFP vector. The transduction efficiency ranged between 65 and 70%. The transduced cells were stimulated with a mixture of cytokines for 2 or 10 min. A, cells were subjected to effector domain pulldown assays, and the activities of Rac1 were examined. Relative Rac-GTP levels were quantified by densitometry normalized to the Mock-transduced nonstimulated control (mean ± S.D. (error bars) from three independent experiments). B, Western blotting of phospho-PAK1, phospho-MLC, and relevant controls of the Lin− cells transduced with MSCV-EGFP-R-Ras G38V or MSCV-EGFP-vector stimulated with a mixture of cytokines for 2 or 10 min is shown. The expression of endogenous R-Ras and overexpressed EGFP-fused R-Ras G38V was detected by anti-EGFP antibody (mean ± S.D. from three independent experiments). *, p < 0.05.
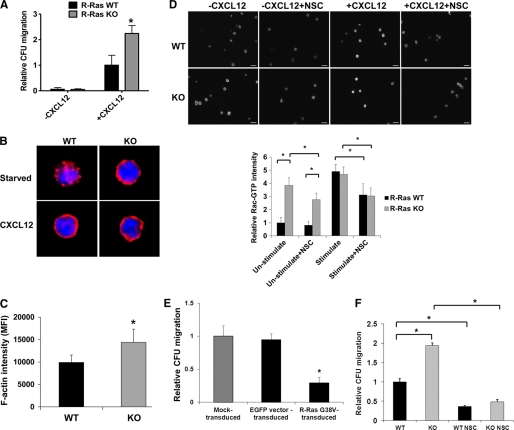
R-Ras Regulates HPC Migration and Actin Organization
Rac GTPases are important regulators of HPC migration and actin cytoskeleton reorganization (13). To investigate whether R-Ras regulates HSPC migration and actin reorganization by modulating Rac1 activity, WT and R-Ras mutant Lin−/c-Kit+ cells were analyzed for their abilities to migrate across a transwell toward a CXCL12 gradient. Although the basal migration activities of WT and R-Ras-KO cells were similarly low, R-Ras-deficient HPCs showed a significantly increased migration ability compared with WT cells in response to CXCL12 (Fig. 4A). Isolated WT and R-Ras-KO Lin−/c-Kit+ cells were also analyzed for F-actin organization upon CXCL12 stimulation. In WT cells, upon CXCL12 stimulation, patched F-actin was reorganized to form a cortical actin network, as revealed by immunofluorescence (Fig. 4B). Even prior to stimulation the F-actin of R-Ras-KO cells was assembled at the cell cortex, indicating that signals regulating cortical actin polymerization, which are dependent on Rac, show higher basal level in R-Ras-deficient cells (Fig. 4B). To quantify the R-Ras deficiency-induced assembly of F-actin, flow cytometry was performed to test the F-actin intensity of both R-Ras WT and KO cells. In the population of Lin−/c-Kit+ cells, without any stimulation, R-Ras-KO cells showed higher F-actin intensity than the R-Ras WT cells, which further suggested that R-Ras deficiency induced higher Rac activity (Fig. 4C). To measure the activation of Rac1 GTPase in R-Ras-KO cells more directly, we stained the unstimulated R-Ras WT and KO Lin−/c-Kit+ cells with a Rac1-GTP-specific antibody. Fig. 4D shows that compared with the WT cells, the intensity of Rac1-GTP in the R-Ras-KO cells was significantly increased. The Rac-specific inhibitor, NSC23766 (12), could inhibit the R-Ras deficiency-induced Rac-GTP elevation (Fig. 4D). To determine further the role of R-Ras activity on HPC migration, R-Ras G38V or a mock vector (control) was retrovirally transduced into Lin− HPCs, and additional transwell migration assays coupled with a cfu activity assay were performed. R-Ras G38V-expressing cells had a 60% lower migration than mock-transduced cells (Fig. 4E), further indicating that R-Ras activity negatively regulates HPC migration.
FIGURE 4.
R-Ras regulates migration and cortical actin assembly in HPCs. A, WT or R-Ras KO LDBM (n = 6 for each group) cells were subjected to migration assays in vitro. B, isolated Lin−/c-Kit+ (LK) BM cells were stimulated with CXCL12 and stained with rhodamine-phalloidin for actin and DAPI for nucleus. Images shown are representative of >100 cells examined for each genotype. C, total BM cells from the WT or KO R-Ras mice without CXCL12 stimulation were stained with rhodamine-phalloidin for F-actin. The mean fluorescence intensity (MFI) of F-actin was subsequently examined through flow cytometry. The LK population was selected for the mean fluorescence intensity calculation. D, LK cells isolated from WT or KO mice were serum-starved overnight in the presence or absence of 50 μm NSC23766. The cells were then stimulated with CXCL12, fixed, and stained with Rac-GTP antibody. The images of the cells were taken under microscope. The relative Rac-GTP fluorescence intensities were measured by ImageJ (1.44p) software. E, Lin− cells from WT mice were transduced with retroviruses containing the MSCV-EGFP-R-Ras G38V vector, the empty MSCV vector (Mock), or the empty EGFP-expressing vector. Transduction efficiency was ∼65–70%. The transduced cells were subjected to transwell migration assays in vitro. F, WT or KO LDBM (n = 6 for each group) cells treated with 50 μm NSC23766 or without treatment were subjected to migration assays in vitro. *, p < 0.05. The data are representative of three independent experiments. Error bars, S.D.
To examine whether R-Ras-mediated directional migration is associated with Rac activity, we applied the Rac-specific inhibitor, NSC23766, to the isolated LDBM cells and carried out transwell migration assay coupled with cfu activity assay of the migrated cells. Our results show that Rac inhibition reduced migration of both WT and R-Ras-deficient HPCs and completely reversed the hypermigratory phenotype of R-Ras-deficient HPCs to that of WT HPCs (Fig. 4F). These data indicate that Rac1 activity is required for the increased cell migration observed in R-Ras mutant cells and further support the role of R-Ras as a key upstream regulator for Rac-mediated HPC migration.
R-Ras Deficiency Increases Progenitor Mobilization to PB
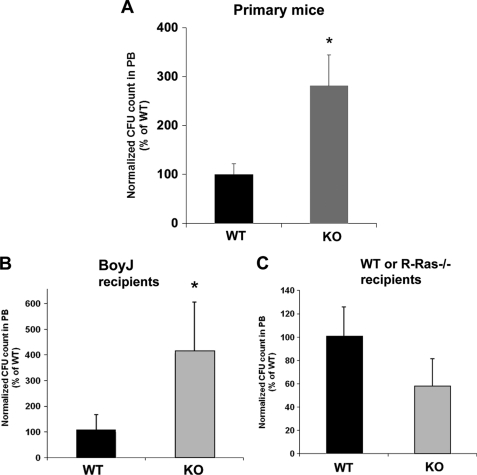
To examine whether the increased in vitro migratory activity of R-Ras KO HPCs could alter in vivo mobilization, the mobilization of WT and mutant R-Ras HSPCs from the BM to PB and spleen after G-CSF stimulation was measured. We administered G-CSF, a cytokine reported to activate migration of HPCs out of BM by inverting the CXCL12 gradient between PB and BM (35), to WT and R-Ras-deficient mice. After 6 days of daily administration, we harvested PB, spleen, and BM cells and assayed their cfu activity. There were no differences in the overall cellularity of these tissues between WT and R-Ras-deficient mice (data not shown). Consistent with the hypermigratory phenotype of R-Ras-deficient HPCs in response to CXCL12, the number of HPCs in PB of R-Ras-KO mice was ∼3-fold higher than in WT mice (p < 0.01, Fig. 5A). We did not detect any significant difference in the colony numbers in the spleen and BM cells of the G-CSF-treated WT versus R-Ras-KO mice, as is the case for basal PB cfu activities (data not shown). Further, BM cells from the WT and the R-Ras-KO mice yielded similar cfu mix activities (Fig. 2F). These data suggest that the increase in HPC count in PB of R-Ras-KO mice was not a result of increased proliferation of HPCs in hematopoietic organs. To investigate further whether R-Ras deficiency-induced HPC mobilization is due to HPC intrinsic property or a change of microenvironment, R-Ras-deficient hematopoietic and nonhematopoietic chimeric mice were generated, followed by CXCL12-induced mobilization assay. In response to the CXCL12, R-Ras-KO BM cells that transplanted to BoyJ mice showed ∼3-fold higher mobilization than the WT BM cells (Fig. 5B). On the contrary, BoyJ mice BM cells that transplanted to WT or KO R-Ras mice showed no significant difference in the mobilization activity (Fig. 5C). These data indicate that R-Ras-KO-induced BM mobilization was a result of HPC intrinsic property. Together, the results confirm the role of R-Ras-mediated signaling in the regulation of HPC mobilization in vivo.
FIGURE 5.
R-Ras deficiency increases hematopoietic progenitor, but not stem cell, mobilization in response to G-CSF administration. A–C, mobilization of hematopoietic progenitors (cfu-C) in PB of primary and chimeric R-Ras−/− mice. Mice submitted to G-CSF administration were compared for their ability to mobilize HSC/P to PB. Results represent normalized PB cfu-C counts in primary WT and R-Ras−/− mice (A), WT and R-Ras−/− hematopoietic chimeras (B), and WT and R-Ras−/− reverse transplantation recipients of WT bone marrow (C). Data represent the average and S.D. (error bars) of the percentage of Lin− PB cells and are representative of three independent experiments. *, p < 0.05.
R-Ras Deficiency Leads to a Homing Defect of HPCs
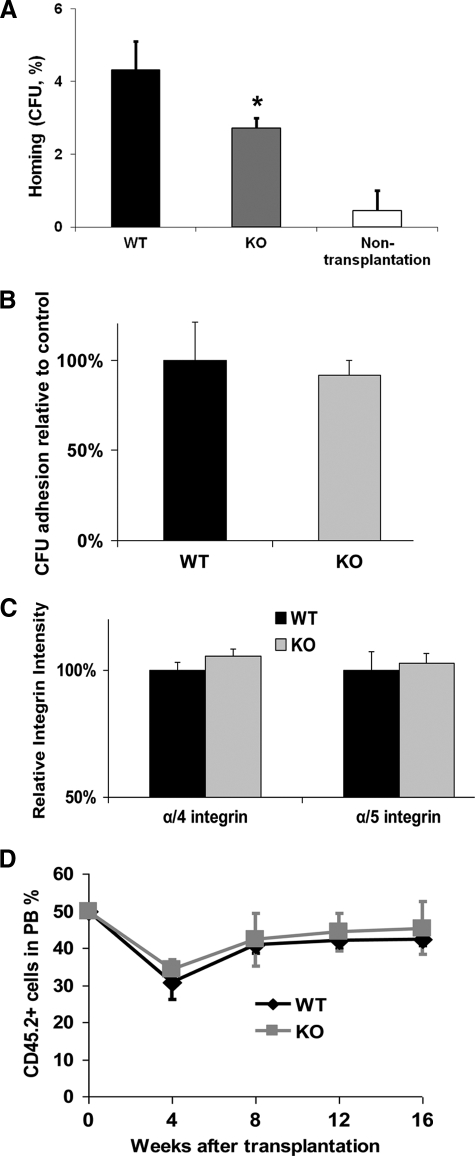
A corollary of the hypothesized activity of R-Ras in HPC migration as a crucial signal transducer is that deficiency of R-Ras may reduce the ability of progenitor cells to home BM after transplantation. WT and R-Ras mutant BM cells were transplanted into lethally irradiated syngeneic mice. After 16 h, BM cells were harvested from the recipients and assayed for cfu activity. The results in Fig. 6A show that R-Ras deficiency led to reduced progenitor homing as indicated by decreased donor-derived cfu activities in BM of recipient mice (WT 4.3% versus KO 2.8%, p < 0.05).
FIGURE 6.
R-Ras-deficient HPCs show defective homing activity. A, the homing ability of BM cells into lethally irradiated hosts was determined. Data are presented as the percentage of cfu residing in BM at 16 h after transplantation into lethally irradiated recipients (basal level is 0) and calculated as indicated under “Experimental Procedures.” B, WT or KO LDBM (n = 6 for each group) cells were harvested and applied for adhesion assay on the CH296-coated 96-well plate. C, total BM cells from the WT or KO R-Ras mice were stained with anti-α4β1 and α5β1 integrin antibody. The α4β1and α5β1 integrin intensities were subsequently examined through flow cytometry. Lin−/c-Kit+ population was selected for the relative intensity calculation. D, CD45.2+ chimera level of WT or R-Ras mutant cells after competitive BM transplantation into lethally irradiated CD45.1+ B6.SJL-Ptprc Pepc/BoyJ recipient mice. Transplanted cells were mixed at 1:1 ratio with CD45.12+ WT competitor cells. The level of chimera was determined in PB by analysis of CD45.2+ cell content by flow cytometry at 16 weeks after transplantation. Data are representative of three independent experiments. *, p < 0.05.
To investigate the mechanism for the defective homing activity of R-Ras-KO HPCs, we examined the expression of β1-integrin, the receptor contributing to the attachment of HPCs in the BM through flow cytometry. Consistent with the in vitro cfu adhesion assay data of the R-Ras WT and KO HPCs on the fibronectin-coated surface (Fig. 6B), no significant difference of β1-integrin expression between WT and KO was detected (Fig. 6C).
To characterize further whether R-Ras deficiency also affects hematopoietic stem cell (HSC) engraftment, a competitive repopulation assay by transplanting lethally irradiated syngeneic BoyJ recipient mice with CD45.1+ WT and CD45.2+ WT or R-Ras-deficient BM cells at 1:1 ratio was performed. No significant difference in the chimerism of R-Ras-deficient hematopoietic cells compared with WT controls, 16 weeks after transplantation, was detected (Fig. 6D), indicating that R-Ras deficiency does not impair HSC engraftment. These data are consistent with the apparently normal blood counts of R-Ras-KO mice (Table 1) and further suggest that R-Ras selectively regulates the traffic of hematopoietic progenitors, but not HSC, between BM and PB, under stress which is distinct from previous observed effects by loss of Rac1/Rac2 function.
TABLE 1.
Hematologic parameters of wild-type (WT) and knock-out (KO) R-Ras mice
Peripheral blood samples were collected from R-Ras WT (n = 6) and KO (n = 6) mice when they were 20 weeks old. Blood counts were performed with a Hemavet 850 hematology analyzer. Data are mean ± S.E.
| Parameters | WT | KO |
|---|---|---|
| White blood cell (109/liter) | 9.784 ± 3.11 | 11.265 ± 3.11 |
| Absolute neutrophil count (109/liter) | 1.702 ± 0.37 | 2.1025 ± 0.72 |
| Red blood cell (1012/liter) | 7.486 ± 0.59 | 7.64 ± 1.00 |
| Hemoglobin (g/liter) | 122 ± 10.7 | 124 ± 12.4 |
| Hematocrit (%) | 38.4 ± 2.72 | 40.975 ± 4.03 |
| Mean corpuscular volume (fl) | 51.34 ± 2.08 | 53.95 ± 5.25 |
| Red blood cell distribution width (%) | 19.58 ± 0.88 | 20.575 ± 0.68 |
| Platelet count (109/liter) | 1,314 ± 102.90 | 1,524.75 ± 126.29 |
| Monocytes (109/liter) | 0.546 ± 0.28 | 0.85250 ± 34 |
| Lymphocytes (109/liter) | 7.516 ± 2.59 | 8.2475 ± 2.21 |
DISCUSSION
How diverse extracellular stimuli available in the BM are transduced in HPCs resulting in their functional retention in the BM and their mobilization to PB is an important question in hematopoiesis. Rac1 and Rac2, members of the Rho family GTPases, are key regulators of these functions in hematopoietic cells. We have previously examined the role of Rac1 and Rac2 in HSPC engraftment and mobilization (12, 13). The deletion of both Rac1 and Rac2 led to a massive egress of HPCs into the blood from the marrow, whereas Rac1−/− but not Rac2−/− HSPCs failed to engraft in the BM of irradiated recipient mice due to impaired homing (12, 13).
In our current study, R-Ras is identified as an intracellular regulator of HPC migration and mobilization. R-Ras may serve as a negative signal mediator required specifically for Rac activation, as deficiency of R-Ras causes an increased activity of Rac1 in HPCs, which, in turn, is involved in controlling HPC migration, homing, and mobilization. R-Ras regulation of Rac1 activity has been studied in cell lines by R-Ras mutant overexpression, and the effect of R-Ras on cell adhesion and migration appears to be cell system-dependent. Our work has now demonstrated that in primary HPCs, under various cytokine/chemokine stimulation conditions, activation of R-Ras correlates with down-regulation of Rac1 activity, whereas deletion of R-Ras induces Rac1/Rac hyperactivation without affecting other GTPases like Cdc42 or H-Ras activity. Thus, R-Ras activity translates into Rac1/Rac2-dependent actin polymerization and migration, impacting on HPC trafficking in vitro and in vivo but does not influence cell proliferation. The data indicate that in HPCs Rac1/Rac2 and R-Ras establish a cross-talk, demonstrated by the influence on Rac1 activity by genetic deletion of R-Ras, constitutively active R-Ras mutant expression, as well as pharmacological inhibition of Rac1/Rac2 on the R-Ras negative background.
Postnatal HPCs reside in the BM in equilibrium with a small circulating fraction residing in the PB. During mobilization, HPCs exit their microenvironment in the BM, presumably migrate through an endothelial barrier and circulate in the bloodstream, without immediately returning (homing) to the BM or any other organ. Mobilization may rely on stem/progenitor cell-intrinsic or -extrinsic pathways; indeed, multiple cellular or local soluble factors from the microenvironment have been considered to be determinants for HPC retention and homing. Our results indicate that BM homing of R-Ras-deficient HPCs is defective whereas mobilization of R-Ras-deficient progenitors in response to G-CSF administration is increased. However, the deficiency of R-Ras only seems to impair HPCs trafficking but not the engraftment or retention after G-CSF administration of the primitive HSC population, suggesting that although R-Ras controls the migratory properties of progenitor cells, it seems not to play a crucial role in the homing, engraftment, or mobilization of stem cells. Several possibilities arise from this observation. First, because it is not clear whether most cell types express R-Ras (21, 36), R-Ras may be differentially expressed or functionally available in different HSPC subpopulations. We identified that LK cells do express and can activate R-Ras upon appropriate stimulation, and upon Rac1 inhibition, R-Ras activation significantly increases. The low frequency of HSC prevented us from performing biochemical assays in further purified HSC populations. Second, despite the use of different agonists that are capable of stimulating chemotaxis and chemokinesis in HSC and HPC, it is possible that R-Ras cross-talks to Rac1 are only preserved or especially relevant in HPCs while being less relevant in HSC. Finally, there is cumulative evidence that HSCs and HPCs share but also differ in the signals dependent on cell-to-cell and paracrine signals from the neighboring microenvironment in relation to different microanatomical locations (37–39) or specific differences in their receptor expression or downstream signaling (40), which may be responsible for a different level of R-Ras activation between HSCs and HPCs. Other pathways, like H-Ras and Cdc42, seem not to be affected by R-Ras deficiency, suggesting that in hematopoietic progenitors, the cross-talk between Rac1 and R-Ras is specifically relevant in the migratory phenotype.
The deletion of Rac2 results in elevated egress of HPCs into the blood from the marrow, and Rac1−/− HPCs failed to engraft in the bone marrow of irradiated recipient mice due to impaired homing (12, 13). These observations might seem to be counterintuitive to our current data in which hyperactivation of Rac1/Rac2 is associated with the same phenotypes. The increased mobilization of Rac2−/− HPCs may be due to the defective adhesion activity, even though they completely lost the transwell migration activity (11–13). However, the reason for R-Ras deficiency-induced mobilization effect might be different because R-Ras−/− HPCs show increased migration activity whereas the adhesion activity is not affected. Together, though, they support a model in which absence or hypoactivity and hyperactivity of Rac GTPase activities disturb similar cell intrinsic signaling pathways, resulting in similar cellular phenotypes. This is not unlike what is observed in the case of Cdc42 gain or loss activity mouse models (33, 41).
In summary, our study using the combined approaches of a R-Ras-KO mouse model and constitutively active R-Ras mutant expression identifies R-Ras as an upstream, negative regulator of Rac1/Rac2 GTPases in HPC migration and retention. The R-Ras and Rac cross-talk controls the actomyosin machinery of hematopoietic progenitors that affects their migration and homing activities and is involved in mobilization of hematopoietic progenitors from BM to PB stimulated by G-CSF.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA125658, R01 CA141341, and P30 DK090971 (to Y. Z.) and R01 HL087159 (to J. A. C.).
- HSPC
- hematopoietic stem and progenitor cell
- BM
- bone marrow
- cfu-C
- colony-forming unit in culture
- HMVEC
- human vascular endothelial cell
- HPC
- hematopoietic progenitor cell
- HSC
- hematopoietic stem cell
- IMDM
- Iscove's DMEM
- LDBM
- low density BM
- Lin−
- lineage-negative
- PB
- peripheral blood
- TRITC
- tetramethylrhodamine isothiocyanate
- SCF
- stem cell factor
- CXCL12
- chemokine ligand 12
- CXCR4
- C-X-C chemokine receptor type 4
- VCAM-1
- vascular cell adhesion molecule 1.
REFERENCES
- 1. Adams G. B., Scadden D. T. (2006) Nat. Immunol. 7, 333–337 [DOI] [PubMed] [Google Scholar]
- 2. Wilson A., Trumpp A. (2006) Nat. Rev. Immunol. 6, 93–106 [DOI] [PubMed] [Google Scholar]
- 3. Morrison S. J., Spradling A. C. (2008) Cell 132, 598–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cancelas J. A., Williams D. A. (2009) Curr. Opin. Hematol. 16, 249–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mulloy J. C., Cancelas J. A., Filippi M. D., Kalfa T. A., Guo F., Zheng Y. (2010) Blood 115, 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bosco E. E., Mulloy J. C., Zheng Y. (2009) Cell. Mol. Life Sci. 66, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Etienne-Manneville S., Hall A. (2002) Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 8. Moon S. Y., Zheng Y. (2003) Trends Cell Biol. 13, 13–22 [DOI] [PubMed] [Google Scholar]
- 9. Zheng Y. (2001) Trends Biochem. Sci. 26, 724–732 [DOI] [PubMed] [Google Scholar]
- 10. Van Aelst L., Joneson T., Bar-Sagi D. (1996) EMBO J. 15, 3778–3786 [PMC free article] [PubMed] [Google Scholar]
- 11. Cancelas J. A., Jansen M., Williams D. A. (2006) Exp. Hematol. 34, 976–985 [DOI] [PubMed] [Google Scholar]
- 12. Cancelas J. A., Lee A. W., Prabhakar R., Stringer K. F., Zheng Y., Williams D. A. (2005) Nat. Med. 11, 886–891 [DOI] [PubMed] [Google Scholar]
- 13. Gu Y., Filippi M. D., Cancelas J. A., Siefring J. E., Williams E. P., Jasti A. C., Harris C. E., Lee A. W., Prabhakar R., Atkinson S. J., Kwiatkowski D. J., Williams D. A. (2003) Science 302, 445–449 [DOI] [PubMed] [Google Scholar]
- 14. Goldfinger L. E., Ptak C., Jeffery E. D., Shabanowitz J., Hunt D. F., Ginsberg M. H. (2006) J. Cell Biol. 174, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lowe D. G., Goeddel D. V. (1987) Mol. Cell. Biol. 7, 2845–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Self A. J., Paterson H. F., Hall A. (1993) Oncogene 8, 655–661 [PubMed] [Google Scholar]
- 17. Sethi T., Ginsberg M. H., Downward J., Hughes P. E. (1999) Mol. Biol. Cell 10, 1799–1809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hughes P. E., Renshaw M. W., Pfaff M., Forsyth J., Keivens V. M., Schwartz M. A., Ginsberg M. H. (1997) Cell 88, 521–530 [DOI] [PubMed] [Google Scholar]
- 19. Kinbara K., Goldfinger L. E., Hansen M., Chou F. L., Ginsberg M. H. (2003) Nat. Rev. Mol. Cell Biol. 4, 767–776 [DOI] [PubMed] [Google Scholar]
- 20. Jeong H. W., Nam J. O., Kim I. S. (2005) Cancer Res. 65, 507–515 [PubMed] [Google Scholar]
- 21. Komatsu M., Ruoslahti E. (2005) Nat. Med. 11, 1346–1350 [DOI] [PubMed] [Google Scholar]
- 22. Ada-Nguema A. S., Xenias H., Hofman J. M., Wiggins C. H., Sheetz M. P., Keely P. J. (2006) J. Cell Sci. 119, 1307–1319 [DOI] [PubMed] [Google Scholar]
- 23. Holly S. P., Larson M. K., Parise L. V. (2005) Mol. Biol. Cell 16, 2458–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keely P. J., Rusyn E. V., Cox A. D., Parise L. V. (1999) J. Cell Biol. 145, 1077–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wozniak M. A., Kwong L., Chodniewicz D., Klemke R. L., Keely P. J. (2005) Mol. Biol. Cell 16, 84–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ginsberg M. H., Du X., Plow E. F. (1992) Curr. Opin. Cell Biol. 4, 766–771 [DOI] [PubMed] [Google Scholar]
- 27. Ginsberg M. H., O'Toole T. E., Loftus J. C., Plow E. F. (1992) Cold Spring Harbor Symp. Quant. Biol. 57, 221–231 [DOI] [PubMed] [Google Scholar]
- 28. Zhang Z., Vuori K., Wang H., Reed J. C., Ruoslahti E. (1996) Cell 85, 61–69 [DOI] [PubMed] [Google Scholar]
- 29. Nakada M., Niska J. A., Tran N. L., McDonough W. S., Berens M. E. (2005) Am. J. Pathol. 167, 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zambrowicz B. P., Friedrich G. A., Buxton E. C., Lilleberg S. L., Person C., Sands A. T. (1998) Nature 392, 608–611 [DOI] [PubMed] [Google Scholar]
- 31. Straub A. C., Klei L. R., Stolz D. B., Barchowsky A. (2009) Am. J. Pathol. 174, 1949–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang L., Yang L., Debidda M., Witte D., Zheng Y. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang L., Yang L., Filippi M. D., Williams D. A., Zheng Y. (2006) Blood 107, 98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boggs D. R. (1984) Am. J. Hematol. 16, 277–286 [DOI] [PubMed] [Google Scholar]
- 35. Petit I., Szyper-Kravitz M., Nagler A., Lahav M., Peled A., Habler L., Ponomaryov T., Taichman R. S., Arenzana-Seisdedos F., Fujii N., Sandbank J., Zipori D., Lapidot T. (2002) Nat. Immunol. 3, 687–694 [DOI] [PubMed] [Google Scholar]
- 36. Saez R., Chan A. M., Miki T., Aaronson S. A. (1994) Oncogene 9, 2977–2982 [PubMed] [Google Scholar]
- 37. Kiel M. J., Morrison S. J. (2006) Immunity 25, 862–864 [DOI] [PubMed] [Google Scholar]
- 38. Lo Celso C., Fleming H. E., Wu J. W., Zhao C. X., Miake-Lye S., Fujisaki J., Côté D., Rowe D. W., Lin C. P., Scadden D. T. (2009) Nature 457, 92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xie Y., Yin T., Wiegraebe W., He X. C., Miller D., Stark D., Perko K., Alexander R., Schwartz J., Grindley J. C., Park J., Haug J. S., Wunderlich J. P., Li H., Zhang S., Johnson T., Feldman R. A., Li L. (2009) Nature 457, 97–101 [DOI] [PubMed] [Google Scholar]
- 40. Mizutani K., Yoon K., Dang L., Tokunaga A., Gaiano N. (2007) Nature 449, 351–355 [DOI] [PubMed] [Google Scholar]
- 41. Yang L., Wang L., Geiger H., Cancelas J. A., Mo J., Zheng Y. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 5091–5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ryan M. A., Nattamai K. J., Xing E., Schleimer D., Daria D., Sengupta A., Köhler A., Liu W., Gunzer M., Jansen M., Ratner N., Le Cras T. D., Waterstrat A., Van Zant G., Cancelas J. A., Zheng Y., Geiger H. (2010) Nat. Med. 16, 1141–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]