Abstract
Species-specific sex pheromones released by female moths to attract conspecific male moths are synthesized de novo in the pheromone gland (PG) via the fatty acid biosynthetic pathway. This pathway is regulated by a neurohormone termed pheromone biosynthesis activating neuropeptide (PBAN), a 33-amino acid peptide that originates in the subesophageal ganglion. In the silkmoth, Bombyx mori, cytoplasmic lipid droplets, which store the sex pheromone (bombykol) precursor fatty acid, accumulate in PG cells. PBAN stimulates lipolysis of the stored lipid droplet triacylglycerols (TAGs) and releases the precursor for final modification. PBAN exerts its physiological function via the PG cell-surface PBAN receptor, a G protein-coupled receptor that belongs to the neuromedin U receptor family. The PBAN receptor-mediated signal is transmitted via a canonical store-operated channel activation pathway utilizing Gq-mediated phospholipase C activation (Hull, J. J., Kajigaya, R., Imai, K., and Matsumoto, S. (2007) Biosci. Biotechnol. Biochem. 71, 1993–2001; Hull, J. J., Lee, J. M., Kajigaya, R., and Matsumoto, S. (2009) J. Biol. Chem. 284, 31200–31213; Hull, J. J., Lee, J. M., and Matsumoto, S. (2010) Insect Mol. Biol. 19, 553–566). Little, however, is known about the molecular components regulating TAG lipolysis in PG cells. In the current study we found that PBAN signaling involves phosphorylation of an insect PAT family protein named B. mori lipid storage droplet protein-1 (BmLsd1) and that BmLsd1 plays an essential role in the TAG lipolysis associated with bombykol production. Unlike mammalian PAT family perilipins, however, BmLsd1 activation is dependent on phosphorylation by B. mori Ca2+/calmodulin-dependent protein kinase II rather than protein kinase A.
Keywords: G Protein-coupled Receptors (GPCR), Insect, Lipid Droplet, Lipolysis, Peptide Hormones, Signal Transduction, Bombyx mori, Sex Pheromone
Introduction
Mating in moths is limited to a specific phase of the photoperiod and developmental stage. Accordingly, the biochemical processes that comprise sex pheromone biosynthesis in female moths must be precisely regulated. In most moth species these processes are regulated by a neurohormone termed pheromone biosynthesis activating neuropeptide (PBAN),4 a 33-amino acid peptide that originates in the subesophageal ganglion and that is characterized by a core C-terminal FSPRLamide sequence (1, 2). After adult emergence, PBAN is released into the hemolymph during a species-specific period and acts on the pheromone gland (PG) to trigger the production and release of species-specific sex pheromones (3, 4).
PG is a functionally differentiated organ in close proximity to the terminal abdominal tip that originates in the intersegmental membrane between the 8th and 9th abdominal segments (5–7). In the silkmoth, Bombyx mori, the sex pheromone, E,Z-10,12-hexadecadien-1-ol, commonly known as bombykol, is synthesized de novo within PG cells from acetyl-CoA via the conventional long chain fatty acid biosynthetic pathway (8, 9). The straight chain fatty acyl intermediate, palmitate, is converted stepwise to bombykol by the actions of a bifunctional Z11–10/12 fatty acyl desaturase, Bmpgdesat1, and a PG-specific fatty acyl reductase, pgFAR (10–12). On the day before adult emergence, B. mori PG cells rapidly accumulate numerous lipid droplets (LDs) within the cytoplasm (13). These LDs play an essential role in bombykol biosynthesis by acting as a reservoir for the de novo synthesized bombykol precursor, Δ10,12-hexadecadienoate, which is deposited in the LDs in the form of triacylglycerols (TAGs) with the precursor predominantly sequestered at the sn-1 and sn-3 positions of the glycerides (14). After adult emergence, the stored fatty acid is cleaved and converted to bombykol in response to PBAN (7, 15).
The pheromonotropic effects of PBAN are dependent on extracellular Ca2+ (3, 4) and are mediated by the PG cell-surface PBAN receptor, a G protein-coupled receptor that belongs to the neuromedin U receptor family (16–18). Biochemical analyses in conjunction with in vivo RNAi-mediated knockdown studies in B. mori have shown that the PBAN signal is transmitted via a canonical store-operated channel activation pathway utilizing Gq-mediated phospholipase C (PLC) activation in which BmGq1, BmPLCβ1, BmIP3R, BmSTIM1, BmOrai1, and BmPLCγ are necessary components (19–21). The precise role of BmPLCγ, however, remains to be elucidated. Although it has been well documented that PBAN stimulation accelerates both lipolysis of the cytoplasmic LDs and subsequent fatty acyl reduction to generate the final product of bombykol (7, 15), the molecular components regulating both steps have yet to be determined. Here we report that several distinct PG cell proteins are phosphorylated in response to PBAN stimulation and that we identified one such protein, B. mori lipid storage droplet protein-1 (BmLsd1), from the PG cytoplasmic fat-cake fraction. We further report that BmLsd1 is a LD-associated insect PAT family protein that plays an essential role in bombykol biosynthetic TAG lipolysis after phosphorylation by B. mori Ca2+/calmodulin-dependent protein kinase II (BmCaMKII).
EXPERIMENTAL PROCEDURES
Insects
Larvae of the inbred p50 strain of B. mori, kindly provided by T. Shimada of the University of Tokyo, were reared on mulberry leaves or an artificial diet and maintained under a 16L:8D photoperiod at 25 °C (22). Pupal age was determined based on morphological characteristics as described (14).
Sample Preparation
PGs were dissected into insect Ringer's solution (35 mm NaCl, 36 mm KCl, 12 mm CaCl2, 16 mm MgCl2, 274 mm glucose, and 5 mm Tris (pH 7.5)) containing phosphatase inhibitors (50 mm NaF, 10 nm okadaic acid, 0.1 mm Na3VO4) and mechanically trimmed as described (23). Trimmed PGs were homogenized in a cytoplasmic fraction buffer (25 mm Hepes (pH 7.5), 50 mm NaCl, 50 mm NaF, 5 mm EDTA, 10 nm okadaic acid, 0.1 mm Na3VO4, 1 mm p-amidinophenylmethanesulfonyl fluoride hydrochloride (p-APMSF, Wako Chemicals, Osaka, Japan), and 10 μg/ml each of aprotinin and leupeptin) for 1 min on ice. PG cell homogenates were centrifuged at 15,000 × g for 10 min to separate soluble and insoluble fractions. The insoluble fraction was washed twice with PBS (137 mm NaCl, 2.7 mm KCl, 8 mm Na2HPO4, 1.5 mm KH2PO4, (pH 7.2)) containing phosphatase inhibitors (50 mm NaF, 10 nm okadaic acid, 0.1 mm Na3VO4) and re-solubilized with a membrane fraction buffer (25 mm Hepes (pH 7.5), 1% Triton X-100, 50 mm NaCl, 50 mm NaF, 5 mm EDTA, 10 nm okadaic acid, 0.1 mm Na3VO4, 1 mm p-APMSF, and 10 μg/ml each of aprotinin and leupeptin). The soluble and insoluble fractions were used as cytoplasmic and membrane fractions, respectively.
Separation of LDs
Forty trimmed PGs in a 1.5-ml tube were gently homogenized in 700 μl of Tris-sucrose buffer (0.25 m sucrose, 20 mm Tris (pH 7.4), 1 mm EDTA, 0.1 mm benzamidine, 10 nm okadaic acid, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mm p-APMSF, 0.1% 2-mercaptoethanol, 2 mm imidazole, 2 mm NaF, 1.5 mm Na2MoO4, 1 mm Na3VO4, 4 mm sodium potassium tartrate, and 1 tablet of PhosSTOP/10 ml (Roche Applied Science)) using a pellet disruptor. The mixture was briefly centrifuged at 2,300 × g at 4 °C for 10 min. The supernatant was transferred to another tube, and the pellet was again homogenized. To achieve complete extraction, a third homogenization was performed in a glass/Teflon homogenizer using 800 μl of Tris-sucrose buffer and added to the pooled supernatants. For sucrose gradient ultracentrifugation, the pooled samples were overlaid with 2 ml of Tris-sucrose buffer lacking sucrose and centrifuged at 400,000 × g for 60 min at 4 °C, which resulted in the formation of a “fat-cake” cap. For efficient harvesting of LDs, the tube was frozen, and the fat cake was cut off and preserved for further investigations (24).
Analysis of PG Protein Phosphorylations
For SDS-PAGE, cytoplasmic, membrane, and fat-cake fractions were incubated with 80 mm Tris buffer (pH 8.8) containing 1% SDS and 2.5% 2-mercaptoethanol in boiling water for 5 min and developed by the method of Laemmli (25). Protein bands from the SDS-PAGE gel were electrically transferred to Immobilon-P Transfer Membrane (Millipore) essentially according to Burnette (26). Membranes were blocked with BSA and then probed with a primary phosphoserine polyclonal antibody (catalog #KAP-ST210, Stressgen), a phosphothreonine polyclonal antibody (catalog #KAP-ST211, Stressgen), or a mouse anti-phosphotyrosine antibody (catalog #03-7700, Zymed Laboratories Inc.). Development was performed using an ECL plus Western Detection System (GE Healthcare) according to the manufacturer's instructions.
Identification of BmLsd1
The PG proteins in the fat-cake fraction were subjected to SDS-PAGE. The resultant SDS-PAGE gel was stained using either a Silver Stain MS Kit (Wako Chemicals, Osaka, Japan) or a Pro-Q Diamond Phosphoprotein Gel Stain kit (Molecular Probes Inc. Eugene, OR). After staining, the silver-stained 44-kDa band was excised, reduced with dithiothreitol, and carboxymethylated by iodoacetic acid. After washing, the protein band was digested with trypsin at 37 °C overnight. An aliquot of the digest was analyzed by nano-LC-MS/MS using an LCQ Deca XP (Finnigan). The peptides were separated on a nano-ESI spray column (100-μm inner diameter × 375 μm outer diameter) packed with reversed-phase material (Inertsil ODS-3, 3 μm, GL Science) using a flow rate of 200 nl/min. The mass spectrometer was operated in the positive-ion mode with spectra acquired in the data-dependent MS/MS mode. The MS/MS spectra were BLASTed against the SwissProt data base (Version 51.6) using an in-house MASCOT server (Version: 2.2.1, Matrix Sciences).
RT-PCR Analysis
PGs and other tissues were dissected into insect Ringer's solution and mechanically trimmed as described (23). Total RNA was isolated from the trimmed PGs by the method of Chomczynski and Sacchi (27). First-strand cDNA synthesis was performed using an RNA PCR kit (Takara Bio Inc., Japan) according to the manufacturer's instructions with 500 ng of total RNA. The specific oligonucleotide primer sets used are shown in Table 1. These primers were designed based on deposited sequences (GenBankTM accession numbers DQ311207, AB563704, EF540759, AB184961, and X05185). PCR was performed using thermacycler conditions consisting of 30 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 60 s. PCR products (5 μl) were electrophoresed on a 1.5% agarose gel in Tris acetate-EDTA buffer and stained with ethidium bromide.
TABLE 1.
Primer sets used to RT-PCR analyses
| Gene | F/Ra | Primer sequence | Nucleotides |
|---|---|---|---|
| BmLsd1-1 | F | ATGCCGAATCTTGAAGTGGTCT | 1098 |
| R | TTAATTGACACCGTTAATGGAA | ||
| BmLsd1-2 | F | AAGGGAGTCCAACCCTCTGT | 491 |
| R | CTTGCCTGGTGAGTCTCCTC | ||
| BmLsd1-3 | F | AGAATCAAGCGAGGCCTGTA | 300 |
| R | CGGCCAAGAATATGGCTAAA | ||
| BmCaMKII-1 | F | TCCGAAGCTCTGAAACATCC | 312 |
| R | CAGGGCTTCTTCGTCATGTT | ||
| BmCaMKII-2 | F | CGCCTTCTCGATTGTTAGGA | 251 |
| R | ATTCTCGCGCCACTATGTCT | ||
| BmPKA-1 | F | CTCGCACCTCCCTTAAGTTG | 250 |
| R | GCCGTGCCTTCAACTATGAT | ||
| BmPKA-2 | F | TAAGTGCAGAGCCCGTAACC | 301 |
| R | AAGCTACCCCCTTCACCAAT | ||
| BmcPKC-1 | F | CCTCTCGTTCGACCTCAAAG | 250 |
| R | CACCGTTGTAGCCCTCATCT | ||
| BmcPKC-2 | F | CTGTGGCTCTCTGCTGTACG | 300 |
| R | GTGCTACGGATGGTCTTGGT | ||
| Actin | F | AGATGACCCAGATCATGTTTG | 721 |
| R | GAGATCCACATCTGCTGGAAG |
a F, forward primer; R, reverse primer.
Synthesis and Injection of Double-stranded RNA (dsRNA)
The templates for synthesis of dsRNAs corresponding to the proteins examined were prepared using gene-specific primers containing T7 polymerase sites. The primer sets used are shown in Table 2. PCR was performed using thermacycler conditions consisting of 6 cycles at 94 °C for 30 s, 56.5 °C for 30 s, 68 °C for 90 s followed by 30 cycles at 94 °C for 30 s, 66 °C for 30 s, and 68 °C for 90 s using KOD-Plus- (Toyobo, Osaka, Japan) with the resulting products purified (Wizard SV Gel and PCR Clean-Up kit, Promega, Madison, WI) and used as templates to generate dsRNAs using the AmpliScribeTM T7 High Yield Transcription kit (Epicentre Technologies, Madison, WI) according to the manufacturer's instructions. After synthesis, dsRNAs were diluted with diethyl pyrocarbonate (DEPC)-treated H2O, the RNA concentrations were measured (Abs260), and the products were analyzed by gel electrophoresis to confirm annealing. Samples were diluted to the desired concentration (final volume 2 μl) and injected into 1-day-old pupae (i.e. pupae 1 day removed from the larval-pupal molt) or newly emerged female moths using a 10-μl microsyringe (Hamilton) as described (12, 20, 21). Control females were injected with 2 μl of DEPC-treated H2O alone. After injection, pupae were maintained under normal conditions until adult emergence.
TABLE 2.
Primer sets used to produce T7 RNA polymerase templates
| Gene | F/Ra | Primer sequenceb | Nucleotides |
|---|---|---|---|
| BmLsd1-1 | F | CCGGATCCTAATACGACTCACTAATAGGGCGATGCCGAATCTTGA | 1160 |
| R | CCGGATCCTAATACGACTCACTAATAGGGCGTTAATTGACACCGT | ||
| BmLsd1-2 | F | CCGGATCCTAATACGACTCACTAATAGGGCGAAGGGAGTCCAACC | 553 |
| R | CCGGATCCTAATACGACTCACTAATAGGGCGCTTGCCTGGTGAGT | ||
| BmLsd1-3 | F | CCGGATCCTAATACGACTCACTAATAGGGCGAGAATCAAGCGAGG | 362 |
| R | CCGGATCCTAATACGACTCACTAATAGGGCGCGGCCAAGAATATG | ||
| BmCaMKII-1 | F | CCGGATCCTAATACGACTCACTAATAGGGCGTCCGAAGCTCTGAAAC | 374 |
| R | CCGGATCCTAATACGACTCACTAATAGGGCGCAGGGCTTCTTCGTCA | ||
| BmCaMKII-2 | F | CCGGATCCTAATACGACTCACTAATAGGGCGCGCCTTCTCGATTG | 313 |
| R | CCGGATCCTAATACGACTCACTAATAGGGCGATTCTCGCGCCACT | ||
| BmPKA-1 | F | CCGGATCCTAATACGACTCACTAATAGGGCGCTCGCACCTCCCTT | 312 |
| R | CCGGATCCTAATACGACTCACTAATAGGGCGGCCGTGCCTTCAAC | ||
| BmPKA-2 | F | CCGGATCCTAATACGACTCACTAATAGGGCGTAAGTGCAGAGCCC | 363 |
| R | CCGGATCCTAATACGACTCACTAATAGGGCGAAGCTACCCCCTTC | ||
| BmcPKC-1 | F | CCGGATCCTAATACGACTCACTAATAGGGCGCCTCTCGTTCGACC | 312 |
| R | CCGGATCCTAATACGACTCACTAATAGGGCGCACCGTTGTAGCCC | ||
| BmcPKC-2 | F | CCGGATCCTAATACGACTCACTAATAGGGCGCTGTGGCTCTCTGC | 362 |
| R | CCGGATCCTAATACGACTCACTAATAGGGCGGTGCTACGGATGGT |
a F, forward primer; R, reverse primer.
b Nucleotide sequences corresponding to the T7 promoter region are underlined.
In Vivo Bombykol Analysis
Adult females were decapitated within 3 h of emergence and maintained at 25 °C for 24 h. They were then injected with either 5 pmol (2 μl) of B. mori PBAN in PBS or PBS alone. Abdominal tips were dissected 90 min after injection, and bombykol production was measured by HPLC as described (28) using a Senshu-Pak NO2 column (Senshu Scientific Co., Tokyo, Japan).
Microscopic Examination of Cytoplasmic Lipid Droplets
Abdominal tips were dissected and mechanically trimmed from normal, decapitated, and RNAi-treated females. The excised glands were fixed with a 4% formalin-PBS solution and stained with Nile Red (a fluorescent probe for intracellular neutral lipids; Molecular Probes Inc.) as described (13). Fluorescence microscopy was performed with an OLYMPUS BX-60 system equipped with a PM-30 exposure unit and a BH20-RFL-T3 light source (400× magnification). Nile Red imaging was done with a 330–385-nm band pass excitation filter, a 400-nm dichroic mirror, and a 420-nm long pass barrier filter (Olympus cube WU). Images were processed and merged using Photoshop CS (Adobe Systems Inc., San Jose, CA).
Analysis of LD TAGs
LD TAGs were analyzed as described previously (14). Five trimmed PGs were prepared from the desired stages of female moths and dipped in 100 μl of acetone for 10 min at room temperature. The dried acetone extracts were dissolved in n-hexane and loaded on a Senshu-Pak PEGASIL-Silica 120–5 column (Senshu Scientific Co.; 4.6-mm inner diameter × 250 mm, pore size 120 Å) equilibrated with n-hexane/acetic acid (99/1). Because various TAG components comprise the cytoplasmic LDs, the entire TAG fraction was separated and detected using an Evaporative Light Scattering Detector (SEDEX model 75, Sedere, France).
Anti-BmLsd1 Antibody
Rabbit polyclonal antibodies for BmLsd1 were commercially generated (Operon Biotechnologies, Tokyo, Japan) by using a mixture of two peptides (HGARFKRKLQRRLT and YEQRDDVSSINGVN) coupled to keyhole limpet hemocyanin. A specific antibody recognizing the HGARFKRKLQRRLT sequence was isolated via affinity chromatography using an antigen-immobilized Thiopropyl Sepharose 6B (GE Healthcare) column prepared after reaction with the thiol-containing peptide, CHGARFKRKLQRRLT, according to the manufacturer's instructions.
Analysis of BmLsd1 Phosphorylation
The cytoplasmic fraction was incubated with a BmLsd1-specific antibody for 2 h at 4 °C, then mixed with 20 μl of rProtein A SepharoseTM Fast Flow (GE Healthcare) and left for 2 h at 4 °C. The resulting immunoprecipitate-bound Sepharose beads were washed 3 times with PBS containing PhosSTOP (1 tablet/10 ml, Roche Applied Science). Bound proteins were eluted from the beads with 45 μl of 0.1 m glycine (pH 4.0), neutralized with 5 μl of 1 m Tris buffer (pH 9.0), and then subjected to SDS-PAGE. Phosphorylation analyses using anti-phosphoamino acid antibodies were performed as described above.
Rapid Amplification of cDNA Ends
Rapid amplification of cDNA ends was performed from about 1 μg of PG total RNA by using a SMARTerTM RACE cDNA amplification kit (Clontech, Palo Alto, CA) according to the manufacturer's instructions. Computer-assisted sequence analyses were performed using GENETYX-MAC Version 15.0.5 (Software Development Co., Tokyo, Japan).
Immunocytochemistry
Mechanically trimmed PGs prepared from abdominal tips of normal and decapitated females were fixed in 4% (w/v) paraformaldehyde and 0.5% (v/v) Triton X-100 in PBS for 20 min at room temperature. After washing 4 times with PBST (0.1% Tween 20 in PBS), PGs were blocked with 5% (v/v) normal goat serum in PBST for 1 h at room temperature and then incubated with primary antibody diluted 1:250 overnight at 4 °C. For detection, PGs were subsequently incubated with Alexa Fluor® 488-conjugated secondary antibody (Molecular Probes) diluted 1:1000 in 5% (v/v) normal goat serum in PBST for 1 h at room temperature and then mounted in a solution containing 90% glycerol, 1% (w/v) n-propyl gallate (Sigma), 2% (w/v) 1,4-diazabicyclo(2.2.2)-octane (Sigma), and PBS. Nile Red was used to visualize neutral lipids.
Confocal images were obtained with a Leica TCS NT instrument using a 488-nm laser line for Alexa Fluor® 488 and a 568-nm laser line for Nile Red. For double-labeling experiments, only one laser line was used for any single-channel recording to avoid cross-talk between channels. Images were processed and merged using Photoshop CS (Adobe Systems).
RESULTS
Detection of PG Proteins Phosphorylated in Response to PBAN Stimulation
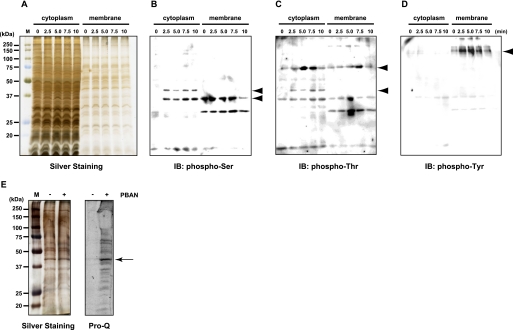
To examine the role of PBAN-mediated phosphorylation in the bombykol biosynthetic pathway, we performed immunoblot analyses of PG homogenates using anti-phosphoamino acid antibodies. We found that a number of PG proteins undergo rapid PBAN-mediated phosphorylation (Fig. 1). Anti-phosphoserine immunoreactive bands corresponding to 35- and 44-kDa (Fig. 1B, arrowheads) and anti-phosphothreonine immunoreactive bands corresponding to 44- and 72-kDa (Fig. 1C, arrowheads) were detected in the cytoplasmic fraction within minutes of PBAN stimulation. A 35-kDa anti-phosphothreonine band was also detected in the cytoplasmic fraction; however, phosphorylation of this band was PBAN-independent, as it was present in non-stimulated samples. In addition, a single 160-kDa anti-phosphotyrosine immunoreactive band that undergoes PBAN-induced phosphorylation was detected in the PG membrane fraction (Fig. 1D, arrowhead).
FIGURE 1.
Detection of PG proteins phosphorylated after PBAN stimulation. A–D, cytoplasmic and membrane fractions of PG homogenates (5 PG equivalents) were prepared from B. mori (p50 strain) female moths that had been decapitated 3 h after eclosion and injected with 5 pmol PBAN 24 h after decapitation. Fractions prepared at the indicated times after PBAN injection were subjected to SDS-PAGE in a 12% acrylamide gel and immunostained with anti-phosphoamino acid antibodies. A, silver staining is shown. B, immunostaining (IB) with an anti-phosphoserine antibody is shown. C, immunostaining with an anti-phosphothreonine antibody is shown. D, immunostaining with an anti-phosphotyrosine antibody is shown. E, the lipid rich fat-cake fraction (40 PG eq) was prepared from B. mori (p50 strain) female moths that had been decapitated 3 h after eclosion and injected with 0 pmol (−) or 5 pmol (+) PBAN 24 h after decapitation. The fat-cake fractions prepared 15 min after PBAN injection were subjected to SDS-PAGE in 12% acrylamide gels and stained with Pro-Q Diamond phosphoprotein stain. Left, silver staining. Right, Pro-Q Diamond staining.
Identification and Characterization of a 44-kDa Protein Phosphorylated by PBAN Stimulation
To facilitate the detection of LD-associated proteins that undergo PBAN-mediated phosphorylation, we used sucrose density gradient ultracentrifugation to prepare a lipid-rich fat-cake fraction from PG cells. After SDS-PAGE, the gel was stained with Pro-Q Diamond phosphoprotein stain, which allows non-selective detection of phosphorylated amino acids depending on the number of phosphate groups in the band. Although the Pro-Q Diamond staining of the PG fat-cake fraction detected more proteins than the immunoblots using the anti-phosphoamino acid antibodies, it clearly indicated the presence of a 44-kDa protein phosphorylated in response to PBAN stimulation (Fig. 1E, arrow). To identify the protein sequence, the 44-kDa band was gel-excised and digested with trypsin. LC/MS analysis of the resulting fragment peptides generated two distinctive fragment sequences, LGTAVLDSR and SKLEVLLQQLQATSK, both of which completely matched sequences found in a B. mori cDNA clone (GenBankTM accession number DQ311207) predicted to encode perilipin. A search of the public B. mori EST data base (SilkBase) (29), which contains expressed sequence tags (EST) derived from a normalized inbred B. mori p50 strain PG cDNA library, identified a PG-expressed perilipin clone (NRPG0891).
Sequence analysis of NRPG0891, in conjunction with both 3′- and 5′-rapid amplification of cDNA ends, revealed that the perilipin gene contains a 1,119-nucleotide open reading frame (ORF) predicted to yield a 373-amino acid PAT family protein (Fig. 2A) with 82.8% sequence identity to the Manduca sexta fat body (FB) Lsd1 (30, 31). Accordingly, we have termed the protein encoded on NRPG0891 as B. mori Lsd1 (BmLsd1). Both BmLsd1 and MsLsd1 contain a PAT domain, an N-terminal sequence of ∼100 residues common to LD-associated proteins including perilipins, adipophilin/adipocyte differentiation-related protein (ADRP), and tail-interacting protein of 47 kilodaltons (TIP47) (32). RT-PCR expression analysis revealed that BmLsd1 expression is limited to the PG, unfertilized egg (Eg), and FB (Fig. 2B). In addition, we found that BmLsd1 expressed in the PG undergoes significant up-regulation 1 day before adult emergence (Fig. 2C). This expression profile is reminiscent of PG proteins crucial to bombykol biosynthesis (i.e. pgFAR, Bmpgdesat1, pgACBP, mgACBP, PBAN receptor, and BmFATP (B. mori fatty acid transport protein)) (10–12, 17, 29, 33), suggesting a potential functional role for BmLsd1 in B. mori pheromonogenesis as well.
FIGURE 2.
Characterization of BmLsd1. A, cDNA and deduced amino acid sequence of BmLsd1 is shown. The PAT domain is shown in boxes. Amino acid sequences used for anti-BmLsd1 antibody production are underlined. Although the rabbit for immunization was injected with a mixture of the two underlined peptides coupled to keyhole limpet hemocyanin, only the affinity-purified antibody recognizing the HGARFKRKLQRRLT sequence was used in the experiments. Amino acid sequences determined by LC-MS/MS analysis are underscored with a dotted line. B, BmLsd1 transcript expression in adult female tissues is shown. Adult tissues were dissected from newly emerged (day 0) p50 female moths. Br, brain; FM, flight muscle; Eg, unfertilized egg; MT, Malpighian tubule; Mg, midgut. C, shown is BmLsd1 transcript expression in the PG at different developmental stages. PGs were dissected from p50 pupae at 3 days (−3) and 1 day (−1) before eclosion and from p50 adults at 0 day (0), 1 day (+1), 2 days (+2), and 3 days (+3) after eclosion. cDNAs were normalized to actin expression levels.
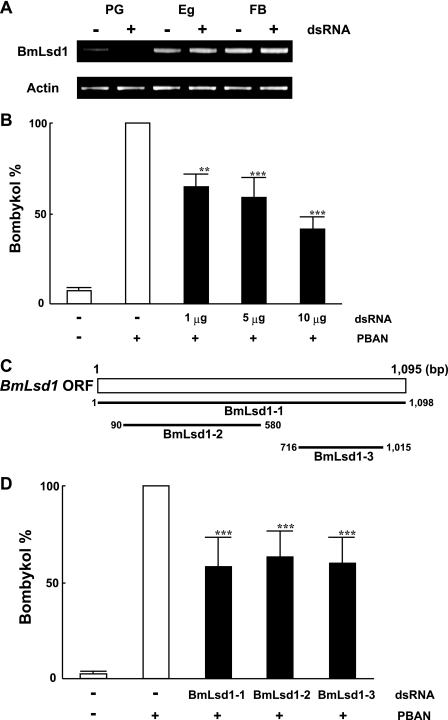
In Vivo Effect of RNAi-mediated BmLsd1 Knockdown on B. mori Pheromonogenesis
To confirm the in vivo function of BmLsd1, we used dsRNA-mediated RNAi knockdown of BmLsd1 and assayed for the predicted inhibitory effect on bombykol production. When we injected 10 μg of BmLsd1 dsRNA into 1-day-old p50 female pupae, BmLsd1 mRNA levels in the PG after eclosion were selectively reduced compared with those in unfertilized egg and FB (Fig. 3A) as well as control pupae injected with DEPC-treated H2O (data not shown). Although we do not know the reason for the PG-selective knockdown of BmLsd1, a similar PG-selective decrease in transcript levels was observed after knockdown of B. mori fatty acid transport protein (29) and B. mori acyl carrier protein.5 Because BmLsd1 dsRNA injections had no effect on pupal development or adult emergence, we next injected varying concentrations (1, 5, and 10 μg) of BmLsd1 dsRNA into 1-day-old female pupae. A dose-dependent reduction in bombykol production was observed (Fig. 3B) with 1, 5, and 10 μg of dsRNA injections resulting in bombykol levels 65, 59, and 42% that of control levels. To confirm the knockdown effect by BmLsd1, we also examined two other dsRNAs corresponding to non-overlapping 5′ and 3′ regions of the BmLsd1 gene; these dsRNAs had a similar knockdown effect on bombykol production (Fig. 3, C and D). Because no decrease in bombykol production was observed in control pupae injected with dsRNAs for unrelated proteins including enhanced green fluorescent protein (data not shown), these results taken together indicate that disruption of bombykol production is related to specific knockdown of the BmLsd1 dsRNA sequence.
FIGURE 3.
Effects of RNAi treatment on in vivo bombykol production. A, RNAi-induced suppression of BmLsd1 transcripts is shown. RT-PCR was performed using cDNAs generated from total RNA prepared from pupae injected with 0 μg (−) or 10 μg (+) dsRNAs for BmLsd1. Tissues were dissected from newly emerged (day 0) p50 female moths. Eg, unfertilized egg. B, effects of RNAi treatment on in vivo bombykol production are shown. 1-day-old pupae were injected with 1, 5, and 10 μg of dsRNA for BmLsd1. Results are expressed relative to the mean value for bombykol levels in non-RNAi-treated females after injection with 5 pmol of PBAN (**, p < 0.01; ***, p < 0.001). Bars represent mean values ± S.D. from independent experimental animals (n = >9). C, shown is a schematic diagram depicting the location of three dsRNAs specific to the BmLsd1 ORF. D, 1-day-old pupae were injected with dsRNAs (5 μg) corresponding to the different BmLsd1 regions. The primer sets used for dsRNA synthesis are indicated in Table 2. Results are expressed relative to the mean value for bombykol levels in non-RNAi-treated females after injection with 5 pmol of PBAN (***, p < 0.001). Bars represent the mean values ± S.D. from independent experimental animals (n = >9).
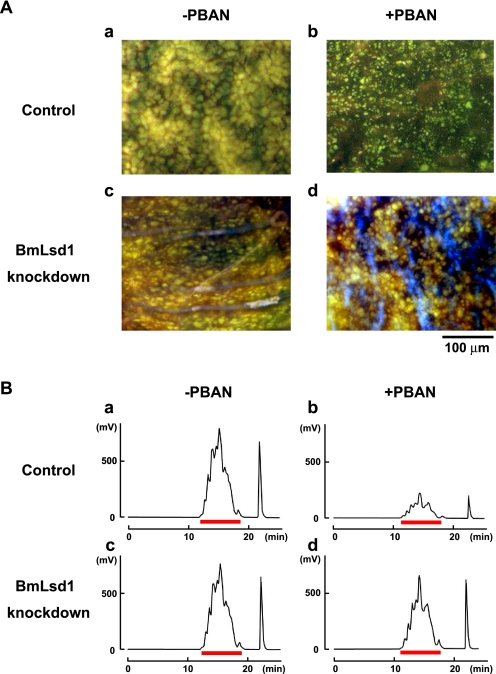
We further examined the knockdown effect of BmLsd1 on LD dynamics during pheromonogenesis (Fig. 4A). In RNAi-untreated controls, female moths accumulated numerous large LDs in the PG, which underwent a striking reduction after PBAN-induced LD lipolysis (Fig. 4A, a and b, also see Ohnishi et al. (29)). In contrast, although RNAi-mediated knockdown of BmLsd1 had no effect on LD accumulation, PBAN-induced LD lipolysis was significantly suppressed compared with the control (Fig. 4A, c and d). Because these LDs store the bombykol precursor in the form of TAGs (14), we further examined the effects of BmLsd1 knockdown on the fluctuation of total TAGs. After extraction with acetone from dissected PGs, total TAG levels were measured by normal-phase HPLC on a PEGASIL-Silica column (12). In BmLsd1 knocked-down PGs, TAG accumulation was comparable with that of the non-RNAi-treated control (Fig. 4B, a and c). In contrast, PBAN-induced lipolysis of TAG content was significantly reduced (20%) in BmLsd1-knockdown PGs (Fig. 4Bd) compared with the 75% reduction observed in the RNAi-untreated control (Fig. 4Bb). Taken together, these results demonstrate that BmLsd1 is integral to the bombykol biosynthetic pathway in that it plays an essential role in liberating the bombykol precursor from TAG storage in the cytoplasmic LDs.
FIGURE 4.
Effects of RNAi-mediated knockdown of BmLsd1 on in vivo LD lipolysis. A, shown are the effects of RNAi treatment on lipolysis of cytoplasmic LDs. PG cells were taken from 1-day-old decapitated female moths previously injected with either DEPC-treated H2O (Control) or 10 μg of BmLsd1 dsRNA (BmLsd1 knockdown) shortly after pupation. After eclosion, females were decapitated within 3 h of emergence, maintained at 25 °C for 24 h, and then injected with PBS alone (−PBAN) or 5 pmol of B. mori PBAN (+PBAN). PBAN was injected 3 times at 3-h intervals. Cytoplasmic LDs were stained with the fluorescent lipid marker, Nile Red. Autofluorescence (blue fluorescence in Figs. 4A, c and d) is not due to Nile Red staining. Magnifications, 400 X. B, effects of RNAi treatment on total TAG contents in the cytoplasmic LDs are shown. 1-Day-old pupae were injected with DEPC-treated H2O (Control) or 10 μg BmLsd1 dsRNA (BmLsd1 knockdown). After eclosion, females were decapitated within 3 h of emergence, maintained at 25 °C for 24 h, and then injected with PBS alone (−PBAN) or 5 pmol of B. mori PBAN (+PBAN). PBAN was injected 3 times at 3-h intervals. Five PGs were extracted with acetone 90 min after PBAN injection, and the extract was subjected to HPLC on a PEGASIL-Silica column (14). The relevant TAG fractions are underlined in red; TAGs were detected using an evaporative light scattering detector.
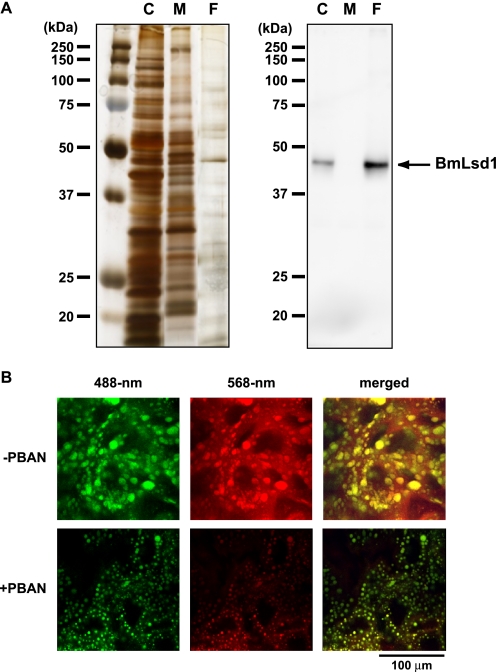
Localization of BmLsd1 in PG Cells
To further characterize BmLsd1, we generated polyclonal anti-BmLsd1 antibodies against residues 186–199 (HGARFKRKLQRRLT) and used the affinity-purified antibody in the following experiments. Because BmLsd1 is a PAT family protein that would be expected to be LD-associated, we sought to use Western blots to examine the spatial distribution of BmLsd1 within PG cells. PG homogenates from newly emerged female moths were fractionated into three subcellular fractions (i.e. cytoplasmic, membrane, and fat-cake fractions). Anti-BmLsd1 immunoblots revealed that although BmLsd1 is predominantly localized to the fat-cake fraction, a small portion is present in the cytoplasm as well (Fig. 5A). Because the fat-cake fraction was separated from the cytoplasmic fraction after ultracentrifugation, these results are consistent with a putative role for BmLsd1 as a PAT family protein that targets cytoplasmic LDs (32). To further examine the subcellular localization of endogenous BmLsd1, PG cells were immunostained using the anti-BmLsd1 antibody in conjunction with selective fluorescent Nile Red-based LD staining. Using this approach, we found that regardless of PBAN stimulation, BmLsd1 predominantly localized to the LDs (Fig. 5B).
FIGURE 5.
Localization of BmLsd1 using a specific anti-BmLsd1 antibody. A, subcellular localization of BmLsd1 in PG cells is shown. After eclosion, females were decapitated within 3 h of emergence and maintained at 25 °C for 24 h, and the cytoplasmic (C), membrane (M), and fat-cake (F) fractions of the PG homogenates were prepared as described under “Experimental Procedures.” Each sample was subjected to SDS-PAGE on a 12% acrylamide gel. Left, subcellular fractions were silver stained. Right, immunostaining was performed with an anti-BmLsd1 antibody. B, localization of endogenous BmLsd1 in the PG cells is shown. After eclosion, females were decapitated within 3 h of emergence, maintained at 25 °C for 24 h, and then injected with PBS alone (−PBAN) or 5 pmol of B. mori PBAN (+PBAN). PBAN was injected 3 times at 3-h intervals. PG cells were stained for endogenous BmLsd1 using an affinity-purified anti-BmLsd1 antibody (488-nm) and for LDs with Nile Red (568-nm). Fluorescent images were obtained by confocal laser microscopy.
Analysis of BmLsd1 Phosphorylation Using an Anti-BmLsd1 Antibody
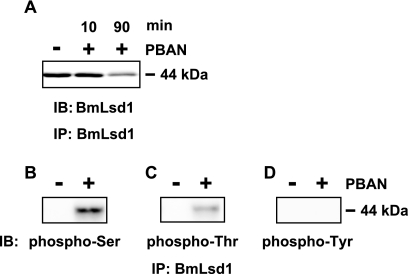
To analyze BmLsd1 phosphorylation in response to PBAN stimulation, PG homogenates were immunoprecipitated with the anti-BmLsd1 antibody. First, to examine BmLsd1 levels after PBAN stimulation, we performed immunoblots using the anti-BmLsd1 antibody (Fig. 6A). BmLsd1 levels appeared to be reduced when PGs were dissected and analyzed 90 min after PBAN injection. In contrast, BmLsd1 levels between non-PBAN-stimulated PG homogenates and 10 min after PBAN injection were comparable (Fig. 6A), indicating that PBAN stimulation has no effect on BmLsd1 protein synthesis. To better characterize PBAN-mediated BmLsd1 phosphorylation, which occurs rapidly within the first 10 min after PBAN injection (Fig. 1), we prepared PGs 10 min after PBAN injection. Using anti-phosphoamino acid antibodies, anti-phosphoserine and anti-phosphothreonine immuno-positive bands in BmLsd1 immunoprecipitated PG homogenates were only observed after PBAN stimulation (Fig. 6, B and C). No anti-phosphotyrosine immuno-positive bands were detected (Fig. 6D), suggesting that BmLsd1 is specifically phosphorylated on Ser and Thr residues in response to PBAN stimulation.
FIGURE 6.
Phosphorylation of BmLsd1 in response to PBAN stimulation. After eclosion, females were decapitated within 3 h of emergence, maintained at 25 °C for 24 h, and then injected with PBS alone (−) or 5 pmol of B. mori PBAN (+). PGs were dissected 10 and 90 min (A) or 10 min (B–D) after PBAN injection, and cytoplasmic fractions were prepared from 10 PGs. After immunoprecipitation (IP) with an anti-BmLsd1 antibody, immunostainings (IB) were performed with the following antibodies: anti-BmLsd1 (A), anti-phosphoserine (B), anti-phosphothreonine (C), and anti-phosphotyrosine (D).
RNAi-mediated Knockdown of Protein Kinases Expressed in the PG
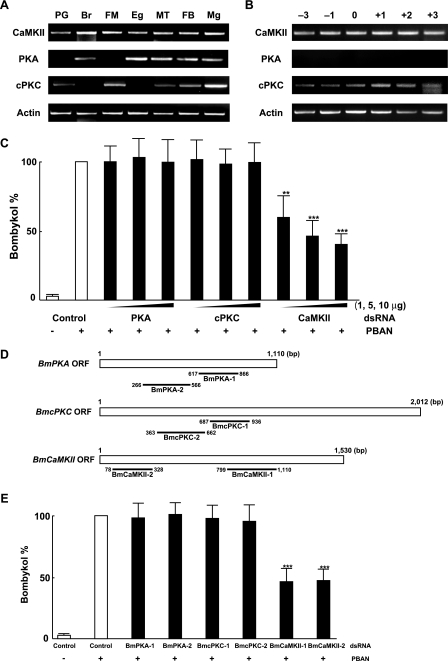
To identify the protein kinase responsible for PBAN-induced phosphorylation, we searched our PG EST data base for protein kinases expressed in the PG. We identified a cDNA clone (NRPG1028) that encodes the B. mori homolog of Ca2+/calmodulin-dependent protein kinase II (CaMKII), which we designated as BmCaMKII. To examine the tissue distribution of the BmCaMKII transcript, we performed RT-PCR analyses using various tissues prepared from p50 female adults. For control purposes, we also examined the tissue expression of two other B. mori protein kinase genes, protein kinase A (BmPKA) and conventional protein kinase C (BmcPKC), which were not identified in our PG EST data base but are present in the public EST data base and described in the literature (34). In adult tissues prepared 3 h after eclosion, BmCaMKII expression was ubiquitous, whereas the BmPKA and BmcPKC transcripts exhibited varied expression patterns (Fig. 7A). A weak but steady signal for BmcPKC was detected in the PG but not for BmPKA (Fig. 7, A and B). To further elucidate involvement of these B. mori protein kinases in PBAN signaling, we performed RNAi-mediated knockdown and examined their effects on in vivo bombykol production. When we injected 10 μg of dsRNA for BmCaMKII into newly emerged adult p50 females, BmCaMKII mRNA levels in the PG 2 days after injection were reduced compared with control females injected with DEPC-treated H2O alone or with dsRNA corresponding to enhanced green fluorescent protein (data not shown). We next injected varying concentrations (1, 5, and 10 μg) of dsRNAs corresponding to BmCaMKII, BmPKA, or BmcPKC. A significant dose-dependent reduction in bombykol production (40, 52, and 60%) was only observed after BmCaMKII knockdown (Fig. 7C). No decrease in bombykol production was observed in females injected with dsRNA corresponding to BmPKA or BmcPKC. To confirm the knockdown effect by BmCaMKII, we also examined dsRNAs corresponding to different regions of the BmCaMKII, BmPKA, and BmcPKC genes. Again, only BmCaMKII dsRNAs had an effect on bombykol production (Fig. 7, D and E). These results suggest that among the protein kinases examined, only BmCaMKII is involved in positive-regulation of PBAN signal transduction cascade.
FIGURE 7.
Characterization of B. mori protein kinases. A, expression of BmCaMKII, BmPKA, and BmcPKC transcripts in various tissues is shown. Tissues examined were dissected from newly emerged (day 0) p50 female moths: Br, brain; FM, flight muscle; Eg, unfertilized egg; MT, Malpighian tubule; Mg, midgut. B, expression of BmCaMKII, BmPKA, and BmcPKC transcripts in the PG at different developmental stages is shown. PGs were dissected from p50 pupae at 3 days (−3) and 1 day (−1) before eclosion and from p50 adults at 0 day (0), 1 day (+1), 2 days (+2), and 3 days (+3) after eclosion. cDNAs were normalized to actin expression levels. C, effects of RNAi-mediated knockdown of protein kinases on in vivo bombykol production are shown. dsRNAs (1, 5, and 10 μg) corresponding to BmPKA, BmcPKC, or BmCaMKII were directly injected into the PG of newly emerged female moths. Results are expressed relative to the mean value for bombykol levels in untreated females after 5 pmol of PBAN injections (**, p < 0.01; ***, p < 0.001). Bars represent mean values ± S.D. from independent experimental animals (n = >9). D, shown is a schematic diagram depicting the location of dsRNAs for the three kinase ORFs. E, effects of RNAi-mediated knockdown of protein kinases on in vivo bombykol production. dsRNAs (5 μg) corresponding to two different regions of BmPKA, BmcPKC, and BmCaMKII were directly injected into the PG of newly emerged female moths. The primer sets used for dsRNA synthesis are indicated in Table 2. Results are expressed relative to the mean value for bombykol levels in untreated females after 5 pmol of PBAN injections (***, p < 0.001). Bars represent mean values ± S.D. from independent experimental animals (n = >9).
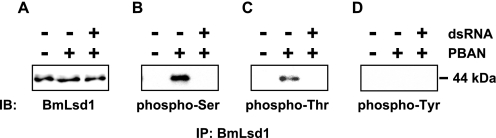
Effect of BmCaMKII Knockdown on BmLsd1 Phosphorylation
We next sought to assess the role of BmCaMKII in PBAN-mediated phosphorylation of BmLsd1. BmCaMKII dsRNAs were injected into the PGs of newly emerged adult p50 females, which were then decapitated 24 h after dsRNA injection and injected with synthetic PBAN 24 h after decapitation. PG homogenates prepared 10 min after PBAN injection were immunoprecipitated with our anti-BmLsd1 antibody and subjected to Western blot analyses (Fig. 8). In the immunoprecipitate prepared from the RNAi-untreated control, an anti-phosphoserine immunoreactive band for BmLsd1 was clearly observed after PBAN stimulation, whereas the intensity of the positive band was greatly diminished in the immunoprecipitate prepared from PGs injected with BmCaMKII dsRNAs (Fig. 8B). Likewise, an anti-phosphothreonine immunoreactive band for BmLsd1 was also greatly diminished in the immunoprecipitate prepared from the BmCaMKII-knockdown PG homogenate (Fig. 8C). Consistent with previous results (i.e. Fig. 6D), no anti-tyrosine immunoreactive band was observed (Fig. 8D).
FIGURE 8.
Effect of BmCaMKII knockdown on BmLsd1 phosphorylation. dsRNAs (10 μg) corresponding to BmCaMKII were directly injected into the PG of newly emerged female moths, which were decapitated 24 h after dsRNA injection, and then injected with PBS alone (−) or 5 pmol of B. mori PBAN (+) 24 h after decapitation. Ten minutes later, PGs were dissected, and the cytoplasmic fractions corresponding to 10 PGs were prepared according to “Experimental Procedures.” After anti-BmLsd1 immunoprecipitation (IP), immunostainings (IB) were performed using anti-BmLsd1 (A), anti-phosphoserine (B), anti-phosphothreonine (C), and anti-phosphotyrosine (D) antibodies.
DISCUSSION
Before adult emergence, female B. mori moths rapidly accumulate numerous LDs within the cytoplasm of PG cells (13). These LDs are composed of various TAGs that play an essential role in providing the bombykol precursor fatty acid (6, 13, 14). To stimulate bombykol production, the external signal of PBAN coincidently activates both steps essential for bombykol biosynthesis, i.e. lipolysis of the cytoplasmic LDs and subsequent fatty acyl reduction of the released precursor fatty acid (7, 15). It is apparent that robust lipolysis proceeds in the PG on the day of eclosion in response to PBAN stimulation; however, the underlying mechanism of lipolysis activation has yet to be elucidated. Because numerous biological processes are regulated by specific modifications of functional molecules in which phosphorylation and/or dephosphorylation often play an essential role in modulating protein functions in the cascade (35), we sought to determine whether PBAN signaling utilizes regulatory mechanisms mediated by phosphorylation. In the current study, we confirmed that PBAN binding results in the phosphorylation of several distinct PG proteins (Fig. 1). In addition, we have identified a 44-kDa protein that is specifically phosphorylated in response to PBAN as BmLsd1 (Fig. 2). Furthermore, LD dynamics in conjunction with dsRNA-mediated knockdown of BmLsd1 demonstrated that BmLsd1 is essential for PBAN-mediated LD lipolysis (Fig. 4) and that PBAN-induced phosphorylation of Ser and Thr residues is BmCaMKII-dependent (Figs. 6 and 8).
BmLsd1 is a member of the structurally related PAT family proteins, which are so name based on similarity among perilipin, adipophilin/adipocyte differentiation-related protein (ADRP) and tail-interacting protein of 47 kilodaltons (TIP47). Members of the PAT family proteins are cytoplasmic LD-associated proteins characterized by the presence of the PAT domain, an N-terminal ∼100-residue sequence (32). Gene cloning (Fig. 2) and localization analysis using a specific antibody (Fig. 5) confirmed these characteristics for BmLsd1. To provide metabolic energy through the oxidation of fatty acids, neutral lipids in LDs are mobilized by lipases. In mammalian adipocytes, the PAT family protein perilipin plays an essential role in catecholamine-regulated TAG lipolysis (32, 36). Catecholamine interaction with the cell surface β-adrenergic receptor results in the generation of cAMP, which in turn activates PKA and the subsequent phosphorylation of both perilipin and hormone-sensitive lipase (37). Phosphorylation of perilipin further promotes release of the lipase activator CGI-58 from the CGI-58-perilipin complex, resulting in the activation of adipose triglyceride lipase (ATGL) (38). Thus, the hormone-induced signal transduction cascade that stimulates LD lipolysis in mammalian adipose tissues requires cAMP-dependent PKA phosphorylation of the LD-surface PAT family protein perilipin. Similar to mammalian adipocytes, PG cells of the silkmoth, B. mori, accumulate cytoplasmic LDs and undergo TAG lipolysis, which releases the sex pheromone (bombykol) precursor fatty acid required for pheromone biosynthesis. As in mammalian adipocytes, TAG lipolysis in PG cells is also under hormonal control and requires the involvement of a perilipin-like protein, BmLsd1 (Figs. 3 and 4). Unlike catecholamine signaling, however, we found that phosphorylation of BmLsd1 is mediated in vivo by BmCaMKII rather than BmPKA (Figs. 7 and 8). We have previously demonstrated that the pheromonotropic effects of PBAN are mediated by the PG cell-surface PBAN receptor, a G protein-coupled receptor that belongs to the neuromedin U receptor family (17). PBAN binding triggers Gq-mediated PLC activation and the subsequent opening of STIM- and Orai-dependent store-operated Ca2+ channels (19–21). The ensuing influx of extracellular Ca2+ drives both cytoplasmic TAG lipolysis and subsequent fatty acyl reduction. In addition, we have reported that the calmodulin inhibitors W-7 and trifluoperazine inhibit the pheromonotropic action of PBAN (39). Furthermore, B. mori does not utilize the cAMP-dependent PKA cascade in PBAN signaling (40). Taken together, these observations are consistent with our findings that BmCaMKII, and not PKA, mediates BmLsd1 phosphorylation and suggest that BmCaMKII plays a key regulatory role in PBAN signaling through phosphorylation of PG proteins crucial to the bombykol biosynthetic pathway. Among the PG proteins phosphorylated in response to PBAN (Fig. 1), we found that other cytoplasmic proteins in addition to BmLsd1 also exhibit diminished phosphorylation in the BmCaMKII-knockdown PG homogenates (data not shown); what role, if any, they play in bombykol production remains to be determined. It is possible that the specific lipase activated in response to PBAN stimulation is among these BmCaMKII-phosphorylated proteins. It has been shown that ERK, a CaMKII substrate, can phosphorylate and activate hormone sensitive lipase (37). The functional significance of BmLsd1 phosphorylation is currently unknown. We speculate that phosphorylation likely triggers a conformational change in BmLsd1 similar to that reported for mammalian perilipins that exposes LD surfaces for lipase binding in conjunction with release of co-activators that have been sequestered (41, 42). Alternatively, the phosphorylation-dependent conformational changes may promote direct interactions with the lipase itself as reported for hormone-sensitive lipase (43). Recently, using an RNAi-mediated gene knockdown screen, we have identified three lipase genes from our PG EST data base that affect LD lipolysis in PG cells.5 Analysis of their role in relation to BmLsd1 phosphorylation and the underlying mechanisms of PBAN-mediated LD lipolysis is expected to further advance our understanding of sex pheromone biosynthetic pathways in moths.
It has been reported that MsLsd1, a lepidopteran homolog of BmLsd1 identified in the M. sexta FB, functions in energy homeostasis and plays an essential role in FB lipid mobilization. In insects, FB is the primary organ for storage of both lipids, in the form of TAGs, and carbohydrates, as glycogen, and thus functionally, FB plays roles that in vertebrates are carried out by both the liver and adipose tissue (44). For energy requirements, the mobilization of energy reserves from the FB is controlled by the neuropeptide adipokinetic hormone (AKH), a small peptide composed of 8–10 amino acids (45). Thus, similar to PBAN, AKH stimulates TAG lipolysis, albeit lipid mobilization in FB cells. AKH promotes a rapid activation of FB cAMP-dependent PKA and a sustained increase in Ca2+ influx (46). Recent studies on AKH-induced phosphorylation identified MsLsd1 as a target of PKA, and phosphorylation of MsLsd1 was proven to be crucial for AKH-induced lipolysis in the FB (30, 47). As mentioned above, because B. mori does not utilize the prevalent cAMP-dependent PKA cascade in PBAN signaling (40), it is interesting that PBAN and AKH utilize distinctive phosphorylation mechanisms despite both regulating the same physiological event of cytoplasmic LD lipolysis in insects.
Despite the anatomical and physiological differences between mammals and insects, a remarkable number of lipolytic factors and their associated signaling mechanisms have been evolutionarily conserved. However, the molecular mechanisms underlying the process by which the substrate TAGs in the LD core are ultimately hydrolyzed after the phosphorylation of PAT family proteins have yet to be fully understood. Elucidation and comparison of the mechanisms underlying TAG lipolysis in insects as well as mammalian systems may facilitate our understandings in this field.
Acknowledgments
We thank Shinji Atsusawa and Masaaki Kurihara for maintaining the B. mori colony.
This work was supported by the Lipid Dynamics Research Program from RIKEN and the Targeted Proteins Research Program and Grants-in-aid for Scientific Research (B) 20380040 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
A. Ohnishi, M. Kaji, K. Hashimoto, and S. Matsumoto, unpublished data.
- PBAN
- pheromone biosynthesis activating neuropeptide
- AKH
- adipokinetic hormone
- CaMKII
- Ca2+/calmodulin-dependent protein kinase II
- cPKC
- conventional PKC
- DEPC
- diethyl pyrocarbonate
- EST
- expressed sequence tag
- FB
- fat body
- LD
- lipid droplet
- Lsd1
- lipid storage droplet protein-1
- PG
- pheromone gland
- TAG
- triacylglycerol
- p-APMSF
- p-amidinophenylmethanesulfonyl fluoride hydrochloride
- BmLsd1
- B. mori Lsd1.
REFERENCES
- 1. Raina A. K., Jaffe H., Kempe T. G., Keim P., Blacher R. W., Fales H. M., Riley C. T., Klun J. A., Ridgway R. L., Hayes D. K. (1989) Science 244, 796–798 [DOI] [PubMed] [Google Scholar]
- 2. Kitamura A., Nagasawa H., Kataoka H., Inoue T., Matsumoto S., Ando T., Suzuki A. (1989) Biochem. Biophys. Res. Commun. 163, 520–526 [DOI] [PubMed] [Google Scholar]
- 3. Rafaeli A. (2002) Int. Rev. Cytol. 213, 49–91 [DOI] [PubMed] [Google Scholar]
- 4. Rafaeli A. (2009) Gen. Comp. Endocrinol. 162, 69–78 [DOI] [PubMed] [Google Scholar]
- 5. Percy J. E., Weatherston J. (1974) in Pheromones (Birch M. C. ed) pp. 11–34, North-Holland Publishing Co., Amsterdam [Google Scholar]
- 6. Fónagy A., Yokoyama N., Okano K., Tatsuki S., Maeda S., Matsumoto S. (2000) J. Insect Physiol. 46, 735–744 [DOI] [PubMed] [Google Scholar]
- 7. Matsumoto S., Hull J. J., Ohnishi A., Moto K., Fónagy A. (2007) J. Insect Physiol. 53, 752–759 [DOI] [PubMed] [Google Scholar]
- 8. Bjostad L. B., Wolf W. A., Roelofs W. L. (1987) in Pheromone Biochemistry (Prestwich G. D., Blomquist G. J. eds) pp. 77–120, Academic Press, Inc., Orlando, FL [Google Scholar]
- 9. Jurenka R. A. (2003) Insect Pheromone Biochemistry and Molecular Biology, pp. 53–80, Elsevier Academic Press, Oxford, UK [Google Scholar]
- 10. Moto K., Suzuki M. G., Hull J. J., Kurata R., Takahashi S., Yamamoto M., Okano K., Imai K., Ando T., Matsumoto S. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8631–8636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moto K., Yoshiga T., Yamamoto M., Takahashi S., Okano K., Ando T., Nakata T., Matsumoto S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9156–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohnishi A., Hull J. J., Matsumoto S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4398–4403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fónagy A., Yokoyama N., Matsumoto S. (2001) Arthropod. Struct. Dev. 30, 113–123 [DOI] [PubMed] [Google Scholar]
- 14. Matsumoto S., Fónagy A., Yamamoto M., Wang F., Yokoyama N., Esumi Y., Suzuki Y. (2002) Insect Biochem. Mol. Biol. 32, 1447–1455 [DOI] [PubMed] [Google Scholar]
- 15. Matsumoto S., Ohnishi A., Lee J. M., Hull J. J. (2010) Vitam. Horm. 83, 425–445 [DOI] [PubMed] [Google Scholar]
- 16. Choi M. Y., Fuerst E. J., Rafaeli A., Jurenka R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 9721–9726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hull J. J., Ohnishi A., Moto K., Kawasaki Y., Kurata R., Suzuki M. G., Matsumoto S. (2004) J. Biol. Chem. 279, 51500–51507 [DOI] [PubMed] [Google Scholar]
- 18. Kim Y. J., Nachman R. J., Aimanova K., Gill S., Adams M. E. (2008) Peptides 29, 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hull J. J., Kajigaya R., Imai K., Matsumoto S. (2007) Biosci. Biotechnol. Biochem. 71, 1993–2001 [DOI] [PubMed] [Google Scholar]
- 20. Hull J. J., Lee J. M., Kajigaya R., Matsumoto S. (2009) J. Biol. Chem. 284, 31200–31213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hull J. J., Lee J. M., Matsumoto S. (2010) Insect Mol. Biol. 19, 553–566 [DOI] [PubMed] [Google Scholar]
- 22. Fónagy A., Matsumoto S., Uchiumi K., Orikasa C., Mitsui T. (1992) J. Pest. Sci. 17, 47–54 [Google Scholar]
- 23. Ozawa R., Matsumoto S. (1996) Insect Biochem. Mol. Biol. 26, 259–265 [DOI] [PubMed] [Google Scholar]
- 24. Fónagy A., Ohnishi A., Esumi Y., Suzuki Y., Matsumoto S. (2005) Ann. N.Y. Acad. Sci. 1040, 310–314 [DOI] [PubMed] [Google Scholar]
- 25. Laemmli U. K. (1970) Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 26. Burnette W. N. (1981) Analyt. Biochem. 112, 195–203 [DOI] [PubMed] [Google Scholar]
- 27. Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 28. Matsumoto S., Kitamura A., Nagasawa H., Kataoka H., Orikasa C., Mitsui T., Suzuki A. (1990) J. Insect Physiol. 36, 427–432 [Google Scholar]
- 29. Ohnishi A., Hashimoto K., Imai K., Matsumoto S. (2009) J. Biol. Chem. 284, 5128–5136 [DOI] [PubMed] [Google Scholar]
- 30. Patel R. T., Soulages J. L., Hariharasundaram B., Arrese E. L. (2005) J. Biol. Chem. 280, 22624–22631 [DOI] [PubMed] [Google Scholar]
- 31. Arrese E. L., Mirza S, Rivera L, Howard A. D., Chetty P. S., Soulages J. L. (2008) Insect Biochem. Mol. Biol. 38, 993–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lafontan M., Langin D. (2009) Prog. Lipid Res. 48, 275–297 [DOI] [PubMed] [Google Scholar]
- 33. Matsumoto S., Yoshiga T., Yokoyama N., Iwanaga M., Koshiba S., Kigawa T., Hirota H., Yokoyama S., Okano K., Mita K., Shimada T., Tatsuki S. (2001) Insect Biochem. Mol. Biol. 31, 603–609 [DOI] [PubMed] [Google Scholar]
- 34. Uno T., Nakao A., Fujiwara Y., Katsurauma C., Nakada T., Itoh O. (2006) Arch. Insect Biochem. Physiol. 61, 65–76 [DOI] [PubMed] [Google Scholar]
- 35. Pawson T., Scott J. D. (2005) Trends Biochem. Sci. 30, 286–290 [DOI] [PubMed] [Google Scholar]
- 36. Miyoshi H., Perfield J. W., 2nd, Souza S. C., Shen W. J., Zhang H. H., Stancheva Z. S., Kraemer F. B., Obin M. S., Greenberg A. S. (2007) J. Biol. Chem. 282, 996–1002 [DOI] [PubMed] [Google Scholar]
- 37. Carmen G. Y., Víctor S. M. (2006) Cell. Signal. 18, 401–408 [DOI] [PubMed] [Google Scholar]
- 38. Guo Y., Cordes K. R., Farese R. V., Jr., Walther T. C. (2009) J. Cell Sci. 122, 749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matsumoto S., Ozawa R., Nagamine T., Kim G. H., Uchiumi K., Shono T., Mitsui T. (1995) Biosci. Biotechnol. Biochem. 59, 560–562 [DOI] [PubMed] [Google Scholar]
- 40. Hull J. J., Kajigaya R., Imai K., Matsumoto S. (2007) J. Insect Physiol. 53, 782–793 [DOI] [PubMed] [Google Scholar]
- 41. Granneman J. G., Moore H. P., Krishnamoorthy R., Rathod M. (2009) J. Biol. Chem. 284, 34538–34544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang H., Bell M., Sreenevasan U., Hu H., Liu J., Dalen K., Londos C., Yamaguchi T., Rizzo M. A., Coleman R., Gong D., Brasaemle D., Sztalryd C. (2011) J. Biol. Chem. 286, 15707–15715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang H., Hu L., Dalen K., Dorward H., Marcinkiewicz A., Russell D., Gong D., Londos C., Yamaguchi T., Holm C., Rizzo M. A., Brasaemle D, Sztalryd C. (2009) J. Biol. Chem. 284, 32116–32125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Law J. H., Wells M. A. (1989) J. Biol. Chem. 264, 16335–16338 [PubMed] [Google Scholar]
- 45. Gäde G., Auerswald L. (2003) Gen. Comp. Endocrinol. 132, 10–20 [DOI] [PubMed] [Google Scholar]
- 46. Van der Horst D. J., Van Marrewijk W. J., Diederen J. H. (2001) Int. Rev. Cytol. 211, 179–240 [DOI] [PubMed] [Google Scholar]
- 47. Patel R. T., Soulages J. L., Arrese E. L. (2006) Arch. Insect Biochem. Physiol. 63, 73–81 [DOI] [PubMed] [Google Scholar]