Abstract
Loss-of-function mutations in the proton-coupled folate transporter (PCFT, SLC46A1) result in the autosomal recessive disorder, hereditary folate malabsorption (HFM). Identification and characterization of HFM mutations provide a wealth of information on the structure-function relationship of this transporter. In the current study, PCR-based random mutagenesis was employed to generate unbiased loss-of-function mutations of PCFT, simulating the spectrum of alterations that might occur in the human disorder. A total of 26 mutations were generated and 4 were identical to HFM mutations. Eleven were base deletion or insertion mutations that led to a frameshift and, along with similar HFM mutations, are predominantly localized to two narrow regions of the pcft gene at the 5′-end. Base substitution mutations identified in the current study and HFM patients were largely distributed across the pcft gene. Elimination of the ATG initiation codon by a one-base substitution (G > A) did not result in a complete lack of translation at the same codon consistent with rare non-ATG translation initiation. Among six missense mutants evaluated, three mutant PCFTs were not detected at the plasma membrane, one mutation resulted in decreased binding to folate substrate, and one had a reduced rate of conformational change associated with substrate translocation. The remaining PCFT mutant had defects in both processes. These results broaden understanding of the regions of the pcft gene prone to base insertion and deletion and inform further approaches to the analysis of the structure-function of PCFT.
Keywords: Drug Transport, Folate, Membrane Proteins, Mutant, Transport Drugs, Hereditary Folate Malabsorption, Random Mutagenesis, Proton-couped Transporter
Introduction
The proton-coupled folate transporter (PCFT)2 (SLC46A1) plays a key role in intestinal folate absorption and folate transport into the central nervous system (1). Loss-of-function mutations in the pcft gene lead to the rare autosomal recessive disorder, hereditary folate malabsorption (HFM) characterized by markedly reduced folate levels in blood and cerebrospinal fluid (1–4). A homozygous mutation in most cases or two compound heterozygous mutations in two cases have been identified in all subjects with the clinical diagnosis of HFM indicating that this disease is caused solely by alterations of the pcft gene (1, 2, 5–12).
Sixteen different loss-of-function pcft mutations have been identified to date in HFM patients. Six result in drastic changes in predicted protein sequences (nonsense). p.Y362_G398del, occurred multiple times in unrelated families and is the result of skipping of exon 3 during RNA splicing (1, 9, 13). C66X introduces a stop codon at position 66 due to a two-base substitution (5). Four frameshift mutations, p. E9Gfs, p.G65Afs, C66Lfs and N68Kfs, are due to base deletions or insertions (2, 8, 10, 12). Ten remaining mutations resulted in a single amino acid substitution in the PCFT protein (missense). Five mutations occurred at charged resides (p.R113C, p.R113S, p.R376W, p.R376Q, and p.D156Y) (2, 6, 7, 11), whereas the other five, p.G147R, p.S318R, p.A335D, p.G338R, and p.P425R, involved substitutions of a non-charged, with a charged, residue (2, 12). There appear to be hot spots for both nonsense and missense mutations. Four of six nonsense mutations occurred between Gly-65 and Gln-68, whereas 40% of the missense mutations occurred at two Arg residues (Arg-113 and Arg-376).
Detailed studies of three residues, Arg-113, Arg-376, and Asp-156, that were mutated in HFM patients provided valuable information on PCFT structure-function. Arg-113 is essential with only a low level of residual activity when it was replaced with like-charged histidine or lysine (R113H and R113K) (14). Although R376W was completely inactive despite the presence of protein accessible at the cell membrane, R376Q retained residual activity, in a substrate-specific manner, with less loss of activity for the antifolate pemetrexed than the reduced folates and folic acid (6). The Asp-156 residue appears to play a key role in protein stability; many mutations at this site resulted in the absence of protein. However, when a mutant protein was expressed, the transporter was fully functional (7).
The current study was designed to identify additional residues required for PCFT function, and vulnerable regions of the carrier, using a random mutagenesis strategy. The mutagenesis rate was adjusted to generate clones with less than an average of 4 mutations per pcft open reading frame following which the specific mutations responsible for the loss-of-function were identified. Using this approach, 144 PCFT mutants were generated; 25 lost function completely or had markedly reduced function. Twenty-six loss-of-function mutations were identified, at least one in each PCFT mutant. Seventeen were nonsense mutations. Molecular mechanisms underlying nine missense mutations ranged from the lack of protein expression, decreased binding of the mutated PCFT to folate substrate, to a reduced rate of conformational change associated with substrate translocation.
MATERIALS AND METHODS
Cell Line and Chemicals
HeLa R1–11 cells were derived from HeLa cells and have lost expression of both the reduced folate carrier and PCFT, due to deletion of the former gene (15) and methylation of the latter promoter (16). HeLa R1–11 cells served as transfection recipients and were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 100 units/ml of penicillin, and 100 μg/ml of streptomycin. HeLa R1–11 cells were thawed regularly from liquid nitrogen to ensure that PCFT expression was absent (17). [3′,5′,7-3H]MTX was purchased from Moravek Biochemicals (Brea, CA).
Random and Site-directed Mutagenesis to Generate pcft Mutants
A random mutagenesis protocol (Diversify PCR Random Mutagenesis Kit, Clontech, Mountain View, CA) was used to generate a large number of PCFT mutant clones containing loss-of-function mutations. This protocol allows modulation of the extent of random mutations induced by independently varying the amount of manganese and dGTP in the PCR. Higher concentrations of both manganese and dGTP result in a higher mutation rate. A 1.35-kb fragment of the pcft gene, which spans from the initiation codon to the XhoI restriction site and encodes the first 451 of 459 amino acid residues, were PCR-amplified with forward primer, TATAAGCTTCACCATGGAGGGGAGCGCGAGC, and reverse primer, AACTCGAGGTGAGGATCAGCCTTTTCC, according to conditions recommended in the kit. The fragment that contained random pcft mutations was restricted with HindIII and XhoI, and cloned into the HA (hemagglutinin)-tagged PCFT expression vector treated with the same restriction enzymes. The resulting plasmids (pcft mutant clones) that contained the correct insert were used for transient transfections. For generation of a PCFT expression vector in which translation was started at the second ATG initiation codon, a pcft fragment spanning from the second ATG codon to the XhoI restriction site was amplified using error-proof Pfu Turbo DNA Polymerase (Stratagene, La Jolla, CA) and a pair of primers, CCCAAGCTTCACCATGCAGGAAGTGGAGACC and AACTCGAGGTGAGGATCAGCCTTTTCC. This fragment was processed and subcloned into the vector in the same way as the fragments for random mutagenesis described above. Specific individual mutations were produced using the QuikChange® II XL Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA). In this case, the HA-tagged PCFT expression vector was used as the template. The coding region of all final expression vectors was sequenced in the Albert Einstein Cancer Center Genomics Shared Resource.
Transient Transfection
HeLa R1–11 cells were seeded into 20-ml Low Background glass scintillation vials (Research Products International Corp., Prospect, IL) at 0.35 million cells/vial for transport studies and grown in 6-well plates at a density of 0.6 million cells/well for cell surface biotinylation experiments. In both cases, transfections were conducted 2 days later with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Transport and labeling assays were conducted 2 days after transfection.
Membrane Transport of MTX
Influx of [3H]MTX (an inexpensive surrogate for folates) was used to assay PCFT function. Transient transfectants were washed twice with HBS (20 mm Hepes, 5 mm dextrose, 140 mm NaCl, 5 mm KCl, 2 mm MgCl2, pH 7.4) and incubated in the same buffer at 37 °C for 20 min. The incubation buffer was then aspirated and transport was initiated by the addition of 0.5 ml of pre-warmed (37 °C) MBS (20 mm MES, 140 mm NaCl, 5 mm KCl, 2 mm MgCl2, pH 5.5) containing 0.5 or 50 μm [3H]MTX. Uptake was carried out at 37 °C over 1 min and stopped by the addition of 5 ml of ice-cold HBS. Cells were washed three times with ice-cold HBS and dissolved in 0.5 ml of 0.2 m NaOH at 65 °C for 40 min. Radioactivity in 0.4 ml of lysate was measured on a liquid scintillation counter and normalized to protein levels obtained with the BCA Protein Assay (Pierce). In most cases, the transport rate was expressed as percentage of wild-type PCFT activity. Otherwise, it was expressed in units of picomole/mg of protein/min.
Cell Surface Biotinylation Assay
This assay was described previously (18). Briefly, cells were washed twice with PBS (pH 8.0) and labeled at room temperature with EZ-Link Sulfo-NHS-LC-Biotin (Pierce Biotechnology), which reacts specifically with primary amine groups, at a concentration of 1 mg/ml in PBS (pH 8.0) for 30 min. Cells were washed twice in PBS and treated with 0.7 ml of hypotonic buffer (0.5 mm Na2HPO4, 0.1 mm EDTA, pH 7.0) containing protease inhibitor mixture (Roche Applied Science) on ice for 30 min. The cells were then detached from the plates with disposable cell lifters and centrifuged at 16,000 × g and 4 °C for 5 min. The pellet was then resuspended in 0.4 ml of lysis buffer (50 mm Tris base, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, pH 7.4) and mixed on a rotator for 1 h at 4 °C. A 25-μl portion (identified as “crude membrane”) was collected and stored in a −20 °C freezer. The remaining crude membrane fraction was centrifuged at 16,000 × g and 4 °C for 15 min and the supernatant was mixed on a rotator overnight at 4 °C with 50 μl of streptavidin-agarose beads (Fisher Scientific) pre-washed three times with the lysis buffer. The agarose beads were washed four times with 0.5 ml of lysis buffer, each with a 20-min mix on a rotator at 4 °C. The precipitated proteins were released from the beads by heating at 95 °C for 5 min in 2× SDS-PAGE sample loading buffer containing dithiothreitol.
Western Blot Analysis
Samples were analyzed on standard 12.5% SDS-PAGE. The precipitated proteins (released from beads as described in the previous section) were loaded directly on gels, whereas the crude membrane fraction was mixed (1:1) with dithiothreitol-containing 2× SDS-PAGE sample loading buffer at room temperature before loading on the gels. After SDS-PAGE, proteins were transferred to Amersham Biosciences Hybond membranes (GE Healthcare) and blocked with 10% dry milk in TBST (20 mm Tris, 135 mm NaCl, 1% Tween 20, pH 7.6) overnight at 4 °C. The blots with crude membrane samples were probed with anti-actin antibody (Cell Signaling Technology, Danvers, MD, 1:2000 in TBST, 0.1% milk) (where shown), then stripped in 100 mm β-mercaptoethanol, 2% SDS, 62.5 mm Tris-Cl (pH 6.7) and re-probed with anti-HA antibody (Sigma, 1:4000 in TBST, 0.1% milk). For precipitated samples, blots were probed directly with anti-HA antibody. After application of the first antibody, the blots were incubated with anti-rabbit IgG-HRP conjugate (Cell Signaling Technology, Danvers, MD, 1:5000 in TBST). The blots were developed with Amersham Biosciences ECL Plus reagent (GE Healthcare) and quantified by Image J.
RESULTS
Optimizing the Conditions for Random Mutagenesis of the pcft Gene
The goal of this study was to generate and identify inactivating PCFT mutations by PCR-based random mutagenesis. The key to this approach is achieving a proper mutagenesis rate. Low rates of mutagenesis generate only a small percentage of PCFT clones that contain an inactive mutant, whereas a high rate of mutagenesis produces clones that contain multiple mutations complicating identification of the inactivating mutation. Initial random mutagenesis was performed under five different conditions with a spectrum of mutation rates. Multiple clones from each condition were sequenced and the function of the encoded mutant PCFTs assessed. As indicated in Table 1, the actual mutations generated per 1 kb of the pcft gene approximated the expected rate of mutagenesis indicative of the accuracy of the random mutagenesis protocol. The more mutations in a pcft clone, the higher the percentage of those clones that lost function. To achieve the desired rate of less than an average of 4 mutations per pcft coding region, further studies were pursued only under conditions “1” (clones with 1 as prefix) and condition “2” (clones with 5 as prefix). However, mutants generated initially under condition “3” (clones with 4 as prefix) were also included in this study.
TABLE 1.
Correlation between the number of mutations in the PCFT coding region and function of mutants generated by random mutagenesis
The pcft coding region was amplified by PCR and random mutations were generated in this reaction. The fragments were cloned into an expression vector and transiently transfected into HeLa R1–11 cells. PCFT function was assessed by measuring MTX influx (0.5 μm) at pH 5.5 over 1 min and was normalized to wild-type PCFT. The values represent the average from three different experiments.
| Mutagenesis condition | Expected mutations/1 kba | Number of clones studied | Actual mutations/1 kb | Mutations/ pcft geneb | PCFT function |
||
|---|---|---|---|---|---|---|---|
| <5% | 5–30% | >30% | |||||
| 1 | 2.0 | 8 | 0.97 | 1.3 ± 0.3 | 1 | 0 | 7 |
| 2 | 2.3 | 10 | 2.4 | 3.3 ± 0.5 | 4 | 3 | 3 |
| 3 | 2.7 | 9 | 3.8 | 5.1 ± 0.7 | 4 | 3 | 2 |
| 4 | 4.6 | 6 | 5.3 | 7.2 ± 1,4 | 5 | 1 | 0 |
| 5 | 8.1 | 5 | 11.7 | 15.8 ± 2.8 | 4 | 0 | 1 |
a Data provided by the manufacturer.
b The mutated pcft region encodes the whole PCFT protein except the last eight amino acid residues at the C-terminus, which does not have any functional role (28).
Identification of pcft Clones with Severely Impaired Function
A total of 144 pcft clones were screened initially for MTX transport activity. As indicated in Fig. 1, transport function of 25 PCFT mutants at pH 5.5, the optimal pH for this carrier (1), was reduced to <10% of that of wild-type PCFT; MTX influx in all but two mutants was not different from the mock transfected cells consistent with a complete loss of activity. MTX influx in clones 1–70 and 5–28 was ∼2 and 6% of the wild-type rate, respectively, significantly higher than that in the mock transfectants (p = 0.010 and 0.015, respectively) and was also different from each other (p = 0.024) based upon three independent experiments.
FIGURE 1.
Identification of PCFT mutant clones with impaired transport function. MTX influx was assessed at 0.5 μm and pH 5.5 over 1 min in HeLa R1–11 cell transient transfectants expressing mutant and wild-type PCFTs. The data are presented as percentage of wild-type PCFT activity. p values are derived from paired t tests comparing mutants and mock transfectants. Data are the mean ± S.E. from three independent experiments.
Identification of Loss-of-function pcft Mutations
The entire pcft coding region of all 25 mutant clones was sequenced. All had more than one mutation in the coding region (silent mutations were not counted). In 19, a pcft mutation in each clone could be identified that either prevented synthesis of full-length PCFT or encoded a known inactive PCFT as indicated in Table 2. Eleven involved base deletion(s) or insertion(s) that led to a frameshift, whereas eight involved base substitutions that resulted in elimination of the initiation codon, introduction of a stop codon, or inactivating amino acid substitutions in the PCFT protein. In one instance, there was a four-base deletion (clone 5-3). Among these deletion and insertion mutations, two, G65Afs and C66Lfs, were previously identified in HFM patients (2, 10). Two point mutations, S318R and R376Q, found in clones 1–46 and 1–74, respectively, were identified in HFM patients and resulted in a complete or substantial loss of activity, respectively (2, 6). Two base substitutions in clones 1–70 and 5–28 resulted in mutation of the ATG initiation codon. Interestingly, there was a low level of residual PCFT activity in both clones, indicating that some PCFT protein was synthesized in the absence of an intact initiation codon.
TABLE 2.
PCFT clones in which a single loss-of-function mutation could be readily identified
| Clonea | Number of mutations in each clone | Inactive mutation at cDNA level | Corresponding mutation at protein level |
|---|---|---|---|
| 1–70 | 2 | c. 1A>C | M1L (no initiation codon) |
| 5–28 | 3 | c. 3G>A | M1I (no initiation codon) |
| 5–46 | 7 | c. 4delGb | p. E2Rfs |
| 5–34 | 4 | c. 4–5insGb | p. E2Gfs |
| 5–136 | 3 | c. 9–10InsGb | p. S4Efs |
| 5–22 | 3 | c. 10delA | p. S4Afs |
| 5–3 | 5 | c. 177–181delCACCC | p. T60Pfs |
| 1–52 | 5 | c. 194delGb | p. G65Afsc |
| 5–76 | 2 | c. 194–195insGb | p. C66Lfsc |
| 5–11 | 6 | c. 275delGb | p. G92Afs |
| 5–133 | 7 | c. 335delGb | p. G112Afs |
| 5–124 | 4 | c. 639G>A | p. W213X |
| 5–78 | 2 | c. 690delTb | p. F230Lfs |
| 5–58 | 4 | c. 918T>A | p. Y306X |
| 1–46 | 4 | c. 952A>C | p. S318Rc |
| 5–33 | 5 | c.1020delCb | p. F341Sfs |
| 1–9 | 2 | c.1091T>A | p. L364X |
| 1–74 | 3 | c. 1127G>A | p. R376Qc |
| 5–88 | 3 | c. 1264A>T | p. K422X |
Non-AUG (or ATG for cDNA) Translation Initiation of the pcft Gene
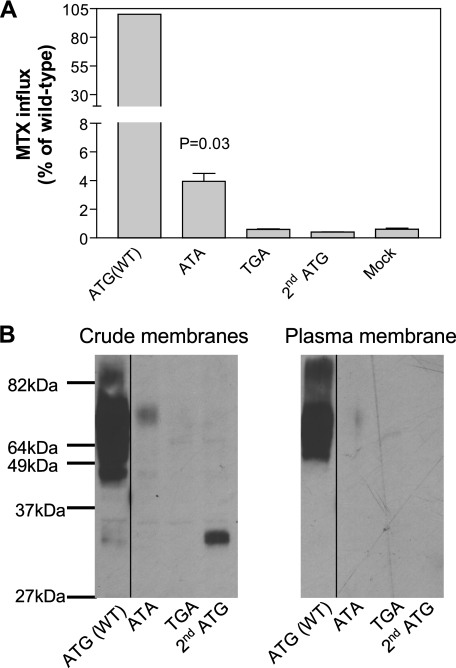
Two possibilities could account for the residual activity present in clones 1–70 and 5–28 despite mutation of the translational initiation codon ATG. First, the mutant could provide an alternative initiation of translation at a non-AUG codon (19). Second, initiation of translation of the pcft gene could occur at the next or second AUG when the first AUG is mutated, a process that would result in a truncated PCFT lacking the first 74 amino acid residues at the N terminus. To test these possibilities, the ATG initiation codon of the pcft gene was changed to ATA (identified in 5–28), or to TGA (stop codon) in the expression vector. In addition, a separate expression vector was generated by deletion of part of the 5′ coding sequence (up to c. 218C) so that translation could only occur at the second ATG codon. As indicated in Fig. 2A, 4% of wild-type PCFT activity was retained when the initiation ATG codon was replaced with ATA. This activity was greater (p = 0.03) than in the mock transfected cells. There was no increase in activity above the mock transfectant level when the ATG codon was substituted with TGA. Also, no activity was detected when translation was started at the second ATG codon.
FIGURE 2.
Comparison of the function and expression of PCFT encoded by cDNAs with different initiation codons. The initiation codon, ATG, of wild-type pcft was changed to ATA or TGA by site-directed mutagenesis. For pcft gene translation initiated by the second ATG codon, all but four nucleotides upstream of the second ATG were deleted. Panel A, PCFT function was assayed with MTX influx (0.5 μm) at pH 5.5 over 1 min and expressed as percentage of wild-type PCFT activity. p values were derived from paired t tests comparing mutants and mock transfectants. Data are the mean ± S.E. from three independent experiments. Panel B, Western blot analysis of PCFT expression in the crude membrane preparation and accessible to biotinylation at the plasma membrane. The selected molecular weight of the protein ladder is indicated. The lines on the images indicate lanes that have been repositioned. The graph is representative of two independent experiments.
Western blot analyses were performed to assess PCFT expression in the crude membrane or biotinylated at the plasma membrane. As indicated in Fig. 2B, the cDNA with ATA as the initiation codon encoded a protein with a molecular weight similar to wild-type PCFT indicating that this protein remained full-length. However, protein expression with this construct was so low it was barely detected at the plasma membrane, consistent with the markedly reduced transport activity. Hence, PCFT synthesized with ATA as the initiation codon is largely functional. Also consistent with the lack of function, there was no protein detected in the crude membrane preparation when the initiation codon ATG was changed to TGA. However, there was protein detected at ∼33 kDa in the crude membrane fraction when the cDNA was initiated with the second ATG initiation codon reflecting a deletion of 74 N-terminal amino acids, including two N-glycosylation sites (Asn-58 and -68), consistent with the absence of function. Therefore, the residual activity detected when the initiation codon is mutated is attributed to non-AUG translation initiation of the pcft gene.
Loss of Transport Activity Due to Point Mutations in PCFT
In the remaining six mutant clones only point mutations were identified, as indicated in Table 3. Because up to five mutations were detected in each clone, a strategy was designed to pinpoint the mutation(s) responsible for the inactivation of PCFT. Based upon the observation that a PCFT point mutation identified in HFM always involves substitution of a charged amino acid residue with a non-charged, or a non-charged residue replaced with a charged residue, one or two point mutations with these characteristics were chosen from each clone for initial assessment. These included: L161R in clone 1–45, L432H in clone 1–60, M222R in clone 4–4, E232G and W333R in clone 4–8, E387K in clone 5–74, as well as V134E and H247P in clone 5–114. These mutants were individually generated by site-directed mutagenesis, transfected into R1–11 cells, and their impact on transport function assessed.
TABLE 3.
PCFT clones that contain multiple base-substitution mutations with unknown functional consequences
| Clone | Number of mutations in each clone | Point mutations indicated at protein and cDNA levels |
|---|---|---|
| 1–45 | 3 | p. L81I (c. 241C>A) |
| p. L161R (c. 482T>G)a | ||
| p. P211S (c. 631C>T) | ||
| 1–60 | 2 | p. F426V (c. 1276T>G) |
| p. L432H (c. 1295T>A) | ||
| 4–4 | 4 | p. P7L (c. 19C>T) |
| p. L194Q (c. 581T>A) | ||
| p. M222R (c. 656T>G)a | ||
| 4–8 | 5 | p. T43A (c. 127A>G) |
| p. V132I (c. 394G>A) | ||
| p. E232G (c. 695A>G)a | ||
| p. W333R (c. 997T>C) | ||
| p. L357P (c. 1070T>C) | ||
| 5–74 | 3 | p. L31P (c. 92T>C)a |
| p I304F (c. 910A>T)a | ||
| p. E387K (c. 1159G>A) | ||
| 5–114 | 5 | p. G40S (c. 118G>A) |
| p. V134E (c. 401 T>A)a | ||
| p. H247P (c. 740A>C)a | ||
| p. A268T (c. 802G>A) | ||
| p. I435T (c. 1304T>C) |
a Mutations that contribute to the loss of PCFT function.
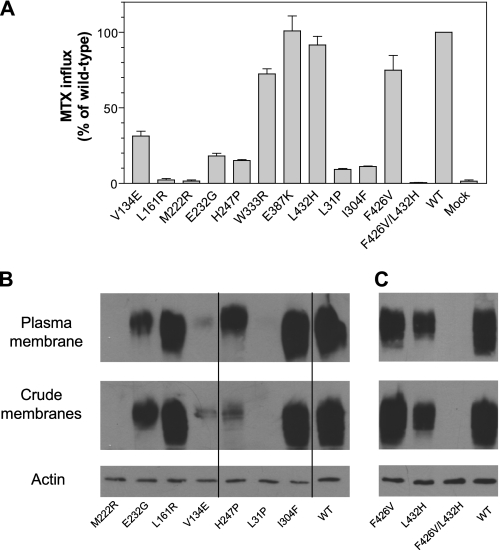
As indicated in Fig. 3A, the L161R and M222R PCFTs resulted in a complete loss of PCFT activity. However, this does not rule out the possibility of another inactivating mutation in these clones. V134E and H247P mutations, both of which were detected in clone 5–114, resulted in reductions in function by ∼70 and 80%, respectively, as compared with wild-type PCFT; both contributed to PCFT inactivation in clone 5–114. The E232G mutant, detected in clone 4–8 had ∼20% the activity of wild-type PCFT; another mutation, W333R in this clone was largely active. Because the E387K and L432H mutants were fully active as compared with wild-type PCFT, the remaining two mutations in clone 5–74, and one remaining mutation in 1–60, were studied further. As indicated in Fig. 3A, both L31P and I304F mutants in clone 5–74 retained only 10% of wild-type activity and thus both contributed to the lack of PCFT function in this clone. Unexpected was the observation that the F426V mutant was largely active, indicating that both mutations (along with L432H) were required for inactivation of PCFT. To exclude any possible defects in the expression vector beyond the PCFT coding region that had been sequenced, the F426V mutation was introduced into the expression vector harboring the functional L432H mutation. All independent clones of the double mutant PCFT lost all transport function (data not shown).
FIGURE 3.
Function and expression of PCFT point mutants. Panel A, analysis of function of PCFT mutants. Transport was measured as described in the legends of Figs. 1 and 2. Data are the mean ± S.E. from three independent experiments. Panels B and C, PCFT expression in both crude membrane preparations and accessible at the plasma membrane. Expression of actin served as the loading control for the crude membrane samples. The lines on the images indicate lanes that have been repositioned. The graphs shown in panels B and C represent different experiments; both are representative of two independent analyses. The relative PCFT levels at the plasma membrane were derived from quantifying band intensity and are the average of two independent analyses.
To examine whether the marked decreases in transport activity resulting from the PCFT mutations were associated with alterations in PCFT expression or trafficking to the cell membrane, Western blot and surface biotinylation analyses were performed for the M222R, E232G, L161R, V134E, H247P, L31P, and I304F PCFT mutants (Fig. 3B). Because the F426V and L432H mutants were individually active, but inactive in combination, they were also included (Fig. 3C). PCFT expression for both the M222R and L31P mutants was not detected at the plasma membrane nor in the crude membrane preparation. There was no expression of the F426V/L432H double-mutant although expression of the individual F426V or L432H PCFT mutants at the plasma membrane was 86, or 61% of wild-type PCFT, respectively. Expression of the V134E mutant at the plasma membrane was only 6% that of wild-type PCFT. There was substantial reduction in H247P expression in the crude membrane preparation, whereas there was only a modest reduction (38%) in expression at the plasma membrane as compared with wild-type PCFT. A reduction (65%) in protein expression at the plasma membrane, as compared with wild-type PCFT, was also observed for E232G. Expression of the L161R and I304F mutant proteins at the plasma membrane was 75 and 102% of wild-type PCFT, respectively.
Functional Analysis of the E232G, L161R, and I304F PCFT Mutants
Because the E232G, L161R, and I304F mutants were expressed, studies were conducted to examine their function and to explore the kinetic basis for their decrease in transport activity. Although the H247P mutant was expressed well at the plasma membrane, it was not included because the role of His-247 was characterized in detail previously (20). MTX influx at neutral pH was markedly decreased in these mutants as compared with the wild-type PCFT excluding the possibility that the mutations disrupt proton coupling as observed for the E185A mutation (18) (data not shown). The extent of increase in MTX influx at pH 4.5 as compared with pH 5.5 for these mutants was comparable with what was observed for wild-type PCFT, suggesting that protonation of these PCFT mutants was not altered in contrast to the H281A mutant (20) (data not shown).
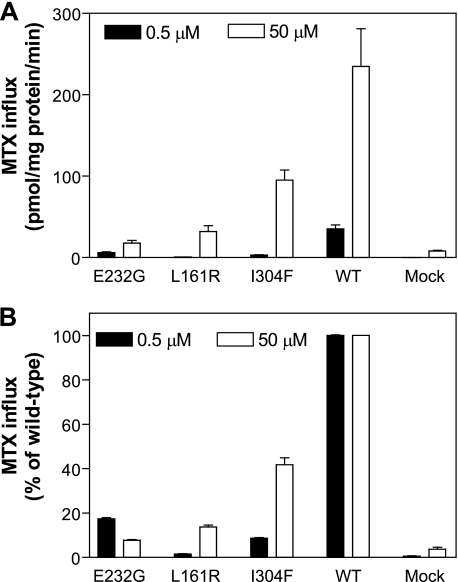
Finally, influx was assessed at MTX concentrations of 0.5 and 50 μm and is indicated in Fig. 4A as absolute rates and in Fig. 4B as percentage of wild-type activity. In this assay, when the percent of activity increases with an increase in concentration, the influx Kt of the mutant is increased. When the opposite occurs, the influx Kt and Vmax are decreased. Hence, the influx Kt for the E232G mutant was decreased and the influx Kt for the I304F mutant, and particularly for the L161R mutant, was increased as compared with the wild-type PCFT. Based on these observations, a full analysis of influx kinetics was undertaken for these mutants. As seen from Table 4, the MTX influx Kt for E232G was decreased to 15% of the wild-type level, consistent with a marked increase in affinity of the mutant carrier for the substrate, whereas MTX influx Kt for the I304F and L161R mutants was increased by a factor of 4.3 and 37, respectively, as compared with wild-type PCFT. The MTX influx Vmax was decreased for all three mutants as compared with the wild-type PCFT with a marked reduction (by 82%) for the E232G mutant. Therefore, the major contributing factor for the decrease in activities appears to be a markedly impaired rate of conformational alteration of the carrier during transport mediated by the E232G mutant and decreased substrate affinities for the S304F and L161R mutants, particularly profound for the latter.
FIGURE 4.
Concentration dependence of the PCFT mutant transport function. MTX influx was performed in transient transfectants under standard conditions (pH 5.5 and over 1 min) at concentrations of 0.5 and 50 μm. MTX influx is expressed in units of picomole/mg of protein (panel A) or as a percentage of wild-type PCFT activity (panel B) at each substrate concentration. Data are the mean ± S.E. from three independent experiments.
TABLE 4.
Comparison of MTX influx kinetics between mutated and wild-type PCFTs
MTX influx was measured at pH 5.5 over 1 min. Kt and Vmax values are the mean ± S.E. from three independent experiments determined from nonlinear regression to the Michaelis-Menten equation. Due to low MTX influx in the L161R mutant, background uptake in mock transfectants was subtracted from uptake in L161R transfectants. Modified Vmax was obtained by normalizing to the protein levels at the plasma membrane.
|
Kt |
Vmax |
|||||
|---|---|---|---|---|---|---|
| μm | Mutant/wild-type | Unmodified | Relative plasma membrane expression | Modified | Mutant/wild-type | |
| pmol/mg protein/min | pmol/mg protein/min | |||||
| Wild-type | 2.3 ± 0.3 | 361 ± 15 | 1.00 | 361 | ||
| E232G | 0.33 ± 0.03 | 0.15 | 23.1 ± 1.5 | 0.35 | 66 | 0.18 |
| I304F | 9.8 ± 1.3 | 4.3 | 199 ± 44 | 1.02 | 195 | 0.54 |
| L161R | 84 ± 8 | 37 | 73 ± 10 | 0.75 | 97 | 0.27 |
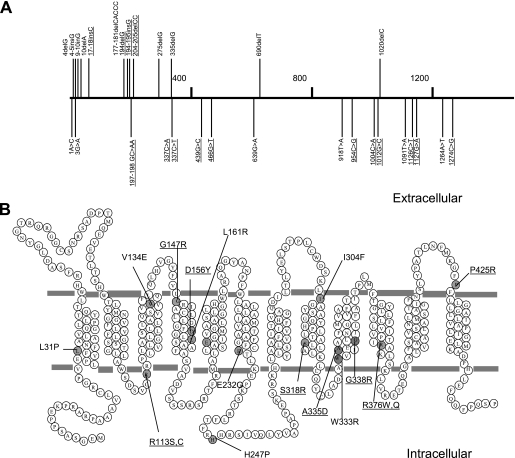
Locations of Random pcft Mutations That Lead to Complete or Partial Losses in PCFT Transport Activities
One aim of the current study was to determine whether there are hot spots (regions) for loss-of-function mutations. The mutations identified in this study by random mutagenesis and in patients with HFM were examined as indicated in Fig. 5A. Base deletion and insertion mutations, indicated above the line, are not evenly distributed. Nine of the 13 mutations are located in the first 205 base pairs and 11 of the 13 mutations are in the first 335 base pairs, a portion representing only 25% of the entire pcft cDNA. In contrast, base substitution mutations are largely evenly distributed, although, in three instances, two mutations are located within 3 base pairs. Focusing on the missense mutations that produce a defective protein, most residues are located within transmembrane domains or close to the boundaries of the membranes except His-247 as indicated in Fig. 5B. Three residues were mutated in the fourth and ninth transmembrane domains, suggesting that these transmembrane domains play important structural and functional roles.
FIGURE 5.
Localization of loss-of-function PCFT mutations. Panel A, pcft mutations are indicated in the cDNA region subjected to random mutagenesis. The horizontal numbers denote the positions of nucleotides in the pcft coding region. Base deletions and insertions are depicted above the line, whereas base substitutions are indicated below the line. The mutations underlined were previously reported in subjects with HFM. Panel B, PCFT missense mutations are indicated on the topological structure of PCFT (27). Point mutations identified in the current study, as well as in a subject with HFM, are indicated by gray circles. Mutations identified in subjects with HFM are underlined.
DISCUSSION
Random mutagenesis has been employed to identify important residues in membrane transporters and channels (21–23). Any part of a gene, or an entire gene, can be subjected to mutagenesis. The hallmark of this method is the generation of mutations in an unbiased manner. A library of mutants can be readily generated; subsequent screening and characterization of the mutant proteins are the limiting steps. In the current study, 144 PCFT mutant clones generated by random mutagenesis were screened. However, in only 25 (17%) was PCFT activity decreased or abolished. Based upon an average of four pcft mutations per clone, as indicated in Tables 2 and 3, a total of 576 pcft mutations would be generated in 144 clones. Yet, only 26 mutations (4.5%) were identified that impacted on PCFT function. The actual percentage may be higher because a second loss-of-function mutation could have been present, but not characterized once the first loss-of-function mutation was identified in a clone. These results indicate that a random mutation in the pcft gene has a very low chance (∼5%) of inactivating PCFT. This finding is consistent with the fact that HFM is a rare autosomal recessive disorder in which both pcft gene copies must be mutated to produce sufficient folate deficiency to result in the clinical syndrome.
Four pcft mutations identified in the current study were the same as found in patients with HFM. This represents 15% of the mutations identified in the current study and 25% of all HFM mutations reported to date. p.G65Afs was identified in a male child of African-American parents (2), whereas p.C66Lfs was found in a subject with consanguineous parents of Pakistani origin (10). One of the missense mutations, p.S318R, was identified as a compound heterozygous mutation in a child of Mexican origin (2). The other missense mutation, p.R376Q, was identified in a child of consanguineous Chinese parents living in Australia (6). The significant overlap between mutations found in the current study and subjects with HFM indicates a high mutation susceptibility at these sites raising the possibility that mutations identified in the current study may be found associated with HFM as more cases are identified and characterized in the future.
With the addition of nine new base deletion or insertion mutations, it is possible to re-examine whether there are mutational hot spots in the pcft gene. Deletion or insertion of bases with a number other than three always leads to a frameshift in translation, independent of the composition of flanking nucleotides. However, 11 of 13 deletion or insertion mutations were localized in the first 335 of 1350 bases subjected to random mutagenesis, clearly indicating a non-random event. There were two regions dense in these mutations (Fig. 5). One is between bases 177 and 205 (c.177–205 region), which contains four such mutations and one base substitution. The other is an even narrower region between bases 4 and 10 (c.4–17 region), which contains five deletion or insertion mutations and is next to the two-base substitution mutations at the initiation codon. Repetitive DNA sequences may play an important role in the propensity of base deletions or insertions (24). Hence, c. 4delG, c. 4–5insG, c. 9–10insG, and c. 10delA mutations are located in a region rich in G (GGAGGGGAG), whereas c. 17–18insC occurs in a poly(C)6 stretch. Similarly, both c. 194delG and c. 194–195insG take place at a poly(G) (7) sequence. However, a repetitive sequence is unlikely to be the only factor because there are two five-base repeats, GGGGG (c.495–499) and TTTTT (c.395–399), in the other part of the pcft gene, and neither a deletion nor insertion mutation was identified at these spots.
Under normal circumstances, translation of proteins starts at the first ATG codon (AUG in mRNA) of the open reading frame. However, a growing number of mammalian genes have been characterized that initiate translation at non-ATG sites (19). One example is another folate-related protein, the enzyme dihydrofolate reductase. When one nucleotide of the ATG-codon was substituted, translation still started at the same site, although at lower efficiency, with methionine incorporated at the first amino acid (25). Very low protein expression, but comparably low function, in the case of ATA initiation of transcription suggests that wild-type PCFT has been synthesized under these conditions, similar to what has been observed for dihydrofolate reductase. It is interesting to note that non-ATG translation did not take place at all when the ATG initiation codon was replaced with a stop codon TGA (Fig. 2A). However, it is unclear as to why the second or downstream ATG codon of pcft was not utilized in this situation since no protein at lower molecular weight was detected (Fig. 2B). Only when the nucleotides upstream of the second AUG (including the first ATG codon) were deleted was the second AUG initiation codon used. However, the encoded protein lacking the first 74 amino acid residues was not functional. These results suggest that translational machinery of the pcft gene can recognize a slightly mutated ATG codon (to ATA or CTG in clone 1–70) but stops at this location if ATG is replaced with a stop codon (TGA).
A spectrum of molecular mechanisms underlying loss-of-function missense mutations were elucidated in the current study. Three mutant PCFTs, M222R, V134E, and L31P resulted in the lack of, or markedly decreased, protein expression. Among the mutants that were expressed and trafficked to the plasma membrane, E232G appeared to have a defect in the ability of the carrier to undergo the required conformational changes associated with substrate transport. In fact, the affinity of this mutant for MTX was markedly increased, a pattern that has been observed with other mutant PCFTs usually accompanied by a fall in Vmax (20). The I304F mutant had a defect in MTX binding (slightly decreased influx Vmax and a larger increase in influx Kt). L161R PCFT had an even greater defect in binding and also a substantial reduction in influx Vmax. The double mutants, F426V/L432H, represent a unique case in which each single mutation was functional but the double mutation resulted in the loss of protein. Based upon the location of Phe-426 and Leu-432 within the helical structure of the 12th transmembrane domain, it is unlikely that these residues are in proximity or interact. More likely, a single mutation results in a transporter that, whereas fully functional, lacks the capacity to sustain the stress of a second mutation. This is similar to what is observed for the Cys-less PCFT, which is also fully functional, but is vulnerable to an additional mutation that, alone, is silent (26).
Results obtained in the current study provide a basis for further analysis of PCFT structure-function relationships. For example, when all the missense mutations are indicated on the topological structure of PCFT (27)(Fig. 5B), it can be seen that the fourth and ninth transmembrane domains contain the most (three) mutations. This suggests that these regions play an important role in PCFT transport function and warrant further structure-function analyses. PCFT mutants, E232G, L161R, and I304F, were all expressed on the plasma membrane and have altered affinity for MTX. Future studies will determine whether the Glu-232, Leu-161, and Ile-304 residues are in, or near, the folate binding pocket.
This work was supported, in whole or in part, by National Institutes of Health Grant CA-82621.
- PCFT
- proton-coupled folate transporter
- HFM
- hereditary folate malabsorption
- MTX
- methotrexate.
REFERENCES
- 1. Qiu A., Jansen M., Sakaris A., Min S. H., Chattopadhyay S., Tsai E., Sandoval C., Zhao R., Akabas M. H., Goldman I. D. (2006) Cell 127, 917–928 [DOI] [PubMed] [Google Scholar]
- 2. Zhao R., Min S. H., Qiu A., Sakaris A., Goldberg G. L., Sandoval C., Malatack J. J., Rosenblatt D. S., Goldman I. D. (2007) Blood 110, 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Geller J., Kronn D., Jayabose S., Sandoval C. (2002) Medicine 81, 51–68 [DOI] [PubMed] [Google Scholar]
- 4. Mahadeo K., Min S. H., Diop-Bove N. K., Kronn D., Goldman I. D. (2010) GeneReviews [Internet], updated 2010 May 06, University of Washington, Seattle, WA [Google Scholar]
- 5. Min S. H., Oh S. Y., Karp G. I., Poncz M., Zhao R., Goldman I. D. (2008) J. Pediatr. 153, 435–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mahadeo K., Diop-Bove N., Shin D., Unal E. S., Teo J., Zhao R., Chang M. H., Fulterer A., Romero M. F., Goldman I. D. (2010) Am. J. Physiol. Cell Physiol. 299, C1153–C1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shin D. S., Min S. H., Russell L., Zhao R., Fiser A., Goldman I. D. (2010) Blood 116, 5162–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Atabay B., Turker M., Ozer E. A., Mahadeo K., Diop-Bove N., Goldman I. D. (2010) Pediatr. Hematol. Oncol. 27, 614–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Borzutzky A., Crompton B., Bergmann A. K., Giliani S., Baxi S., Martin M., Neufeld E. J., Notarangelo L. D. (2009) Clin. Immunol. 133, 287–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meyer E., Kurian M. A., Pasha S., Trembath R. C., Cole T., Maher E. R. (2010) Mol. Genet. Metab. 99, 325–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lasry I., Berman B., Straussberg R., Sofer Y., Bessler H., Sharkia M., Glaser F., Jansen G., Drori S., Assaraf Y. G. (2008) Blood 112, 2055–2061 [DOI] [PubMed] [Google Scholar]
- 12. Shin D. S., Mahadeo K., Min S. H., Diop-Bove N., Clayton P., Zhao R., Goldman I. D. (2011) Mol. Genet. Metab. 103, 33–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mahadeo K., Diop-Bove N., Ramirez S. I., Cadilla C. L., Rivera E., Martin M., Lerner N. B., DiAntonio L., Duva S., Santiago-Borrero P. J., Goldman I. D. (2011) J. Pediatr., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lasry I., Berman B., Glaser F., Jansen G., Assaraf Y. G. (2009) Biochem. Biophys. Res. Commun. 386, 426–431 [DOI] [PubMed] [Google Scholar]
- 15. Zhao R., Gao F., Hanscom M., Goldman I. D. (2004) Clin. Cancer Res. 10, 718–727 [DOI] [PubMed] [Google Scholar]
- 16. Diop-Bove N. K., Wu J., Zhao R., Locker J., Goldman I. D. (2009) Mol. Cancer Ther. 8, 2424–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao R., Chattopadhyay S., Hanscom M., Goldman I. D. (2004) Clin. Cancer Res. 10, 8735–8742 [DOI] [PubMed] [Google Scholar]
- 18. Unal E. S., Zhao R., Goldman I. D. (2009) Am. J. Physiol. Cell Physiol. 297, C66–C74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Touriol C., Bornes S., Bonnal S., Audigier S., Prats H., Prats A. C., Vagner S. (2003) Biol. Cell 95, 169–178 [DOI] [PubMed] [Google Scholar]
- 20. Unal E. S., Zhao R., Chang M. H., Fiser A., Romero M. F., Goldman I. D. (2009) J. Biol. Chem. 284, 17846–17857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Penado K. M., Rudnick G., Stephan M. M. (1998) J. Biol. Chem. 273, 28098–28106 [DOI] [PubMed] [Google Scholar]
- 22. Shibagaki N., Grossman A. R. (2006) J. Biol. Chem. 281, 22964–22973 [DOI] [PubMed] [Google Scholar]
- 23. Loukin S. H., Vaillant B., Zhou X. L., Spalding E. P., Kung C., Saimi Y. (1997) EMBO J. 16, 4817–4825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lovett S. T. (2004) Mol. Microbiol. 52, 1243–1253 [DOI] [PubMed] [Google Scholar]
- 25. Peabody D. S. (1989) J. Biol. Chem. 264, 5031–5035 [PubMed] [Google Scholar]
- 26. Zhao R., Shin D. S., Goldman I. D. (2011) Biochim. Biophys. Acta 1808, 1140–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhao R., Unal E. S., Shin D. S., Goldman I. D. (2010) Biochemistry 49, 2925–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Subramanian V. S., Marchant J. S., Said H. M. (2008) Am. J. Physiol. Cell Physiol. 294, C233–C240 [DOI] [PubMed] [Google Scholar]