Abstract
Structural studies showed that Dnmt3a has two interfaces for protein-protein interaction in the heterotetrameric Dnmt3a/3L C-terminal domain complex: the RD interface (mediating the Dnmt3a-3a contact) and the FF interface (mediating the Dnmt3a-3L contact). Here, we demonstrate that Dnmt3a-C forms dimers via the FF interface as well, which further oligomerize via their RD interfaces. Each RD interface of the Dnmt3a-C oligomer creates an independent DNA binding site, which allows for binding of separate DNA molecules oriented in parallel. Because Dnmt3L does not have an RD interface, it prevents Dnmt3a oligomerization and binding of more than one DNA molecule. Both interfaces of Dnmt3a are necessary for the heterochromatic localization of the enzyme in cells. Overexpression of Dnmt3L in cells leads to the release of Dnmt3a from heterochromatic regions, which may increase its activity for methylation of euchromatic targets like the differentially methylated regions involved in imprinting.
Keywords: Chromatin, DNA Enzymes, DNA Methylation, Protein DNA-interaction, Protein-Protein Interactions
Introduction
The coordinated gene expression during development and cell differentiation is regulated by epigenetic signals comprising DNA methylation, post-translational modification of histone tails, and non-coding RNAs (1). Among them, DNA methylation is a key epigenetic process involved in the control of gene activity (2–3), parental imprinting (4–6), X-chromosome inactivation (7–10), and maintenance of the genome integrity and stability through protection against endogenous retroviruses and transposons (11). Aberrant DNA methylation patterns are associated with several human diseases, including cancer (12–17). In mammals, DNA methylation patterns are established during embryogenesis by the action of the de novo DNA methyltransferases Dnmt3a and Dnmt3b, together with their regulatory factor, Dnmt3L (18–19). In addition, the Dnmt3a and Dnmt3b enzymes have a role in the preservation of DNA methylation at heterochromatin regions (20–21). Disruption of the Dnmt3a or Dnmt3b gene in mice is lethal (22), whereas Dnmt3L knock-out mice are viable and do not show discernable morphological abnormalities (23–25), indicating that Dnmt3a and Dnmt3b are functional in the absence of Dnmt3L.
Dnmt3a and Dnmt3b are nuclear proteins, which stably associate with chromatin containing methylated DNA (21), including mitotic chromosomes, and localize to pericentromeric heterochromatin (26–28). Dnmt3 proteins comprise two parts: a large multidomain N-terminal part of variable size, which has regulatory and targeting functions and a C-terminal catalytic part. The N-terminal part of Dnmt3a and Dnmt3b contains two defined domains: a cysteine-rich ADD (ATRX–DNMT3–DNMT3L) domain, which binds to H3 tails unmodified at K4 (29–30) and a PWWP domain, which binds to H3 trimethylated at K36 (31) and is essential for the heterochromatin localization of the enzymes (27–28, 31). The C-terminal parts of Dnmt3 enzymes resemble prokaryotic DNA-(cytosine C5)-methyltransferases and harbor the active centers of the enzymes and are active in an isolated form (32). Dnmt3L is catalytically inactive, directly interacts with the catalytic domains of Dnmt3a and Dnmt3b and stimulates their activity both in vivo and in vitro (25, 33–37).
The structure of a complex consisting of the C-terminal parts of Dnmt3a and Dnmt3L (Dnmt3a-C/Dnmt3L-C) revealed a linear heterotetramer, in which the two Dnmt3L subunits are positioned at the edges and two Dnmt3a molecules in the center (Fig. 1A) (38–39). The Dnmt3a C-terminal domain provides two interfaces for protein-protein contacts: one hydrophobic FF interface at the Dnmt3a-C/3L-C contact formed by the stacking interaction of four phenylalanine residues (two from each subunit) and one polar RD interface at the Dnmt3a-C/3a-C contact mediated by a hydrogen bonding network between arginine and aspartate residues. The RD interface forms the DNA binding site, while both interfaces are essential for cofactor S-adenosyl-l-methionine binding and catalytic activity of the enzyme (38, 40). We demonstrate here that Dnmt3a-C oligomerizes in a reversible manner. Initial dimer formation occurs through the FF interface, further oligomerization via the RD interface of the FF dimers. Because each RD interface constitutes an independent DNA binding site, Dnmt3a-C oligomers can bind to two or more DNA molecules, oriented next to each other, roughly in parallel. Dnmt3L prevents this oligomerization, because it lacks the RD interface, and it therefore precludes binding of Dnmt3a to more than one DNA molecule. The formation of Dnmt3a oligomers is required for the localization of the enzyme to heterochromatin, and binding of Dnmt3L changes the subnuclear localization of Dnmt3a, which could be one mechanism describing how Dnmt3L stimulates the methylation of imprinted loci.
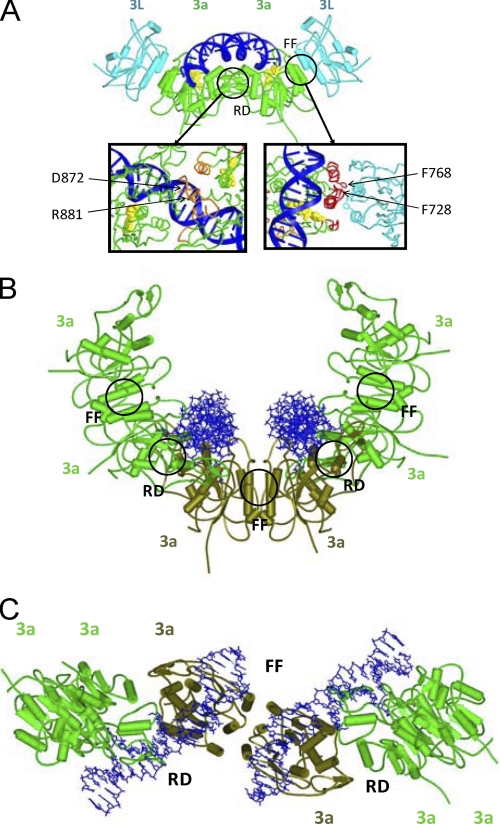
FIGURE 1.
Models of Dnmt3a-C/3L-C heterotetramer and Dnmt3a-C oligomers in complex with DNA. A, model of the Dnmt3a/Dnmt3L C-terminal domain tetramer, colored in green for Dnmt3a-C and in cyan for Dnmt3L-C. The cofactor S-adenosyl-l-methionine is shown in yellow and the modeled DNA in blue. The close up of the FF and RD interfaces is shown with the relevant amino acid residues labeled. B and C, modeling of Dnmt3a-C oligomers formed by the interaction of FF-dimers via their RD interface. The figure shows a hexameric complex as an example; the three FF-dimeric building blocks are colored light green and dark olive green. The two DNA molecules bound at the RD interfaces in the Dnmt3a-C hexamer are oriented roughly in parallel. Panel B shows a view along the axis of the bound DNA molecules, panel C shows a view perpendicular to the axis of the Dnmt3a oligomer.
EXPERIMENTAL PROCEDURES
Mutagenesis, Protein Expression, and Protein Purification
The sequences encoding C-terminal domains of the murine Dnmt3a (residues 608–908) and Dnmt3L (residues 208–421) were subcloned as N-terminal His6 fusion proteins into pET-28a vector (Novagen) (32, 36). Selected Dnmt3a-C and Dnmt3L-C variants were generated using the megaprimer site-directed mutagenesis method as described previously (41). Introduction of the mutations was confirmed by restriction marker site analysis and DNA sequencing. Protein expression was carried out in Escherichia coli BL21 (λDE3) pLysS (Novagen). Cells were grown at 32 °C, induced with 1 mm IPTG at A600 nm of approximately 0.5 and harvested after 3 h. The proteins were purified at high μm concentrations using Ni-NTA-agarose. Each protein was purified at least twice, and the purity of the preparations was estimated to be greater than 98% (supplemental Fig. S4A). The concentrations of the proteins were determined by UV spectrophotometry using extinction coefficients of 39290 m−1 cm−1 and 40330 m−1 cm−1 for Dnmt3a-C and Dnmt3L-C, respectively. Concentration determinations were confirmed by densitometric analysis of Coomassie-stained SDS-polyacrylamide gels. The folding of the mutant proteins was verified by circular dichroism spectroscopy as described (42), indicating that the secondary structure of the R881A variant was indistinguishable from wild-type Dnmt3a-C. The F728 variants were both folded as demonstrated by the strong negative ellipticity at 220 nm, which is indicative of α-helix content (supplemental Fig. S4B). Both F728 variants displayed a small but significant shift in the ascending limb of the spectrum around 200 nm, suggesting some deviations from the wt folding. The aggregation of Dnmt3a-C shown in Fig. 2A and supplemental Fig. S1A and recovery of enzyme activity from aggregated Dnmt3a-C was only done for illustration. All other experiments reported in this study were conducted with Dnmt3a-C proteins that never went through aggregation, because they were purified at lower protein concentration.
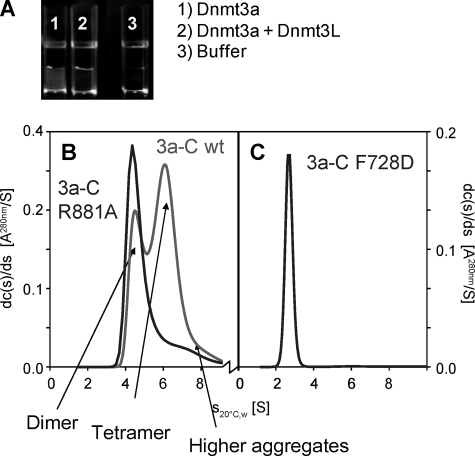
FIGURE 2.
Oligomerization of Dnmt3a-C. A, reversible aggregation of Dnmt3a-C during protein purification is prevented in the presence of Dnmt3L-C. The figures show samples of Dnmt3a-C (40 μm) or Dnmt3a-C/3L-C (40 μm each) after reduction of salt concentration from 500 mm KCl to 200 mm KCl by 2 h of dialysis. B and C, differential sedimentation coefficient (S20 °C,W) distributions obtained from sedimentation velocity experiments in the analytical ultracentrifuge with Dnmt3a-C wt (2B, red), R881A (2B, blue) and F728D (2C) mutants.
Analytical Ultracentrifugation
Analytical ultracentrifugation was done with an An50-Ti 8-place rotor in a Beckman-Coulter model XL-A centrifuge equipped with UV absorption optics. Sedimentation velocity experiments were run at 4 °C and 50 krpm in a buffer containing 10 mm HEPES pH 7.2, 0.2 m KCl, 1.34 m glycerol (10%), 1 mm EDTA, and 0.2 mm DTT. They were evaluated using the program SEDFIT, which transforms the measured data into a diffusion corrected differential sedimentation coefficient distribution (43–44). For every sedimenting species the sedimentation constant was read off the corresponding maximum of this distribution. All distributions were corrected to water at 20 °C using density and viscosity data and partial specific volumes incorporated into SEDNTERP.4
Fluorescence Spectroscopy
Fluorescence spectroscopy was carried out in a Cary Eclipse fluorimeter (Varian) using a double-labeled two flank oligonucleotide substrate obtained by annealing of three individual oligonucleotides, one 50mer that could hybridize with two 20mers (one of them labeled by Cy3, one by Cy5): 50mer: 5′-TGC ACT CTC CTC CCG GAA GTG TTC TTC GTA GAC TGC GGC ACA GTT TTTTG-3′; Cy3–20-L: 5′-ACT TCC GGG AGG AGA GTG CA-3′-Cy3; Cy5–20-R: Cy5–5′-CAA AAA ACT GTG CCG CAG TC-3′. Control substrates only containing the Cy3 or Cy5 probe were generated using unlabeled versions of the 20mers. 20-L: 5′-ACT TCC GGG AGG AGA GTG CA-3′; 20-R: 5′-CAA AAA ACT GTG CCG CAG TC-3′.
The DNA substrate used for Fluorescence Resonance Energy Transfer (FRET)5 experiments was obtained by annealing of the 50mer with one 20-L and one 20-R oligonucleotide. It provides two double-stranded 20 bps stretches, which are separated by a 10 nucleotide single-stranded part, and contain either two (Cy3 and Cy5) or one fluorophore (only Cy3 or only Cy5) (Sequence 1).
SEQUENCE 1.

FRET DNA Substrate.
The FRET DNA was annealed by mixing all three individual strands at equal concentration (20 μm), heating to 86 °C for 5 min, and slowly cooling to ambient temperature. The annealed substrate was purified using Qiagen nucleotide purification kit. FRET experiments were carried out in the presence and absence of Dnmt3a-C proteins in buffer (20 mm HEPES pH 7.5, 1 mm EDTA, 100 mm KCl) containing 100 nm FRET DNA. For the protein-bound sample, 200 nm Dnmt3a-C proteins complex were added and incubated with the DNA at ambient temperature for 20 min to allow for complex formation. The 3a/3L complex was prepared by incubation of Dnmt3a-C and Dnmt3L-C (final concentration of 200 nm each) on ice for 20 min prior to the incubation with DNA. FRET was studied by using Cy3 excitation at 540 nm. Cy5 excitation in control reaction was at 640 nm. Emission and excitation slits were always set at 5 nm. In all experiments, 5 separate scans were taken and averaged. All experiments were carried out at least in duplicate; usually 3–4 repeats were performed.
Scanning-force Microscopy
Scanning force microscopy (SFM) experiments were carried out using a 509mer DNA fragment derived from the human CG island upstream of the human SUHW1 gene and purified catalytic domains of Dnmt3a. DNA-protein filaments were generated in a total volume of 30 μl by incubating 12 nm DNA with 200 nm Dnmt3a protein in 50 mm HEPES (pH 7.5), 250 mm NaCl, 1 mm EDTA, and 100 μm of sinefungin (Sigma). Samples were incubated for 1 h at room temperature to allow for DNA binding. 1 μl of the complex solution was mixed with 9 μl of 10 mm MgCl2 solution and deposited on freshly cleaved mica (Plano, Wetzlar), allowed to adhere for 40 s and then washed with 1 ml of bi-distilled water. The sample was then dried using compressed air. Protein-DNA filaments where observed by tapping mode in air using a Multimode AFM with a Nanoscope III controller (Digital instruments, Santa Barbara, CA). We used NSTNCHF silicon cantilevers (Nascatec, Stuttgart) with a nominal spring constant of 50 N/m and a resonance frequency of ∼150 kHz. All images were obtained with a scanning speed of 0.75–1 Hz and a resolution of 512 × 512 pixels. Experiments were carried out blinded, i.e. the operator of the microscope did not know the data obtained in prior experiments. To remove background slope, raw images were flattened using the Nanoscope software. DNA protein complexes were regarded as a filament if the height exceeded 150% of the height observed for DNA molecule alone and were at least 20 nm wide. Filaments were evaluated using the section tool of the Nanoscope V6r12 software.
Cell Culture and Laser Scanning Microscopy
The expression construct for murine full length Dnmt3a fused to yellow fluorescent protein (YFP) was obtained from Dr. G. Xu (Shanghai) (45). To prepare a cyan fluorescent protein (CFP) fusion of Dnmt3L, human full-length Dnmt3L (AP001753) was cloned into the pECFP-C1 vector (Invitrogen), using XhoI and EcoRI sites. The interface variants of full length Dnmt3a and Dnmt3L were created in a similar way as the interface variants of the C-terminal domains (see above). NIH3T3 cells were grown in DMEM with 10% (v/v) fetal calf serum and 2 mm l-glutamine at 37 °C in 5% (v/v) CO2. Cells (1–2 × 105) were transfected in 6-well plates using FuGENE 6 (Roche, Basel, Switzerland; 1 μg of total plasmid DNA per well). In co-transfection experiments equal amounts of Dnmt3a-YFP and Dnmt3L-CFP encoding plasmids were used. Transfected NIH3T3 cells were fixed in 4% (w/v) paraformaldehyde. Confocal images were taken using a Carl Zeiss LSM510 (Jena, Germany; software version 3.0). For the analysis of the co-transfection experiments, the observed localization pattern of Dnmt3a was divided into three categories: spotty, spotty plus diffused and diffused; 100, 54, or 73 cells were analyzed for the Dnmt3a wt, Dnmt3a-Dnmt3L wt, and Dnmt3a-Dnmt3L F261D mutant, respectively.
RESULTS
Oligomerization of Dnmt3a-C Depends on Both Interfaces and Is Disrupted by Dnmt3L-C
We have noticed for several years, that Dnmt3a-C tends to aggregate during purification at the dialysis step when the salt concentration is reduced from 500 to 200 mm KCl if the protein concentration is too high (Fig. 2A and supplemental Fig. S1A). Recently, we observed that the aggregated enzyme preparations could be re-dissolved after dilution and then display almost 80% of the catalytic activity of an enzyme preparation that never went through aggregation (supplemental Fig. S1B). The observation that recovery of enzyme activity did not require refolding indicates that the aggregation process is reversible, and it probably occurs through native protein/protein interfaces. The structure of the Dnmt3a-C/3L-C tetramer shows that Dnmt3a-C has two such interfaces (Fig. 1A), the FF and RD interface. The conservation of key residues of the FF interface between all members of the Dnmt3 family (supplemental Fig. S2), as well as modeling, suggested that Dnmt3a could replace Dnmt3L at the Dnmt3a-3L interface (the FF interface) to form another Dnmt3a/3a contact. Hence, the C-terminal domain of Dnmt3a possesses two putative interfaces for self-interaction, leading to the possibility of forming linear Dnmt3a-C oligomers (Fig. 1, B and C and supplemental Fig. S2B). Dnmt3a-C precipitation experiments in the presence of Dnmt3L-C showed that Dnmt3L-C considerably reduced aggregation of Dnmt3a-C (Fig. 2A and supplemental Fig. S1A). The amino acid sequence alignment of Dnmt3a and Dnmt3L shows that Dnmt3L-C lacks an RD interface (supplemental Fig. S2A), suggesting that the RD interface plays an important role in the reversible aggregation of Dnmt3a-C. This conclusion is also in agreement with the salt dependence of the aggregation, because the RD interface is dominated by ionic contacts which strengthen during reduction of salt concentration.
Using analytical ultracentrifugation, we investigated in detail the oligomeric state of the purified wild type Dnmt3a-C that never went through aggregation. The experiment revealed one fraction sedimenting with an S20 °C,water-value of 4.5–4.6 S (Fig. 2B), which corresponds to a dimer with a frictional ratio of 1.35–1.4. The frictional ratio of a folded protein lies between ∼1.2 for a globular structure and 1.5–1.7 for extended or elongated structures (46). A second, major fraction of Dnmt3a-C sedimented at higher S values (∼6–6.2 S) corresponding to a Dnmt3a-C tetramer with elongated structure (frictional ratio 1.6 ± 0.3). Higher aggregates were also observed, depending on the initial concentration of the preparation. In contrast, in ultracentrifugation experiments with purified Dnmt3a-C/3L-C complex at comparable concentrations only the 2:2 heterotetramer of ∼5.4 S was detected (40), indicating that Dnmt3L-C prevents oligomerization of Dnmt3a-C. These observations are in agreement with published data obtained with Dnmt3a2, an isoform of Dnmt3a also containing most of the N-terminal part, showing that Dnmt3A2 forms large structures of heterogeneous sizes, although binding of Dnmt3L to Dnmt3A2 leads to the formation of specific heterotetrameric complexes (37). To analyze the potential self-interaction of Dnmt3L-C via its FF interface, purified Dnmt3L-C was also studied by analytical ultracentrifugation and showed a sedimentation constant of 2.35 S, which corresponds to a monomeric state with a frictional ratio of 1.35 (supplemental Fig. S3).
To study the role of the interfaces in Dnmt3a function, we mutated the Arg-881 at the RD interface to alanine (R881A) and the Phe-728 at the FF interface to alanine (F728A) and to aspartic acid (F728D), assuming that the F to D exchange, which introduces a charged residue instead of a hydrophobic one, would result in a stronger phenotype than the F to A mutation. The protein variants were expressed, purified and their folding confirmed by circular dichroism spectroscopy (supplemental Fig. S4). As expected, DNA binding experiments showed that the RD interface constitutes the DNA binding site of Dnmt3a-C homooligomers, because the R881A exchange abrogated DNA binding, while the F728A and F728D variants bound DNA with even slightly better affinity than the wild type enzyme (supplemental Fig. S5). These results replicate previous observations made with the corresponding heterotetrameric Dnmt3a-C/3L-C complexes (40). We have also shown before that all three Dnmt3a-C variants are catalytically inactive (40).
In the ultracentrifugation experiments, the Dnmt3a-C R881A mutant retained its ability to dimerize (S20 °C,water = 4.3–4.4 S) (Fig. 2B). However, tetramers as observed for the wild type protein were not detected. The dimeric state of the R881A mutant demonstrates that the FF interface is fully functional and mediates dimerization of Dnmt3a. In contrast, the F728D mutation converted the protein into a monomer (S20 °C,water = 2.6–2.7 S, frictional ratio 1.5, Fig. 2C), indicating that the intact RD interface does not support self-interaction in absence of DNA. The efficient DNA binding of the FF variants suggests that dimerization across the RD interface can occur in presence of DNA, because DNA binding requires the cooperation of both subunits at the RD interface (38). The Dnmt3a-C F728A variant behaved as a dimer (data not shown), indicating that the F to A exchange did not fully disrupt the FF interface, such that dimerization was still possible under the conditions of the analytical ultracentrifugation, which was performed at relatively high protein concentrations.
Taken together, we show that the FF interface is the primary interface for Dnmt3a-C homodimerization, because the R881A mutant retained its ability to dimerize (via the FF interface), whereas the F728D mutation converted the dimer into a monomer. This finding is in agreement with the structure, because the FF interface is hydrophobic while the RD interface is hydrophilic in nature, and hydrophobic protein/protein interfaces are known to be more stable (47–48). Further reversible oligomerization of Dnmt3a-C is mediated by the salt-dependent interaction of Dnmt3a-C FF dimers via their RD interfaces. This process is prevented by Dnmt3L-C, because it does not contain an RD interface. The requirement for the Dnmt3a-C homodimer to first dissociate before Dnmt3L-C/3a-C complexes can form also explains the long preincubation times of separately purified Dnmt3a-C and Dnmt3L-C needed to reach a maximum stimulation of Dnmt3a-C activity (37). The formation of Dnmt3a-C/3L-C complexes is supported by the monomeric state of Dnmt3L-C.
Binding of Parallel DNA Molecules by Dnmt3a-C Oligomers
We modeled the structure of a Dnmt3a-C/3a-C contact via the FF interface by superimposing a Dnmt3a-C subunit onto one of the Dnmt3L-C molecules of the Dnmt3a-C/3L-C heterotetramer structure. This superposition can be done with good confidence, because of the strong structural and amino acid sequence similarity between Dnmt3a-C and Dnmt3L-C. By combining these Dnmt3a-C/3a-C FF dimers obtained by modeling with Dnmt3a-C/3a-C dimers formed via the RD interface taken from the Dnmt3a-C/3L-C structure, a model of a Dnmt3a-C hexamer containing three FF and two RD interfaces could be generated. In principle, this hexamer could be further extended. Because each RD interface constitutes an independent DNA binding site, such a hexamer could bind two DNA molecules (Fig. 1, B and C). Interestingly, the modeling suggests that the DNA binding sites are oriented roughly in parallel to each other in a distance of ∼3–4 nm, which would not allow for one DNA molecule to bind side by side to both sites simultaneously. Instead, binding of two DNA molecules oriented roughly in parallel to each other could occur (Fig. 1, B and C and supplemental Fig. S2B).
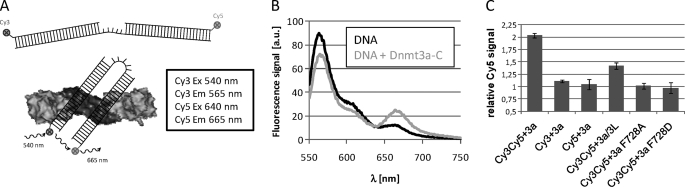
To study the potential binding of two parallel DNA molecules by the Dnmt3a-C oligomer in solution, we developed a FRET assay, using a DNA substrate that consists of two 20 base pair double-stranded regions separated by a 10 nucleotide single-stranded region, which are flexible. The ends of the DNA were labeled with fluorophores that constitute a FRET donor/acceptor pair (Cy3/Cy5 with a Förster radius of about 56 Å). In the annealed oligonucleotide, the average distance of both ends is large and does not permit FRET. However, after binding of the FRET substrate to adjacent RD interfaces of a Dnmt3a-C oligomer, the ends should approach each other, and FRET could be established (Fig. 3A). Indeed, with the free DNA only a small Cy5 signal was observed after excitation of Cy3 at 540 nm (Fig. 3B). This signal does not correspond to FRET, but could be attributed to direct excitation of Cy5 (data not shown). However, after addition of Dnmt3a-C to the FRET substrate, we observed a strong increase in Cy5 fluorescence after Cy3 excitation, which is indicative of FRET (Fig. 3B). Control experiments confirmed that an increase in Cy5 fluorescence after addition of Dnmt3a-C was not observed after direct excitation of Cy5 (data not shown) and with identical substrates that only contained the Cy3 or Cy5 probe (Fig. 3C). These controls confirm that FRET is detected and parallel DNA binding occurs. Next we investigated the effect of Dnmt3L-C on parallel DNA binding of Dnmt3a-C. A strongly reduced FRET signal was seen after addition of a Dnmt3a-C/3L-C complex when compared with Dnmt3a-C at same concentration. The Dnmt3a-C/3L-C complex was generated by preincubation of Dnmt3a-C and Dnmt3L-C. Under these conditions, it is unlikely that all Dnmt3a-C oligomers are resolved, which explains the residual FRET effect that was still observed. The FRET signal was also lost with the Dnmt3a-C mutants that disrupt the FF interface, such that DNA binding is possible, but no oligomerization of Dnmt3a-C occurs (Fig. 3C). These results confirm that Dnmt3a-C oligomers can bind to two DNA molecules oriented in parallel. Dnmt3L-C or mutations at the FF interface of Dnmt3a-C disrupt the oligomers and, thereby, prevent parallel binding of more than one DNA molecule.
FIGURE 3.
Analysis of binding of parallel DNA molecules by Dnmt3a-C oligomers. A, schematic setup of the FRET assay. In free form, the distance of both ends of the FRET substrate is too large for FRET to occur. After binding of the two double-stranded regions to adjacent DNA binding sites in a Dnmt3a-C oligomer, the ends approach each other and FRET can be established. B, fluorescence emission spectra of the free FRET DNA (black line) and of FRET DNA-Dnmt3a-C complexes (grey line) after Cy3 excitation. FRET is indicated by the strong increase in Cy5 emission at 665 nm in the presence of Dnmt3a-C. C, Cy5 emission of the FRET DNA after addition of Dnmt3a-C, Dnmt3a-C interface variants or Dnmt3a-C/3L-C complex divided by the corresponding Cy5 emission of the FRET obtained for free DNA. In addition, as a control, experiments with single labeled FRET DNA (either only carrying Cy3 or Cy5 label) were carried out, but no signal change was observed after addition of Dnmt3a-C. Excitation was at 540 nm. Error bars represent the error of the mean derived from the variation of repeated experiments.
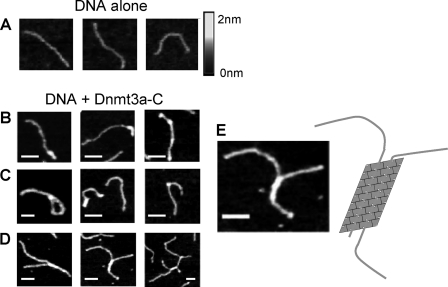
Next, we studied the binding of long DNA molecules to Dnmt3a-C oligomers by SFM. We have shown previously that the Dnmt3a-C/3L-C complex polymerizes on DNA, using purified Dnmt3a-C and a 509 bp PCR product, which can be visualized accurately by SFM (40). We now observed that Dnmt3a-C forms DNA-nucleoprotein filaments in the absence of Dnmt3L as well (Fig. 4). However, additional structural features like sharp curvatures and lariats were also seen with Dnmt3a-C, indicating that protein bound stretches from different parts of a long single DNA molecule can interact after forming DNA loops (Fig. 4C). Interestingly, about one third of the observed complexes contained two DNA molecules connected by more or less extended protein-DNA filaments (Fig. 4D). These connected synaptonemal-like complexes were probably formed by a direct interaction between Dnmt3a-C molecules bound to different DNA molecules oriented in parallel (Fig. 4E). Such complexes were not observed in the previous Dnmt3a-C/3L-C experiments. They document the ability of Dnmt3a-C oligomers to bind simultaneously to two DNA molecules oriented in parallel.
FIGURE 4.
Scanning force microscopy of Dnmt3a-C-DNA filaments. A, images showing free DNA. B–D, images showing examples of Dnmt3a-DNA filaments. Regular filaments (B), structures like lariats and sharp curvatures (C), as well as branched, synaptonemal-like complexes (D) can be observed. E, magnification of an exemplary picture from D and a schematic representation of a DNA oligomer bound to two parallel DNA molecules. The scale bars correspond to 50 nm.
Subnuclear Localization of Dnmt3a Depends on Both Interfaces and Is Altered by Dnmt3L
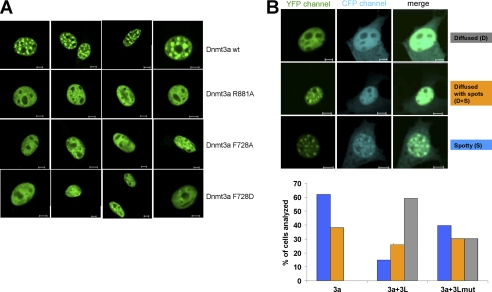
It had been shown before that Dnmt3a localizes to heterochromatic foci in mouse cell lines (26–28, 31) and that this effect depends on both the PWWP and the catalytic domain of Dnmt3a (27–28, 31). To investigate if oligomerization of Dnmt3a has an effect on its cellular and subnuclear localization, we studied the cellular distribution of YFP-tagged full-length Dnmt3a and its interface mutants R881A, F728A, and F728D in NIH 3T3 cells. As expected, wild type Dnmt3a was located exclusively in subnuclear spots (Fig. 5 and supplemental Figs. S6 and S7), which were previously shown to co-localize with pericentromeric heterochromatin and reflect the localization of the endogenous enzyme (26–28, 31). The F728A, F728D, and R881A variant fusion proteins showed similar expression levels as the wild type protein (as judged by the intensity of the fluorescence signal) and exclusive nuclear localization as well. However, all variants displayed a great change in the sub-nuclear localization pattern. Instead of the enrichment in distinct heterochromatic spots characteristic of the wild-type protein, a diffuse staining was observed, with the fluorescence signal almost uniformly distributed within the nucleus (Fig. 5A). These results indicate that the ability of Dnmt3a to bind to heterochromatic regions was lost or greatly reduced with the interface mutants indicating that both interfaces are required for the stable localization of Dnmt3a to the heterochromatic foci. A similar change in localization was observed after complete removal of the catalytic domain of Dnmt3a (27).
FIGURE 5.
Subnuclear localization of Dnmt3a and Dnmt3L. A, localization of the YFP-tagged full-length Dnmt3a wt and its interface variants R881A, F728A and F728D in NIH3T3 cells. B, localization of the YFP-tagged full-length Dnmt3a wt in NIH3T3 cells in the presence and absence of Dnmt3L. Three patterns of nuclear localization were observed: diffused, spotty plus diffused and spotty. The bar diagram shows a quantification of the Dnmt3a localization in the presence and absence of Dnmt3L wt. Additional experiments were conducted with the Dnmt3L F261D interface mutant that partially lost its ability to interact with Dnmt3a. The blue bars represent cells with spotty Dnmt3a localization, the orange bars spotty plus diffused, and the gray bars correspond to diffused localization pattern. The scale bars represent 5 μm.
We next investigated the subuclear localization of CFP-fused Dnmt3L, which showed a homogenous nuclear and some weak cytoplasmic localization (supplemental Fig. S6). We confirmed that the CFP and YFP channels showed no cross-talk in these experiments (supplemental Fig. S7) and studied nuclear localization of Dnmt3a and Dnmt3L after co-transfection of Dnmt3L-CFP with Dnmt3a-YFP. Dnmt3L showed weak localization to heterochromatic spots in some cells, indicating some targeting influence of Dnmt3a (Fig. 5B). This finding reproduces results of an earlier study (49). Interestingly, we observed a major change in the distribution of Dnmt3a in the nucleus in the absence and presence of Dnmt3L (Fig. 5B). When Dnmt3L was not present, Dnmt3a was localized to the heterochromatic foci in the great majority of cells (62%). Only a minority of cells showed spotty Dnmt3a localization overlaid with some diffuse background (38%), but we never observed cells with uniformly diffused nuclear Dnmt3a localization without any spots. In contrast, in the presence of Dnmt3L, the majority of cells showed a diffused nuclear Dnmt3a localization pattern (60%) and only a minor fraction of cells displayed the spotty localization characteristic for the wild-type Dnmt3a (15 and 25% for spotty and spotty plus diffused, respectively). We performed similar experiments with the Dnmt3L F261D interface mutant, which showed strongly reduced interaction with Dnmt3a in vitro (40) and observed a much reduced effect of the Dnmt3L variant on the localization of Dnmt3a. In the presence of Dnmt3L F261D the majority of cells retained the spotty or spotty and diffused Dnmt3a localization pattern and only in a minor fraction of cells Dnmt3a was found diffusely distributed in the nucleus. These results indicate that Dnmt3L interaction reduces the heterochromatic localization of Dnmt3a and redistributes it into euchromatin.
DISCUSSION
The Dnmt3a C-terminal domain employs two interfaces for protein/protein contacts in the structure of the Dnmt3a/Dnmt3L heterotetrameric complex: one FF interface and one RD interface. We show here that the FF interface also supports self-interaction of Dnmt3a-C and that Dnmt3a-C dimers can form protein oligomers using their RD interfaces. Since each RD interface constitutes a potential DNA binding site, oligomerization of Dnmt3a creates several DNA binding sites. Our modeling of a Dnmt3a oligomer predicted binding to several DNA molecules oriented next to each other roughly in parallel (Fig. 1 and supplemental Fig. S2B). We document this unusual mode of DNA binding to Dnmt3a-C oligomers by binding experiments performed in solution (FRET) and imaging (SFM). Binding of two DNA molecules in parallel orientation so far has been observed only with few proteins (one noticeable example being RecA (50)) and such DNA binding mode was not anticipated for Dnmt3a.
Dnmt3a has been shown to bind tightly to chromatin (21). It is engaged in two interactions with the H3 tail, mediated by its ADD domain binding to the end of the H3 tail unmodified at K4 (29–30), and by its PWWP domain binding to H3K36me3, which was found to be essential for heterochromatic localization (31). In addition, Dnmt3a polymerizes on the DNA (38, 40) and we show here that Dnmt3a oligomers bind to several DNA molecules oriented in parallel. Since heterochromatin is defined as a region of very high DNA density, the ability of Dnmt3a oligomers to bind to parallel DNA close to each other may contributes to the targeting of the enzyme to such DNA dense regions. This hypothesis is supported by our finding that heterochromatic localization of Dnmt3a was lost with non-oligomerizing Dnmt3a mutants affected at the FF interface and in the presence of Dnmt3L, which disrupts oligomerization and multiple DNA binding because it lacks an RD interface. The structure of heterochromatic DNA is not known. However, at least one model of the 30 nm chromatin fiber, the helical ribbon model (51), proposes an almost parallel arrangement of the linker DNA regions in a spacing that would fit to the model of the Dnmt3a oligomers binding to parallel DNA molecules. Hence, the unusual DNA binding mode of Dnmt3a could represent a novel mechanism of heterochromatic targeting.
We have shown that Dnmt3L disrupts the reversible oligomerization of Dnmt3a-C and it interferes with the simultaneous binding to several DNA molecules to Dnmt3a. We investigated the influence of Dnmt3L on the subnuclear localization of Dnmt3a. Strikingly, more than 60% of the cells lost the heterochromatic localization of Dnmt3a in the presence of Dnmt3L, but not when Dnmt3a was co-expressed with the Dnmt3L F261D mutant, which partially lost its ability to interact with Dnmt3a. This observation suggests a novel mode of action of Dnmt3L in setting DNA methylation imprints. In addition to stimulating the activity of Dnmt3a, it might be involved in the release of Dnmt3a from dense heterochromatin to make it available to act at imprinted differentially methylated regions, which are generally located in euchromatin. The expression of Dnmt3L in germ line cells may redistribute Dnmt3a from heterochromatin to euchromatin and, thereby, increase Dnmt3a availability and DNA methylation activity for the generation of DNA methylation imprints.
Supplementary Material
Acknowledgment
We thank Dr. Xiadong Cheng for insightful discussions and remarks.
This work was supported by the DFG (JE 252/6).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
D. B. Hayes, T. Laue, and J. Philo, unpublished data.
- FRET
- fluorescence resonance energy transfer
- YFP
- yellow fluorescent protein
- CFP
- cyan fluorescent protein
- SFM
- scanning force microscopy.
REFERENCES
- 1. Allis C. D., Jenuwein T., Reinberg D. (2007) Epigenetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2. Miranda T. B., Jones P. A. (2007) J. Cell. Physiol. 213, 384–390 [DOI] [PubMed] [Google Scholar]
- 3. Lande-Diner L., Zhang J., Ben-Porath I., Amariglio N., Keshet I., Hecht M., Azuara V., Fisher A. G., Rechavi G., Cedar H. (2007) J. Biol. Chem. 282, 12194–12200 [DOI] [PubMed] [Google Scholar]
- 4. Delaval K., Feil R. (2004) Curr. Opin. Genet. Dev. 14, 188–195 [DOI] [PubMed] [Google Scholar]
- 5. Hore T. A., Rapkins R. W., Graves J. A. (2007) Trends Genet. 23, 440–448 [DOI] [PubMed] [Google Scholar]
- 6. Sha K. (2008) Annu Rev. Genomics Hum. Genet. 9, 197–216 [DOI] [PubMed] [Google Scholar]
- 7. Heard E. (2004) Curr. Opin. Cell Biol. 16, 247–255 [DOI] [PubMed] [Google Scholar]
- 8. Chang S. C., Tucker T., Thorogood N. P., Brown C. J. (2006) Front Biosci. 11, 852–866 [DOI] [PubMed] [Google Scholar]
- 9. Yen Z. C., Meyer I. M., Karalic S., Brown C. J. (2007) Genomics 90, 453–463 [DOI] [PubMed] [Google Scholar]
- 10. Straub T., Becker P. B. (2007) Nat. Rev. Genet. 8, 47–57 [DOI] [PubMed] [Google Scholar]
- 11. Howard G., Eiges R., Gaudet F., Jaenisch R., Eden A. (2008) Oncogene 27, 404–408 [DOI] [PubMed] [Google Scholar]
- 12. Jones P. A., Baylin S. B. (2007) Cell 128, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Egger G., Liang G., Aparicio A., Jones P. A. (2004) Nature 429, 457–463 [DOI] [PubMed] [Google Scholar]
- 14. Feinberg A. P., Tycko B. (2004) Nat. Rev. Cancer 4, 143–153 [DOI] [PubMed] [Google Scholar]
- 15. Rodenhiser D., Mann M. (2006) Cmaj 174, 341–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Robertson K. D. (2005) Nat. Rev. Genet. 6, 597–610 [DOI] [PubMed] [Google Scholar]
- 17. Esteller M. (2007) Nat. Rev. Genet. 8, 286–298 [DOI] [PubMed] [Google Scholar]
- 18. Jurkowska R., Jurkowski T. P., Jeltsch A. (2011) Chembiochem 12, 206–222 [DOI] [PubMed] [Google Scholar]
- 19. Klose R. J., Bird A. P. (2006) Trends Biochem. Sci. 31, 89–97 [DOI] [PubMed] [Google Scholar]
- 20. Dodge J. E., Okano M., Dick F., Tsujimoto N., Chen T., Wang S., Ueda Y., Dyson N., Li E. (2005) J. Biol. Chem. 280, 17986–17991 [DOI] [PubMed] [Google Scholar]
- 21. Jeong S., Liang G., Sharma S., Lin J. C., Choi S. H., Han H., Yoo C. B., Egger G., Yang A. S., Jones P. A. (2009) Mol. Cell. Biol. 29, 5366–5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Okano M., Bell D. W., Haber D. A., Li E. (1999) Cell 99, 247–257 [DOI] [PubMed] [Google Scholar]
- 23. Bourc'his D., Xu G. L., Lin C. S., Bollman B., Bestor T. H. (2001) Science 294, 2536–2539 [DOI] [PubMed] [Google Scholar]
- 24. Bourc'his D., Bestor T. H. (2004) Nature 431, 96–99 [DOI] [PubMed] [Google Scholar]
- 25. Hata K., Okano M., Lei H., Li E. (2002) Development 129, 1983–1993 [DOI] [PubMed] [Google Scholar]
- 26. Bachman K. E., Rountree M. R., Baylin S. B. (2001) J. Biol. Chem. 276, 32282–32287 [DOI] [PubMed] [Google Scholar]
- 27. Chen T., Tsujimoto N., Li E. (2004) Mol. Cell. Biol. 24, 9048–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ge Y. Z., Pu M. T., Gowher H., Wu H. P., Ding J. P., Jeltsch A., Xu G. L. (2004) J. Biol. Chem. 279, 25447–25454 [DOI] [PubMed] [Google Scholar]
- 29. Otani J., Nankumo T., Arita K., Inamoto S., Ariyoshi M., Shirakawa M. (2009) EMBO Rep 10, 1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang Y., Jurkowska R., Soeroes S., Rajavelu A., Dhayalan A., Bock I., Rathert P., Brandt O., Reinhardt R., Fischle W., Jeltsch A. (2010) Nucleic Acids Res. 38, 4246–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dhayalan A., Rajavelu A., Rathert P., Tamas R., Jurkowska R. Z., Ragozin S., Jeltsch A. (2010) J. Biol. Chem. 285, 26114–26120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gowher H., Jeltsch A. (2002) J. Biol. Chem. 277, 20409–20414 [DOI] [PubMed] [Google Scholar]
- 33. Chedin F., Lieber M. R., Hsieh C. L. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 16916–16921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Z. X., Mann J. R., Hsieh C. L., Riggs A. D., Chédin F. (2005) J. Cell. Biochem. 95, 902–917 [DOI] [PubMed] [Google Scholar]
- 35. Suetake I., Shinozaki F., Miyagawa J., Takeshima H., Tajima S. (2004) J. Biol. Chem. 279, 27816–27823 [DOI] [PubMed] [Google Scholar]
- 36. Gowher H., Liebert K., Hermann A., Xu G., Jeltsch A. (2005) J. Biol. Chem. 280, 13341–13348 [DOI] [PubMed] [Google Scholar]
- 37. Kareta M. S., Botello Z. M., Ennis J. J., Chou C., Chédin F. (2006) J. Biol. Chem. 281, 25893–25902 [DOI] [PubMed] [Google Scholar]
- 38. Jia D., Jurkowska R. Z., Zhang X., Jeltsch A., Cheng X. (2007) Nature 449, 248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheng X., Blumenthal R. M. (2008) Structure 16, 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jurkowska R. Z., Anspach N., Urbanke C., Jia D., Reinhardt R., Nellen W., Cheng X., Jeltsch A. (2008) Nucleic Acids Res. 36, 6656–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jeltsch A., Lanio T. (2002) Methods Mol. Biol. 182, 85–94 [DOI] [PubMed] [Google Scholar]
- 42. Gowher H., Loutchanwoot P., Vorobjeva O., Handa V., Jurkowska R. Z., Jurkowski T. P., Jeltsch A. (2006) J. Mol. Biol. 357, 928–941 [DOI] [PubMed] [Google Scholar]
- 43. Schuck P. (2000) Biophys J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schuck P., Rossmanith P. (2000) Biopolymers 54, 328–341 [DOI] [PubMed] [Google Scholar]
- 45. Li J. Y., Pu M. T., Hirasawa R., Li B. Z., Huang Y. N., Zeng R., Jing N. H., Chen T., Li E., Sasaki H., Xu G. L. (2007) Mol. Cell. Biol. 27, 8748–8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lebowitz J., Lewis M. S., Schuck P. (2002) Protein Sci. 11, 2067–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chothia C., Janin J. (1975) Nature 256, 705–708 [DOI] [PubMed] [Google Scholar]
- 48. Bahadur R. P., Chakrabarti P., Rodier F., Janin J. (2003) Proteins 53, 708–719 [DOI] [PubMed] [Google Scholar]
- 49. Nimura K., Ishida C., Koriyama H., Hata K., Yamanaka S., Li E., Ura K., Kaneda Y. (2006) Genes Cells 11, 1225–1237 [DOI] [PubMed] [Google Scholar]
- 50. Bell C. E. (2005) Mol. Microbiol. 58, 358–366 [DOI] [PubMed] [Google Scholar]
- 51. Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R. R., Richmond T. J. (2004) Science 306, 1571–1573 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.