Abstract
The human C-type lectin-like molecule CLEC5A is a critical macrophage receptor for dengue virus. The binding of dengue virus to CLEC5A triggers signaling through the associated adapter molecule DAP12, stimulating proinflammatory cytokine release. We have crystallized an informative ensemble of CLEC5A structural conformers at 1.9-Å resolution and demonstrate how an on-off extension to a β-sheet acts as a binary switch regulating the flexibility of the molecule. This structural information together with molecular dynamics simulations suggests a mechanism whereby extracellular events may be transmitted through the membrane and influence DAP12 signaling. We demonstrate that CLEC5A is homodimeric at the cell surface and binds to dengue virus serotypes 1–4. We used blotting experiments, surface analyses, glycan microarray, and docking studies to investigate the ligand binding potential of CLEC5A with particular respect to dengue virus. This study provides a rational foundation for understanding the dengue virus-macrophage interaction and the role of CLEC5A in dengue virus-induced lethal disease.
Keywords: Immunology, Innate Immunity, Molecular Dynamics, Viral Immunology, X-ray Crystallography, C-type Lectin-like, CLEC5A, Dengue, MDL-1
Introduction
Macrophage effector functions can be triggered by the interactions of activating receptors on the macrophage surface with appropriate ligands. The activation of macrophages is beneficial in the immune response against many infectious pathogens, but dengue virus infection can trigger excessive cytokine release, which contributes to mortality by various mechanisms, including increased capillary permeability (1). There is no vaccine or effective antiviral treatment against dengue virus, which is the most common arboviral (or arthropod-borne) cause of disease worldwide and a major cause of mortality in endemic regions (1). The World Health Organization estimates that there are 50 million dengue virus infections each year with 890,000 cases reported in the Americas alone in 2007 (2). The C-type lectin-like protein CLEC5A has recently been shown to be a macrophage receptor for dengue virus, and the CLEC5A-virus interaction triggers macrophage activation with a marked proinflammatory cytokine release (3). CLEC5A contributes substantially to the mortality associated with dengue virus-induced infection by triggering excessive macrophage activation (3). Blockade of CLEC5A improves survival and shows promise as a therapeutic approach in dengue virus disease (3).
CLEC5A is expressed on the surface of monocytes and macrophages and was identified by its interaction with DNAX-activating protein of molecular mass 12 kilodaltons (DAP12)3; it is also known as myeloid DAP12-associated lectin (MDL-1) (4). CLEC5A contains a C-type lectin-like domain in its C-terminal extracellular region and has only four amino acids in its predicted N-terminal cytoplasmic region. Signal transmission occurs through the interaction with DAP12, which has a cytoplasmic immunoreceptor tyrosine-based activation motif (4, 5). The CLEC5A-DAP12 interaction depends upon a charge-charge interaction between a lysine residue and an aspartate residue in the transmembrane domains of CLEC5A and DAP12, respectively (4, 6). Ligand binding by CLEC5A induces the phosphorylation of DAP12 by Src kinases and an interaction with Syk, which triggers further downstream signaling events (6). CLEC5A has been shown to associate with both DAP12 and DAP10 in bone marrow-derived osteoclasts, triggering osteoclastogenesis and bone remodeling (7).
Human CLEC5A is expressed in monocyte/macrophage cell lines but not in non-myeloid cells (5). CLEC5A plays an important role in the activation of macrophages; CLEC5A stimulation of mouse myeloid cells up-regulates expression of the inflammatory mediator CD11b, inducing significant chemokine production and concurrent signaling through Toll-like receptors (4, 8). Both human and mouse CLEC5A have been reported to bind to dengue virions, and the interaction is inhibited in vitro by fucose and mannan (3). However, the precise ligand on the virions bound by CLEC5A has not yet been identified. To better understand how host immune cells interact with dengue virus and to explore the role of CLEC5A in dengue virus-induced lethal disease, we have solved the crystal structure of the extracellular, C-type lectin-like domain of CLEC5A and undertaken functional and computational analyses. We examined the binding of CLEC5A to different serotypes of dengue virus and investigated carbohydrate binding to the protein using glycan microarrays and computational docking experiments to map putative ligand binding sites on the structure. These studies demonstrate that CLEC5A is a dimer capable of undergoing a clear conformational switch, which may occur in the presence of a ligand. CLEC5A is capable of binding the different serotypes of dengue virus, although the precise ligand is not known. Collectively, these findings suggest a model whereby a conformational switch may mediate downstream signaling through DAP12, ultimately causing cytokine release and stimulating an inflammatory response.
EXPERIMENTAL PROCEDURES
CLEC5A Protein Expression, Purification, Crystallization, and Structural Determination
Recombinant human CLEC5A protein constructs were expressed in Escherichia coli as insoluble inclusion bodies, refolded, and purified by size exclusion chromatography as described previously (9). Protein purity and molecular weight were assessed by SDS-PAGE and mass spectroscopy, respectively. Liquid chromatography-electrospray ionization-mass spectrometry was performed using a reversed-phase C4 column on an Ultima HPLC system (Dionex, Sunnyvale, CA) connected to a quadrupole time-of-flight Micromass spectrometer (Waters, Milford, MA). For microarray analyses, BirA enzyme was produced and used to biotinylate CLEC5A protein, which had a BirA recognition tag engineered onto its N terminus using the approach described previously (10). Biotinylated CLEC5A protein was conjugated to FITC-labeled neutravidin (Sigma) or Alexa Fluor 647-labeled streptavidin (Invitrogen). Recombinant CLEC5A lacking an N-terminal BirA recognition sequence was crystallized as reported (9). The structure of CLEC5A was solved from a twinned crystal with twin fractions of 0.79 and 0.21 using a multistep molecular replacement protocol (see supplemental data) using the program MOLREP and human CD94 (Protein Data Bank code 3BDW) as a search model (11, 12). Subsequent manual rebuilding and refinement cycles used Coot and Refmac5, and the twin refinement function of Refmac was used (13, 14). The structure of CLEC5A was subjected to validation using the programs PROCHECK and WHATIF before Protein Data Bank deposition under code 2YHF (15, 16).
Structural Analyses
The Dynamite package was used to infer, analyze, and graphically represent the likely modes of motion of CLEC5A (17). Electrostatic surface potentials and isocontour representations were calculated using Adaptive Poisson-Boltzmann Solver (APBS) software scaled between −5 and +5, −7 and +7, or −10 and +10 kTe, and images were created using Visual Molecular Dynamics and PyMol (Schrödinger, Portland, OR) and rendered with Raster3D (18).
Modeling of Dimeric CLEC5A
The model of dimeric CLEC5A was assembled by manual superimposition of two copies of the structure of monomeric CLEC5A onto the dimeric structure of the related C-type lectin-like receptor NKG2D (Protein Data Bank code 1KCG) using Coot. Subsequent energy minimization was performed using algorithms implemented by the WHATIF server (16).
Transmembrane Helical Simulations of CLEC5A, DAP12, and DAP10
Coarse-grained molecular dynamics simulations of the transmembrane helices of these proteins were performed following the approach described by Psachoulia et al. (19). Briefly, simulations were set up with helices preinserted into a 1,2-dipalmitoyl-sn-glycero-3-phosphocholine bilayer and separated from one another. Simulations lasted 5 μs, and the helices were allowed to freely diffuse through the bilayer and interact with one another. Simulations were performed with Gromacs 4 at 323 K. Temperature and pressure were coupled using a Berendsen thermostat (tau_p = 10 ps and tau_t = 1 ps). Pressure was coupled semi-isotropically with a compressibility of 3 × 10−5 bar−1. The force field used was the same derivative of the MARTINI force field used by Psachoulia et al. (19). Each simulation was repeated five times with different starting velocities.
Flow Cytometry and Confocal Microscopy
HEK 293T cells were washed in cold PBS and incubated with either mouse anti-CLEC5A antibody (R&D Systems, Minneapolis, MN) or the appropriate isotype control for 20 min on ice in PBS, 0.5% FCS. Cells were then washed three times and incubated with an allophycocyanin-conjugated anti-mouse IgG secondary antibody for 20 min on ice. Cells were then washed and analyzed on a FACSCanto machine (BD Biosciences) and subsequently with FlowJo software (Treestar, Ashland, OR). For confocal microscopy, HEK 293T cells were grown on glass-bottomed dishes suitable for live cell microscopy (MatTek Corp., Ashland, MA) and imaged 24 h following transfection. A 488 nm argon laser was used to excite fluorophores, and live cells were visualized under an oil-immersed objective with a Zeiss LSM510 confocal microscope and a Zeiss XL3 live cell incubator system (Carl Zeiss MicroImaging, Munich, Germany).
Bioluminescence Resonance Energy Transfer (BRET)
60% confluent HEK 293T cells were transfected with varying ratios of plasmids encoding full-length human CLEC5A protein tagged at its cytoplasmic N terminus with either green fluorescent protein 2 (GFP2) or luciferase together with either an empty pcDNA3 plasmid or one encoding full-length DAP12 using GeneJuice (Novagen, Gibbstown, NJ). These CLEC5A fusion proteins contain the CLEC-2 cytoplasmic domain (residues 1–32) in place of the very short CLEC5A cytoplasmic domain (residues 1–4). Cells were harvested 24 h after transfection. For each transfection, 10 μm DeepBlueC coelenterazine (PerkinElmer Life Sciences) was added to 100 μl of cells in a 96-well plate, and light emission in the 410 ± 40- and 515 ± 15-nm wavelength ranges was collected immediately as described previously using a Fusion microplate analyzer (PerkinElmer Life Sciences) (20). GFP2 and luciferase expression was measured in a separate well and converted to a ratio of concentrations. BRET efficiency (BRETeff) and GFP:luciferase ratio values were calculated by comparison with those obtained for a positive BRET control consisting of a soluble, fused form of luciferase and GFP, which were taken as 1 and 1:1, respectively.
Dengue Virus Binding
CLEC5A or CLEC-2 protein samples were spotted onto nitrocellulose membrane. The membranes were blocked with 5% skimmed milk in PBS and then incubated with supernatants from UV light-treated dengue virus serotype DEN1 (Hawaii), DEN2 (16681), DEN3 (H87), DEN4 (H241), or mock-infected C6/36 cells for 2 h. After washing three times with PBS, anti-dengue virus mouse monoclonal antibody (clone 4G2, Armed Forces Research Institute of Medical Sciences, Bangkok, Thailand) was added to the membrane and incubated for an additional hour followed by a secondary HRP-labeled anti-mouse IgG (Dako, Glostrup, Denmark) at a 1:1000 dilution. After washing, the membranes were developed with enhancement chemiluminescence substrate (Amersham Biosciences, GE Healthcare). Recombinant CLEC-2 protein was used as a negative control and was prepared as reported previously (21, 22). Human anti-dengue virus antibody was purified from hyperimmune serum. The assay was repeated four times.
Glycan Binding Microarray Analyses
Recombinant CLEC5A protein was tested for its capacity to bind to a wide range of glycan sequences displayed on two different microarray systems as described in detail in the supplemental data.
Docking of Potential Carbohydrate Ligands onto CLEC5A
The software GOLD, a program for calculating the docking modes of molecules in protein binding sites, was used to predict the binding of the putative carbohydrate ligands to the crystal structure of CLEC5A (23, 24). The carbohydrates included the monosaccharides fucose and mannose and the N-glycan core tetrasaccharide (α1,6-fucosylated chitobiose core with a mannose residue), which was used in modeling of the core region of the insect N-glycans visualized in the crystal structure of the dengue virus serotype 2 envelope (E) protein dimer (Protein Data Bank code 1OAN) (25).
RESULTS
CLEC5A Displays Conformational Flexibility
Recombinant protein encoding the extracellular domain of human CLEC5A (Fig. 1A) was purified and crystallized (9). Crystals with maximum dimensions of 0.63 mm diffracted synchrotron radiation to 1.9-Å resolution (Table 1). These crystals exhibited partial merohedral twinning with twin fractions of 0.79 and 0.21 and were additionally found to possess a non-crystallographic symmetry axis aligned with the twin axis (see supplemental data). Therefore, a multistep molecular replacement approach was used to assign the precise three-dimensional structure of CLEC5A from these pseudosymmetric crystals. There are nine copies of CLEC5A within the asymmetric unit, representing a range of structural conformers and so providing a valuable insight into the range of possible conformations that this receptor may adopt in vivo.
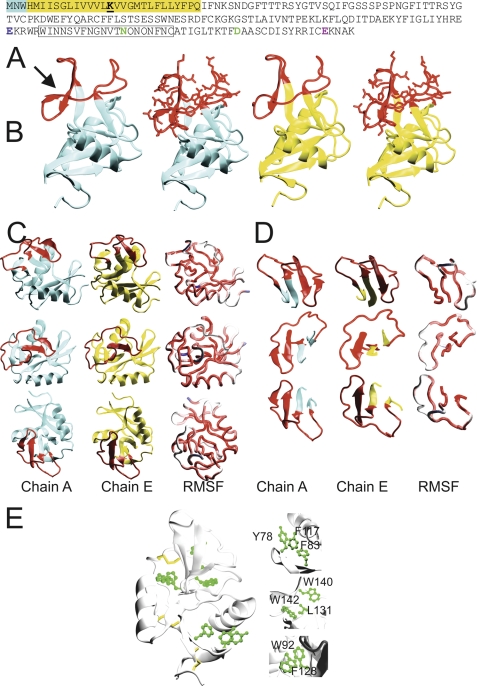
FIGURE 1.
CLEC5A exists as different structural conformers. A, the protein sequence of CLEC5A with intracellular and transmembrane portions highlighted blue and yellow, respectively. Lysine 16 (bold and underlined) in the transmembrane region interacts with an aspartate in the transmembrane helix of DAP12. The vestiges of canonical carbohydrate/Ca2+ binding sites 1, 2, and 4 are colored blue, green, and pink, respectively. The long loop region is boxed. B, overview of the structure of molecules A (cyan) and E (yellow) of CLEC5A showing the long loop region in red. In the second and fourth panels from the left, the side chains of the long loop region are also displayed. Molecule A contains the additional β-strand (indicated by an arrow), whereas molecule E does not. C, different orientations of molecules A (cyan) and E (yellow) are shown with the long loop region in red. In the far right panel, the average structure over all nine molecules in the asymmetric unit is represented as a tube that is colored according to the relative mean square fluctuation (RMSF) of Cα atoms calculated over all nine unique conformations in the asymmetric unit (the range is from red (low fluctuation) to blue (high fluctuation)). D, different orientations of a section of CLEC5A from amino acids 134–161 containing the long loop region are displayed with the long loop region in red. In the far right panels of C and D, the average structure over all nine molecules in the asymmetric unit is represented as a tube colored according to the relative mean square fluctuation as in C. E, CLEC5A core stability is based on three disulfide bonds (yellow) and three hydrophobic regions (green). The internal hydrophobic cores of CLEC5A include the following residues: Tyr-78, Phe-117, Phe-83, Trp-92, Phe-128, Trp-140, Trp-142, and Leu-131.
TABLE 1.
Crystallographic data collection for CLEC5A
Values for the highest resolution shell are indicated in parentheses. Rmerge = ΣhklΣi|Ii − <I>|/|<I>| where Ii is the intensity for the ith measurement of an equivalent reflection with indices h, k, and l. Rcryst = Σ‖Fo| − |Fc‖/Σ|Fo| using all data except 5.1%, which were used for the Rfree calculation. r.m.s.d., root mean square deviation.
| Data collection | |
| Space group | P31 |
| Cell dimensions | a = 109.109, b = 109.109, c = 84.879, α = β = 90.000, γ = 120 |
| Wavelength (Å) | 1.117 |
| Resolution (Å) | 50.00-1.90 (1.97-1.90) |
| Rmerge | 0.072 (0.495) |
| I/σ(I) | 12.90 (1.63) |
| Completeness (%) | 87.1 (76.4) |
| Twin operators, twin fraction | h, k, l, 0.79; −h, h + k, −l, 0.21 |
| Refinement | |
| Resolution (Å) | 50.00-1.90 |
| No. protein atoms | 8560 |
| No. solvent atoms | 1134 |
| Average B-factor (Å2) | 22.408 |
| r.m.s.d. bond lengths (Å) | 0.01 |
| r.m.s.d. bond angles (°) | 0.947 |
| Rcryst/Rfree | 0.216/0.267 |
| Ramachandran analysis | |
| Favored (%) | 92.91 |
| Allowed (%) | 6.32 |
| Outliers (%) | 0.77 |
The overall architecture of CLEC5A is that of a C-type lectin-like molecule based on two core antiparallel β-sheets flanked by two α-helices. Typically, the long loop region of these proteins plays a key role in ligand binding (22, 26, 27). The CLEC5A long loop is unusual in two respects. First, of the 20 loop residues, 10 are glutamines or asparagines (Fig. 1A). Regions of high glutamine/asparagine content often act as molecular polar zippers, influencing ligand binding. Such polar zipper regions influence the interactions between hemoglobin subunits and between several transcription factors and specific DNA segments (28). Second and most significantly, two different long loop region conformations exist in the crystal. In four of the nine CLEC5A molecules within the asymmetric unit, the long loop region contains an extra β-strand, which is positioned such that it forms an extension of the antiparallel β-sheet beneath it, thus simultaneously reducing loop flexibility and stabilizing the protein core (Fig. 1, B–D, molecule A). However, in the remaining molecules (Fig. 1, B–D, molecule E), this section is disordered, suggesting that in the absence of a stabilizing crystal lattice ligand binding may promote a conformational switch between these two states. This arrangement has not been documented hitherto for members of this family of proteins. Although the long loop regions of related molecules NKG2D and LOX-1 also contain short, β-strand-like elements, these features are considerably smaller than the CLEC5A long loop region β-strand and do not form part of a greater β-sheet assembly; in contrast, the CLEC-2 long loop region contains a short 310 helix (22, 29, 30). Therefore, this β-sheet can potentially function as a “tether,” attenuating the flexibility of the long loop region at its N terminus, and as an enhanced internal scaffold, imparting greater central stability to CLEC5A. This raises the likelihood that CLEC5A has substantial potential dynamic flexibility compared with related molecules and is capable of undergoing a conformational change on binding to a ligand at the cell surface.
As is typical of C-type lectin-like proteins, the core of CLEC5A is maintained by three intramolecular disulfide bonds. Non-covalent mechanisms also contribute to the core stability of CLEC5A with three internal hydrophobic centers drawing together the secondary structural elements of the C-type lectin-like domain (Fig. 1E). These hydrophobic cores play an important role in the internal stability of CLEC5A through adhesive interactions between the delocalized electrons of aromatic rings.
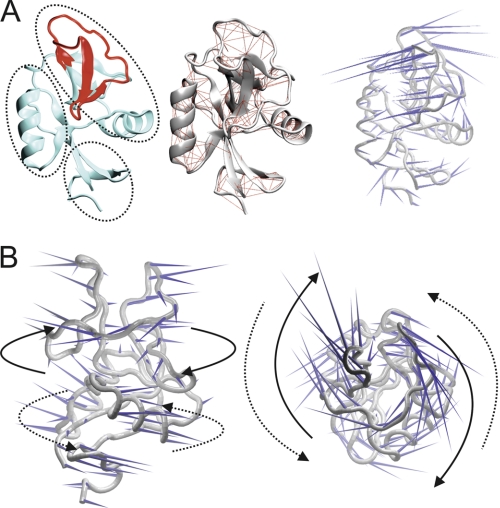
To explore the possible roles of these structural features of CLEC5A in ligand binding and subsequent signal transduction, we performed computational dynamics analyses of the crystal structural data. The analyses incorporated information from all of the nine structural conformers present in the unit cell. A covariance web for CLEC5A indicates the presence of three distinct structural subdomains, which appear to move independently from one another, centering on the unique third β-sheet as a fulcrum (Fig. 2A). Interestingly, a porcupine plot reveals that the “distal” and “proximal” portions of CLEC5A (defined in terms of relative distance from the cell membrane) are predicted to be capable of moving in opposing, twisting motions around a rigid, central core (Fig. 2B). Although it is unclear how this putative twisting motion might affect the ligand binding capabilities of CLEC5A, such twisting may influence downstream signaling via the adapter molecule DAP12. A twisting motion in the extracellular domain of a membrane protein can result in a change in the orientation of a transmembrane helix in terms of its tilt angle and/or rotation; this is well demonstrated for the Tar chemoreceptor of E. coli (31). Such a change in the CLEC5A transmembrane helix could bring the CLEC5A and DAP12 pairs of charged residues into closer proximity, which may influence subsequent signaling through the activating motifs of DAP12. This may occur by reciprocal twisting of the signaling region into a more accessible position perhaps less sterically inhibited by the membrane.
FIGURE 2.
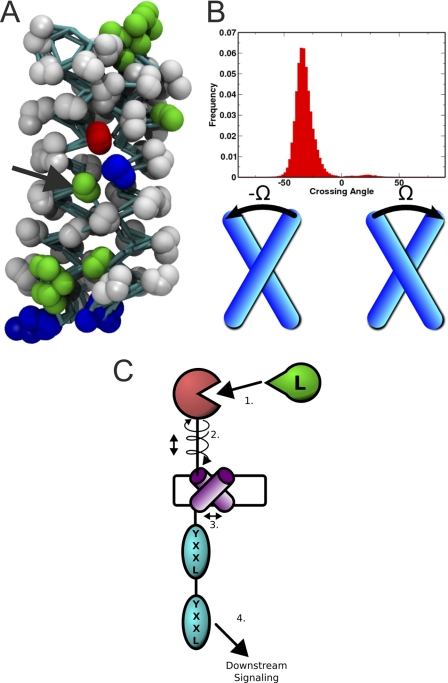
CLEC5A is capable of distinct twisting in its upper and lower lobes. A, in the left panel, the long loop region of chain E is displayed in red, and the rest of the molecule is colored cyan. The three subdomains that are predicted to move independently around the unique third β-sheet are indicated by dotted circles. The middle panel shows a CLEC5A covariance web; red lines link pairs of Cα atoms that are likely to undergo concerted motions. The right panel shows a porcupine plot of the principal mode of conformational variability of the Cα atoms calculated from a CONCOORD ensemble. The orientation of the blue cones indicates the direction of motion of the atoms and their length is proportional to the amplitude of the motion. B, in the left panel, a CLEC5A porcupine plot is oriented with its predicted binding surface seen en face. The right image is a 90° anticlockwise rotation about the x axis. Arrows indicate flexible twisting motions of the upper membrane-distal (solid lines) relative to the lower membrane-proximal (dotted lines) CLEC5A subdomain.
CLEC5A Forms Homodimers At Cell Surface
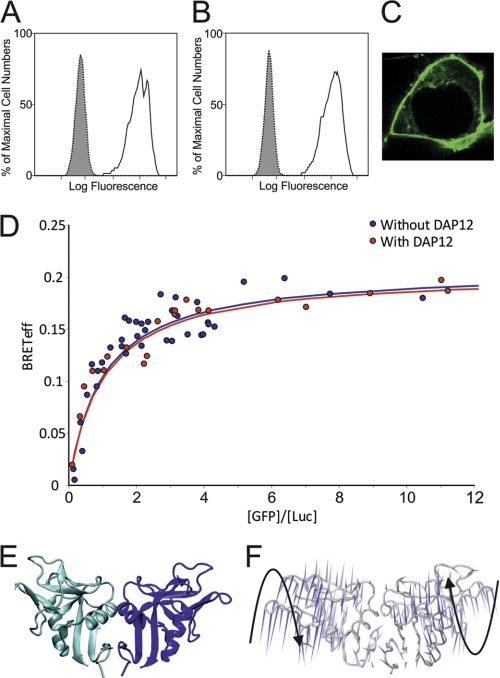
Many C-type lectin-like molecules bind their ligands as dimers (26, 29, 30, 32, 33). Additionally, all DAP12-associated C-type lectin-like molecules are dimeric (34). CLEC5A crystallized as a monomer, and the site on its surface equivalent to the homodimerization site of many C-type lectin-like proteins does not possess a surface distribution of hydrophobic patches sufficient to dictate constitutive dimerization as observed in some family members, such as the immune receptor NKG2D and members of the Ly49 receptor family (26, 29). However, we have recently shown that the related receptor CLEC-2, which similarly lacks a prominent “dimerization patch,” does indeed form dimers in the plasma membrane, and this dimerization is central to its function (32). This and the homodimeric nature of DAP12 prompted us to investigate whether CLEC5A can similarly homodimerize in the membrane. BRET assays determine the BRETeff between luciferase (a light donor) and GFP2 (a light acceptor) fusion proteins. As this efficiency depends upon the distance between the donor and acceptor, this provides a quantitative method for distinguishing between monomers and dimers at the cell surface (20). Cells were transiently cotransfected with a “BRET pair” of plasmids encoding full-length CLEC5A (one tagged with GFP2 and the other tagged with luciferase) and with either an empty control vector or one expressing DAP12 (see supplemental data). Surface expression of these CLEC5A constructs was confirmed by confocal microscopy and flow cytometric analysis (Fig. 3, A–C). Quantitative analysis of this type 1 BRET data demonstrates that CLEC5A oligomerizes in the absence and presence of DAP12, and the dependence of BRETeff on the acceptor:donor ratio fits best to a dimer model (Fig. 3D). The BRETeff maxima for the CLEC5A BRET pairs reside between the documented BRETeff maxima for known monomers and for disulfide-linked homodimers, and the curve shape is typical of nonconstitutive homodimerization (20). These curves resemble those we previously reported for the nonconstitutive homodimer and related C-type lectin-like protein CLEC-2 and indicate that human CLEC5A homodimerizes at the cell surface and that this dimerization occurs independently of DAP12 (20, 32). A model of dimeric CLEC5A based on the CLEC5A crystal structure and standard dimerization arrangement for related molecules (Fig. 3E) indicates that there will be a large potential binding surface area for ligand interactions. Importantly, the features of dimeric CLEC5A are consistent with the composite capacity to undergo a pronounced twisting motion wherein the upper and lower segments of each subunit of CLEC5A twist away from one another relative to their central core regions, extending the surface area available for accommodating a large ligand, such as dengue virus (Fig. 3F). This will have implications for a twisting/signaling mechanism as a larger, more pronounced motion may be more readily translated across the plasma membrane.
FIGURE 3.
CLEC5A is a homodimer at the cell surface. A and B, flow cytometry of HEK 293T cells cotransfected with GFP-tagged CLEC5A and either an empty control pcDNA3 plasmid (A) or a plasmid expressing DAP12 (B) using an anti-human CLEC5A antibody (unfilled histogram) or an IgG2b isotype control antibody (filled histogram) followed by an allophycocyanin-conjugated secondary antibody. C, confocal microscopy of HEK 293T cells transfected with GFP-tagged CLEC5A demonstrating GFP localization to the cell surface. D, BRET analysis. HEK 293T cells were cotransfected with the GFP2-CLEC5A/luciferase (Luc)-CLEC5A pair plus either DAP12 (red dots) or an empty control vector (blue dots) at varying DNA concentration ratios of the BRET pair constructs. The BRETeff trace for the CLEC5A BRET pair in both cases is consistent with that of a non-disulfide-linked dimer. E, dimeric CLEC5A is represented as a schematic where the individual monomer chains are colored blue and cyan. F, a porcupine plot of the principal mode of conformational variability of the Cα atoms calculated from a CONCOORD ensemble of model dimeric CLEC5A is shown where the orientation of the blue cones indicates the direction of motion of the atom, and their length is proportional to the amplitude of the motion. Black arrows indicate the relative dynamic twisting motions of each CLEC5A monomer.
Binding of Dengue Virus Serotypes by CLEC5A
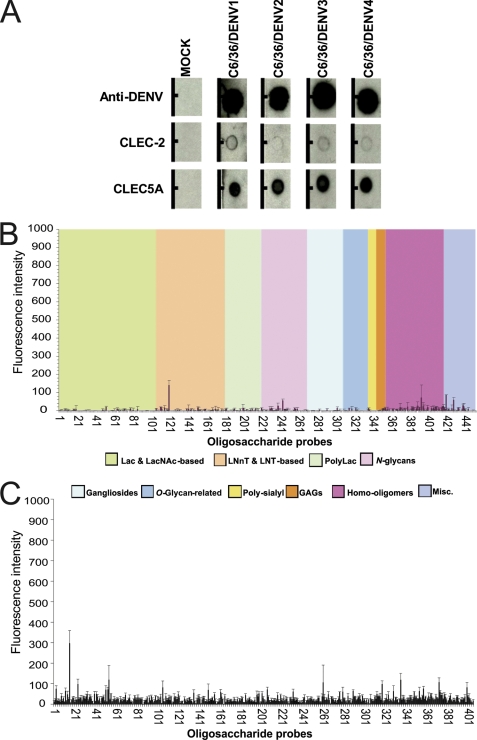
Chen et al. (3) demonstrated that CLEC5A interacts with dengue virus serotype 2. There are also data suggesting that different dengue virus serotypes may interact with different immune receptors (35). Therefore, we sought to investigate whether CLEC5A is capable of binding to each of the four dengue virus serotypes. Using a dot blot approach, binding of recombinant CLEC5A to dengue virus was detected for all four serotypes (Fig. 4A). After background subtraction, there was a suggestion that CLEC5A bound most strongly to dengue virus serotypes 1 and 4.
FIGURE 4.
Analyses of binding of CLEC5A to dengue viruses and glycan microarrays. A, dot blot binding assays. Representative blots showing binding of dengue virus serotypes 1–4 by recombinant CLEC5A and CLEC-2. In the lower two rows, proteins were spotted onto a nitrocellulose membrane and incubated with supernatants from UV light-treated dengue virus (DENV) serotype DENV1 (Hawaii), DENV2 (16681), DENV3 (H87), or DENV4 (H241) or from mock-infected C6/36 cells and then with the monoclonal mouse anti-dengue antibody 4G2 and subsequently detected with the anti-mouse IgG antibody. In the upper row, designated Anti-DENV, purified polyclonal human antibody to dengue virus was blotted onto the membrane and incubated in a similar manner with virus and then with 4G2 and anti-mouse IgG for detection of viral binding. B and C, glycan microarray binding studies. In B, recombinant CLEC5A protein was tested for binding to a panel of 452 lipid-linked oligosaccharide probes. Signals are expressed as the means of duplicates of fluorescence intensities with error bars representing half of the difference between the two values. Probes are sorted into 10 groups according to their backbone-type sequences. Lac, lactose; LacNAc, N-acetyllactosamine; LNnT, lacto-N-neotetraose; LNT, lacto-N-tetraose; PolyLac, polylactosamine; GAGs, glycosaminoglycans; Misc., miscellaneous. In C, recombinant CLEC5A protein was tested for binding to a panel of 406 oligosaccharide probes developed by the Consortium for Functional Glycomics. The mean is shown with error bars representing the S.E. The probes and data for B and C are tabulated in supplemental Tables 1 and 2, respectively.
Carbohydrate Microarray Analyses of CLEC5A
The dengue virus E protein contains two potential N-linked glycosylation sites at Asn-67 and Asn-153. The presence of glycosylation at Asn-67 is a unique signature for dengue virus among the flaviviruses and is conserved in all the strains of the four dengue serotypes (36). The cryoelectron microscopy model of dengue virus in complex with DC-SIGN, a C-type lectin known to mediate dengue virus attachment to dendritic cells, suggested the N-glycan at Asn-67 on the E protein as a receptor binding site, and the interaction of DC-SIGN with dengue virus glycans has been corroborated biochemically (37, 38). The E protein N-glycans have also been reported as ligands for the carbohydrate recognition domain of the mannose receptor (39). These findings together with the report that fucose and mannan inhibit the CLEC5A-dengue virus interaction have raised the possibility that the N-glycans on the virus E protein play a role in the CLEC5A-dengue virus interaction (3).
To examine directly the ability of CLEC5A to bind to oligosaccharide sequences, CLEC5A was analyzed in two glycan microarray platforms. Both microarrays included oligosaccharide probes recognized by C-type lectins, such as DC-SIGN, SIGN-R1, SIGN-R3, and Langerin (40–42). One array included a diverse set of 452 glycan probes based on the neoglycolipid-based technology (43, 44) (supplemental Table 1). This array included a wide range of N-glycan-related probes. Among these were paucimannose N-glycans with or without the α1,6-linked core fucose typically found on insect-derived glycoproteins and an N-glycan with the alternative α1,3-linked core fucosylation found in insects (45). Also included were high mannose-type N-glycans, many of which are common to mammals and insects. Complex-type N-glycans with or without α1,6-linked core fucose whose core region sequences are common to insects and mammals were also in the arrays. None of the N-glycan probes nor any of the other probes in this array gave binding signals with CLEC5A (Fig. 4B). The other array included a set of 406 glycan probes assembled by the Consortium for Functional Glycomics (supplemental Table 2) (42). This array also included an extensive repertoire of high mannose- and paucimannose-type N-glycans as well as complex- and hybrid-type N-glycans with or without α1,6 core fucosylation. There were replicate forms of N-glycan sequences having a variety of spacers or linkers, including linked asparagines and also flanking short peptide sequences at their core regions. No specific binding signals were observed with CLEC5A (Fig. 4C).
Structural Influences on Ligand Binding by CLEC5A
The lack of binding signals in the current carbohydrate microarrays does not formally rule out carbohydrate recognition by CLEC5A. To explore this, we analyzed the crystal structure of CLEC5A using computational calculations of docking modes for the monosaccharides fucose and mannose and the dengue virus N-linked tetrasaccharide observed in the crystal structure of dengue E protein (the chitobiose core with α1,6-linked fucose and a mannose residue) (25).
There are four canonical carbohydrate binding sites reported for C-type lectins, which typically bind carbohydrates through coordinated calcium ions (27, 46). The protein sequence of CLEC5A contains an incomplete set of residues from canonical sites 1, 2, and 4 but none from canonical site 3. It is unclear how CLEC5A might recognize a carbohydrate ligand in the absence of a full set of C-type lectin calcium-coordinating and carbohydrate binding sites.
Three discrete potential predicted carbohydrate binding patches (distinct from the canonical sites) were identified and are illustrated in Fig. 5. The most favorable patch (Predicted Patch 1) comprises asparagine residues 110, 144, 145, and 149. Predicted Patch 1 contains none of the conserved calcium binding/carbohydrate-coordinating residues in the C-type lectin canonical sites 1–3, but the proximity of the conserved canonical site 4 residue glutamate 184 could help to stabilize a sugar binding interaction (Fig. 5A). Predicted Patch 2 superimposes onto the canonical site 2 calcium- and carbohydrate-recognizing residues in CLEC5A (Fig. 5A). This site is composed of asparagines 154, 156, and 158 and glutamine 157. Predicted Patch 3 consists of a set of acidic amino acid side chains (glutamates 89 and 125 and aspartate 123), none of which correspond to the canonical site 1–4 sets of conserved residues (Fig. 5A). However, although Predicted Patch 3 was identified as a possible carbohydrate binding patch for fucose and mannose, this patch was not identified as a binding site for the dengue virus E protein N-glycan core tetrasaccharide in our docking experiments. Predicted Patch 3 could in principle be involved in interactions with the more peripheral glycan moieties distal to the core tetrasaccharide. Thus, these predictions propose three possible sites for sugar binding by CLEC5A: Predicted Patch 1 (near canonical site 4), Predicted Patch 2 (canonical site 2), and Predicted Patch 3 (not corresponding to a canonical site).
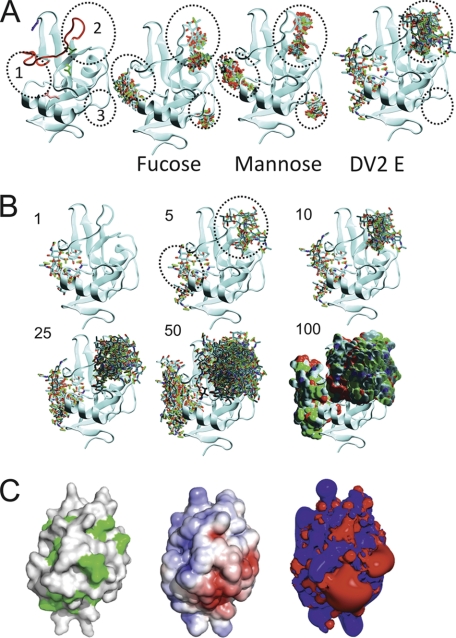
FIGURE 5.
Docking of possible glycan ligands on structure of CLEC5A. A, Predicted Patches 1–3 are encircled by numbered, black dotted lines in all panels. In the far left panel only, the side chains of the vestigial canonical sites 1, 2 and 4 carbohydrate/calcium-binding residues conserved with other C-type lectins are indicated on the CLEC5A structure in blue, green, and pink, respectively. In the following images, the predicted modes of binding of the 10 most favorable binding sites for fucose, mannose, and dengue virus serotype 2 (DV2) E protein sugars are shown. B, images of the top one, five, 10, 25, 50, and 100 conformations of dengue virus E sugars docked onto CLEC5A as ranked by the GOLD algorithms. For clarity, Predicted Patches 1 and 2 are indicated (black dotted circles as in A) on the panel showing the top five conformations of dengue virus E sugars. C, molecular surface representations of the predicted binding face of CLEC5A are shown in the left and middle panels. In the left panel, surface hydrophobic patches are green. In the middle panel, electrostatic charge is illustrated with negatively and positively charged regions colored red and blue, respectively. The right panel is an electrostatic isocontour representation. The surface electrostatic potential scale is from −5 to +5 kTe.
Calcium ions usually play a crucial role in the coordination of carbohydrate ligands on the surface of C-type lectins (46). However, no calcium ions are present in the crystal structure of CLEC5A. Surface representations and subsequent analyses of the electrostatic potential and presentation of non-polar regions of CLEC5A indicate that the surface of CLEC5A is predominantly basic. However, three acidic regions proximal to Predicted Patches 2 and 3 might feasibly accommodate calcium ions (Fig. 5C). The dimeric nature of CLEC5A will increase the potential surface area for ligand binding sites.
Implications of Conformational Flexibility of CLEC5A for Transmembrane Signaling
Given the different structural conformations seen in the CLEC5A crystal structure and the potential for dimeric CLEC5A to twist in response to ligand binding, we analyzed whether it was feasible for such conformational changes to be translated through the membrane and so influence signaling through adapter molecules. DAP12 has no extracellular domain and interacts with CLEC5A through an ionic interaction between polar residues in the transmembrane regions of both proteins (a lysine in CLEC5A and an aspartate in DAP12). The presence of this transmembrane ionic interaction has been shown to be critical for both DAP12 and DAP10 signaling (4, 7, 34). However, it is unclear how this charge-mediated association within the membrane could transmit information about ligand binding from the extracellular domain of CLEC5A to the intracellular signaling domain of DAP12 as there are no structural studies of the transmembrane interactions of DAP12 or DAP10 with CLEC5A. Therefore, to investigate this association and to suggest a mechanism for ligand-induced signaling, we performed coarse-grained molecular dynamics simulations of the transmembrane helices of these proteins in a membrane environment.
After ⅘ of the simulations, the helices adopt a consensus structural arrangement in which they are held together via the CLEC5A lysine and DAP12 aspartate side chain interaction stabilized by a neighboring threonine side chain (Fig. 6A). This structure is right-handed and remains stable in each simulation where it occurs (Fig. 6, A and B). An alternative structure is observed in ⅕ of the simulations where a distinct right-handed CLEC5A-DAP12 heterodimer forms through the same triad of residues but by interacting via an alternative face. In the light of the recent data suggesting a possible CLEC5A-DAP12-DAP10 interaction in osteoclasts, this alternative structure may represent a binding mode that accommodates an additional DAP10 helix (7). Such a structure would represent a right-handed CLEC5A-DAP12-DAP10 heterotrimer, which insulates the polar residues from the hydrophobic core of the membrane. Simulations of the consensus CLEC5A-DAP12 heterodimeric structure with an additional DAP10 helix present produced a range of CLEC5A-DAP12-DAP10 heterotrimeric structures with a right-handed bundle. However, these heterotrimeric structures did not pack as tightly or form as discrete a consensus structure as the CLEC5A-DAP12 heterodimer, which is likely to be the predominant configuration.
FIGURE 6.
Computational simulations of interactions between transmembrane helices of CLEC5A and DAP12. A, results from four coarse-grained molecular dynamics simulations of the transmembrane helices of CLEC5A and DAP12 are superimposed; non-hydrogen atoms are represented as spheres, and covalent bonds are represented as cyan sticks. The helices conform to a stable structure with a clear interaction between a CLEC5A lysine (right) and a DAP12 aspartate (left). Polar residue side chains are colored green, basic residues are blue, and acidic residues are red. The stabilizing threonine residue (green) is indicated with a black arrow. B, the crossing angle (Ω) is defined as the counterclockwise screw angle relating two helices in their plane of contact. The direction of the two helices is ignored. The angle is negative if the near helix is rotated clockwise relative to the far helix. The distribution reflects a tight right-handed dimer of helices across the simulations. C, a model of how ligand binding could trigger signaling. 1, a ligand binding event (“L”) promotes a twisting conformational change. 2, a twisting movement promotes movement of the transmembrane helix of CLEC5A within the membrane. 3, this movement alters the interaction with DAP12, possibly by changing the screw angle between the two transmembrane helices. 4, this change might promote downstream signaling through the immunoreceptor tyrosine-based activation motif domain by altering its disposition in relation to the membrane and possibly its accessibility to other signaling molecules.
Our data are in close agreement with, but independent of, recent nuclear magnetic resonance structures of the membrane-embedded, heterotrimeric assembly of DAP12 with the natural killer cell-activating receptor NKG2C (47). Notably, a threonine residue plays a similar role in stabilizing the association of NKG2C with DAP12.
DISCUSSION
This study presents the crystal structure and functional analysis of the human C-type lectin-like dengue virus receptor CLEC5A. The structure was solved from twinned, pseudosymmetric crystals using a multistep molecular replacement strategy. The structural data offer a valuable opportunity to analyze the conformational flexibility of the CLEC5A in detail because of the presence of an ensemble of structural conformers in the asymmetric unit. This analysis provides molecular insight into the potential flexibility of this protein and the likely effects of this on ligand binding and signaling. CLEC5A displays the typical architecture of the C-type lectin-like domain, but interestingly, one conformer also contains an extra β-strand in its otherwise unstructured long loop region (Fig. 1). This strand contributes to a unique three-stranded antiparallel β-sheet located directly beneath the long loop region itself, an arrangement that has not been seen previously for this family of proteins. This β-sheet will provide a central scaffold to CLEC5A that could function as a hinge region supporting flexible twisting motions of the subdomains on either side of it. This is especially relevant as the long loop region is typically centrally involved in ligand binding in this class of molecules (22, 26, 27). The range of conformational variations found in the crystal structure was further explored using molecular dynamics approaches. Computational dynamics analyses of CLEC5A indicate how the membrane-distal and membrane-proximal lobes of CLEC5A are capable of moving in opposing, twisting motions around a rigid, central core (Fig. 2B). These data suggest that a ligand for CLEC5A may induce an extended β-sheet formation, thus stabilizing the ligand-bound structure.
The long loop region β-strand can be considered to have properties that allow it to serve as a molecular binary switch. In the absence of a stabilizing influence, this portion of the long loop region is relatively disordered and flexible, which may maintain CLEC5A in an “open” state for ligand interactions. However, the presence of an appropriate stabilizing environment, here arising from the crystal lattice but in vivo feasibly arising from ligand binding, can trigger a stable “closed” state. In this closed state, the stabilizing influence triggers a cascade of increased stability with local β-strand formation, which promotes tethering of this part of the long loop region to form an extended β-sheet, which in turn is associated with increased core rigidity of the C-type lectin-like domain of CLEC5A. This switch between two conformational states with differing flexibility might also have consequences for the propagation of signaling.
Twisting movements within the C-type lectin-like domains of the CLEC5A dimer may translate across the transmembrane helical regions and influence signaling through DAP12. Our molecular simulations indicate that relative sliding motions of the transmembrane domains may result in piston-like movements within the membrane and that these may influence the signaling capacity of DAP12 (Fig. 6C). This may occur in a manner similar to that described for two-component signaling in bacterial receptors (31, 48–50). Motions within this CLEC5A-DAP12 heterodimeric bundle may offer insight into possible signaling mechanisms. In particular, our molecular dynamics studies reveal a sliding of the transmembrane helix of CLEC5A along the GXXXGXXXG motif of DAP12. DAP12 is itself a disulfide-linked homodimer, and therefore each of its transmembrane helices is capable of interacting with one CLEC5A helix (34). We have shown that CLEC5A is dimeric at the cell surface (Fig. 3D). Therefore, each DAP12 helix could associate with each CLEC5A transmembrane helix via the Lys-Asp-Thr interfacial triad in an arrangement similar to that reported for the assembly of DAP12 and the heterodimeric NKG2C and CD94 immune receptor complex (47). Such an arrangement may allow piston-type displacements between the CLEC5A and DAP12 helices to facilitate downstream signaling.
We have demonstrated that the extracellular domain of CLEC5A directly and specifically interacts with dengue virus serotypes 1–4 made in mosquito cells with a likely preference for serotypes 1 and 4. This is consistent with studies comparing RNA genomes from the different dengue virus serotypes that indicate a close genetic relationship between serotypes 1 and 4 (51). However, complete structural data on the E protein of all four serotypes and detailed analysis of glycosylation of the E protein grown in insect cells are not yet available. These experiments demonstrate that the recombinant CLEC5A protein used is biologically active as it binds to dengue virus.
The structure of CLEC5A demonstrates that it is not a canonical C-type lectin as it does not have a full set of calcium-coordinating and carbohydrate binding sites. This is also the case for C-type lectin-like molecules, such as CLEC-2 and NKG2D, that bind to unglycosylated protein ligands (22, 52). Therefore, from a structural perspective, CLEC5A might be expected to bind to a protein ligand but is reported to bind to dengue virus in a manner that is inhibited by sugars (3). However, there is a precedent for non-canonical sugar binding in Dectin-1, which is a C-type lectin that binds β-glucan oligosaccharides in the absence of calcium (43), although the molecular basis for this interaction is incompletely understood (53). We therefore analyzed the structure of CLEC5A for sites with potential for interactions with known dengue virus surface carbohydrate species and investigated whether CLEC5A bound to a wide range of microarrayed glycans. The surface of CLEC5A has regions that might contribute to carbohydrate binding, but no binding was seen to the wide spectrum of glycans investigated. There are several possible explanations for the negative glycan binding results. (a) The ligand may not be a carbohydrate. (b) The ligand may be a glycan not included in the arrays. (c) The ligand may be a composite of a glycan and a protein component.
We have shown that CLEC5A is homodimeric at the cell surface (Fig. 3D). This oligomerization may enhance the capacity of CLEC5A to bind to a dengue virus glycoprotein ligand at the cell surface by providing a larger surface for ligand binding. Our data are consistent with the view that the ligand of CLEC5A could be a protein with acidic residues surrounding a number of small non-polar regions on its surface and that this ligand may display carbohydrate modifications. However, it is unlikely that CLEC5A binds carbohydrate in the manner of canonical C-type lectins.
This structural and functional characterization of CLEC5A presents a rational basis for exploring the interaction of dengue virus with host immune cells and provides insight into the role of CLEC5A in dengue virus-induced lethal disease. The structure provides a secure foundation for exploration of the viral ligand and for the design of inhibitory compounds that may be of value in the treatment of dengue virus disease.
Supplementary Material
Acknowledgments
We thank Mark Ellis at the Synchrotron Radiation Source, Daresbury, UK and Keith Morris, Paul Young, Aleksandra Flanagan, Jon Grimes, Geoff Sutton, and Mark Sansom for useful conversations. We acknowledge colleagues in the Glycosciences Laboratory: Wengang Chai, Yibing Zhang, Maria Campanero-Rhodes, and Robert Childs for help with microarrays. We acknowledge The Protein-Glycan Interaction Core (H) of The Consortium for Functional Glycomics (funded by National Institutes of Health Grant GM62116 from the NIGMS) for the glycan array analysis.
This work was supported by the UK Medical Research Council, National Institute for Health Research Oxford Comprehensive Biomedical Research Centre Program, UK Research Councils Basic Technology Grant GR/S79268 (“Glycoarrays”), Engineering and Physical Research Councils Translational Grant EP/G037604/1, and the Portuguese Foundation for Science and Technology.

The on-line version of this article (available at http://www.jbc.org) contains supplemental experimental procedures, Figs. 1 and 2, and Tables 1 and 2.
The atomic coordinates and structure factors (code 2YHF) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- DAP
- DNAX-activating protein
- BRET
- bioluminescence resonance energy transfer
- BRETeff
- BRET efficiency
- E
- envelope.
REFERENCES
- 1. Wilder-Smith A., Schwartz E. (2005) N. Engl. J. Med. 353, 924–932 [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization (2009) Dengue and Dengue Haemorrhagic Fever, Fact Sheet Number 117, World Health Organization, Geneva [Google Scholar]
- 3. Chen S. T., Lin Y. L., Huang M. T., Wu M. F., Cheng S. C., Lei H. Y., Lee C. K., Chiou T. W., Wong C. H., Hsieh S. L. (2008) Nature 453, 672–676 [DOI] [PubMed] [Google Scholar]
- 4. Bakker A. B., Baker E., Sutherland G. R., Phillips J. H., Lanier L. L. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9792–9796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yim D., Jie H. B., Sotiriadis J., Kim Y. S., Kim Y. B. (2001) Cell. Immunol. 209, 42–48 [DOI] [PubMed] [Google Scholar]
- 6. Gingras M. C., Lapillonne H., Margolin J. F. (2002) Mol. Immunol. 38, 817–824 [DOI] [PubMed] [Google Scholar]
- 7. Inui M., Kikuchi Y., Aoki N., Endo S., Maeda T., Sugahara-Tobinai A., Fujimura S., Nakamura A., Kumanogoh A., Colonna M., Takai T. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 4816–4821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aoki N., Kimura Y., Kimura S., Nagato T., Azumi M., Kobayashi H., Sato K., Tateno M. (2009) J. Leukoc. Biol. 85, 508–517 [DOI] [PubMed] [Google Scholar]
- 9. Watson A. A., O'Callaghan C. A. (2010) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 29–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O'Callaghan C. A., Byford M. F., Wyer J. R., Willcox B. E., Jakobsen B. K., McMichael A. J., Bell J. I. (1999) Anal. Biochem. 266, 9–15 [DOI] [PubMed] [Google Scholar]
- 11. Vagin A., Teplyakov A. (2000) Acta Crystallogr. D Biol. Crystallogr. 56, 1622–1624 [DOI] [PubMed] [Google Scholar]
- 12. Sullivan L. C., Clements C. S., Beddoe T., Johnson D., Hoare H. L., Lin J., Huyton T., Hopkins E. J., Reid H. H., Wilce M. C., Kabat J., Borrego F., Coligan J. E., Rossjohn J., Brooks A. G. (2007) Immunity 27, 900–911 [DOI] [PubMed] [Google Scholar]
- 13. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 14. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 15. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 16. Vriend G. (1990) J. Mol. Graph. 8, 52–56, 29 [DOI] [PubMed] [Google Scholar]
- 17. Barrett C. P., Hall B. A., Noble M. E. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2280–2287 [DOI] [PubMed] [Google Scholar]
- 18. Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 19. Psachoulia E., Fowler P. W., Bond P. J., Sansom M. S. (2008) Biochemistry 47, 10503–10512 [DOI] [PubMed] [Google Scholar]
- 20. James J. R., Oliveira M. I., Carmo A. M., Iaboni A., Davis S. J. (2006) Nat. Methods 3, 1001–1006 [DOI] [PubMed] [Google Scholar]
- 21. Watson A. A., O'Callaghan C. A. (2005) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61, 1094–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Watson A. A., Brown J., Harlos K., Eble J. A., Walter T. S., O'Callaghan C. A. (2007) J. Biol. Chem. 282, 3165–3172 [DOI] [PubMed] [Google Scholar]
- 23. Jones G., Willett P., Glen R. C. (1995) J. Mol. Biol. 245, 43–53 [DOI] [PubMed] [Google Scholar]
- 24. Verdonk M. L., Cole J. C., Hartshorn M. J., Murray C. W., Taylor R. D. (2003) Proteins 52, 609–623 [DOI] [PubMed] [Google Scholar]
- 25. Modis Y., Ogata S., Clements D., Harrison S. C. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dimasi N., Moretta L., Biassoni R. (2004) Immunol. Res. 30, 95–104 [DOI] [PubMed] [Google Scholar]
- 27. Zelensky A. N., Gready J. E. (2003) Proteins 52, 466–477 [DOI] [PubMed] [Google Scholar]
- 28. Perutz M. (1994) Protein Sci. 3, 1629–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Radaev S., Rostro B., Brooks A. G., Colonna M., Sun P. D. (2001) Immunity 15, 1039–1049 [DOI] [PubMed] [Google Scholar]
- 30. Ohki I., Ishigaki T., Oyama T., Matsunaga S., Xie Q., Ohnishi-Kameyama M., Murata T., Tsuchiya D., Machida S., Morikawa K., Tate S. (2005) Structure 13, 905–917 [DOI] [PubMed] [Google Scholar]
- 31. Moore J. O., Hendrickson W. A. (2009) Structure 17, 1195–1204 [DOI] [PubMed] [Google Scholar]
- 32. Watson A. A., Christou C. M., James J. R., Fenton-May A. E., Moncayo G. E., Mistry A. R., Davis S. J., Gilbert R. J., Chakera A., O'Callaghan C. A. (2009) Biochemistry 48, 10988–10996 [DOI] [PubMed] [Google Scholar]
- 33. Hughes C. E., Pollitt A. Y., Mori J., Eble J. A., Tomlinson M. G., Hartwig J. H., O'Callaghan C. A., Fütterer K., Watson S. P. (2010) Blood 115, 2947–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feng J., Call M. E., Wucherpfennig K. W. (2006) PLoS Biol. 4, e142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thepparit C., Smith D. R. (2004) J. Virol. 78, 12647–12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mondotte J. A., Lozach P. Y., Amara A., Gamarnik A. V. (2007) J. Virol. 81, 7136–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pokidysheva E., Zhang Y., Battisti A. J., Bator-Kelly C. M., Chipman P. R., Xiao C., Gregorio G. G., Hendrickson W. A., Kuhn R. J., Rossmann M. G. (2006) Cell 124, 485–493 [DOI] [PubMed] [Google Scholar]
- 38. Hacker K., White L., de Silva A. M. (2009) J. Gen. Virol. 90, 2097–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miller J. L., de Wet B. J., Martinez-Pomares L., Radcliffe C. M., Dwek R. A., Rudd P. M., Gordon S. (2008) PLoS Pathog. 4, e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fukui S., Feizi T., Galustian C., Lawson A. M., Chai W. (2002) Nat. Biotechnol. 20, 1011–1017 [DOI] [PubMed] [Google Scholar]
- 41. Galustian C., Park C. G., Chai W., Kiso M., Bruening S. A., Kang Y. S., Steinman R. M., Feizi T. (2004) Int. Immunol. 16, 853–866 [DOI] [PubMed] [Google Scholar]
- 42. Blixt O., Head S., Mondala T., Scanlan C., Huflejt M. E., Alvarez R., Bryan M. C., Fazio F., Calarese D., Stevens J., Razi N., Stevens D. J., Skehel J. J., van Die I., Burton D. R., Wilson I. A., Cummings R., Bovin N., Wong C. H., Paulson J. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 17033–17038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Palma A. S., Feizi T., Zhang Y., Stoll M. S., Lawson A. M., Díaz-Rodríguez E., Campanero-Rhodes M. A., Costa J., Gordon S., Brown G. D., Chai W. (2006) J. Biol. Chem. 281, 5771–5779 [DOI] [PubMed] [Google Scholar]
- 44. Feizi T., Chai W. (2004) Nat. Rev. Mol. Cell Biol. 5, 582–588 [DOI] [PubMed] [Google Scholar]
- 45. Tomiya N., Narang S., Lee Y. C., Betenbaugh M. J. (2004) Glycoconj. J. 21, 343–360 [DOI] [PubMed] [Google Scholar]
- 46. Drickamer K. (1989) Biochem. Soc. Trans. 17, 13–15 [DOI] [PubMed] [Google Scholar]
- 47. Call M. E., Wucherpfennig K. W., Chou J. J. (2010) Nat. Immunol. 11, 1023–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Falke J. J., Erbse A. H. (2009) Structure 17, 1149–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Laub M. T., Goulian M. (2007) Annu. Rev. Genet. 41, 121–145 [DOI] [PubMed] [Google Scholar]
- 50. Szurmant H., White R. A., Hoch J. A. (2007) Curr. Opin. Struct. Biol. 17, 706–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blok J. (1985) J. Gen. Virol. 66, 1323–1325 [DOI] [PubMed] [Google Scholar]
- 52. O'Callaghan C. A., Cerwenka A., Willcox B. E., Lanier L. L., Bjorkman P. J. (2001) Immunity 15, 201–211 [DOI] [PubMed] [Google Scholar]
- 53. Brown J., O'Callaghan C. A., Marshall A. S., Gilbert R. J., Siebold C., Gordon S., Brown G. D., Jones E. Y. (2007) Protein Sci. 16, 1042–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.