Abstract
Cyclotides, a large family of cyclic peptides from plants, have a broad range of biological activities, including insecticidal, cytotoxic, and anti-HIV activities. In all of these activities, cell membranes seem likely to be the primary target for cyclotides. However, the mechanistic role of lipid membranes in the activity of cyclotides remains unclear. To determine the role of lipid organization in the activity of the prototypic cyclotide, kalata B1 (kB1), and synthetic analogs, their bioactivities and affinities for model membranes were evaluated. We found that the bioactivity of kB1 is dependent on the lipid composition of target cell membranes. In particular, the activity of kB1 requires specific interactions with phospholipids containing phosphatidylethanolamine (PE) headgroups but is further modulated by nonspecific peptide-lipid hydrophobic interactions, which are favored in raft-like membranes. Negatively charged phospholipids do not favor high kB1 affinity. This lipid selectivity explains trends in antimicrobial and hemolytic activities of kB1; it does not target bacterial cell walls, which are negatively charged and lacking PE-phospholipids but can insert in the membranes of red blood cells, which have a low PE content and raft domains in their outer layer. We further show that the anti-HIV activity of kB1 is the result of its ability to target and disrupt the membranes of HIV particles, which are raft-like membranes very rich in PE-phospholipids.
Keywords: Antimicrobial Peptides, Antiviral Agents, Phosphatidylethanolamine, Phospholipid Vesicle, Plasma Membrane, Cyclic Peptides, Hemolytic Activity, Cyclotides, Model Membranes, Peptide-Membrane Interactions
Introduction
Cyclotides are plant-derived peptides characterized by a cyclic backbone and three disulfide bonds forming a cystine knot motif (1) (Fig. 1) that confers them with exceptional stability (2). Their natural function appears to be as host defense insecticidal agents (3), but many other activities have also been reported, including uterotonic (4), anti-HIV (5), hemolytic (6), antibacterial (7), anti-cancer (8), and pesticidal action (9). These activities reflect the ability of cyclotides to target cells of different compositions. More than 150 cyclotides have been characterized (10), and their diverse sequences, remarkable stability, and various bioactivities suggest that they have exciting potential as molecular frameworks in drug design (11).
FIGURE 1.
Sequence and structure of kB1. A, the sequence of kB1 is shown. Backbone cyclization between Gly-1 and Asn-29 is illustrated with a black line, and the disulfide connectivity is shown in yellow. The segments between Cys residues are termed loops. B, a three-dimensional structure of kB1 (PDB ID 1nb1) shows the cyclic knotted nature of cyclotides. The six Cys are labeled, and backbone loops are identified. Blue and green circles show the position of Gly-1 and Trp-23 residues, respectively. The black arrow indicates the direction of the peptide chain. C, surface representation of the molecule is shown in two views, illustrating the hydrophobic (green), bioactive (red), and amendable faces (blue). The same color code is used in the primary sequence shown in panel A.
It is believed that the activity of cyclotides is mediated by their ability to target and disrupt cell membranes. Such a mechanism of action is consistent with their hemolytic properties (6) and ability to disrupt gut epithelial cells in lepidopteron larvae (3) and is further supported by a range of biophysical studies (12–15). The prototypic cyclotide kalata B1 (kB1)7 is the most well studied cyclotide, and it has been shown to bind to (12) and disrupt phospholipid bilayers by a pore-forming mechanism (15). Furthermore, in a previous study we showed that the all-d enantiomer (d-kB1) is bioactive, suggesting that activity does not require recognition by a chiral protein receptor (9). In combination, these reports suggest that cyclotides target cell membranes by interacting directly with lipids.
Differences in bioactivity are observed when different targets (16) and/or different cyclotides are compared (17), suggesting that a range of factors in membrane and cyclotide structures contribute to bioactivity. As different organisms have distinct membrane characteristics (e.g. phospholipid composition and asymmetric distribution, the presence/absence of sterol and raft-like domains or the presence of charge), we were interested in determining the key-properties of cell membranes that dictate the activity of cyclotides.
In this study the membrane properties that modulate cyclotide bioactivity were identified by correlating biophysical studies in model membranes with bioassays conducted using red blood cells (RBCs), bacteria, and HIV particles. Specifically, kB1 and several analogs of varying potencies were assessed for their affinities for several phospholipid mixtures. The results were compared with those obtained in RBCs, bacteria, and HIV, each having its own characteristic membrane composition. Our results show a clear correlation between membrane affinity/selectivity and biological potency.
EXPERIMENTAL PROCEDURES
Peptide Extraction
Native kB1 was extracted and isolated from the above ground parts of Oldenlandia affinis and purified as described previously (18). The concentrations of kB1 samples were determined before each assay by absorbance at 280 nm (ϵ280 = 5875 m−1 cm−1).
Peptide Synthesis
Cyclic kB1 mutants were synthesized, cyclized, and oxidized as described previously (16, 19). The names of the mutants are abbreviated for simplicity; for instance, W23K refers to substitution of the Trp residue at position 23 with a Lys residue. The single Lys-kB1 mutants T8K, V10K, N15K, T16K, T20K, W23K, V25K, and N29K, the double Lys-kB1 mutant T20K/N29K, and the Asp mutant E7D were used in this study. An acyclic form of kB1 opened in loop 6, leaving Gly-1 at the C terminus and Asn-29 at the N terminus, was synthesized using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry and oxidized as previously described (16). The purity of the peptides was assessed using analytical RP-HPLC, and the correct folding of the peptides was confirmed by 1H NMR spectrometry, as described previously (20, 21). All peptides had ≥95% purity.
Preparation of Lipid Vesicles
Large unilamellar vesicles (LUVs) of diameter of 100 nm were used in fluorescence studies, and small unilamellar vesicles of diameter of 50 nm were used for surface plasmon resonance (SPR) studies, prepared by extrusion as described earlier (22). Synthetic lipids palmitoyloleoylphosphatidylcholine (POPC), palmitoyloleoylphosphatidylglycerol (POPG), palmitoyloleoylphosphatidylethanolamine (POPE), palmitoyloleoylphosphatidylserine (POPS), dipalmitoylglycerophosphatidylcholine (DPPC), and cholesterol (Chol) or extracted lipids (octadecanoyl sphingomyelin (SM) extracted from porcine brain, palmitoyl-linoleoylphosphatidylinositol (PLPI) from soy, lipopolysaccharide (LPS) from Escherichia coli (outer membrane)) were used to prepare model membranes. Different lipid mixtures were prepared, and the molar percentage of each lipid component except POPC is indicated for each mixture in the text; for instance, POPC/Chol/SM (20/40%) refers to a mixture with 40% POPC, 20% Chol, and 40% SM in molar ratio). Unless otherwise stated, peptide and lipid samples were prepared in buffer 10 mm HEPES, pH 7.4, containing 150 mm NaCl to represent the physiological pH and ionic strength.
Preparation of Vesicles from Spheroplasts
To remove the outer membrane of E. coli, spheroplasts were prepared by the osmotic shock procedure, as described previously (23, 24). Vesicles obtained with spheroplasts were used for SPR studies. For these studies the spheroplasts were resuspended in HEPES buffer and sonicated for lysis of membrane and release of internal contents. Homogenization of the vesicles to a 50-nm diameter was obtained by extrusion.
Surface Plasmon Resonance Studies
The binding affinity of kB1 and analogs to lipid bilayers was evaluated by SPR. Solutions were freshly prepared and filtered (0.22-μm pore size). Several lipid mixtures were used to determine how membrane properties modulate kB1 membrane binding. L1 sensor chips and a BIAcore 3000 system (Biacore, GE Healthcare) were used. Lipids were deposited on the L1 chip, as described previously (22). Association to the lipid bilayer was evaluated by injection of peptide sample over the lipid surface for 180 s (5 μl/min), whereas the dissociation was followed for 600 s. The chip surface was regenerated after each injection cycle, as described previously (22). All measurements were conducted at 25 °C. Association and dissociation rates were calculated by fitting with BIAeval software (Version 4.1).
Unless otherwise stated, the lipid mixtures studied were in the homogenous liquid disordered (ld) phase at 25 °C. The amount of lipid deposited on the chip surface depends on the lipid mass density of the packing of the mixture components (see Table 1); it should be stressed that both the amount of kB1 and the total amount of lipid deposited on the chip surface are important for an accurate comparison of the binding affinity of kB1 for the different lipid mixtures. Thus, the peptide/lipid ratio (P/L) at a reporting point, at the end of the association phase (t = 170 s), was calculated to estimate the affinity for the different lipid mixtures (25).
TABLE 1.
Affinity of kB1 for membranes containing POPE
| Lipid composition | Lipid phase | Lipid depositiona | kB1/PEb | kac | kdd |
|---|---|---|---|---|---|
| RU | mol/mol | ×103 M.s−1 | ×10−3s−1 | ||
| POPC/POPE (10%) | ld | 7594 | 1.53 | 4.20 ± 0.09 | 13.00 ± 0.09 |
| POPC/POPE/Chol (10/20%) | ld + lo | 8649 | 1.91 | 3.98 ± 0.08 | 8.45 ± 0.07 |
| POPC/POPE/Chol (10/33%) | ld + lo | 8486 | 2.63 | 3.50 ± 0.11 | 8.10 ± 0.06 |
| POPC/POPE/SM (10/40%) | ld + so | 8650 | 1.76 | 5.02 ± 0.08 | 6.37 ± 0.02 |
| POPC/POPE/Chol/SM (10/33/40%) | lo | 9019 | 2.38 | 3.90 ± 0.15 | 6.45 ± 0.04 |
| POPC/POPE/Chol/SM (10/20/40%) | ld + lo + so | 8492 | 1.62 | 4.53 ± 0.07 | 4.61 ± 0.01 |
| POPC/POPE/POPG/Chol/SM (10/10/20/40%) | ld + lo + so | 7867 | 1.55 | 4.30 ± 0.07 | 5.23 ± 0.01 |
a Lipid was deposited on an L1 chip, reaching a plateau, which confirms coverage of the chip surface. The total amount of lipid was converted to mass (1 response unit (RU) = 1 pg/mm2 of peptide or lipid).
b The amount of kB1 bound to the membrane was estimated after 170 s of peptide sample injection. The total number of lipid moles was estimated taking into account the average mass of the lipid mixture being PE 10% of the lipid.
c The kinetic of kB1-membrane binding is a complex event, and globally fitting was not possible. For comparison of the initial association rate the apparent association rate, ka, was estimated in the beginning of injection (0–40 s).
d Apparent dissociation rate, kd, was estimated in the dissociation phase considering a peptide-lipid Langmuir kinetic model.
NMR Spectroscopy
Samples of kB1 and E7D were prepared in 450 μl of 50 mm sodium phosphate buffer, pH 7.0, with 100 mm NaCl and 50 μl of D2O to a final concentration of 0.58 and 0.46 mm, respectively. A 0.2 m stock solution of PE was prepared in buffer with 10% (v/w) D2O. One-dimensional 1H NMR and two-dimensional 1H,1H two-dimensional total correlation spectra were recorded for each sample of pure peptide and upon titration with PE stock solution (0–67 mm final concentration). Resonance assignments were confirmed from two-dimensional NOESY spectra. Chemical shifts were monitored throughout the titration, and each resonance that experienced a shift of more than 0.02 ppm in the studied range was analyzed using a one-site-specific binding model in GraphPad prism. All data points could be fitted to a one-site binding with a dissociation constant Kd of 56 mm (95% confidence interval 47–65 mm).
Leakage Studies
Vesicle leakage induced by kB1 or by its analogs was quantified by carboxyfluorescein (CF) dequenching, as detailed previously (15). Several concentrations (2-fold dilution starting from 10 μm) of kB1, N29K, or V25K were incubated during 10 min with LUVs (5 μm lipid). The fluorescence intensity (λexcitation = 489 nm and λemission = 515 nm) of the released CF solutions was followed after 10 min of incubation with the peptide and read using a PerkinElmer LS50B fluorescence spectrophotometer. The percentage of leakage was calculated as before (15).
Transbilayer Movement of Phospholipids
The flip-flop of phospholipids labeled with a nitrobenzoxadiazole (NBD) moiety was evaluated by quenching of the NBD fluorescence by Co2+, as described previously (26). To maintain the properties of the headgroup, phospholipids with the NBD moiety attached to the acyl chain were used. Briefly, LUVs doped with 1% 1-palmitoyl-2-{6-(NBD)-hexanoyl}-sn-glycero-3-phosphocholine (C6-NBD-PC) or with C6-NBD-PE were compared to identify possible differences in outward movement of these two headgroups. 20 mm Co2+ was added to vesicle suspensions (100 μm lipid concentration) to quench the fluorescence of NBD moieties in the outer layer, and the transbilayer movement was evaluated in the presence of kB1 or its analogs by titration with a stock solution. The fluorescence emission spectrum (λexcitation = 460 nm) before and after peptide addition was scanned using a PerkinElmer LS50B fluorescence spectrophotometer. POPC and POPC/POPE/Chol/SM (10/33/40%) vesicles were compared. It is worth mentioning that with these lipid systems the leakage induced by kB1 is negligible up to P/L = 2; therefore, variations in quenching are not due to permeabilization of the vesicles.
Hemolytic Studies
Fresh human blood was used to quantify hemolysis induced by kB1 and its analogs. Assay and data analysis were conducted, as described earlier (16). All peptide solutions were assayed in triplicate and prepared in HEPES buffer, pH 7.4, containing 150 mm NaCl.
Antibacterial Studies
Bacterial growth inhibition induced by kB1 was evaluated to examine its antimicrobial activity. Growth of Gram-negative E. coli (DH5α) and Gram-positive Staphylococcus aureus (RN4220) was tested in the presence of 100 μm of kB1 and compared with controls as previously described (27).
Atomic Force Microscopy Imaging
Direct observation of the effects induced by kB1 or its analogs V25K and N29K in bacterial cells and eukaryotic cells was obtained by atomic force microscopy. E. coli was used as a bacterial model, whereas RBCs were used as a model for eukaryotic cells. E. coli was grown as for antibacterial studies; incubation with peptide and sample preparation was as previously described (28). Final peptide concentrations were 1, 10, or 100 μm kB1, V25K, or N29K. Fresh blood was incubated with kB1 (final concentrations, 1, 5, 10, or 50 μm) for 2 h at room temperature to evaluate the effect on RBCs. The number of RBCs in the blood, as determined using Cell-Dyn 1600, was 5.16 × 106 cells/ml. After incubation, a red suspension revealing hemolysis was visible for the two highest concentrations. A drop of blood was smeared on a glass microscope slide and left to dry at room temperature. Samples were scanned in intermittent contact mode on NanoWizard II equipment (JPK Instruments, Berlin, Germany) mounted on an Axiovert 200 inverted microscope (Carl Zeiss, Jena, Germany) with oxidized sharpened silicon tips (ACL tips from Applied Nanostructures with a tip radius of 6 nm, resonant frequency of about 190 kHz, and spring constant of 45 newtons/m). Scanning speed was set to 0.8–1 and 0.3 Hz for E. coli and RBC imaging, respectively. Images were acquired with a typical 512 × 512 resolution. Data were analyzed using JPK image processing software Version 3 (JPK Instruments) and Gwyddion Version 2.19 (Czech Metrology Institute).
Anti-HIV Studies
HIV-1 particles (NL4.3 strain (29) or 92RW016 and a clade A strain, obtained from the NIH AIDS Research and Reference Reagent Program) were immobilized onto a 96-well plate by HLA-DR antibody capture, as previously described (30). Virucidal activity of kB1 or its analogs V25K and N29K was evaluated by treatment of the immobilized virus with different concentrations of kB1 or its analogs. After 1 h of incubation at 37 °C, peptides were thoroughly washed with culture medium to remove unbound cyclotide followed by the addition of 2.5 × 104 TZM-bl indicator cells per well (31) to measure the infectiveness of HIV particles. HIV infection of TZM-bl cells was measured after 48 h of incubation at 37 °C and 5% CO2 by measuring luciferase activity using the Steady Glo Luciferase Assay System (Promega) according to the manufacturer's instructions. The concentration of cyclotide leading to a 50% reduction in viral infectivity (VC50) compared with untreated virus was calculated by nonlinear regression analysis of the log10 concentration of cyclotide versus the percentage inhibition of luciferase activity using GraphPad PRISM software, as published (32). Data were obtained from three independent assays.
HIV Particle Disruption
To evaluate if kB1 induces disruption of HIV-1 particles, NL4.3 virus stock was incubated with 50 μg/ml kB1 (∼17 μm), V25K, or N29K for 1 h at 37 °C. Samples were centrifuged through a 20% w/v sucrose cushion (100,000 × g, 1 h 4 °C) to pellet viral particles. The pellet was lysed, and virion proteins were separated by SDS-PAGE and detected by Western blot analysis using the Odyssey Infrared Imaging System (LI-COR, Lincoln, NE) and HIV human immune serum as described (33).
RESULTS
kB1 Has Selectivity for Membranes Containing Phosphatidylethanolamine Phospholipids
The affinity of kB1 for different lipid mixtures was evaluated using SPR (22). Membranes of eukaryotic cells are fluid and rich in lipids having PC; thus, POPC, which forms bilayers with ld properties (i.e. fluid phase) at room temperature, was chosen to mimic the bulk phase in eukaryotic cell membranes. The selectivity of kB1 was discerned by comparing pure neutral POPC bilayers with more complex lipid mixtures (Fig. 2A) that mimicked varying properties of cell membranes.
FIGURE 2.
Binding of kB1 to lipid membranes requires the presence of PE-phospholipids. A, the affinity of kB1 for the different lipid systems was studied using SPR and is compared by the amount of kB1 bound to the membrane at the end of association phase (after injection of 25 μm kB1 over the membrane surface during 170 s). The P/L ratio was calculated to normalize the response to the total amount of lipid deposited in the chip (1 response unit (RU) = 1 pg/mm2 of peptide or lipid). B, sensorgrams were obtained upon injection of 25 μm kB1 over POPC or POPC/POPE (5%; 10 or 20%) during 180 s (association phase) and dissociation from the membranes followed for 600 s (dissociation phase). C, shown is the amount of the kB1 bound to POPC or POPC/POPE (5%; 10 or 20%) at the end of association phase versus the amount of POPE in the chip surface (half of the total amount of POPE was considered, as only half of the lipid is in the outer leaflet). D, sensorgrams were obtained upon injection of 25 μm kB1 onto membranes that mimic the outer membrane (LPS and POPC/LPS (20%)) or the inner (POPG/POPE (40/60%)) membrane of E. coli as well as native inner membrane.
kB1 was found to bind weakly to neutral POPC bilayers and negatively charged bilayers, as shown by studies increasing the amount of the anionic phospholipid POPG in POPC matrix (Fig. 2A). This finding was confirmed by the use of another anionic phospholipid, POPS, which revealed that negative headgroups in the bilayer do not significantly increase the affinity of kB1.
When membranes with different fluidities (e.g. POPC at ld, POPC/Chol/SM (33/40%) at liquid-ordered phase (lo) and DPPC at solid phase (so)) were compared, no effect on the affinity of kB1 for the lipid bilayer was detected (Fig. 2A). The same trend was observed when homogenous ld (POPC) was compared with heterogeneous mixtures (i.e. with lateral segregation of clustered lipids, forming rigid domains in the bulk fluid matrix; POPC/Chol (20%) (ld + lo), POPC/Chol (33%) (ld + lo), POPC/SM (40%) (ld + so), and POPC/Chol/SM (20/40%) (ld + lo + so) (34). These results show that the existence of phase boundaries does not increase the affinity of kB1 for membranes. Moreover, Chol or SM by itself does not facilitate the binding of kB1 to membranes.
The effects of other biologically relevant phospholipid headgroups, such as phosphatidylinositol (PI) and PE were also evaluated. The presence of the carbohydrate-containing and negatively charged PI did not increase kB1 binding, as shown with POPC/PLPI (20%) bilayers, whereas the presence of the neutral, but rather small, PE headgroup had a clear effect (Fig. 2, A and B). Plotting the amount of kB1 bound as a function of the amount of POPE in the membrane (Fig. 2C) revealed a strong linear correlation, suggesting that PE headgroups are necessary for membrane targeting by kB1. Thus, kB1 binding to membranes is specific (i.e. not simply dependent on electrostatics or surface adsorption) and stems from PE targeting.
When POPE Is Present, kB1 Prefers Liquid-ordered Domains Over the Liquid-disordered Phase
It was of interest to evaluate whether the lipid environment (i.e. ld, lo, ld + lo, ld + so, or ld + lo + so) influences the binding of kB1 when PE is present. The data indeed showed that the lipid environment surrounding PE significantly influences the extent and kinetics of kB1 binding to membranes in which 10% of POPE was included (Fig. 2A and supplemental Fig. S1). A comparison (Table 1) of the ratio kB1/PE for homogeneous ld (POPC/POPE (10%)) and homogeneous lo (POPC/POPE/Chol/SM (10/33/40%)) revealed that when a similar percentage of POPE is available, kB1 has greater affinity for membranes in the lo phase, rich in Chol/SM, than in the ld phase. The kinetic data (Table 1) show similar apparent association rates (ka) and a decrease in the apparent dissociation rate (kd). Similar ka values suggest similar initial binding rates, whereas a slower kd reveals a tighter binding of kB1 to lo membranes. Heterogeneous membrane systems showed intermediate behavior. These results suggest that after initial membrane binding, through specific interactions with PE headgroups, nonspecific lipid-peptide hydrophobic interactions mediate insertion of the cyclotide into bilayers. Overall, the data showed that PE-phospholipids are responsible for the membrane targeting by kB1, whereas hydrophobic interactions appear to modulate the insertion of the cyclotide into the membrane.
kB1 Does Not Target the Negatively Charged Outer Membrane of Bacteria
kB1 binds weakly to negatively charged bilayers, as shown by studies increasing the amount of the anionic phospholipid POPG in POPC matrix (Fig. 2A). The addition of POPG to membranes containing PE (see POPC/POPE/Chol/SM (10/20/40%) versus POPC/POPE/POPG/Chol/SM (10/10/20/40%), Fig. 2A, Table 1) revealed that negatively charged headgroups do not favor the binding of kB1 to POPE-containing membranes, supporting the hypothesis that kB1 membrane-binding is not primarily governed by electrostatic interactions.
The ability of kB1 to bind to bacteria-like membranes was further investigated with model membranes of compositions similar to the E. coli outer membrane (35) or inner membrane (36, 37). The outer membrane contains LPS in the outer layer and phospholipids in the inner layer, whereas the inner membrane comprises a phospholipid bilayer composed mainly of PE and PG phospholipids (37). Vesicles containing 100% LPS from E. coli were compared with POPC/LPS (20%) (Fig. 2D). The results revealed that kB1 does not bind to LPS. An inner membrane mimic system was prepared using vesicles from E. coli spheroplast and compared with POPG/POPE (40/60%) mixture. Similar binding efficiency for these membranes was observed (Fig. 2D) showing not only that membranes obtained from synthetic lipids can be correlated with those from biological extracts (i.e. POPG/POPE versus E. coli spheroplasts) but also confirming the importance of PE for the binding of kB1 to biological membranes. These results show that kB1 can target the inner membrane of E. coli, which is very rich in PE headgroups but does not target the LPS layer in the outer membrane.
The Bioactivity of kB1 Is Dependent on Membrane Affinity
Previous Ala-scan and Lys-scan studies show that a single mutation can ablate (16, 38) or increase (16) the hemolytic or pesticidal activities of kB1, depending on where the mutation occurs. Three patches in the molecule were identified: a bioactive patch, a hydrophobic patch, in which substitutions may ablate the activity, and an “amendable” face, where certain substitutions increase activity (Fig. 1C). Although the mutants showed differences in activity, they have similar three-dimensional folds to native kB1 (16) and can, therefore, be used to establish a comparison between activity and membrane binding properties. Inactive analogs with a mutation in the bioactive patch (i.e. E7D, T8K, N15K, and T16K) or in the hydrophobic patch (i.e. V4K, V10K, W23K, and V25K) and more potent analogs with mutations in the amendable face (i.e. T20K, N29K, and T20K/N29K) were compared with native kB1. Additionally, reduced kB1 (disulfide bonds reduced but the circular backbone maintained) and an acyclic form of kB1 (opened in loop 6, Gly-1 at the C terminus and Asn-29 at the N terminus, in which disulfide bonds, bioactive patch, and hydrophobic patch were kept) were also compared to establish if structural features are important for the binding.
The SPR data obtained for the kB1 mutants injected over POPE-containing bilayers revealed that inactive mutants have weak binding, whereas more active mutants bind better than native kB1, as shown with POPC/POPE/Chol/SM (10/33/40%) bilayers (Fig. 3A) and confirmed with other lipid mixtures (e.g. POPC/POPE (10 or 20%), POPC/POPE/Chol/SM (10/20/40%), and POPC/POPE/POPG/Chol/SM (10/10/20/40%)). Hemolytic potency correlates well with the amount of peptide bound to the membrane (Fig. 3B), indicating that the hemolytic efficiency kB1 and ability to bind to the lipid bilayer are intimately related. As expected, both inactive and active mutants had weak binding to bilayers that lack POPE. Reduced and acyclic forms of kB1 did not bind to lipid bilayers even when 20% POPE was present, indicating that both the cystine knot and cyclic backbone are important for membrane binding.
FIGURE 3.
Activity of kB1 is correlated with affinity for the lipid bilayer. Native kB1, active mutants (R28K, T20K, N29K, and T20K/N29K), and inactive mutants (V4K, E7D, T8K, V10K, N15K, V16K, W23K, and V25K) are compared in their affinity for lipid membranes. A, sensorgrams obtained upon injection of 25 μm of kB1 or analogs onto POPC/POPE/Chol/SM (10/33/40%) membranes are shown. B, correlation between the amount of peptide bound to the membrane and the hemolytic activity of the different analogs are shown. The values were normalized for the response obtained with the native kB1 (see supplemental Fig. S2 and Table S1). RU, response unit.
The Affinity of kB1 for POPE Is Due to a Specific Interaction with the Head Group
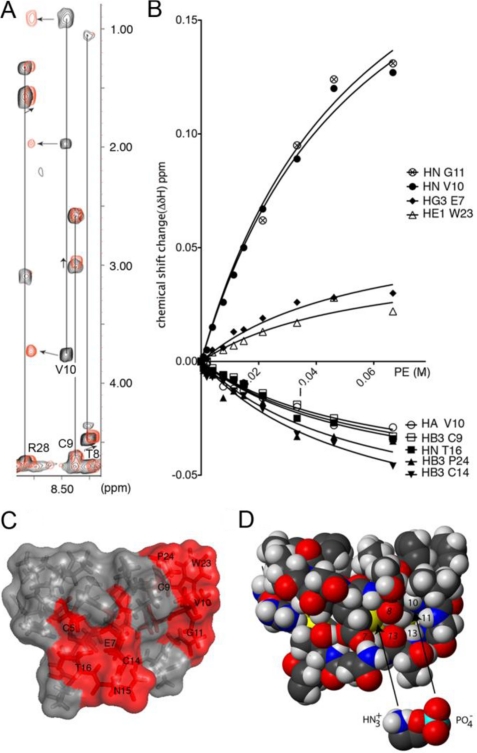
NMR was used to further investigate whether the selectivity of kB1 for membranes containing POPE is dependent on a specific interaction involving the lipid head group. On the addition of PE, chemical shift changes were observed for a number of kB1 residues, as shown in Fig. 4. Fitting of the data yielded a dissociation constant of 56 mm. The observed chemical shift changes are clustered to a large extent in the bioactive patch of kB1, as shown in Fig. 4C. To confirm the specificity of this interaction, the experiment was repeated with the highly conservative, but inactive, mutant E7D. No chemical shift differences above 0.005 ppm were seen for this mutant, confirming that the interaction between kB1 and PE is specific and that its disruption accounts for the poor affinity for PE-containing membranes and consequent lack of bioactivity of E7D (see Fig. 3). Analysis of the region in kB1 central for binding to PE reveals the presence of a distinct cluster of atoms with negatively (carboxyl of Glu-7 and the carbonyls of Thr-8 and Thr-13) and positively (NH of Val-10, Gly-1, and Thr-13) charged character. We propose a putative binding mode in which these regions coordinate with the PE zwitterionic structure (Fig. 4C).
FIGURE 4.
NMR monitored titration of kalata B1 with PE. A, two-dimensional total correlation spectra of kB1 (black) and kB1 after the addition of 67 mm PE (red) show examples of observed chemical shift changes. B, binding curves for selected resonances show an overall chemical shift change of >0.02 ppm within the studied range. Curves were fitted to the data using a one-to-one specific binding model. C, the PE binding surface of kalata B1 is shown. Residues with at least one resonance show a chemical shift change of >0.02 ppm upon binding to PE are shown in red. D, shown is the putative binding mode of PE to kB1.
NMR monitored studies of lipid-binding proteins with isolated soluble lipid headgroups have been reported for profilin I and inositol 1,4,5-triphosphate (39). Specific binding between human profilin I and inositol 1,4,5-triphosphate of similar affinity (10–50 mm) and chemical shift changes of the same order of magnitude to those observed here for kB1 and PE were reported.
Leakage Efficiency of kB1 Correlates with Its Membrane Selectivity
It was previously reported that kB1 can induce leakage of phospholipid vesicles (15). In the present study, leakage of LUVs of different lipid compositions (POPC, POPC/Chol/SM (33/40%), POPC/POPE (10%), and POPC/POPE/Chol/SM (10/33/40%)) induced by kB1 was studied to determine whether membrane affinity correlates with leakage efficiency. Vesicles loaded with CF were incubated with varying concentrations of kB1 (up to a P/L of two), and the leakage was quantified by the increase in CF fluorescence, as described previously (15). N29K, a membrane-active analog, and V25K, an inactive analog, were compared with native kB1.
Fig. 5A shows that kB1 induces leakage in POPC/POPE and POPC/POPE/Chol/SM vesicles with an effective EC50 of 2.55 μm (P/L = 0.51) and 0.65 μm (P/L = 0.13), respectively. By contrast, there is no detectable leakage of content from vesicles without POPE (i.e. POPC and POPC/Chol/SM, Fig. 5A). These results confirm that PE is required for membrane targeting by kB1 and show that membrane affinity correlates with leakage efficiency, with higher efficiency against more rigid vesicles. Studies with N29K and V25K (Fig. 4B) showed that the active mutant induced leakage, whereas the inactive analog did not.
FIGURE 5.
Membrane leakage and flip-flop induced by kB1. A, the fluorescence of CF was converted to the percentage of leakage and plotted as a function of P/L ratio. The efficiency of vesicle leakage induced by kB1 is compared for membrane with different lipid compositions. B, leakage from POPC/POPE/Chol/SM (10/33/40%) vesicles induced by kB1 is compared with the membrane-inactive V25K and the membrane-active N29K. C, outward movement of C6-NBD-PE and C6-NDB-PC phospholipids was compared for POPC and POPC/Chol/SM. NBD fluorescence quenching by Co2+ was followed upon titration with kB1. Quenching efficiency (represented by I0/I, where I0 is the initial fluorescence intensity, and I is the fluorescence obtained upon addition of peptide) is plotted as a function of P/L.
Transbilayer Movement of Phospholipids Is Facilitated in the Presence of kB1
Flip-flop, or transbilayer movement, is a process in which the polar headgroup of a lipid passes through the hydrophobic bilayer to locate in the opposing bilayer interface. In practice, lipids are symmetrically distributed in artificial membranes, and the passive spontaneous transbilayer movement is very slow due to the unfavorable passage of a hydrophilic headgroup across the hydrophobic membrane core; the transbilayer movement that occurs in biological membranes is an event that requires consumption of energy and lipid transporters to catalyze the process. Nevertheless, it has been reported that incorporation of peptides or proteins into a pure lipid membrane may accelerate flip-flop of phospholipids between both leaflets (40, 41).
Transbilayer movement of phospholipids induced by kB1 was evaluated by fluorescence quenching of NBD-labeled phospholipids induced by Co2+ (26). In POPC membranes, neither C6-NDB-PC nor C6-NBD-PE undergoes detectable transbilayer movement induced by kB1 (Fig. 5C). On the other hand, in POPC/Chol/SM vesicles, there is a strong increase of C6-NBD-PE fluorescence quenching upon titration with kB1; in contrast, the increase in quenching is weak for C6-NBD-PC. These results show that kB1 can promote net transbilayer movement of phospholipids and suggest that kB1 inserts close to, and recruits, phospholipids containing PE headgroups. In addition, the difference in the NBD-PE fluorescence quenching efficiency for POPC and POPC/Chol/SM bilayers suggests that at a low percentage of PE headgroups (1% of C6-NDB-PE), the presence of Chol and SM are determinants for the binding/insertion of kB1 in the membrane.
kB1 Has Hemolytic Properties but Not Antimicrobial Activity
The hemolytic activity of kB1 determined here is comparable with previous reports (16). The acyclic permutant had no hemolytic activity, consistent with other acyclic permutants (42) (see supplemental Fig. S1 and Table S1 for comparison of hemolysis efficiency obtained for kB1 and its analogs).
The antimicrobial activity of kB1 was tested in growth inhibition assays against E. coli (Gram-negative) and S. aureus (Gram-positive). No activity was observed against either of the organisms with 100 μm kB1 at physiologically relevant salt conditions (150 mm NaCl). The lack of antimicrobial activity at physiological salt conditions is in agreement with previous studies (7, 43).
Further information on the effects induced by kB1 in bacterial and eukaryotic cells was obtained by atomic force microscopy, in which direct observation of cell shape is possible. E. coli was used as a bacterial model, and RBCs were used as a model for eukaryotic cells. The active analog N29K and the inactive V25K were compared with kB1.
kB1 induces significant changes in the shape of RBCs (Fig. 6). In the presence of 1 μm kB1, RBCs lose their typical concave shape and exhibit a flat surface. With increasing kB1 concentration, the changes are even more evident; RBCs lose their disc-like shape and acquire a crenate shape. Such alterations in the RBC membrane are concomitant with an alteration in membrane permeability (44, 45), which eventually results in hemolysis. A similar concentration effect was observed for N29K. With V25K, alterations in the RBC shape are only evident at the highest concentration tested (see supplemental Fig. S3).
FIGURE 6.
kB1 binds to the RBCs membrane and induces changes in the RBCs shape. Images of RBCs were generated by atomic force microscopy for both control samples without the addition of peptide and after incubation with different kB1 concentrations.
Alteration in the shape of E. coli in the presence of antimicrobial peptides (AMPs) has recently been observed by atomic force microscopy (28) whereby E. coli loses shape with roughness at low micromolar concentration (e.g. 5 μm). In the presence of kB1, no effect was detected up to 10 μm (Fig. 7), suggesting weak antimicrobial activity. When 100 μm of kB1 was added to bacteria, alterations were visible but were distinct from the severe effects observed with other AMPs (28). In addition, the membrane-active N29K and the inactive V25K show a similar effect to kB1 (Fig. 7), showing that the membrane activity of kB1 does not produce antimicrobial activity against E. coli.
FIGURE 7.
The effect of kB1 and its analogs on the shape of E. coli followed by atomic force microscopy. Three different concentrations of kB1, N29K, or V25K were incubated with E. coli during 2 h. The images show that at low micromolar concentrations, neither kB1 nor its analogs induce effect on E. coli. Effects on E. coli shape are only evident when a high concentration of peptide is incubated with the bacteria. No differences among the three peptides were detected.
kB1 Has Direct Virucidal Activity against HIV Particles
Anti-HIV activity of kB1 has been reported (46), but little is known about the mechanism of the antiviral effects. We recently hypothesized that the protective effect of cyclotides is exerted before the entry of virus into cells (47). To test this hypothesis, HIV virions were captured by HLA-DR antibodies on the surface of 96-well plates incubated with kB1 at different concentrations and then washed before being cultured with TZM-bl reporter cells. With this protocol it is possible to evaluate whether kB1 demonstrates virucidal activity against HIV. kB1 was tested against two different HIV strains: NL4.3, which uses CXCR4 as co-receptor for entry into cells, and a clade A strain, which uses the CCR5 co-receptor. V25K and N29K were also compared to evaluate how important the ability to target the viral membrane is for kB1 anti-HIV activity.
Inhibition of virus replication, with a kB1 dose-dependent response (Fig. 8A), is evident for both strains tested, with a VC50 of 2.04 μm for NL4.3 and 4.54 μm for Clade A. Targeting of HIV by kB1 is likely to occur by peptide-membrane interactions, as supported by the activity of N29K (VC50 of 8.06 μm) and lack of activity of V25K (Fig. 8B).
FIGURE 8.
KB1 induces disruption of HIV membrane. A, activity of kB1 against HIV-1 NL4.3 or HIV-1 Clade is shown. A, immobilized virus particles were pretreated with kB1 for 1 h and washed off before being cultured with target TZM-bl cells. The error bars represent the S.D. B, KB1 is compared with the membrane-inactive V25K and the membrane-active N29K for their virucidal activity against NL4.3. The error bars represent S.D. For clarity, only the positive S.D. is represented in this panel. C, after treatment of NL4.3 virus particles with 50 μg/ml kB1, V25K, or N29K, viral proteins were detected by Western blot. A low level of viral capsid protein p24 for kB1- and N29K-treated virus compared with control treatment, but not for V25K, indicates removal of membrane envelope and disruption of the particle.
To further evaluate the membrane-targeting hypothesis, the ability of kB1 to disrupt HIV-1 particles was evaluated. NL4.3 virus particles were treated with kB1, V25K, or N29K. A clear decrease in the viral capsid protein, p24, with both kB1 and N29K, but not with V25K, was observed compared with NL4.3 not incubated with cyclotide (Fig. 8C), indicating perturbation of the viral lipid envelope and disruption of viral particles.
DISCUSSION
In this study we analyzed the binding of kB1 and analogs to model membranes with defined lipid compositions to characterize the nature of the peptide-lipid interactions. These studies were compared with the ability of kB1 to target organisms with distinct biological membranes (i.e. RBCs, bacteria, and HIV particles), and a correlation was found between them. As well as increasing our understanding of cyclotide-membrane interactions, the results provide a mechanistic insight into the ability of kB1 to disrupt HIV particles by targeting the HIV membrane envelope.
The SPR results show that the affinity of kB1 for membranes depends on lipid composition (see Fig. 2). Specifically, the extent of binding to homogenous fluid lipid bilayers is dependent on the amount of PE-phospholipids, with kB1 showing very weak binding to membranes that lack PE-phospholipids. In addition, the environment surrounding PE-phospholipids further modulates the membrane affinity; interestingly, negative charges have no effect, whereas a raft-like environment favors insertion of kB1 into membranes (Table 1). Even a small percentage of PE is enough to allow association of kB1 with the lipid bilayer if in a membrane containing Chol and SM (see Fig. 5C). The results are consistent with a specific interaction between kB1 and PE headgroups being required for efficient membrane targeting, with the hydrophobic surroundings assisting with the insertion.
The importance of an interaction with the lipid bilayer for the bioactivity of kB1 is seen when the membrane affinities of kB1 analogs are correlated with hemolytic potencies (see Fig. 3). Inactive Lys mutants (16) lack affinity for the membrane, whereas more active Lys mutants show higher affinity than native kB1. The reduced form and the acyclic permutant of kB1 are unable to bind to membranes. Overall, the results show that kB1 activity is correlated with membrane binding and that both the sequence and the cyclic knotted structure of kB1 are important for membrane-targeting.
The surface-exposed bioactive face and hydrophobic patch on kB1 (see Fig. 1C) have been reported to be important for activity (16, 38). The current study has shown that both the bioactive face and hydrophobic patch contribute to membrane affinity, as single mutations in either of these patches (e.g. T16K and V25K, respectively) suppress membrane binding (see Fig. 3). Whereas the bioactive patch has a specific interaction with the PE headgroup (see Fig. 4), the hydrophobic patch is necessary for insertion of the kB1 into the membrane core. When the integrity of these regions is compromised either by alteration of the secondary structure (by reduction or linearization) or by residue mutation, the peptide is unable to target membranes and loses activity. Incorporation of Lys residues at key positions (e.g. in the amendable face) increases the binding (see Fig. 3), probably by providing additional favorable interactions with the polar headgroups of the lipid bilayer.
More insights into the mechanism of action of kB1 were obtained with leakage and flip-flop studies (see Fig. 5). That kB1 is able to induce leakage in LUVs containing PE lipids established a correlation between membrane selectivity and membrane leakage. Furthermore, it also supports the hypothesis that the mode of action of kB1 involves membrane targeting followed by insertion, permeabilization, and membrane disruption. These results are consistent with electrophysiological results (15), which show a gradual increase of kB1 on the membrane surface until a critical peptide concentration is reached, leading to pore formation and membrane permeabilization.
The ability of kB1 to induce the outward movement of PE-phospholipids is consistent with the targeting of PE-headgroups and indicates that kB1 is capable of self-promoting its binding by exposing more PE-phospholipids in the outer membrane. This mechanism is very similar to that of cinnamycin, a tetracyclic peptide antibiotic, that recognizes PE in both model and biological membranes (48). Cinnamycin inserts into the lipid bilayer and self-promotes its own binding by inducing the outward movement of PE and causing the exposure of additional PE (49), eventually leading to pore formation and hemolysis (50). Similar to kB1, the PE environment and nonspecific hydrophobic interactions were also shown to be important for cinnamycin affinity for the membrane (51).
The fact that Chol/SM domains in PE-containing membranes improve the binding/insertion of kB1 (Fig. 2A and Table 1) and, consequently, the membrane leakage efficiency (Fig. 5) is consistent with the reported ability of Chol to undergo fast flip-flop (52). Therefore, the presence of Chol might add dynamism and promote the faster exposition of more PE-phospholipids in the outer layer.
With a well known membrane lipid composition, RBCs are excellent models to study kB1-membrane interactions and establish a correlation between model and biological membranes. PC-phospholipids, SM, and Chol are mainly located in the outer layer. PE-phospholipids, although mainly located in the inner layer, have some surface exposition, whereas PS-phospholipids are exclusively sequestered in the inner layer (53). Hemolytic properties of kB1 confirm its ability to target and permeabilize the membrane of RBCs. The crenated shape (Fig. 6) reveals that there is expansion of the outer layer (44) and redistribution of phospholipids whereby PE becomes more exposed to the outer layer, whereas PC and SM are redistributed to the inner layer (45). Although only a small percentage of PE is initially exposed, the presence of lo domains rich in Chol and SM (so-called raft domains) in the outer leaflet (54, 55) seems to facilitate the initial binding and insertion of kB1 into the outer layer. The increased concentration of PE in the outer membrane promotes the binding of more peptide molecules, leading to pore formation and lysis of RBCs.
Bacterial cells are characterized by a negatively charged membrane very rich in PE and lacking sterol but also by the presence of an external cell wall enriched with either the anionic LPS or the anionic peptidoglycan in Gram-negative or Gram-positive bacteria, respectively. Although kB1 could, in principle, target the inner membrane of E. coli rich in PE (Fig. 2D), it does not show any preference for negatively charged membranes (Fig. 2A) and does not bind to model membranes that mimic the outer membrane of E. coli (Fig. 2D). Therefore, a lack of activity against both E. coli and S. aureus is expected, as the inner membrane cannot be reached before the negatively charged outer membrane is overcome. The lack of bacterial membrane targeting was confirmed by atomic force microscopy imaging, as weak activity and no differences between membrane-active and -inactive analogs were observed (Fig. 7). These results are in contrast with the striking effect on bacterial membranes induced by AMPs (28).
Generally, peptides with hemolytic properties insert into the hydrophobic core of the erythrocyte membrane through hydrophobic interactions, and when the hydrophobic patch is disrupted, hemolytic properties are lost (56). By contrast, antibacterial peptides exert their action through electrostatic attractions and adsorption at the polar interface in which the insertion into the hydrophobic core may facilitate the efficiency but is not a requirement (57). Hemolytic and bacterial results with kB1 are of significance as they show a strong correlation between affinity for model lipid bilayers and bioactivity. In summary, kB1 activity is dependent on PE-phospholipids and insertion into the membrane hydrophobic core but is not correlated with electrostatic attractions. When classic antibacterial peptide features are compared with kB1 properties, it is clear that kB1 has distinct properties (47); i.e. kB1 has a neutral global charge, is unable to target negatively charged membranes, and has weak activity against tested Gram-positive and Gram-negative bacteria. Although activity against other bacteria, not tested yet, cannot be ruled out, the antimicrobial activity of kB1 against the tested strains is weak.
Similar to trends observed for hemolytic activity, the anti-HIV activity of cyclotides is dependent on the maintenance of the overall structure (46) and on the hydrophobic patch (58), which suggests that membrane interactions are also involved in the anti-HIV action of cyclotides (47). HIV is an enveloped virus, i.e. it is encased within a membrane, which is mainly composed of viral glycoproteins and a raft-like membrane with a large amount of SM, Chol, and PE-phospholipids (59, 60). Therefore, results with membrane models rich in lo phase can be compared with raft domains in eukaryotic cells but also with the rigid viral membrane.
In this study we have shown that kB1 has selectivity for model membranes that contain PE, but the interaction is further enhanced if Chol/SM domains are present. Leakage studies (see Fig. 5A) with model membranes that mimic HIV membrane properties (POPC/POPE/Chol/SM (10/33/40%)) revealed that kB1 has a more pronounced ability to disrupt these rigid membranes than membranes in fluid phase (POPC/POPE 10%). Overall, the model membrane results are consistent with kB1 targeting the HIV membrane via peptide-lipid interactions. To test this hypothesis, NL4.3 and the clade A HIV-1 strain were treated with kB1 before being cultured with host cells. The results show that kB1 decreases the infectivity of both strains in a dose-dependent manner. A similar response was obtained with the membrane-active N29K, whereas the membrane-inactive analog, V25K, had little effect. These results confirm that kB1 has HIV-1 virucidal activity, which is mediated by its membrane binding properties.
Anti-HIV studies of kB1 confirmed the ability of kB1 to destroy the raft-like membrane of the virus. The fact that HIV membranes have more Chol/SM and PE than the host cells (59, 60) supports the notion that kB1 will select HIV virions over eukaryotic cells. These results are in agreement with previous studies (46) in which it was shown that kB1 has anti-HIV activity 25-fold lower than its cytotoxic concentration.
To the best of our knowledge kB1 is the first peptide shown to have virucidal activity against HIV, i.e. destroying directly the viral particle by disrupting the membrane envelope. Other reported anti-HIV peptides inhibit HIV entry into the cell by fusion inhibition; T20 (enfurvitide, the first approved anti-HIV fusion inhibitor drug) and T1249 are two examples (61). Interestingly, human secretory ribonucleases have also been reported to have cytoprotective activity against HIV, and a possible effect on viral particles was suggested (62).
Inactivating HIV through a lipid-targeting mechanism is unlikely to result in the emergence of cyclotide resistant strains of HIV, as replacement of membrane lipids is virtually impossible for the virus. Furthermore, a membrane target mechanism suggests a broad-spectrum HIV virucidal activity and opens the possibility of using cyclotides against other enveloped viruses. Overall, with a long half-life, thermal resistance (2), and high efficacy against HIV (46), kB1 and other cyclotides (17) are promising anti-HIV agents with potential applications in topical microbicidal gels to prevent the sexual transmission of HIV. Delivering the drugs locally by the use of topical gels minimizes systemic toxicity (63) and potentially maximizes the therapeutic index.
In conclusion, the biological activity of kB1 is dependent on membrane affinity, which is governed by lipid composition. Based on biophysical studies and bioassays, the mechanism of kB1 can be summarized as follows. A bioactive patch on the kB1 surface targets membranes through specific interaction with PE; the hydrophobic domain of kB1 inserts into the hydrophobic core through peptide-lipid interactions, which are more favorable in raft-like domains and promotes outward movement of PE-phospholipids; the presence of more PE in the outer leaflet facilitates the binding of more peptide molecules, leading to pore formation and eventual membrane disruption. This model explains the hemolytic properties of kB1 and its lack of antimicrobial activity. In addition, the anti-HIV activity of kB1 is clearly correlated with its ability to target the HIV membrane envelope, a raft-like membrane with a high proportion of PE-phospholipids.
Supplementary Material
Acknowledgments
We thank Dr. Brit Winnen (University of Queensland) for providing the bacterial strains and Laura Cascales (University of Queensland) for help with the bacterial assays. We thank the National Institutes of Health AIDS Research and Reference Reagent Program for providing the HIV-1 isolate 92RW016 and the TZM-bl indicator cell line, contributed by Dr. John C Kappes, Dr. Xiaoyun Wu, and Tranzyme Inc.
This research was supported by Australian Research Council Grant DP0880105 and by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Program (PIOF-GA-2008-220318).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- kB1
- kalata B1
- LUVs
- large unilamellar vesicles
- PE
- phosphatidylethanolamine
- POPC
- palmitoyloleoylphosphatidylcholine
- POPG
- palmitoyloleoylphosphatidylglycerol
- POPE
- palmitoyloleoylphosphatidylethanolamine
- Chol
- cholesterol
- SM
- sphingomyelins
- P/L
- peptide/lipid ratio
- CF
- carboxyfluorescein
- PC
- phosphatidylcholine
- lo
- liquid-ordered phase
- so
- solid phase
- ld
- liquid disordered
- PI
- phosphatidylinositol
- AMP
- antimicrobial peptide
- NBD
- nitrobenzoxadiazole
- C6-NBD-
- 1-palmitoyl-2-{6-(NBD)-hexanoyl}-sn-glycero-3-
- LPS
- lipopolysaccharide.
REFERENCES
- 1. Craik D. J., Daly N. L., Bond T., Waine C. (1999) J. Mol. Biol. 294, 1327–1336 [DOI] [PubMed] [Google Scholar]
- 2. Colgrave M. L., Craik D. J. (2004) Biochemistry 43, 5965–5975 [DOI] [PubMed] [Google Scholar]
- 3. Barbeta B. L., Marshall A. T., Gillon A. D., Craik D. J., Anderson M. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1221–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gran L., Sandberg F., Sletten K. (2000) J. Ethnopharmacol. 70, 197–203 [DOI] [PubMed] [Google Scholar]
- 5. Gustafson K. R., Sowder R. C. I., Henderson L. E., Parsons I. C., Kashman Y., Cardellina J. H. I., McMahon J. B., Buckheit R. W. J., Pannell L. K., Boyd M. R. (1994) J. Am. Chem. Soc. 116, 9337–9338 [Google Scholar]
- 6. Schöpke T., Hasan Agha M. I., Kraft R., Otto A., Hiller K. (1993) Sci. Pharm. 61, 145–153 [Google Scholar]
- 7. Tam J. P., Lu Y. A., Yang J. L., Chiu K. W. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8913–8918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lindholm P., Göransson U., Johansson S., Claeson P., Gullbo J., Larsson R., Bohlin L., Backlund A. (2002) Mol. Cancer Ther. 1, 365–369 [PubMed] [Google Scholar]
- 9. Colgrave M. L., Kotze A. C., Huang Y. H., O'Grady J., Simonsen S. M., Craik D. J. (2008) Biochemistry 47, 5581–5589 [DOI] [PubMed] [Google Scholar]
- 10. Wang C. K., Kaas Q., Chiche L., Craik D. J. (2008) Nucleic Acids Res. 36, D206–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gunasekera S., Foley F. M., Clark R. J., Sando L., Fabri L. J., Craik D. J., Daly N. L. (2008) J. Med. Chem. 51, 7697–7704 [DOI] [PubMed] [Google Scholar]
- 12. Kamimori H., Hall K., Craik D. J., Aguilar M. I. (2005) Anal. Biochem. 337, 149–153 [DOI] [PubMed] [Google Scholar]
- 13. Shenkarev Z. O., Nadezhdin K. D., Sobol V. A., Sobol A. G., Skjeldal L., Arseniev A. S. (2006) FEBS J. 273, 2658–2672 [DOI] [PubMed] [Google Scholar]
- 14. Svangård E., Burman R., Gunasekera S., Lövborg H., Gullbo J., Göransson U. (2007) J. Nat. Prod. 70, 643–647 [DOI] [PubMed] [Google Scholar]
- 15. Huang Y. H., Colgrave M. L., Daly N. L., Keleshian A., Martinac B., Craik D. J. (2009) J. Biol. Chem. 284, 20699–20707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang Y. H., Colgrave M. L., Clark R. J., Kotze A. C., Craik D. J. (2010) J. Biol. Chem. 285, 10797–10805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ireland D. C., Wang C. K., Wilson J. A., Gustafson K. R., Craik D. J. (2008) Biopolymers 90, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Plan M. R., Göransson U., Clark R. J., Daly N. L., Colgrave M. L., Craik D. J. (2007) Chembiochem 8, 1001–1011 [DOI] [PubMed] [Google Scholar]
- 19. Daly N. L., Love S., Alewood P. F., Craik D. J. (1999) Biochemistry 38, 10606–10614 [DOI] [PubMed] [Google Scholar]
- 20. Daly N. L., Clark R. J., Craik D. J. (2003) J. Biol. Chem. 278, 6314–6322 [DOI] [PubMed] [Google Scholar]
- 21. Clark R. J., Daly N. L., Craik D. J. (2006) Biochem. J. 394, 85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Henriques S. T., Pattenden L. K., Aguilar M. I., Castanho M. A. (2008) Biophys. J. 95, 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yanagida N., Uozumi T., Beppu T. (1986) J. Bacteriol. 166, 937–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Papo N., Shai Y. (2005) J. Biol. Chem. 280, 10378–10387 [DOI] [PubMed] [Google Scholar]
- 25. Henriques S. T., Castanho M. A., Pattenden L. K., Aguilar M. I. (2010) Biopolymers 94, 314–322 [DOI] [PubMed] [Google Scholar]
- 26. Henriques S. T., Castanho M. A. (2004) Biochemistry 43, 9716–9724 [DOI] [PubMed] [Google Scholar]
- 27. Henriques S. T., Tan C. C., Craik D. J., Clark R. J. (2010) Chembiochem 11, 2148–2157 [DOI] [PubMed] [Google Scholar]
- 28. Alves C. S., Melo M. N., Franquelim H. G., Ferre R., Planas M., Feliu L., Bardají E., Kowalczyk W., Andreu D., Santos N. C., Fernandes M. X., Castanho M. A. (2010) J. Biol. Chem. 285, 27536–27544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., Martin M. A. (1986) J. Virol. 59, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fletcher P. S., Wallace G. S., Mesquita P. M., Shattock R. J. (2006) Retrovirology 3, 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wei X., Decker J. M., Liu H., Zhang Z., Arani R. B., Kilby J. M., Saag M. S., Wu X., Shaw G. M., Kappes J. C. (2002) Antimicrob. Agents Chemother. 46, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yap S. H., Sheen C. W., Fahey J., Zanin M., Tyssen D., Lima V. D., Wynhoven B., Kuiper M., Sluis-Cremer N., Harrigan P. R., Tachedjian G. (2007) PLoS Med. 4, e335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Figueiredo A., Moore K. L., Mak J., Sluis-Cremer N., de Bethune M. P., Tachedjian G. (2006) PLoS Pathog. 2, e119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Almeida R. F., Fedorov A., Prieto M. (2003) Biophys. J. 85, 2406–2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsuzaki K., Sugishita K., Miyajima K. (1999) FEBS Lett. 449, 221–224 [DOI] [PubMed] [Google Scholar]
- 36. Ahn T., Oh D. B., Kim H., Park C. (2002) J. Biol. Chem. 277, 26157–26162 [DOI] [PubMed] [Google Scholar]
- 37. Murzyn K., Róg T., Pasenkiewicz-Gierula M. (2005) Biophys. J. 88, 1091–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simonsen S. M., Sando L., Rosengren K. J., Wang C. K., Colgrave M. L., Daly N. L., Craik D. J. (2008) J. Biol. Chem. 283, 9805–9813 [DOI] [PubMed] [Google Scholar]
- 39. Richer S. M., Stewart N. K., Tomaszewski J. W., Stone M. J., Oakley M. G. (2008) Biochemistry 47, 13455–13462 [DOI] [PubMed] [Google Scholar]
- 40. Fattal E., Nir S., Parente R. A., Szoka F. C., Jr. (1994) Biochemistry 33, 6721–6731 [DOI] [PubMed] [Google Scholar]
- 41. Kol M. A., van Dalen A., de Kroon A. I., de Kruijff B. (2003) J. Biol. Chem. 278, 24586–24593 [DOI] [PubMed] [Google Scholar]
- 42. Daly N. L., Craik D. J. (2000) J. Biol. Chem. 275, 19068–19075 [DOI] [PubMed] [Google Scholar]
- 43. Pränting M., Lööv C., Burman R., Göransson U., Andersson D. I. (2010) J. Antimicrob. Chemother. 65, 1964–1971 [DOI] [PubMed] [Google Scholar]
- 44. Sheetz M. P., Singer S. J. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 4457–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lin S., Yang E., Huestis W. H. (1994) Biochemistry 33, 7337–7344 [DOI] [PubMed] [Google Scholar]
- 46. Daly N. L., Gustafson K. R., Craik D. J. (2004) FEBS Lett. 574, 69–72 [DOI] [PubMed] [Google Scholar]
- 47. Henriques S. T., Craik D. J. (2010) Drug Discov. Today 15, 57–64 [DOI] [PubMed] [Google Scholar]
- 48. Choung S. Y., Kobayashi T., Inoue J., Takemoto K., Ishitsuka H., Inoue K. (1988) Biochim. Biophys. Acta 940, 171–179 [DOI] [PubMed] [Google Scholar]
- 49. Makino A., Baba T., Fujimoto K., Iwamoto K., Yano Y., Terada N., Ohno S., Sato S. B., Ohta A., Umeda M., Matsuzaki K., Kobayashi T. (2003) J. Biol. Chem. 278, 3204–3209 [DOI] [PubMed] [Google Scholar]
- 50. Choung S. Y., Kobayashi T., Takemoto K., Ishitsuka H., Inoue K. (1988) Biochim. Biophys. Acta 940, 180–187 [DOI] [PubMed] [Google Scholar]
- 51. Machaidze G., Seelig J. (2003) Biochemistry 42, 12570–12576 [DOI] [PubMed] [Google Scholar]
- 52. Hamilton J. A. (2003) Curr. Opin. Lipidol. 14, 263–271 [DOI] [PubMed] [Google Scholar]
- 53. Daleke D. L., Huestis W. H. (1989) J. Cell Biol. 108, 1375–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cheng H. T., Megha, London E. (2009) J. Biol. Chem. 284, 6079–6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kiessling V., Crane J. M., Tamm L. K. (2006) Biophys. J. 91, 3313–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Glukhov E., Burrows L. L., Deber C. M. (2008) Biopolymers 89, 360–371 [DOI] [PubMed] [Google Scholar]
- 57. Chen Y., Guarnieri M. T., Vasil A. I., Vasil M. L., Mant C. T., Hodges R. S. (2007) Antimicrob. Agents. Chemother 51, 1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang C. K., Colgrave M. L., Gustafson K. R., Ireland D. C., Goransson U., Craik D. J. (2008) J. Nat. Prod. 71, 47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Aloia R. C., Tian H., Jensen F. C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5181–5185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brügger B., Glass B., Haberkant P., Leibrecht I., Wieland F. T., Kräusslich H. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rusconi S., Scozzafava A., Mastrolorenzo A., Supuran C. T. (2007) Curr. Top. Med. Chem. 7, 1273–1289 [DOI] [PubMed] [Google Scholar]
- 62. Bedoya V. I., Boasso A., Hardy A. W., Rybak S., Shearer G. M., Rugeles M. T. (2006) AIDS Res. Hum. Retroviruses 22, 897–907 [DOI] [PubMed] [Google Scholar]
- 63. Nuttall J. (2010) Drugs 70, 1231–1243 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.