Abstract
Arrestins bind active phosphorylated forms of G protein-coupled receptors, terminating G protein activation, orchestrating receptor trafficking, and redirecting signaling to alternative pathways. Visual arrestin-1 preferentially binds rhodopsin, whereas the two non-visual arrestins interact with hundreds of G protein-coupled receptor subtypes. Here we show that an extensive surface on the concave side of both arrestin-2 domains is involved in receptor binding. We also identified a small number of residues on the receptor binding surface of the N- and C-domains that largely determine the receptor specificity of arrestins. We show that alanine substitution of these residues blocks the binding of arrestin-1 to rhodopsin in vitro and of arrestin-2 and -3 to β2-adrenergic, M2 muscarinic cholinergic, and D2 dopamine receptors in intact cells, suggesting that these elements critically contribute to the energy of the interaction. Thus, in contrast to arrestin-1, where direct phosphate binding is crucial, the interaction of non-visual arrestins with their cognate receptors depends to a lesser extent on phosphate binding and more on the binding to non-phosphorylated receptor elements.
Keywords: Electron Paramagnetic Resonance (EPR), G Protein-coupled Receptors (GPCR), Protein Structure, Protein-Protein Interactions, Signal Transduction, Arrestin, BRET
Introduction
Arrestins are a small family of signaling regulators that preferentially bind active phosphorylated forms of G protein-coupled receptors (GPCRs),5 blocking G protein coupling, switching the signaling to alternative pathways, and orchestrating receptor trafficking (for review, see Refs. 1 and 2). Mammals have four arrestin subtypes that demonstrate >50% amino acid conservation (1, 3, 4) and very similar elongated two-domain structures in the basal state (5–9) (Fig. 1A). Arrestin-16 (formerly known as visual or rod arrestin) and arrestin-4 (cone arrestin) are predominantly expressed in photoreceptors (10, 11), whereas arrestin-2 and -3 (also know as β-arrestins 1 and 2) are present in virtually every cell in the body (3, 4), with the highest expression levels in mature neurons (12, 13). Broad receptor specificity of both non-visual arrestins in vitro (14–16) and in living cells (17–19) suggests that these two subtypes “serve” the great majority of ∼800–3,400 GPCRs found in different mammalian species (SEVENS database). In contrast, arrestin-1 shows high selectivity for its cognate receptor rhodopsin (8, 14, 20, 21). The precise structural basis of receptor specificity of arrestin proteins remains to be elucidated. Recent studies of arrestin-1 identified an extensive receptor binding surface covering virtually all of the concave sides of both arrestin domains (22–25). Here we demonstrate that a similarly extensive surface of the most abundant non-visual subtype, arrestin-2, is involved in receptor binding and identify surprisingly few residues that play key role in receptor interaction and GPCR preference.
FIGURE 1.
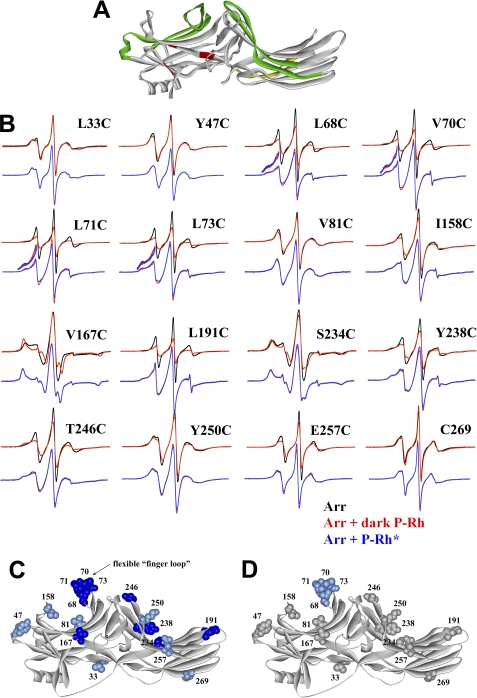
Extensive receptor footprint on arrestin-2 identified by SDSL EPR spectroscopy. A, shown is the structure of arrestin-2 (6) with functionally important elements indicated as follows: phosphate binding residues (red); elements that determine receptor specificity (green); interdomain hinge (yellow) (image generated using ViewerPro 6.0). B, for each spin-labeled arrestin, normalized spectra in the absence (black) or presence of dark P-Rh (red) are compared in the top overlays, and spectra in the presence of dark P-Rh (red) and P-Rh* (blue) are compared in the bottom overlays. The lowfield portion of the bottom overlays for the finger loop spectra are magnified 2-fold in intensity to better illustrate the small changes in line shape. C and D, the magnitude of the detected changes in spin label mobility is color-coded on the arrestin-2 crystal structure as follows: gray, no change; light blue, small decrease in mobility; dark blue, large decrease in mobility. C, changes upon interaction with dark P-Rh are shown. D, additional changes induced by light activation of phosphorylated rhodopsin (P-Rh*) relative to those induced by binding to dark P-Rh are shown.
EXPERIMENTAL PROCEDURES
Materials
[γ-32P]ATP, [14C]leucine, and [3H]leucine were from PerkinElmer Life Sciences. All restriction enzymes were from New England Biolabs (Ipswich, MA). Sepharose 2B and all other chemicals were from sources previously described (26, 27). Rabbit reticulocyte lysate was from Ambion (Austin, TX), and SP6 RNA polymerase was prepared as described (28). 11-Cis-retinal was generously supplied by Dr. R. K. Crouch (Medical University of South Carolina, Charleston, SC).
Construction of Arrestin-1/2 Chimeras and Mutagenesis
Bovine arrestin-1 cDNA (29) was a gift from Dr. T. Shinohara (National Eye Institute, NIH, Bethesda, MD). Plasmids that encode bovine arrestin-1, the prevailing long splice variant of bovine arrestin-2 (3), and the most abundant short splice variant of arrestin-3 (3, 15) with an “idealized” 5′-untranslated region (28) under control of a SP6 promoter were described in detail earlier (14, 15, 21). Mutations were introduced by PCR, as described (23). All mutant constructs were confirmed by dideoxy sequencing.
In Vitro Transcription, Translation, and Evaluation of Mutant Stability
Plasmids were linearized using a unique HindIII site downstream of the coding sequence. In vitro transcription and translation were performed as previously described (21, 27). All arrestin proteins were labeled by incorporation of [3H]leucine and [14C]leucine with a specific activity of the mix of 1.5–3 Ci/mmol, resulting in the specific activity of arrestin proteins within the range of 66–85 Ci/mmol (150–230 dpm/fmol). The translation of every mutant used in this study produced a single labeled protein band with the expected mobility on SDS-PAGE. Two parameters were used for the assessment of mutant relative stability, as described (30); that is, its yield multiplied by the percentage of the protein remaining in the supernatant after incubation for 10 min at 37 °C followed by centrifugation. This integral parameter calculated for a mutant was expressed as a percent of that for corresponding wild type (WT) arrestin. The relative stability of all mutants used in this study exceeds 85% that of the parental WT proteins.
Receptor Preparations
Rhodopsin
Urea-treated rod outer segment membranes containing rhodopsin phosphorylated by endogenous GRK1 (also know as rhodopsin kinase) and regenerated with 11-cis-retinal were prepared as described previously (31). The stoichiometry of phosphorylation for the rhodopsin preparations used in these studies was 3.2 mol of phosphate/mol of rhodopsin, which ensures high affinity binding of arrestin-1 (31) and non-visual arrestins (14).
Muscarinic Receptor
The human M2 muscarinic cholinergic receptor was expressed in Sf9 cells, purified by affinity chromatography, reconstituted into chick heart phospholipids, and phosphorylated by purified GRK2 to a stoichiometry of 3.1–3.7 moles of phosphate/mole of M2 receptor, as described previously (14, 32).
Arrestin Binding to Receptors
Binding was performed, as described (14).
Rhodopsin
Briefly, in vitro translated radiolabeled arrestins (50 fmol) in 50 mm Tris-HCl, pH 7.5, 0.5 mm MgCl2, 1.5 mm dithiothreitol, and 50 mm potassium acetate were incubated with 7.5 pmol (0.3 μg) of phosphorylated light-activated rhodopsin in a final volume of 50 μl for 5 min at 37 °C in room light.
Muscarinic M2 Receptor
Radiolabeled arrestins (50 fmol) were incubated with 50 fmol of phosphorylated receptor in the presence of 100 μm muscarinic agonist carbachol for 35 min at 30 °C. After incubation with either receptor, the samples were immediately cooled on ice and loaded onto 2 ml of Sepharose 2B columns equilibrated with 10 mm Tris-HCl, pH 7.5, 100 mm NaCl. Bound arrestin eluted with receptor-containing membranes in the void volume (between 0.5–1.1 ml). Nonspecific binding determined in the presence of 0.3 μg liposomes (less than 10% of the total binding and less than 0.5% of the arrestin present in the assay) was subtracted.
Arrestin-2 Expression and Purification
Arrestin was expressed in Escherichia coli and purified as described (33). The cysteine-less arrestin-2 (A2-CL) (C59V, C125S, C140L, C150V, C242V, C251V, C269S) was used as a base mutant for the introduction of unique cysteines. The best substituting residue in each position was chosen based on our high resolution crystal structure of arrestin-2 (6) to preserve existing intramolecular interactions as much as possible. We have previously shown that cysteine-less arrestin-2 is fully functional in terms of receptor binding (34).
Electron Paramagnetic Resonance Spectroscopy
Arrestin cysteine mutants were dialyzed into 50 mm MOPS, 100 mm NaCl, pH 7.0, and labeled with a 10-fold molar excess of 2,2,5,5-tetramethylpyrroline-3-yl-methanethiosulfonate spin label (MTSL; Toronto Research Chemicals, Ontario, Canada) overnight at 4 °C to generate the R1 side chain, as described (22, 34). Final protein concentrations were determined by the BCA protein assay (Pierce) using BSA as a standard. EPR samples contained 25 μm spin-labeled arrestin-2 and 250 μm light-activated phosphorhodopsin in native disc membranes (26) in a final volume of 10 μl. X-band EPR spectra were recorded for samples in glass capillaries at room temperature over a 100-gauge range with an incident microwave power of 10 milliwatts on a Bruker ELEXSYS E500 fitted with a super high Q cavity. Spectra shown are typically the average of 16–36 scans, base-line-corrected, and normalized to the same area for each graphic overlay in Fig. 1B. For a qualitative interpretation of EPR spectral line shapes, as used in the present study, the reader is referred to Crane et al. (36) and Kusnetzow et al. (37).
Plasmid Construction for Bioluminescence Energy Transfer (BRET)
The plasmids containing the sequences of the indicated arrestins N-terminal-tagged with Venus (a variant of enhanced yellow fluorescent protein (38); a generous gift from Dr. J. A. Javitch, Columbia University) were constructed using a modified pGEM2 in vitro transcription vector (Promega, Madison, WI) that is under the control of an SP6 promoter idealized 5′-untranslated region (28) with an upstream EcoRI site followed by the coding sequence of arrestins between NcoI and HindIII sites. Venus was amplified by PCR using the 5′-AGTCAGAATTCGCGATCGCGGCCACGATGGTGAGCAAGGGCGA-3′ forward primer that adds EcoRI and AsiSI sites upstream of the start codon and the 5′-TCTCCCCCATGGAGTCGAGCGCTCGCCGAGACTTAAGTCCGGAGGTGGCCT-3′ reverse primer that codes for a short spacer with the SGLKSRRALDS sequence and an in-frame NcoI site. Venus was subcloned between the EcoRI and NcoI restriction sites. The arrestins were subcloned in-frame with the Venus-spacer sequence using NcoI and HindIII sites. The Venus-arrestin fusion proteins were subcloned into a pcDNA3 mammalian expression vector (Invitrogen) modified as described (39, 40) using the EcoRI and HindIII restriction sites. A plasmid encoding Renilla luciferase variant 8 (RLuc8) (41) was a generous gift from Dr. Nevin A. Lambert (Medical College of Georgia, Augusta, GA). RLuc8 was fused in-frame with the sequence of the triple HA-tagged human β2-adrenergic receptor (β2AR), M2 muscarinic cholinergic, and D2 dopamine receptors purchased from the cDNA resource center (Missouri S&T cDNA Resource Center). To this end, the coding sequences of all GPCRs were amplified by PCR using forward primers that introduce EcoRI and AsiSI restriction sites upstream of the receptor start codon and reverse primers that introduce an in-frame SbfI restriction site. These fragments were subcloned using EcoRI and SbfI sites in-frame with a C-terminal RLuc8.
BRET Assays
The well established BRET1 assay (42–44) with Venus as the acceptor and RLuc8 as the donor was used to characterize the binding of Venus-arrestins to β2-adrenergic receptor-RLuc8, M2-RLuc8, and D2-RLuc8. COS-7 cells were transfected with the indicated plasmids using LipofectamineTM 2000 (Invitrogen) according to the manufacturer's protocol (3 μl of LipofectamineTM 2000/1 μg of DNA). Increasing amounts of the indicated Venus-arrestin constructs (0–12 μg) along with 250 ng of the indicated receptor plasmid and empty pcDNA3 to equalize DNA were used to transfect 80–90% confluent COS-7 cells on 60-mm dishes. Twenty-four hours after transfection, cells were trypsinized and re-seeded at 100,000–200,000 cells per well onto white opaque 96-well microplates (Nunc, Rochester, NY) for luminescence measurements or black opaque microplates (Nunc) for fluorescence determination. Forty-eight hours post-transfection, the medium was replaced with PBS with Ca2+ and Mg2+ containing 0.01% glucose (w/v), 36 mg/liter sodium pyruvate, and 25 mmol/liter HEPES, pH 7.2. Coelenterazine-h (DiscoveRx, Fremont, CA) at a final concentration of 5 μm was added 8 min after agonist (25 μm isoproterenol) stimulation, and luminescence was measured immediately using a POLARstar Optima dual channel luminometer and fluorometer microplate reader (BMG Labtech, Cary, NC). The light emitted by coelenterazine-h and Venus in each well was measured simultaneously 5 times through a 465–485-nm and a 522.5–547.5-nm bandpass filter, respectively. The net BRET ratio was calculated as the long wavelength emission divided by the short wavelength emission and expressed as the relative change compared with unstimulated cells. The expression of each Venus-arrestin was evaluated using fluorescence at 535 nm upon excitation at 485 nm. The Venus-arrestin fluorescence, which is directly proportional to the expression levels, was normalized by the basal luminescence from the β2-adrenergic receptor-RLuc8 to account for variations in cell number and expression levels. The curves resulting from the titration of the various amounts of Venus-arrestin were fit by non-linear regression to a one-site hyperbola model using Prism version 5.04 (GraphPad Software, San Diego, CA). To assess the difference between the respective WT and mutant arrestins curves, a global fit algorithm was used.
RESULTS
Mapping the Receptor Binding Surface of Arrestin-2
An extensive surface covering most of the concave sides of both arrestin-1 domains was implicated in rhodopsin binding by several groups using a variety of methods (14, 22–24, 26, 45, 46). Structural similarity within the arrestin family (5–9) as well as the functionality of numerous chimeras with combinations of elements from different arrestins (14, 21, 47) suggests that homologous elements of arrestin-2 may be involved in receptor binding. To test this hypothesis, we used site-directed spin-labeling EPR spectroscopy (48, 49), which worked remarkably well with arrestin-1 (22, 25, 50). The EPR spectra of spin-labeled proteins contain information on the motional dynamics of R1 side chain and thus allow the elucidation of protein-protein interaction sites. To use this method, we introduced unique cysteines in different positions on the background of fully functional cysteine-less arrestin-2 (34) and used receptor binding-induced changes in spin label mobility (Fig. 1B) to determine the receptor “footprint” on arrestin-2 (Fig. 1, C and D). We used light-activated phosphorylated rhodopsin (P-Rh*) as a model receptor because both non-visual arrestins were shown to bind it specifically in an activation- and phosphorylation-dependent fashion (14–16, 47). To determine the functional consequences of mutations and labeling of arrestin-2, we incubated purified spin-labeled arrestin-2 (25 μm) with 250 μm P-Rh* for 5 min at 37 °C (23, 51). Arrestin-2 bound to rhodopsin-containing membranes was pelleted by centrifugation. All 16 spin-labeled cysteine mutants demonstrated similar (>95%) binding to P-Rh* (supplemental Fig. S1). We expected dark P-Rh-arrestin-2 complex to form essentially as well as the P-Rh*-arrestin-2 complex based on our previous findings that non-visual arrestins bind comparably both dark P-Rh and P-Rh* (14–16, 47).
The spin labels at each site evaluated in arrestin-2 show relatively high mobility, with the exception of sites 167 and 234, consistent with their surface-exposed positions on the protein (Fig. 1B). In sharp contrast to arrestin-1 (51–54, 93), at the concentration used in the EPR experiments (25 μm), arrestin-2 does not demonstrate appreciable self-association (56). Therefore, the observed changes in spin label mobility can be directly attributed to the effects of receptor binding. The spin labels attached to arrestin-2 at positions 68, 70, 71, 73, 167, 191, 234, 238, and 246 become substantially immobilized upon binding to dark P-Rh. Strong immobilization of these surface residues suggests that the binding of P-Rh causes the residues to come into direct contact with either the receptor itself, the adjacent membrane, or with other arrestin elements. Although arrestin-2 does not bind membranes with appreciable affinity, as very small fraction associates with liposomes we use to determine nonspecific binding, in the complex with receptor parts of the molecule may come into contact with the membrane. Immobilized residues are localized along the whole span of the concave surfaces of both domains. This distribution resembles that observed for receptor binding to arrestin-1, suggesting a similarly large receptor footprint on all arrestins.
Spin labels at positions 33, 47, 81, 158, 250, 257, and 269 exhibit less dramatic immobilization upon arrestin binding to the receptor (Fig. 1B). Smaller decreases in nitroxide mobility at these positions, which are located on the non-receptor binding surface and on the periphery of the receptor binding surface of both arrestin-2 domains, indicate that these residues are not in the closely packed interaction interface. The relatively subtle reductions in mobility could arise from indirect effects of receptor binding, i.e. small structural rearrangements, protein tumbling changes due to protein-protein docking, changes in local backbone motion, and/or hydration changes due to receptor contact with arrestin-2 residues within close proximity. Irrespective of the origin of the changes, all sites showed some change upon binding to P-Rh, indicating that a large area of arrestin-2 may be affected by receptor binding (Fig. 1C), similar to the case for arrestin-1 (22).
The motional changes observed in the spectra at positions V167R1 and S234R1 could be due to changes in rotational diffusion of the protein upon binding to rhodopsin-containing membranes. However, the motional changes observed in the remaining highly mobile arrestin-2 spectra can be attributed to internal changes in motion rather than rotational diffusion; the small changes in motion observed for L33R1 upon binding to the receptor serves as an internal control showing that rotational diffusion effects are very small at sites with high mobility.
Although the largest changes occur upon binding to P-Rh, small additional decreases in mobility are noted upon light activation, notably in the cluster of residues in the loop between β-strands V and VI (positions 70, 71, 73) (termed the “finger loop” in arrestin-1 (22)), which is also consistent with observations in arrestin-1 (22). The remaining sites examined showed no further significant changes in mobility upon light activation of the receptor (Fig. 1D). The difference in arrestin-2 binding to dark P-Rh and P-Rh* by spin label immobilization (Fig. 1) was much less dramatic than the ∼2-fold difference observed in the direct binding assay (14, 47). The reason for this is likely 4 orders of magnitude difference of arrestin-2 concentrations used; that is, 1 nm radiolabeled arrestin in the binding and 25 μm spin-labeled protein in EPR. As the result, a 10-fold difference in arrestin affinity for these two forms of rhodopsin would translate into large disparity in binding at 1 nm, but in EPR experiments arrestin concentration would be ≫KD in both cases, with essentially the same fraction of bound arrestin. However, further changes induced by rhodopsin activation reflect structural rearrangements within the arrestin-receptor interface.
Identification of Arrestin Residues That Determine Receptor Specificity
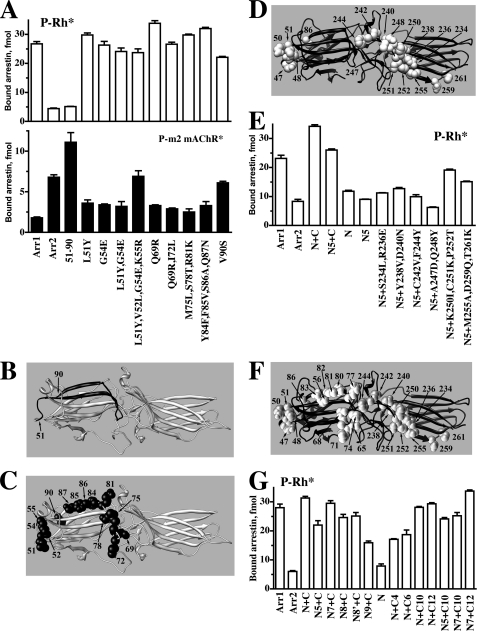
We recently found that the exchange of two elements between arrestin-1 and -2 completely reverses their receptor preference (21). These elements encompass β-strands V and VI in the N-domain and XV and XVI in the C-domain and adjacent loops, which cover almost half of the receptor interaction interface in arrestin-2 (highlighted in green in Fig. 1A). Sequence comparison shows that 15 and 20 residues differ in the N- and C-domain elements, respectively (Fig. 2). According to the crystal structures of both proteins (5, 6), most of these residues are surface-exposed and may directly participate in receptor binding. We have previously shown that “swapping” elements important for receptor specificity between arrestin-1 and -2 has two effects; it reduces the binding to the receptor preferred by the “acceptor” arrestin and increases the binding to the receptor preferred by the “donor” arrestin (21). Therefore, we used a similar approach to identify individual residues that determine receptor preference. The introduction of non-identical arrestin-2 residues, individually and in groups, into the N-domain element of arrestin-1 allowed us to exclude nine that neither significantly reduced the binding to P-Rh* nor increased the binding to carbachol-activated phosphorylated M2 muscarinic acetylcholine receptor (P-m2 mAChR*) to the level comparable with that of WT arrestin-2, leaving relatively few “suspects” in this region (Fig. 3, A–C). Based on these data, we constructed an arrestin-2 mutant (Y47L,L48V,E50G,R51K,S86V), where these five residues were replaced with the corresponding arrestin-1 residues and added groups of arrestin-1 residues to the C-domain element in the context of this base mutant (Fig. 3, D and E). Two triple mutations (K250I,C251K,P252T and M255A,D259Q,T261K) yielded the most dramatic increase in P-Rh* binding, whereas double mutations (S234L,R236E, Y238V,D240N, and C242V,F244Y) were less potent (Fig. 3E). The A247D,Q248Y double mutation had no effect, excluding the role of these two residues in receptor preference. Importantly, the effect of a quintuple mutation (Y47L,L48V,E50G,R51K,S86V) was smaller than that of the whole arrestin-1-derived N-domain element both in the context of arrestin-2 and arrestin-2 with the C-domain element from arrestin-1 (Fig. 3E), suggesting the involvement of additional residues in this element.
FIGURE 2.
Sequence comparison of the receptor discriminator regions in three arrestin subtypes. The residues that differ between bovine arrestin-1 and arrestin-2 are shown in bold, and the residues that are exposed (according to the crystal structure of both proteins) are underlined. Residues unique for arrestin3 are italicized. Ten residues replaced with alanines in Figs. 5 and 6 are shadowed. The positions of the first and last residues are shown in parentheses before and after the sequence.
FIGURE 3.
Identification of arrestin residues that determine receptor specificity. A–C, shown is the binding to P-Rh* (upper panel) and phosphorylated M2 muscarinic acetylcholine receptor (P-m2 mAChR*) of arrestin-1 (Arr1), arrestin-2 (Arr2), arrestin-1 with the N-domain residues 51–90 replaced with arrestin-2 residues 47–86 (51–90) (shown in panel B; arrestin-1- and arrestin-2-derived elements and residues are shown in gray and black, respectively), and arrestin-1 mutants with the indicated residues replaced with the corresponding arrestin-2 residues (shown as black CPK models on gray arrestin-1 molecule in panel C). D, arrestin-2 molecule (black) with arrestin-1 residues introduced in the mutants characterized in panel E are shown as gray CPK models. E, direct binding to P-Rh* of Arr1, Arr2, and Arr2 with elements 47–86 and 233–261 replaced with arrestin-1 elements 51–90 and 239–267, respectively (N+C), Arr2 replaced with a quintuple mutation (Y47L,L48V,E50G,R51K,S86V) in the N-domain, residues 233–261 replaced with the arrestin-1 element 239–267 (N5+C), element 47–86 replaced with arrestin-1 residues 51–90 (N), and with combinations of N5 quintuple mutations in the N-domain and the indicated mutations in the C-domain element. F, arrestin-2 molecule (black) with all arrestin-1 residues introduced in the mutants characterized in panel G are shown as gray CPK models. G, direct binding to P-Rh* of arrestin-2 with the indicated elements replaced with the corresponding residues from arrestin-1: N7, N5+R65Q+L68I; N8, N5+L71M+T74S+K77R; N8, N7+T56S; N9, N5+F80Y+V81F+A82S+N83Q; C4, S234L+R236E+Y238V+D240N; C6, K250I+C251K+P252T+M255A+D259Q+T261K; C10, C4+C6; C12, C10+C242V+F244Y. Binding experiments were performed 2–3 times in duplicate. Means ± S.D. are presented.
Therefore, we then constructed a series of N-domain mutants where additional arrestin-1-derived residues were added to these five substitutions and tested them on the background of arrestin-2 containing the arrestin-1-derived C-domain element (Fig. 3, F and G). The results indicate that the combination of seven mutations (Y47L, L48V, E50G, R51K, R65Q, L68I, and S86V) closely mimics the effect of the N-domain element. Using a similar gain-of-function approach, we combined an increasing number of substitutions in the C-domain element that enhanced P-Rh* binding (Fig. 3E) and tested them in the context of arrestin-2 with the arrestin-1-derived N-domain element. The data show that the combination of 12 point mutations in arrestin-2 most closely mimics the effect of the arrestin-1 C-domain element (Fig. 3G). Moreover, the combination of 7 mutations in the arrestin-2 N-domain with 12 mutations in the C-domain has the same effect as the simultaneous introduction of complete N- and C-domain elements from arrestin-1 (Fig. 3G). Thus, this set of 19 residues is sufficient to switch receptor specificity of arrestin-2 to that of arrestin-1.
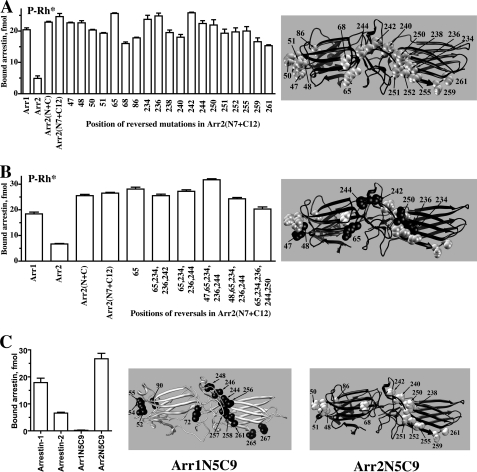
However, because we initially tested (Fig. 3, A and E) and then combined (Fig. 3G) groups of residues rather than individual residues, these data do not indicate that every one of these 19 residues is necessary. To test this, we reversed individual point mutations in the context of the base mutant with 19 substitutions and compared the binding of these proteins to P-Rh* (Fig. 4A). The results indicate that two mutations in the N-domain and three in the C-domain are not required. Based on these data, we then constructed several combinations of reversals with little to no effect on P-Rh* binding (Fig. 4B). These data allowed us to exclude residues Tyr-47, Arg-65, Ser-234, Arg-236, and Phe-244, thereby demonstrating that simultaneous substitution of 5 and 9 residues in the N- and C-domain, respectively, is sufficient to create an arrestin-2 protein with arrestin-1-like binding to P-Rh* (Fig. 4B). In addition, these data identified 2 residues in the N-domain (Leu-68 and Ser-86) and 3 in the C-domain (Asp-240, Asp-259, and Thr-261) that appear to play the most prominent roles in defining receptor preference; the reversal of each of these individual mutations reduced an observed increase in P-Rh* binding by 27–38% (Fig. 4A). Considering the size of the receptor-binding arrestin surface (Fig. 1) and the number of individual residues ensuring receptor specificity (Fig. 4), the magnitude of the effects of these point mutations is quite remarkable. This is in agreement with our EPR data that show large line-shape changes for spin labels at position Leu-68 as well as positions Tyr-238 and Thr-246, which are close to Asp-240. To further test the importance of the 14 residues identified, we introduced five N-domain along with 9 C-domain arrestin-1 residues into arrestin-2 (Arr2N5C9) as well as homologous 14 arrestin-2 residues into arrestin-1 (Arr1N5C9) and found that these sets of substitutions completely reverse receptor preference, at least as far as P-Rh* is concerned (Fig. 4C).
FIGURE 4.
Ranking receptor-discriminator residues by their importance. A, shown is direct binding to P-Rh* of the indicated arrestin-2-based mutants (abbreviations as in the legend to Fig. 3). The mutants designated by a single number are based on Arr2(N7+C12) base mutant with 19 substitutions (shown as gray CPK models on black arrestin-2 molecule on the right) in which individual mutation in the indicated positions were reversed to the residue present in arrestin-2. Note that five reversals of single amino acid substitutions, two in the N-domain (68 and 86) and three in the C-domain (240, 259, and 261), reduce P-Rh* binding by 27–38%. B, binding to P-Rh* of indicated arrestin-2-based mutants is shown. Six mutants on the right carry the reversals of up to five indicated mutations. The arrestin-2 molecule (black) on the right shows CPK models of arrestin-1 residues that do or do not play a role in rhodopsin preference shown in gray and black, respectively. C, the exchange of five and nine residues in the N- and C-domain, respectively, reverses receptor preference of arrestin-1 and -2. Arr1/2-N5C9, arrestin-1-(V52L,G54E,K55R,I72L,V90S,V244Y,N246D,V248C,I256K,K257C,T258P,A261M,Q265D,K267T); Arr2/1-N5C9, arrestin-2-(L48V,E50G,R51K,L68I,S86V,Y238V,D240N,C242V,K250I,C251K,P252T,M255A,D259Q,T261K). Arrestin-2-derived residues in Arr1N5C9 (black on gray molecule) and arrestin-1-derived residues in Arr2N5C9 (gray on black molecule) are shown on the right. Binding experiments were performed two-three times in duplicate. Means ± S.D. are presented.
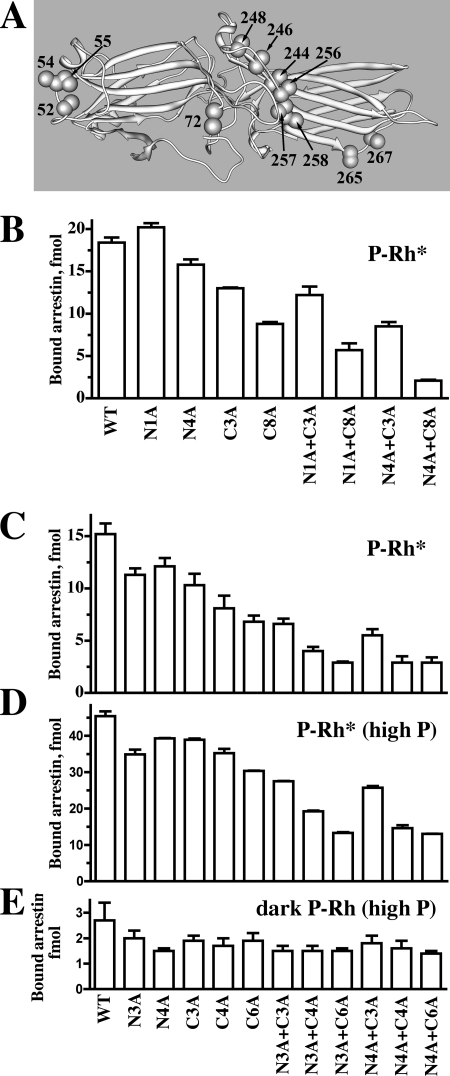
Arrestin elements with different functions contribute to the energy of the arrestin-receptor interaction. Several positively charged residues interact with multiple receptor-attached phosphates (23, 31, 57–60). A least 10 exposed residues mediate receptor subtype-specific binding, contributing to receptor discrimination (Figs. 3 and 4 and Refs. 14 and 21). In addition, a number of conserved non-phosphate binding residues are also engaged by the receptor (Fig. 1 and Ref. 22). Because phosphate binding and other conserved arrestin elements would readily mediate the binding to any GPCR, the contribution of “receptor discriminator” residues determines the extent of receptor subtype specificity that arrestin proteins can achieve. To gauge this contribution quantitatively, we replaced key residues that determine receptor preference with alanines (Fig. 5A) in arrestin-1 and measured the binding of the mutants to its cognate receptor, P-Rh*. These mutations reduce the binding to varying degrees, with the reduction roughly proportional to the number of altered side chains (Fig. 5, B and C). Interestingly, the effect of any mutation strongly depends on the context; it tends to be much greater when combined with other alanine substitutions. For example, the I72A (N1A in panel B; note that position 72 in arrestin-1 corresponds to position 68 in arrestin-2 (cf. Fig. 2)) mutation did not reduce the binding in the context of WT arrestin-1 and slightly reduced it in the context of the protein with three alanine substitutions in the C-domain (compare C3A and N1A/C3A) while reducing it by a third in the context of arrestin-1 with eight alanines in the C-domain (compare C8A and N1A/C8A). Similarly, the quadruple alanine substitution (V52A,G54A,K55A,I72A; N4A) reduced binding by 15% in the context of WT (compare WT and N4A), but in the context of mutants with 3 and 8 alanines in the C-domain it reduced the binding by 35 and 75%, respectively (compare C3A with N4A/C3A and C8A with N4A/C8A). The effect of a triple substitution (V52A,G54A,K55A) in the N-domain (N3A in Fig. 5, panel C) grew from 26% reduction in the context of WT (WT versus N3A) to 43–50% reduction in the context of C4A or C6A mutants (Fig. 5C). The same tendency is apparent if one calculates the effect of C-domain substitutions. Binding decreases due to another triple mutation (N246A,Q265A,K267A) (C3A), from ∼30% in WT to 42–45% in the N3A and N4A backgrounds (compare C3A with WT, N3A/C3A with N3A, and N4A/C3A with N4A in panel C). The effects of C4A and C6A mutations also increased from ∼50% in WT to ∼75% reduction in the context of N3A or N4A mutants (Fig. 5C). Importantly, just 8 appropriately selected alanine substitutions reduce P-Rh* binding ∼5-fold. The same cooperativity was observed when we used highly phosphorylated P-Rh* (4 or more phosphates) instead of P-Rh* with a modest phosphorylation level (2–3 phosphates per molecule), although due to increased contribution of phosphate binding residues, the maximum decrease in P-Rh* binding was reduced from 80 to 70% (Fig. 5, C and D). None of the mutations dramatically reduced the binding to dark P-Rh (Fig. 5E), which is mediated solely by the interactions of highly conserved phosphate-binding elements with receptor-attached phosphates (61). We found that even the most disabling mutations did not reduce arrestin-1 binding to P-Rh* to the level of dark P-Rh binding (Fig. 5, C–E), indicating that other conserved receptor binding residues that were not mutated support ∼20% of the interaction.
FIGURE 5.
Arrestin-1 residues that determine receptor preference are key contributors to receptor binding. A, shown is the arrestin-1 molecule with alanines (CPK models) in all positions substituted in panels B, C, D, and E. Direct binding of arrestin-1-based mutants to P-Rh* (2–3 mol of phosphates/mol of Rh) (B and C) or highly phosphorylated (at least four phosphates/mol of Rh) P-Rh* (D) or dark inactive P-Rh (E) was determined as described under “Experimental Procedures.” The mutants are designated as follows: N1A, point N-domain mutant I72A; N3A, triple N-domain mutant V52A,G54A,K55A; N4A, quadruple N-domain mutant V52A,G54A,K55A,I72A; C3A, triple C-domain mutant N246A,Q265A,K267A; C4A, quadruple C-domain mutant V244A,N246A,Q265A,K267A; C6A, six substitutions in the C-domain V244A,N246A,K257A,I258A,Q265A,K267A; N8A, eight substitutions in the C-domain V244A,N246A,V248A,I256A,K257A,I258A,Q265A,K267A. Binding experiments were performed two-three times in duplicate. Means ± S.D. are presented.
Receptor Discriminator Residues Are Crucial for the Binding of Non-visual Arrestins to Their Cognate GPCRs
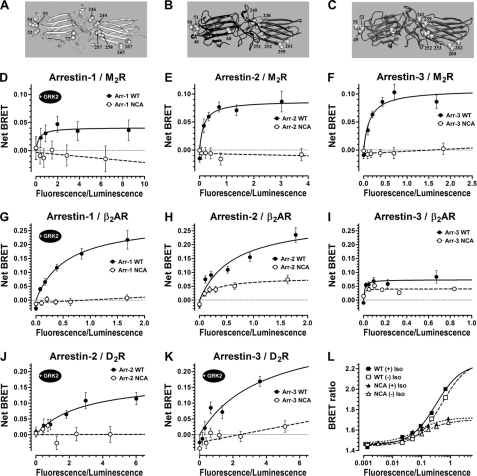
Arrestin-1 has more phosphate binding residues than other members of the family (8, 23, 57) and demonstrates the strictest dependence on receptor phosphorylation, with the binding to P-Rh* requiring at least three receptor-attached phosphates (31, 62) and exceeding that of Rh* by 10–20-fold (26, 59). In contrast, non-visual arrestins show only a ∼3–5-fold binding differential (14–16, 19) and were reported to bind unphosphorylated receptors in response to their activation (42, 63–68). Thus, it appears that non-phosphate binding elements likely contribute more significantly to the binding of non-visual arrestins to their cognate receptors. To determine the contribution of receptor-discriminator residues in non-visual arrestins, we constructed mutants (termed NCA in Fig. 6) with alanine substitutions of key receptor-discriminating residues: four in the N-domain (Val-52, Gly-54, Lys-55, Ile-72 or Leu-48, Glu-50, Arg-51, Leu-68 or Leu-49, Asp-51, Arg-52, Leu-69 in arrestin-1, -2, or -3, respectively) and six in the C-domain (Val-244, Asn-246, Lys-257, Thr-258, Gln-265, Lys-267 or Tyr-238, Asp-240, Cys-251, Pro-252, Asp-259, Thr-261 or Tyr-239, Asp-241, Cys-252, Pro-253, Asp-260, Q262 in arrestin-1, -2, or -3, respectively) (Fig. 6, A–D). WT arrestins and NCA mutants were tagged with Venus (enhanced YFP (38)) on the N terminus and co-expressed with M2 muscarinic cholinergic, β2-adrenergic, and D2 dopamine receptor C-terminally-tagged with Renilla luciferase (41) in COS-7 cells. Arrestin recruitment to the receptor in response to agonist stimulation is reflected in BRET from luciferase to Venus, which saturates with increasing Venus-arrestin expression (42, 43) (Fig. 6). Although this assay is not as quantitative as in vitro direct binding, it has two important advantages; it allows the measurement of arrestin interactions with receptors that are not available in a purified form, and it does so under the more physiological conditions of an intact cell. To validate the assay, we confirmed earlier observations (14, 18, 32, 42, 47, 69, 71, 72) that both non-visual arrestins readily interact with β2-adrenergic and M2 receptors (Fig. 6, E, F, H, I, J, and K). Unexpectedly, we found that the binding of arrestin-2 or -3 to the D2 dopamine receptor could only be detected upon overexpression of GRK2 (Fig. 6, K and L). Arrestin-1 has much lower affinity for β2-adrenergic and M2 receptors (14, 47), as reflected in the absence of detectable BRET signal without GRK2 co-expression (data not shown). Its interactions with β2-adrenergic and M2 receptors in cells can be observed only upon overexpression of GRK2 (Fig. 6, D and G). Arrestin-1 binding to non-cognate receptors appears to be largely driven by the receptor-attached phosphates, because it is undetectable without GRK2 co-expression. However, receptor discriminator residues also contribute, as NCA mutations in arrestin-1 obliterate detectable interaction with both receptors. We also found that NCA mutations in both non-visual arrestins effectively prevented the binding to M2 and D2 receptors and greatly reduced the observed interaction with β2-adrenergic receptor, demonstrating the crucial role of receptor-discriminator residues in these interactions (Fig. 6, E, F, H, I, J, and K). Even in the case of arrestin-3 binding to β2-adrenergic receptor, which yields relatively low BRET signal, NCA mutations reduce agonist-stimulated (Fig. 6I) as well as agonist-independent (Fig. 6L, (−)Iso curves), binding. The latter appears to be specific, as was observed previously in a direct binding assay with purified β2-adrenergic receptor (14–16). Thus, 10 residues in arrestin-2 and -3 that determine their receptor preference critically contribute to the energy of their interaction with non-visual GPCRs.
FIGURE 6.
Receptor-discriminator residues play a decisive role in arrestin interactions with different GPCRs in living cells. A, B, and C, molecules of arrestin-1 (A), -2 (B), and -3 (C) with alanines (shown as CPK models) in all positions substituted in corresponding NCA mutants. BRET assays were used to test the binding of WT (filled circles and solid lines) or NCA mutants (alanine substitutions of Val-52, Gly-54, Lys-55, Ile-72, Val-244, Asn-246, Lys-257, Thr-258, Gln-265, Lys-267 or Leu-48, Glu-50, Arg-51, Leu-68, Tyr-238, Asp-240, Cys-251, Pro-252, Asp-259, Thr-261 or Leu-49, asp-51, Arg-52, Leu-69, Tyr-239, Asp-241, Cys-252, Pro-253, Asp-260, Gln-262 in arrestin-1, -2, or -3, respectively; open circles and dashed lines). N-terminal Venus-tagged arrestin-1 (D and G), arrestin-2 (E, H, and J), and arrestin-3 (F, I, and K) were expressed with the M2 muscarinic (D–F), β2-adrenergic (G, H, I and L), and D2 (J and K) dopamine receptors C-terminal-fused with Renilla luciferase variant 8 (RLuc8). D–K, agonist-induced increase in BRET signal as a function of arrestin expression is shown. Cells expressing M2, β2-adrenergic, and D2 receptors with 25 μm carbamoylcholine, 10 μm isoproterenol, or 10 μm quinpirole, respectively, for 25 min at 37 °C are shown. L, shown is the BRET ratio for the data depicted in panel I from the cells expressing increasing levels of Venus-tagged WT arrestin-3 (squares) or the corresponding NCA mutant (triangles) and a fixed amount of β2-adrenergic receptor-RLuc8. Cells were treated with β-agonist isoproterenol (+Iso; filled symbols) or vehicle (−Iso; open symbols). Although net maximum agonist-stimulated BRET signals for WT and the mutant arrestin-3/β2-adrenergic receptor combination was not dramatically different (I), the actual interaction, as demonstrated by the BRET ratios shown in panel L, is significantly greater for WT arrestin-3. Data points were fitted to a one-site binding hyperbola model or a simple linear regression (D–K) when convergence was not achieved (most of the mutant NCA arrestins) or to a one-site sigmoidal dose-response model (L) using the GraphPad Prism Version 5.04 software. Means ± S.E. of six parallel measurements in a typical experiment (representative of two to four independent experiments for each receptor/arrestin combination) are shown.
DISCUSSION
Arrestin binding to active phosphoreceptor is a complex multi-step process (for review, see Ref. 61). It is driven by direct arrestin interactions with two types of elements: receptor-attached phosphates, which make every receptor look the same and, therefore, are largely responsible for the binding to non-cognate GPCRs, and non-phosphorylated cytoplasmic residues, unique for every receptor that likely determines arrestin preference for particular subtypes. Visual arrestin-1 is the only vertebrate subtype that shows clear specificity toward just one receptor, rhodopsin (73). The disparity in numbers between hundreds of non-visual GPCRs in vertebrates and only two arrestins to regulate their signaling is striking (1).
The receptor binding surface has so far been mapped out only on arrestin-1, where it was shown by a variety of methods to be fairly extensive, covering a large part of the concave sides of both arrestin domains (14, 22–25, 45, 46, 74). Here we demonstrate that an equally extensive surface of arrestin-2 is engaged by the receptor (Fig. 1), suggesting that a large receptor footprint is a common feature of arrestin proteins and likely responsible for the very high affinity of the interaction (14, 71, 75–78). The sheer size of the receptor binding surface suggested an idea that a single arrestin molecule could fit two GPCRs in a dimer (79). However, a series of recent studies clearly demonstrated that the physiologically relevant complex comprises one arrestin bound to a single receptor molecule in vertebrates (51, 78, 80–82) and in Drosophila (83). Apparently, the large area of receptor contact ensures the ability of arrestin to effectively preclude G-protein interaction (84–86).
So far only two structural elements that contribute to the observed selectivity of arrestin-1 and broad specificity of non-visual arrestins were identified. One is Val-90 in arrestin-1, which is buried and participates in stabilization of the β-strand sandwich of the N-domain; its replacement with Ser or Ala found in arrestin-2 and -3, respectively, makes the N-domain more flexible and enhances arrestin-1 binding to non-visual GPCRs (6). The other is a uniquely loose conformation of the receptor binding β-strand XVI in arrestin-3, which is detached from the β-sheet and appears to be responsible for reduced selectivity of arrestin-3 for active phosphoreceptor (9). Here we identified several exposed residues in the N- and C-domains of arrestin proteins that largely determine their preference for particular receptor subtypes. We also showed that these residues critically contribute to receptor interaction in arrestin-1, -2, and -3, as their alanine substitution dramatically decreases or completely prevents receptor binding (Figs. 5 and 6). The deleterious effects of alanine substitutions at the non-phosphate binding residues in arrestin-1 are not as dramatic as in non-visual subtypes, suggesting that the contribution of receptor-attached phosphates to receptor interaction in arrestin-2 and -3 is smaller than in the case of arrestin-1. Based on our data, we cannot exclude the possibility that the some of the residues deduced from substitutions of arrestin-1 residues with those of arrestin-2 and vice versa affect receptor specificity indirectly by altering the structure of the receptor-binding elements and do not necessarily directly contact the receptor. However, in many positions the change of the side chain impacts rhodopsin binding (Figs. 3 and 4), its elimination suppresses that binding of three different arrestins to multiple GPCRs (Figs. 5 and 6), and nearby residues in arrestin-2 are significantly immobilized by P-Rh* interaction. We believe that in these cases direct engagement of these residues by GPCRs is the most parsimonious explanation. In terms of their relative contribution to receptor selectivity and binding energy, receptor discriminator residues fall into two groups: five are absolutely crucial and nine others play a supportive role (Fig. 7). Although in a direct binding assay (Figs. 3 and 4) these were identified essentially as discriminating between rhodopsin and non-visual receptors, their crucial role in arrestin-2 and -3 interaction with three different GPCRs (Fig. 6) strongly suggests that these residues play a more general role in the recognition of particular receptor subtypes. This idea needs to be tested experimentally. Interestingly, several arrestin-2 residues significantly immobilized by receptor binding (Fig. 1) are in the elements that do not play an appreciable role in receptor preference. These likely represent more “generic” elements engaging different GPCRs.
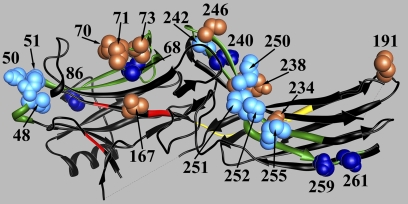
FIGURE 7.
Arrestin residues important for receptor binding. The structure of arrestin-2 (6) with key functional elements shown is as follows: phosphate binding residues (Lys-10, Lys-11, Arg-165, Arg-169, Lys-170), red; two previously identified (21) elements (residues 45–86 and 231–262 in the N- and C-domain, respectively) that determine receptor specificity, green; individual residues in these elements playing a key role in receptor preference (Leu-68, Ser-86, Asp-240, Asp-259, Thr-261), dark blue CPK models; residues playing supporting roles in receptor preference (Leu-48, Glu-50, Arg-51, Tyr-238, Cys-242, Lys-250, Cys-251, Pro-252, Met-255), light blue CPK models; residues in positions that showed significant immobilization by P-Rh* binding (Val-70, Leu-71, Leu-73, Val-167, Leu-191, Ser-234, Tyr-238), light brown CPK models; interdomain hinge (residues 173–184) (34, 39, 40, 50, 35), yellow. The image was generated using ViewerPro 6.0.
Only two non-receptor binding partners, microtubules (34, 50) and calmodulin (87), interact at the same concave sides of the two arrestin domains as GPCRs. Arrestins bind a wide variety of other signaling proteins, most of which engage the convex non-receptor binding side of the molecule (for review, see Refs. 1 and 2), whereas clathrin and AP2, which mediate the trafficking of the arrestin-receptor complex, interact with well defined sites in the C termini of non-visual arrestins (88–91). This spatial arrangement allows independent manipulation of arrestin interactions with these three types of binding partners, with a view of constructing special mutants with therapeutic potential (92). This approach was recently validated by the demonstration that “enhanced” arrestin-1, which binds unphosphorylated active rhodopsin with high affinity, partially compensates for the deficit of rhodopsin phosphorylation in vivo (55). Many human disorders are associated with excessive signaling by various GPCRs (70) that can be dampened by enhanced arrestins. However, most cells express multiple GPCR subtypes, only one of which is the mutant that needs to be restrained. Broad receptor specificity of both non-visual arrestins would prevent selective targeting of a particular receptor. The small number of key receptor discriminator residues identified here opens up the possibility of constructing arrestins with greatly enhanced receptor specificity, which would make selective targeting of a group of receptors, or even an individual subtype, feasible.
Supplementary Material
Acknowledgments
We are grateful to Dr. T. Shinohara for bovine arrestin-1 cDNA, Dr. Rosalie K. Crouch for 11-cis-retinal, Dr. Jonathan A. Javitch for expert advice on receptor-arrestin BRET and the plasmid encoding Venus, Dr. Nevin A. Lambert for plasmid encoding Renilla luciferase variant 8, F. B. Couch for making alanine substitution mutants, and Dr. Carl Johnson for use of POLARstar Optima dual channel luminometer and fluorimeter microplate reader.
This work was supported, in whole or in part, by National Institutes of Health Grants GM081756, GM077561, EY011500 (to V. V. G.), GM070642 (to C. S. K.), and EY005216 (to W. L. H.). This work was also supported by the Jules Stein Professorship Endowment (to W. L. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
We use systematic names for the arrestin proteins: arrestin-1 (historic names: visual or rod arrestin, 48 kDa protein, or S-antigen), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin).
- GPCR
- G protein-coupled receptor
- BRET
- bioluminescence energy transfer s
- P-Rh*
- light-activated phosphorylated rhodopsin
- Arr
- arrestin
- CPK
- Corey-Pauling-Koltun.
REFERENCES
- 1. Gurevich E. V., Gurevich V. V. (2006) Genome Biology 7, 236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. DeWire S. M., Ahn S., Lefkowitz R. J., Shenoy S. K. (2007) Annu. Rev. Physiol. 69, 483–510 [DOI] [PubMed] [Google Scholar]
- 3. Sterne-Marr R., Gurevich V. V., Goldsmith P., Bodine R. C., Sanders C., Donoso L. A., Benovic J. L. (1993) J. Biol. Chem. 268, 15640–15648 [PubMed] [Google Scholar]
- 4. Attramadal H., Arriza J. L., Aoki C., Dawson T. M., Codina J., Kwatra M. M., Snyder S. H., Caron M. G., Lefkowitz R. J. (1992) J. Biol. Chem. 267, 17882–17890 [PubMed] [Google Scholar]
- 5. Hirsch J. A., Schubert C., Gurevich V. V., Sigler P. B. (1999) Cell 97, 257–269 [DOI] [PubMed] [Google Scholar]
- 6. Han M., Gurevich V. V., Vishnivetskiy S. A., Sigler P. B., Schubert C. (2001) Structure 9, 869–880 [DOI] [PubMed] [Google Scholar]
- 7. Milano S. K., Pace H. C., Kim Y. M., Brenner C., Benovic J. L. (2002) Biochemistry 41, 3321–3328 [DOI] [PubMed] [Google Scholar]
- 8. Sutton R. B., Vishnivetskiy S. A., Robert J., Hanson S. M., Raman D., Knox B. E., Kono M., Navarro J., Gurevich V. V. (2005) J. Mol. Biol. 354, 1069–1080 [DOI] [PubMed] [Google Scholar]
- 9. Zhan X., Gimenez L. E., Gurevich V. V., Spiller B. W. (2011) J. Mol. Biol. 406, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gurevich V. V., Hanson S. M., Gurevich E. V., Vishnivetskiy S. A. (2007) in Methods in Signal Transduction Series (Kisselev O., Fliesler S. J. eds) pp. 55–88, CRC Press, Boca Raton, FL [Google Scholar]
- 11. Nikonov S. S., Brown B. M., Davis J. A., Zuniga F. I., Bragin A., Pugh E. N., Jr., Craft C. M. (2008) Neuron 59, 462–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gurevich E. V., Benovic J. L., Gurevich V. V. (2002) Neuroscience 109, 421–436 [DOI] [PubMed] [Google Scholar]
- 13. Gurevich E. V., Benovic J. L., Gurevich V. V. (2004) J. Neurochem. 91, 1404–1416 [DOI] [PubMed] [Google Scholar]
- 14. Gurevich V. V., Dion S. B., Onorato J. J., Ptasienski J., Kim C. M., Sterne-Marr R., Hosey M. M., Benovic J. L. (1995) J. Biol. Chem. 270, 720–731 [DOI] [PubMed] [Google Scholar]
- 15. Celver J., Vishnivetskiy S. A., Chavkin C., Gurevich V. V. (2002) J. Biol. Chem. 277, 9043–9048 [DOI] [PubMed] [Google Scholar]
- 16. Kovoor A., Celver J., Abdryashitov R. I., Chavkin C., Gurevich V. V. (1999) J. Biol. Chem. 274, 6831–6834 [DOI] [PubMed] [Google Scholar]
- 17. Kohout T. A., Lin F. S., Perry S. J., Conner D. A., Lefkowitz R. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 1601–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Barak L. S., Ferguson S. S., Zhang J., Caron M. G. (1997) J. Biol. Chem. 272, 27497–27500 [DOI] [PubMed] [Google Scholar]
- 19. Pan L., Gurevich E. V., Gurevich V. V. (2003) J. Biol. Chem. 278, 11623–11632 [DOI] [PubMed] [Google Scholar]
- 20. Smith W. C., Gurevich E. V., Dugger D. R., Vishnivetskiy S. A., Shelamer C. L., McDowell J. H., Gurevich V. V. (2000) Invest. Ophthalmol. Vis. Sci. 41, 2445–2455 [PubMed] [Google Scholar]
- 21. Vishnivetskiy S. A., Hosey M. M., Benovic J. L., Gurevich V. V. (2004) J. Biol. Chem. 279, 1262–1268 [DOI] [PubMed] [Google Scholar]
- 22. Hanson S. M., Francis D. J., Vishnivetskiy S. A., Kolobova E. A., Hubbell W. L., Klug C. S., Gurevich V. V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4900–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanson S. M., Gurevich V. V. (2006) J. Biol. Chem. 281, 3458–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pulvermüller A., Schroder K., Fischer T., Hofmann K. P. (2000) J. Biol. Chem. 275, 37679–37685 [DOI] [PubMed] [Google Scholar]
- 25. Vishnivetskiy S. A., Francis D., Van Eps N., Kim M., Hanson S. M., Klug C. S., Hubbell W. L., Gurevich V. V. (2010) J. Mol. Biol. 395, 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gurevich V. V., Benovic J. L. (1993) J. Biol. Chem. 268, 11628–11638 [PubMed] [Google Scholar]
- 27. Gurevich V. V., Benovic J. L. (1992) J. Biol. Chem. 267, 21919–21923 [PubMed] [Google Scholar]
- 28. Gurevich V. V. (1996) Methods Enzymol. 275, 382–397 [DOI] [PubMed] [Google Scholar]
- 29. Shinohara T., Dietzschold B., Craft C. M., Wistow G., Early J. J., Donoso L. A., Horwitz J., Tao R. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 6975–6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gurevich V. V. (1998) J. Biol. Chem. 273, 15501–15506 [DOI] [PubMed] [Google Scholar]
- 31. Vishnivetskiy S. A., Raman D., Wei J., Kennedy M. J., Hurley J. B., Gurevich V. V. (2007) J. Biol. Chem. 282, 32075–32083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pals-Rylaarsdam R., Gurevich V. V., Lee K. B., Ptasienski J. A., Benovic J. L., Hosey M. M. (1997) J. Biol. Chem. 272, 23682–23689 [DOI] [PubMed] [Google Scholar]
- 33. Gurevich V. V., Benovic J. L. (2000) Methods Enzymol. 315, 422–437 [DOI] [PubMed] [Google Scholar]
- 34. Hanson S. M., Cleghorn W. M., Francis D. J., Vishnivetskiy S. A., Raman D., Song X., Nair K. S., Slepak V. Z., Klug C. S., Gurevich V. V. (2007) J. Mol. Biol. 368, 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vishnivetskiy S. A., Hirsch J. A., Velez M. G., Gurevich Y. V., Gurevich V. V. (2002) J. Biol. Chem. 277, 43961–43967 [DOI] [PubMed] [Google Scholar]
- 36. Crane J. M., Mao C., Lilly A. A., Smith V. F., Suo Y., Hubbell W. L., Randall L. L. (2005) J. Mol. Biol. 353, 295–307 [DOI] [PubMed] [Google Scholar]
- 37. Kusnetzow A. K., Altenbach C., Hubbell W. L. (2006) Biochemistry 45, 5538–5550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nagai T., Ibata K., Park E. S., Kubota M., Mikoshiba K., Miyawaki A. (2002) Nat. Biotechnol. 20, 87–90 [DOI] [PubMed] [Google Scholar]
- 39. Song X., Raman D., Gurevich E. V., Vishnivetskiy S. A., Gurevich V. V. (2006) J. Biol. Chem. 281, 21491–21499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song X., Gurevich E. V., Gurevich V. V. (2007) J. Neurochem. 103, 1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Loening A. M., Fenn T. D., Wu A. M., Gambhir S. S. (2006) Protein Eng. Des. Sel. 19, 391–400 [DOI] [PubMed] [Google Scholar]
- 42. Namkung Y., Dipace C., Javitch J. A., Sibley D. R. (2009) J. Biol. Chem. 284, 15038–15051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Namkung Y., Dipace C., Urizar E., Javitch J. A., Sibley D. R. (2009) J. Biol. Chem. 284, 34103–34115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuravi S., Lan T. H., Barik A., Lambert N. A. (2010) Biophys. J. 98, 2391–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ohguro H., Palczewski K., Walsh K. A., Johnson R. S. (1994) Protein Sci. 3, 2428–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhuang T., Vishnivetskiy S. A., Gurevich V. V., Sanders C. R. (2010) Biochemistry 49, 10473–10485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gurevich V. V., Richardson R. M., Kim C. M., Hosey M. M., Benovic J. L. (1993) J. Biol. Chem. 268, 16879–16882 [PubMed] [Google Scholar]
- 48. Columbus L., Hubbell W. L. (2002) Trends Biochem. Sci. 27, 288–295 [DOI] [PubMed] [Google Scholar]
- 49. Hubbell W. L., Gross A., Langen R., Lietzow M. A. (1998) Curr. Opin. Struct. Biol. 8, 649–656 [DOI] [PubMed] [Google Scholar]
- 50. Hanson S. M., Francis D. J., Vishnivetskiy S. A., Klug C. S., Gurevich V. V. (2006) J. Biol. Chem. 281, 9765–9772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hanson S. M., Van Eps N., Francis D. J., Altenbach C., Vishnivetskiy S. A., Arshavsky V. Y., Klug C. S., Hubbell W. L., Gurevich V. V. (2007) EMBO J. 26, 1726–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hanson S. M., Dawson E. S., Francis D. J., Van Eps N., Klug C. S., Hubbell W. L., Meiler J., Gurevich V. V. (2008) Structure 16, 924–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schubert C., Hirsch J. A., Gurevich V. V., Engelman D. M., Sigler P. B., Fleming K. G. (1999) J. Biol. Chem. 274, 21186–21190 [DOI] [PubMed] [Google Scholar]
- 54. Imamoto Y., Tamura C., Kamikubo H., Kataoka M. (2003) Biophys. J. 85, 1186–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Song X., Vishnivetskiy S. A., Gross O. P., Emelianoff K., Mendez A., Chen J., Gurevich E. V., Burns M. E., Gurevich V. V. (2009) Curr. Biol. 19, 700–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hanson S. M., Vishnivetskiy S. A., Hubbell W. L., Gurevich V. V. (2008) Biochemistry 47, 1070–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gurevich V. V., Benovic J. L. (1995) J. Biol. Chem. 270, 6010–6016 [DOI] [PubMed] [Google Scholar]
- 58. Vishnivetskiy S. A., Paz C. L., Schubert C., Hirsch J. A., Sigler P. B., Gurevich V. V. (1999) J. Biol. Chem. 274, 11451–11454 [DOI] [PubMed] [Google Scholar]
- 59. Gurevich V. V., Benovic J. L. (1997) Mol. Pharmacol. 51, 161–169 [DOI] [PubMed] [Google Scholar]
- 60. Vishnivetskiy S. A., Schubert C., Climaco G. C., Gurevich Y. V., Velez M. G., Gurevich V. V. (2000) J. Biol. Chem. 275, 41049–41057 [DOI] [PubMed] [Google Scholar]
- 61. Gurevich V. V., Gurevich E. V. (2004) Trends Pharmacol. Sci. 25, 105–111 [DOI] [PubMed] [Google Scholar]
- 62. Mendez A., Burns M. E., Roca A., Lem J., Wu L. W., Simon M. I., Baylor D. A., Chen J. (2000) Neuron 28, 153–164 [DOI] [PubMed] [Google Scholar]
- 63. Whistler J. L., Tsao P., von Zastrow M. (2001) J. Biol. Chem. 276, 34331–34338 [DOI] [PubMed] [Google Scholar]
- 64. Kishi H., Krishnamurthy H., Galet C., Bhaskaran R. S., Ascoli M. (2002) J. Biol. Chem. 277, 21939–21946 [DOI] [PubMed] [Google Scholar]
- 65. Kim O. J., Gardner B. R., Williams D. B., Marinec P. S., Cabrera D. M., Peters J. D., Mak C. C., Kim K. M., Sibley D. R. (2004) J. Biol. Chem. 279, 7999–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mukherjee S., Palczewski K., Gurevich V. V., Hunzicker-Dunn M. (1999) J. Biol. Chem. 274, 12984–12989 [DOI] [PubMed] [Google Scholar]
- 67. Mukherjee S., Gurevich V. V., Preninger A., Hamm H. E., Bader M. F., Fazleabas A. T., Birnbaumer L., Hunzicker-Dunn M. (2002) J. Biol. Chem. 277, 17916–17927 [DOI] [PubMed] [Google Scholar]
- 68. Walther C., Nagel S., Gimenez L. E., Moerl K., Gurevich V. V., Beck-Sickinger A. G. (2010) J. Biol. Chem. 285, 41578–41590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lee K. B., Ptasienski J. A., Pals-Rylaarsdam R., Gurevich V. V., Hosey M. M. (2000) J. Biol. Chem. 275, 9284–9289 [DOI] [PubMed] [Google Scholar]
- 70. Schöneberg T., Schulz A., Biebermann H., Hermsdorf T., Römpler H., Sangkuhl K. (2004) Pharmacol. Ther. 104, 173–206 [DOI] [PubMed] [Google Scholar]
- 71. Gurevich V. V., Pals-Rylaarsdam R., Benovic J. L., Hosey M. M., Onorato J. J. (1997) J. Biol. Chem. 272, 28849–28852 [DOI] [PubMed] [Google Scholar]
- 72. Klewe I. V., Nielsen S. M., Tarpø L., Urizar E., Dipace C., Javitch J. A., Gether U., Egebjerg J., Christensen K. V. (2008) Neuropharmacology 54, 1215–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gurevich V. V., Gurevich E. V. (2006) Pharmacol. Ther. 110, 465–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Dinculescu A., McDowell J. H., Amici S. A., Dugger D. R., Richards N., Hargrave P. A., Smith W. C. (2002) J. Biol. Chem. 277, 11703–11708 [DOI] [PubMed] [Google Scholar]
- 75. Schleicher A., Kühn H., Hofmann K. P. (1989) Biochemistry 28, 1770–1775 [DOI] [PubMed] [Google Scholar]
- 76. Pulvermüller A., Maretzki D., Rudnicka-Nawrot M., Smith W. C., Palczewski K., Hofmann K. P. (1997) Biochemistry 36, 9253–9260 [DOI] [PubMed] [Google Scholar]
- 77. Zhang L., Sports C. D., Osawa S., Weiss E. R. (1997) J. Biol. Chem. 272, 14762–14768 [DOI] [PubMed] [Google Scholar]
- 78. Bayburt T. H., Vishnivetskiy S. A., McLean M., Morizumi T., Huang C. C., Tesmer J. J., Ernst O. P., Sligar S. G., Gurevich V. V. (2011) J. Biol. Chem. 285, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Fotiadis D., Jastrzebska B., Philippsen A., Müller D. J., Palczewski K., Engel A. (2006) Curr. Opin. Struct. Biol. 16, 252–259 [DOI] [PubMed] [Google Scholar]
- 80. Hanson S. M., Gurevich E. V., Vishnivetskiy S. A., Ahmed M. R., Song X., Gurevich V. V. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3125–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tsukamoto H., Sinha A., Dewitt M., Farrens D. L. (2010) J. Mol. Biol. 399, 501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Song X., Vishnivetskiy S. A., Seo J., Chen J., Gurevich E. V., Gurevich V. V. (2011) Neuroscience 174, 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Satoh A. K., Xia H., Yan L., Liu C. H., Hardie R. C., Ready D. F. (2010) Neuron 67, 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Krupnick J. G., Gurevich V. V., Benovic J. L. (1997) J. Biol. Chem. 272, 18125–18131 [DOI] [PubMed] [Google Scholar]
- 85. Wilden U., Hall S. W., Kühn H. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 1174–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lohse M. J., Andexinger S., Pitcher J., Trukawinski S., Codina J., Faure J. P., Caron M. G., Lefkowitz R. J. (1992) J. Biol. Chem. 267, 8558–8564 [PubMed] [Google Scholar]
- 87. Wu N., Hanson S. M., Francis D. J., Vishnivetskiy S. A., Thibonnier M., Klug C. S., Shoham M., Gurevich V. V. (2006) J. Mol. Biol. 364, 955–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Goodman O. B., Jr., Krupnick J. G., Santini F., Gurevich V. V., Penn R. B., Gagnon A. W., Keen J. H., Benovic J. L. (1996) Nature 383, 447–450 [DOI] [PubMed] [Google Scholar]
- 89. Laporte S. A., Oakley R. H., Zhang J., Holt J. A., Ferguson S. S., Caron M. G., Barak L. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3712–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kang D. S., Kern R. C., Puthenveedu M. A., von Zastrow M., Williams J. C., Benovic J. L. (2009) J. Biol. Chem. 284, 29860–29872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kim Y. M., Benovic J. L. (2002) J. Biol. Chem. 277, 30760–30768 [DOI] [PubMed] [Google Scholar]
- 92. Gurevich V. V., Gurevich E. V. (2010) Expert Rev. Mol. Med. 12, e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kim M., Hanson S. M., Vishnivetskiy S. A., Song X., Cleghorn W. M., Hubbell W. L., Gurevich V. V. (2011) Biochemistry 50, 2235–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.