Abstract
In Gram-negative bacteria, type II secretion systems assemble a piston-like structure, called pseudopilus, which expels exoproteins out of the cell. The pseudopilus is constituted by a major pseudopilin that when overproduced multimerizes into a long cell surface structure named hyper-pseudopilus. Pseudomonas aeruginosa possesses two type II secretion systems, Xcp and Hxc. Although major pseudopilins are exchangeable among type II secretion systems, we show that XcpT and HxcT are not. We demonstrate that HxcT does not form a hyper-pseudopilus and is different in amino acid sequence and multimerization properties. Using structure-based mutagenesis, we observe that five mutations are sufficient to revert HxcT into a functional XcpT-like protein, which also becomes capable of forming a hyper-pseudopilus. Phylogenetic and experimental analysis showed that the whole Hxc system was acquired by P. aeruginosa PAO1 and other Pseudomonas species through horizontal gene transfer. We thus identified a new type II secretion subfamily, of which the P. aeruginosa Hxc system is the archetype. This finding demonstrates how similar bacterial machineries evolve toward distinct mechanisms that may contribute specific functions.
Keywords: Bacteria, Membrane Trafficking, Protein Assembly, Protein Secretion, Protein-Protein Interactions, Pseudomonas aeruginosa, Pseudopilus, T2SS, Xcp
Introduction
At least six distinct pathways have been identified in Gram-negative bacteria for the secretion of a diverse pool of extracellular proteins (1). Among them, the type II secretion pathway is a two-step process in which exoproteins are initially exported through the cytoplasmic membrane by either the Sec or Tat translocons (2). Translocation across the outer membrane is then performed by the type II secretion system (T2SS),5 also called secreton. In our model organism, the gammaproteobacterium, Pseudomonas aeruginosa, the secreton is a multiprotein complex involving at least 12 different proteins called XcpA, XcpP–Z (3). In the current model of Xcp secreton assembly, three subcomplexes are defined: the inner membrane platform (XcpRSYZ), required to provide energy during the secretion process; the secretin XcpQ, forming a pore in the outer membrane through which the substrate is transported and further expelled by the third subcomplex of the system called pseudopilus in analogy with the type IV pilus assembly system (4). Five Xcp components (XcpT–X) are, like the major type IV pilin, PilA, all substrates for the prepilin peptidase XcpA/PilD (5, 6). They have consequently been called pseudopilins. The three-dimensional structure of all pseudopilins characterized so far displays the typical αβ fold found in the pilin/pseudopilin family (7–11). XcpT, which is the most abundant pseudopilin (12), is the only pseudopilin able to assemble into a pilus-like structure. We called this structure hyper-pseudopilus (HPP) because it is only observable under conditions of XcpT overproduction (13–15). These observations strongly suggest that the Xcp secreton contains a pilus-like structure and that the main component of this so-called pseudopilus is XcpT. XcpT is then considered as the major pseudopilin, whereas the four other pseudopilins, XcpU–X are called minor pseudopilins.
Due to their high specificity for a given organism and a set of substrates, components of the diverse T2SSs found in Gram-negative bacteria are hardly exchangeable (4). Interestingly, the major pseudopilin, generally called GspG in Gram-negative bacteria, appears much less specific because homologs of GspG from six different T2SSs, including the P. aeruginosa Xcp system, are fully functional in the Klebsiella oxytoca Pul T2SS lacking its major pseudopilin PulG (15). This observation suggests that in type II secretion, substrate- or species-specificity is not related to the major pseudopilin, which therefore probably assembles a pseudopilus exchangeable between most T2SSs.
In addition to the typical Xcp T2SS, the PAO1 P. aeruginosa strain possesses a second T2SS, referred to as Hxc system (for homolog to Xcp) (16). Whereas more than a dozen exoproteins utilize the Xcp T2SS for their secretion, the Hxc T2SS is dedicated to the secretion of one single protein, the low molecular mass alkaline phosphatase LapA (16, 17). We have previously observed in complementation assays the nonredundancy of the two machineries and more particularly that HxcT, which is 60% identical to XcpT, is unable to compensate for the lack of XcpT for Xcp secretion in P. aeruginosa (16). Based on other studies (15), such incompatibility between two major pseudopilins is unexpected, especially between two T2SSs belonging to the same organism.
We show with the present study that, in addition to its inability to compensate for the lack of XcpT, HxcT pseudopilins are also unable to assemble into an HPP even when overproduced. Interestingly, these two phenotypes are restored when using an HxcT variant in which 9 residues have been replaced with corresponding residues in XcpT and other compatible major pseudopilins. Because those key residues are different in HxcT, our data clearly indicate that the second Hxc T2SS of P. aeruginosa is unable to assemble a HPP, which suggests different pseudopilus architecture. We concluded that such differences in the Xcp and Hxc systems probably result in subtle modification of the type II secretion process and highlight the existence of two T2SS subtypes that we propose to name T2aSS and T2bSS for Xcp- and Hxc-type systems, respectively. Interestingly, such HxcT-specific amino acids are found in phylogenetically related GspG proteins, revealing a larger distribution of this alternative T2bSS.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
All bacterial strains, vectors, and plasmids used in this study are listed in supplemental Tables S3 and S4. Escherichia coli strains were grown at 37 °C in Luria broth. P. aeruginosa strains were grown at 30 °C in proteose peptone medium supplemented with glucose 0.4% (w/v). When required, media were supplemented with the following antibiotics at the indicated concentrations: for E. coli, 50 μg/ml ampicillin, 30 μg/ml kanamycin, and 15 μg/ml gentamycin; and for P. aeruginosa, 150 μg/ml carbenicillin and 50 μg/ml gentamycin. The E. coli TG1 strain was used to propagate all plasmids. Plasmids were introduced into P. aeruginosa by triparental mating by use of the conjugative properties of pRK2013. The P. aeruginosa strains used were PAO1 and its derivatives ΔxcpT (in-frame deletion of the xcpT gene in PAO1) and ΔxcpTΔhxcT (in-frame deletions of the xcpT and hxcT genes in PAO1). P. aeruginosa transconjugants were selected on Pseudomonas isolation agar supplemented with antibiotics.
Construction of the P. aeruginosa Mutant
The deletion of the hxcT gene in ΔxcpT, leading to the ΔxcpTΔhxcT strain, was constructed as described previously (16). Briefly, 500-bp sections upstream (hxcV′) and downstream (hxcX′) of the hxcT gene were PCR-amplified using the primers hxcTup1 (5′-AGCGCGGGGTCGTCGGTGCCTCGACG-3′) and hxcTup2 (5′-CTCAGAGCTCATCTCGACACTCCTCGG-3′) for hxcV′ and hxcTdw1 (5′-GTCGAGATAGCTCTAGCGCGCCAT-3′) and hxcTdw2 (5′-GAAGTCTCGCGGCCGCTTCGAAGCCT-3′) for hxcX′. The oligonucleotides were designed for amplifying fragments with overlapping 3′ and 5′ ends. Both fragments were ligated by performing an overlapping PCR, using hxcTup1 and hxcTdw2 in a second run of PCR with a mix of the two fragments as the matrix. The resulting PCR product was cloned into the pCR2.1 vector (TA cloning kit; Invitrogen). A 1,000-bp BamHI/ApaI DNA fragment was then subcloned into the suicide pKNG101 vector. The resulting construct (pKNG-ΔhxcT) was transferred to P. aeruginosa by mobilization with the pRK2013. The strain in which the chromosomal integration occurred was selected on Pseudomonas isolation agar plates containing 2,000 μg of streptomycin/ml. Excision of the plasmid, resulting in the deletion of the chromosomal target gene, was performed after selection on Luria broth plates containing 5% sucrose. Clones that became sucrose-resistant and streptomycin-sensitive were confirmed to contain the hxcT gene deletion by PCR analysis.
Site-directed Mutagenesis of HxcT and Cloning of HxcT Variants
The pCR2.1 vector carrying the hxcT gene (pCR2.1-hxcTwt) (16) was used as DNA template to generate all the hxcT mutant alleles (supplemental Table S1). We used the QuikChange PCR-based targeted mutagenesis approach with a pair of specific primers (supplemental Table S2) carrying both the desired mutations and the Pfu-Turbo (Stratagene) Taq polymerase to amplify the entire pCR-hxcT plasmid. The mutation in the hxcT gene was confirmed by DNA sequencing (GATC biotech). For expression in P. aeruginosa we used two types of broad host range vectors: the pMMB190 and the pJN105. Each hxcT allele from pCR2.1 was cut out using the BamHI/EcoRV restriction sites and subcloned into the BamHI/SmaI sites of the pMMB190 (supplemental Table S3). For the subcloning into the pJN105, XbaI/SacI restriction sites were used for all the hxcT alleles (supplemental Table S3). To generate the C-terminal His6-tagged form of each hxcT mutant allele, we designed a single pair of primers, HxcTh6-F2 (BamHI: 5′-ATTGATCCGAATCGAACCATCCGAGGAGTTCGAGATGATTCGT-3′) and HxcTh6-R2 (HindIII: 5′-ATTGATCCGAATCGAACCATCCGAGGAGTTCGAGATGATTCGT-3′), for amplifying the hxcT alleles and flanking at the same time a C-terminal His6 DNA-encoding sequence. The PCR products were then cloned into the pCR2.1 by TA-cloning (Invitrogen) and the DNA sequence checked by sequencing. Each allele was subcloned into the pMMB190 vector using the BamHI/HindIII restriction sites (supplemental Table S3).
The HxcT homolog from Pseudomonas fluorescens (HxcTPf) was PCR-amplified using the primers HxcTpf5-F1 (5′-ATAGAATTCGCTTGTTTGCGCAGGGCTTGCATGGGCCGCTCG-3′) and HxcTpf5-R1 (5′-ATAGAATTCCGACCGGCCCTATAGTTGCCAGGAGCCGATATC-3′) from the genomic DNA of the P. fluorescens strain Pf-5. Each primer contained an EcoRI restriction site. The resulting PCR product was then digested by EcoRI and subcloned into the pMMB190.
Immunofluorescence (IMF) Microscopy
P. aeruginosa strains were grown overnight at 37 °C on a Luria broth plate supplemented with 250 μg of carbenicillin/ml and 2 mm of isopropyl β-d-1-thiogalactopyranoside. We used the same protocol as described before (18) to prepare glass slides for IMF observation. The primary mouse antibody directed against the His6 tag (Roche Applied Science) was used at a 1:500 dilution, and the secondary fluorescein/anti-mouse (Vector Laboratories) conjugate was used at a 1:400 dilution. Samples were observed on a Zeiss Axio microscope.
Secretion Assays
Protease secretion by P. aeruginosa was tested after the organism was plated on tryptic soy agar (Difco) plates containing 1.5% skim milk. Elastase secretion was analyzed on tryptic soy agar plates containing 1.0% elastine. For the detection of lipase secretion, lipid agar plates were used. Lipid agar is a minimal medium containing olive oil as the sole carbon source (19). For liquid secretion assays, bacterial strains were grown for 24 h at 30 °C with aeration in proteose peptone medium (Difco) containing 0.4% glucose, 50 μg gentamycin/ml, and 0.5% l-arabinose (inducer of the pBAD). Cells and supernatants were separated by centrifugation; proteins contained in the supernatants were precipitated by adding trichloroacetic acid (TCA, 15% final concentration) and incubation for 2 h at 4 °C. Samples were subsequently centrifuged, and the pellets were washed with acetone and resuspended in SDS-PAGE sample buffer (2% SDS, 5% β-mercaptoethanol, 10% glycerol, 0.1 m Tris-HCl, pH 6.8, 0.02% bromphenol blue). The samples were heated for 7 min at 95 °C before electrophoresis. The proteins contained in the supernatant of 1.0 A600 unit equivalent of bacterial cell culture were loaded and separated on SDS gels containing 12% acrylamide. Gels were stained with Coomassie Brilliant Blue.
Purification of Pseudopilin-soluble Domains and Chemical Cross-linking
The His6-tagged periplasmic soluble domain of XcpT (XcpTp) was purified as described by Durand et al. (18). The untagged periplasmic soluble domain of HxcT (HxcTp) was purified following the protocol used for the minor pseudopilins by Douzi et al. (20). Purified proteins were then used for chemical cross-linking. Each protein (0.6 mm) was mixed in phosphate-buffered saline (PBS×1) supplemented with 10 mm ZnSO4 in a total volume of 50 μl. After 30 min of preincubation at room temperature 1% of freshly prepared paraformaldehyde was added to the mix. Incubation was continued during 20 min, after which the reaction was blocked by adding 4-fold concentrated SDS-PAGE sample buffer. 20 μl of each mix was directly analyzed on a 15% SDS-PAGE. The gel was stained with Coomassie Brilliant Blue.
Two-dimensional SDS-PAGE
Each sample was run on two consecutive 15% SDS-PAGEs as follows: after boiling cell samples were electrophoresed on the first gel. Then the entire band was cut out and run without further boiling perpendicular on the second gel. After running, the gel was immunoblotted and revealed with the anti-XcpT (laboratory collection) or anti-HxcT (antipeptide generated by Thermo Fisher scientist) as described in Viarre et al. (17).
Phylogenetic Reconstruction
We built a first dataset containing a subset of 99 pilins and major pseudopilins (dataset D1) representative of major pilins and pseudopilins. Then we built a second dataset with the 145 type II major pseudopilins identified in Alphonse et al. (7) (dataset D2). One of these sequences was highly divergent and was thus removed from the analysis. A survey of the 1,269 complete prokaryotic genomes available at the NCBI using BlastP (21) (default parameters, except the max_target sequences that was fixed to 1,000) allows the identification of 397 homologs of P. aeruginosa PAO1 HxcT and XcpT. The corresponding protein sequences were gathered in a third dataset (D3). Each dataset was aligned using mafft (22) version 6.833 (default parameters). The resulting alignments were visually inspected using the alignment editor (ed) from the MUST package (23) and manually refined when necessary. Regions where the alignment was doubtful were removed prior to phylogenetic analyses. Maximum likelihood phylogenetic trees were inferred using PhyML and the JTT (Jones, Taylor, Thornton) model for dataset D2. For the alignments D1 and D3 we used the Le and Gascuel (LG) model and a gamma distribution (four discrete categories, an estimated alpha parameter) and an estimated proportion of invariable sites, to take into account the heterogeneity of evolutionary rates among sites. The branch robustness of the resulting trees was estimated using the nonparametric bootstrap procedure implemented in PhyML (100 replicates of the original dataset) and the same parameters as for the tree reconstruction.
Logo Construction
Based on the tree inferred from the dataset D3 we have inferred the logo corresponding to major clades using the Phylo_mLogo tool (23).
RESULTS
HxcT Is an Atypical Major Pseudopilin (HxcT Overproduction Does Not Lead to HPP Formation)
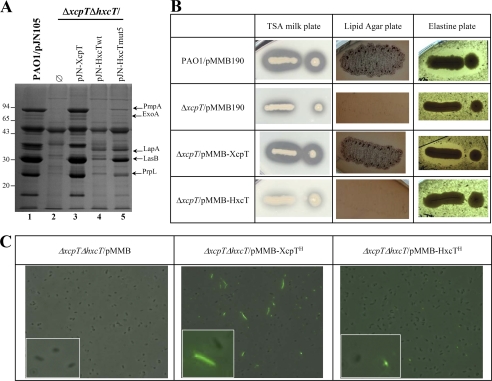
As reported previously, the P. aeruginosa Xcp and Hxc machineries are involved in the secretion of exoproteins in a system-specific manner. In a P. aeruginosa PAO1 mutant strain deleted for xcpT and hxcT genes (referred to as ΔxcpTΔhxcT strain), the introduction of a plasmid carrying either the xcpT or hxcT genes specifically restores Xcp-dependent (LasB, PmpA, aminopeptidase) or Hxc-dependent (LapA) secretion (Fig. 1A, compare lanes 1, 3, and 4). The functional complementation of the Xcp-dependent secretion was tested by three different phenotypic plate assays. In the wild-type PAO1 strain (Fig. 1B, first row), proficient Xcp-dependent secretion is observed by the formation of a halo around the bacterial colony on a skimmed milk plate due to the activity of several proteases including PmpA and LasB, bacterial growth on lipid agar plate is due to the lipase activity of LipA, and formation of a halo on elastin plate is due to the elastolytic activity of LasB. In contrast, in the xcpT mutant strain (Fig. 1B, second row) none of these phenotypes could be observed, indicating the absence of Xcp-dependent secretion of lipase and proteases. Introduction of a plasmid carrying the xcpT gene (Fig. 1B, third row) restores the Xcp-dependent secretion proficiency of the ΔxcpT strain whereas the introduction of a plasmid carrying the hxcT gene had no effect (Fig. 1B, fourth row). This observation confirmed that the HxcT major pseudopilin cannot replace the XcpT function in the Xcp secreton. Moreover, we used His-tagged forms of XcpT (XcpTH) and HxcT (HxcTH) and combined IMF microscopy to demonstrate that HxcT (Fig. 1C, right) is unable to assemble into a HPP in the conditions where XcpT does (Fig. 1C, center). Note that the overproduction of HxcT is similar to XcpT in the IMF experiment (data not shown). Our data clearly confirm that HxcT is an atypical major pseudopilin in the T2SS. Indeed, in addition to its inability to replace XcpT for Xcp-dependent secretion, it is also unable to assemble into an HPP when overproduced.
FIGURE 1.
HxcT and XcpT are not interexchangeable. A, 12% SDS-PAGE stained with Coomassie Brilliant Blue of supernatant fractions. Molecular mass markers are indicated on the left side of the gel. Some of the Xcp- (PmpA, ExoA, LasB, and PrpL) and the Hxc- (LapA) dependent substrates are indicated by arrows on the right side. B, phenotypical plate assays revealing Xcp-dependent secretion and activities of substrates. Proteolytic activity is shown by a halo around the colony on a skimmed milk plate (second column). Secretion of the lipase LipA is revealed by the growth of P. aeruginosa strains on a lipid agar plate (third column). Elastase LasB activity is shown by a clear halo around the bacterial colony on elastine plate (fourth column). The name of the P. aeruginosa strains and plasmids is indicated in the first column. C, IMF microscopy revealing HPP assembly by P. aeruginosa strains grown on plate (see “Experimental Procedures”). HPPs are visible as fluorescent filaments labeled by the anti-His antibody. Insets show detailed views of a part of each panel.
HxcT Reversion by Structure-based Sequential Mutagenesis
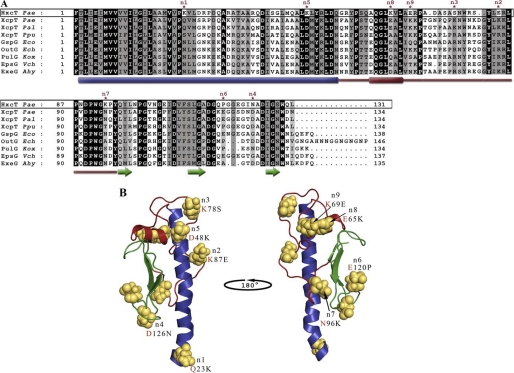
Heterologous complementation studies have shown that the major pseudopilins XcpT from Pseudomonas alcaligenes and Pseudomonas putida could replace XcpT from P. aeruginosa for T2SS-dependent secretion of exoenzymes (24, 25). However, XcpT cannot replace HxcT for LapA secretion in P. aeruginosa (Fig. 1A, lane 3). Importantly, heterologous complementation studies performed in K. oxytoca have shown that the major pseudopilins, XcpT from P. aeruginosa, GspG from E. coli, OutG from Erwinia chrysanthemi, EpsG from Vibrio cholerae, OutG from Erwinia carotovora, and ExeG from Aeromonas hydrophila could replace PulG from K. oxytoca in T2SS-dependent HPP assembly and secretion of exoenzyme (15). To understand the functional difference observed between XcpT and HxcT major pseudopilins, we compared the amino acid sequence of HxcT with the sequences of a set of compatible and HPP-forming major pseudopilins (Fig. 2A). The alignment revealed nine significant and remarkable differences between the HxcT sequence and the consensus sequence of the compatible major pseudopilins (Fig. 2A). In XcpT, the corresponding residues are Gln23, Asp48, Glu65, Lys69, Lys78, Lys87, Asn96, Glu120, and Asp126. These nine residues were mapped on the tri-dimensional structure of XcpT (Fig. 2B) and replaced in the pseudopilus context. To do so, the structure of XcpT (7) was positioned in the Neisseria gonorrhoeae type IV pilus filament (Protein Data Bank ID code 2HIL; data not shown) (26). To allocate each identified residues, a possible role in HPP formation and/or Xcp-dependent secretion, we gradually reintroduced in HxcT the nine corresponding residues found in XcpT. The reconstruction was performed by sequentially introducing each one of the nine substitutions in HxcT. The order by which the substitutions were introduced was driven by the predicted importance of these residues in the pseudopilus assembly. More precisely, in our reconstituted pseudopilus, the N-terminal part of the α-helix of major pseudopilins is nearly parallel to the fiber axis in the center of the filament creating the hydrophobic core crucial for pseudopilus assembly. Therefore, residues located in this α-helix and residues found at the interface between two neighboring XcpT and therefore possibly involved in intersubunit interaction were the first selected for the “gradual” directed mutagenesis. Moreover, mutation n1 was chosen first because it is located in the N-terminal α-helix which has a predominant role in the multimerization process of XcpT. We thus call the HxcT substitutions K23Q, E84K, S77K, N123D, and K48D, n1, n2, n3, n4, and n5 mutations, respectively. The substitution P117E is the n6 mutation and is located in the putative calcium binding site in the β2β3 loop (27), shown to be important in the secretion process. The three last targeted positions did not seem to be involved in intersubunit interactions and were considered as minor in terms of pseudopilus formation and were therefore the last to be introduced. These substitutions, K93N, K65E, and E69K, were named n7, n8, and n9, respectively. We thus design the progressive conversion of HxcT into XcpT by generating nine HxcT variants (HxcTmut1 until HxcTmut9). For example, HxcTmut9 bears all nine substitutions, whereas the HxcTmut2 variant carries the K23Q and E84K substitutions (see supplemental Table S1).
FIGURE 2.
Selection criteria for mutations of HxcT. A, sequence alignment of matured major pseudopilins from T2SSs. Sequence of HxcT from the T2SS of P. aeruginosa PAO1 is boxed. Identical residues are highlighted in black, conserved residues are highlighted in dark gray, and similar residues are highlighted in light gray. The secondary structure elements are indicated below the sequence alignment where cylinders represent the α-helices, arrows represent the β-strand, and the red line indicates the αβ-loop region. The point mutations selected to transform HxcT in XcpT are indicate by asterisks. Pseudopilin gene identification (gi) numbers: Pal, P. alcaligenes M-1(gi 3978481/AAC83358.1); Ppu, P. putida WCS358 (gi 3297910/CAA56982.1); Eco, E. coli SMS-3-5 (gi 170681001/YP_001745224.1); Ech, E. chrysanthemi (gi 259985/AAB24181.1); Kox, K. oxytoca UNF5023 (gi 131595/P15746.1); Vch, V. cholerae MZO-3 (gi 153802200/ZP_01956786); Ahy, A. hydrophila ATCC 7966 (gi 117620343/YP_855105); and Pae, P. aeruginosa PAO1 (gi 15598297/NP_251791 and 15595878/NP_249372) for XcpT and HxcT, respectively. B, ribbon view of the XcpT pseudopilin from P. aeruginosa (PAO1). N-terminal helix is colored in blue, αβ-loop region in red, and the C-terminal three-stranded β-sheet is colored in green. Residues selected to transform HxcT into XcpT are represented in sphere mode. Mutations performed are indicated below the priority tag with a red letter for the XcpT residue.
Successful HxcT Reversion with HxcTmut5
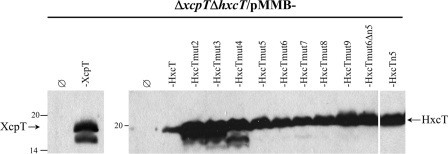
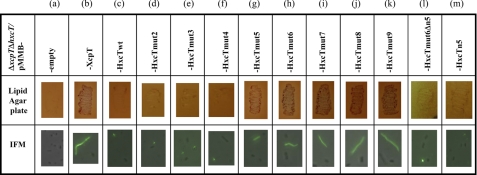
The newly engineered genes encoding the nine HxcT variants were subcloned into the pMMB190 broad-host range vector and expressed in the PAO1ΔxcpTΔhxcT background. All of the HxcT variants are produced by the bacterial strain as seen by SDS-PAGE and Western blot analysis of cell extracts (Fig. 3). To test the functionality of the HxcT variants we checked two different phenotypes, the restoration of Xcp-dependent secretion on a lipid agar plate and XcpT-like HPP formation by IMF microscopy (Fig. 4). All assays were performed using the PAO1ΔxcpTΔhxcT strain carrying either the plasmid pMMB190, the plasmid carrying the xcpT gene, the plasmid carrying the wild-type hxcT gene, or the plasmids carrying either one of the nine hxcTmut alleles. The HxcT variants HxcTmut2 to HxcTmut4 behave as the wild-type HxcT protein and do not provide any of the above-mentioned phenotypes. Interestingly, the HxcTmut5 variant is able to grow on a lipid agar plate, albeit only slightly, and assembles into few and short HPPs. Analysis of the supernatant of a liquid culture of the PAO1ΔxcpTΔhxcT strain producing HxcTmut5 (Fig. 1A, lane 5) confirmed the restoration, albeit partial, of the Xcp-dependent secretion, which is consistent with the growth on lipid agar (Fig. 4g). The HxcTmut6 variant grows better on a lipid agar plate and assembles into more and longer HPPs (Fig. 4h). From HxcTmut5 the more HxcT carries Xcp-like amino acids the more the Xcp phenotypes are striking. Indeed, strains producing HxcTmut7–9 grow increasingly better on a lipid agar plate and assemble more and longer HPPs. However, the presence of an aspartate residue at position n5 appears to be absolutely required for HxcT reversion because its absence in HxcTmut6 variant HxcTmut6Δn5 does not allow growth restoration on lipid agar or HPP formation (Fig. 4l). Substitution of n5 is, however, not sufficient for reversion because HxcTn5 does not allow growth restoration on lipid agar or HPP formation (Fig. 4m).
FIGURE 3.
Production of HxcT variants. 15% SDS-PAGE and Western blot of cell extracts from the ΔxcpTΔhxcT strain producing XcpT, HxcT, and the various HxcT variants and grown on Luria broth plate supplemented with 2 mm isopropyl β-d-1-thiogalactopyranoside. The membrane was revealed by anti-XcpT (left) or anti-HxcT (right) antibodies. An arrow indicates the position of the proteins. Molecular mass markers are indicated on the left side of each panel.
FIGURE 4.
Phenotypical studies of the HxcT variants. The complementation of the Xcp-dependent secretion is shown by the lipid agar plate assay (second row), and the formation of HPP is investigated using IMF microscopy (third row). The name of the constructs used in each case is indicated in the first row.
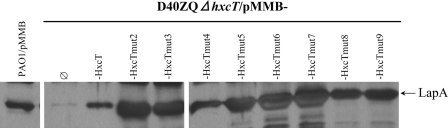
Note that the Hxc-dependent LapA secretion can be achieved by any of the HxcT variants (Fig. 5). The LapA secretion remains, moreover, Xcp-independent because LapA is still secreted in a strain lacking all xcp genes and the hxcT gene (PAO1D40ZQΔhxcT) but producing any of the HxcT variants (Fig. 5). The observation that all HxcTmut variants retained functionality in the Hxc secreton indicates that key residues for Xcp pseudopilus formation and secretion are not crucial for Hxc secretion.
FIGURE 5.
All HxcT variants remain functional in the Hxc secreton. Analysis of the supernatant of various P. aeruginosa strains grown under phosphate-limitation conditions is shown. The equivalent of 1.0 A600 has been loaded in each well. Samples were first run on a 12% SDS-PAGE and then blotted and revealed using an antibody directed against LapA. Two strains of P. aeruginosa were used here: the PAO1 parental strain and its D40ZQΔhxcT derivative, which lacks the entire xcp T2SS operon together with an in-frame deletion of the hxcT gene. The two strains were complemented either with the empty vector or with plasmids expressing the wild-type hxcT gene or its mutant derivatives. The position of LapA is indicated by an arrow on the right side of the gel.
In conclusion, by accumulation of amino acid changes, HxcT was progressively turned into XcpT. Each of the mutations brings an additional “XcpT feature” to HxcT and makes it more efficient at complementing an xcpT deletion mutant strain. Moreover, our observation demonstrates that restoration of both Xcp-dependent secretion and ability to assemble a HPP are coupled and therefore tightly linked during the Xcp secretion process.
XcpT and HxcT Pseudopilin Multimerization Capacity
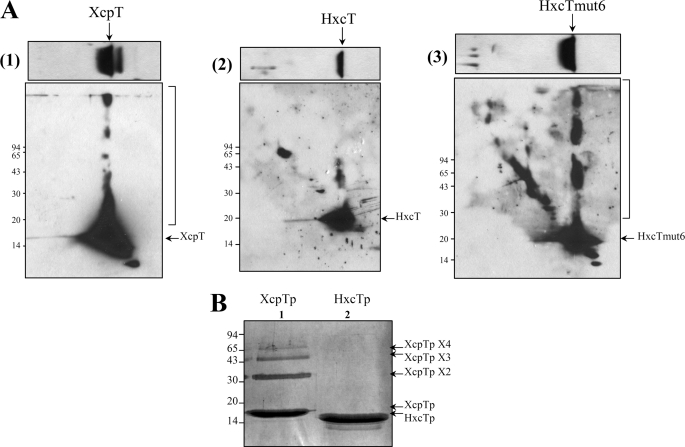
One major functional difference between XcpT and HxcT seems to relate to their ability to multimerize into an HPP when these pseudopilins are overproduced. To confirm this observation, we used two-dimensional SDS-PAGE to investigate the multimerization properties of XcpT and HxcT (Fig. 6A). This technique consists in loading a cell extract containing the full-length pseudopilin on a SDS-PAGE and in proceeding with migration (first dimension). After migration, a band of gel corresponding to one lane of loaded sample is cut out on its entire length, laid down on top of a second SDS-PAGE (second dimension), and run without heat treatment perpendicular to the first dimension. The migration in the second dimension triggered a local increase of the concentration of pseudopilin subunits before they enter in the running gel. This leads to spontaneous multimerization of pseudopilins which are intrinsically capable of doing so. Using this technique we visualized the multimerization of XcpT (Fig. 6A(1)), marked by the bracket) but not that of the wild-type form of HxcT (Fig. 6A(2)). However, HxcTmut6 (Fig. 6A(3)) is clearly able to form multimers to the same extent as XcpT does. We thus found a correlation between the ability of pseudopilins to form multimers in these conditions and their ability to restore the Xcp-related phenotypes (secretion on lipid agar plate and HPP assembly).
FIGURE 6.
Pseudopilin multimerization. A, two-dimensional SDS-PAGE analysis of the multimerization of full-length major pseudopilins. The first dimension is presented in the upper panel, and the second dimension is presented in the lower panel. After running, the gels were immunoblotted and revealed by the anti-XcpT antibody (1) or the anti-HxcT antibody (2) and (3). An arrow indicates the position of the protein of interest. The right bracket on the side of the gel shows the protein multimers. B, paraformaldehyde cross-linking-mediated multimerization of the major pseudopilin-soluble domains. The soluble periplasmic domains of XcpT (XcpTp, lane 1) and of HxcT (HxcTp, lane 2) have both been subjected to chemical cross-linking. Samples were then run on a 15% SDS-PAGE, and the gel was stained with Coomassie Brilliant Blue. Arrows on the side of the gel indicate the proteins and protein multimers. Molecular mass markers are indicated on one side of each panel.
Type IV pili and T2SS-associated pseudopili are formed by the helical organization of pilins and pseudopilins, respectively, stabilized by the hydrophobic contacts between the N-terminal α-helices tightly packed in the filament core. However, the position of the soluble domains of pilin/pseudopilin subunits close to the fiber axis does not exclude their involvement in intersubunits contact. Moreover, the soluble domains of pseudopilins are stable, easy to purify in large amount, and thus amenable to in vitro biochemical assays (20). We therefore purified the XcpT- and HxcT-soluble/periplasmic domains (designated as XcpTp and HxcTp, respectively) and subjected them to chemical cross-linking to assess their multimerization properties. Upon paraformaldehyde treatment, XcpTp forms various multimeric species, from dimer to tetramer (Fig. 6B, lane 1), whereas HxcTp is unable to form any oligomeric forms in the same conditions, and only the monomeric species is detected (Fig. 6B, lane 2). This result confirms that HxcT is unable to multimerize in conditions where XcpT does.
In conclusion, we demonstrated that the intrinsic difference between the T2SS major pseudopilins XcpT and HxcT relies on the ability to multimerize. Therefore, the reason why HxcT in unable to substitute for XcpT in the Xcp-dependent secretion might reside in its limited multimerization capacity, which reflects likely different pseudopilus organization in Xcp and Hxc T2SS.
HxcT and Relatives Form a Subfamily within the Type II Pseudopilin Family
In a recent study (7), we solved the three-dimensional structure of XcpT, revealing that the previously called variable pilin region is in fact highly conserved among major type II pseudopilins and constitutes a specific consensus motif to specifically distinguish them from type IV pilins. This also suggests that both proteins form distinct but homologous families. This observation is in agreement with the maximum likelihood (ML) phylogeny showing that type IV pilins and type II pseudopilins form distinct monophyletic groups (bootstrap value = 100%) separated by an important evolutionary distance (see the length of the branch connecting both groups in supplemental Fig. S1). By contrast, the evolutionary distance separating the two major pseudopilins of P. aeruginosa PAO1 is quite small, and both sequences grouped within the well supported monophyletic group that contains all the major pseudopilins (bootstrap value = 100%; supplemental Fig. S1). To examine in more detail the phylogenetic relationships between P. aeruginosa PAO1 XcpT and HxcT and other major pseudopilins, we performed an analysis involving the 145 major pseudopilins we described previously (7). The resulting ML tree shows some major inconsistencies with organism taxonomy (e.g. some sequences from different taxonomic groups are mixed in the tree; Fig. 7A and supplemental Fig. S2), indicating that horizontal gene transfers occurred during the evolution of type II major pseudopilins. Interestingly, P. aeruginosa PAO1 HxcT is nested together with six other Pseudomonas (two P. aeruginosa, one Pseudomonas entomophila, two P. fluorescens, and one Pseudomonas syringae) sequences within a large cluster of 40 Betaproteobacteria (purple group, bootstrap value = 92%, Fig. 7A and supplemental Fig. S2). By contrast, the P. aeruginosa PAO1 XcpT together with five other Pseudomonas sequences form an isolated monophyletic cluster (Fig. 7A and supplemental Fig. S2). The closer (and well supported) relationship observed between HxcT and Betaproteobacterial sequences (but not XcpT) excludes the possibility that the two P. aeruginosa PAO1 major pseudopilins results from a duplication event. On the contrary, this strongly suggests that P. aeruginosa PAO1 and a few other Pseudomonas species have acquired their HxcT genes from Betaproteobacteria. The same interpretation could be proposed to explain the emergence of the two Strenotrophomonas (Gammaproteobacteria) sequences within Betaproteobacteria (supplemental Fig. S2). The two T2SS major pseudopilins in P. aeruginosa PAO1 have therefore two distinct evolutionary origins and may represent two distinct subfamilies of major pseudopilins. Importantly, the emergence of P. aeruginosa PAO1 HxcT within Betaproteobacteria is recovered when a larger set of major pseudopilin homologs (dataset D3) is considered (supplemental Fig. S3), indicating that our result is not the consequence of a bias in the taxonomic sampling.
FIGURE 7.
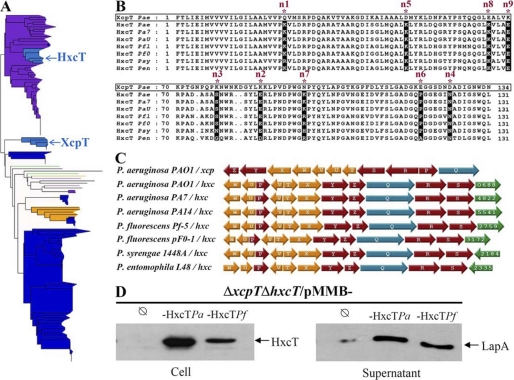
HxcT major pseudopilin phylogeny. A, maximum likelihood unrooted phylogenetic tree based on the D2 alignment (145 sequences, 114 positions). Taxonomic groups are indicated by colors: green, Delta/Epsilonproteobacteria; purple, Betaproteobacteria; dark blue, Gammaproteobacteria; light blue, cluster of Gammaproteobacteria containing P. aeruginosa PAO1 sequences; yellow, Alphaproteobacteria; pink, Firmicutes; red, Aquificae; black, other bacteria). The scale bar represents the inferred average number of substitution per site. This phylogeny is available in expanded format in supplemental Fig. S3A. B, sequences alignment of the seven HxcT-like major pseudopilins found in the Pseudomonas genus. The sequence of XcpT from P. aeruginosa (boxed) was aligned against the seven HxcT isolated in the phylogenetic tree. The nine positions selected in HxcT are indicated by asterisks. Their conservation among HxcT-like pseudopilins is highlighted in black. The gene identification (gi) numbers: Pa7, Pseudomonas aeruginosa PA7 (gi 152986054); PaU, P. aeruginosa PA14 (gi 116048588); Pfl, P. fluorescens Pf-5 (gi 70730120); Pf0, P. fluorescens Pf0–1 (gi 77459401); Psy, P. syringae pv. phaseolicola 1448A (gi 71735284); Pen, P. entomophila L48 (gi 104781443); and Pae, P. aeruginosa PAO1 (gi 15598297 and 15595878) for XcpT and HxcT, respectively. C, genetic organization of the seven T2SS gene clusters involving one of the seven hxcT identified compared with the classical genetic organization found in xcpT-like T2SSs. Pseudopilin-encoding genes are in yellow, inner membrane platform-encoding genes are in red, and secretin-encoding genes in are blue. In green are represented putative substrate-encoding genes (PA numbers labeled inside) systematically found in hxc-type clusters. D, phenotypic complementation of P. aeruginosa PAO1 hxcT (hxcTPa) by P. fluorescens Pf-5 hxcT (hxcTPf). SDS-PAGE and Western blot of cell extracts (cell) and supernatant fractions are shown. Bacteria were grown in phosphate-limiting conditions. The equivalent of 0.2 (cell) or 0.4 (supernatant) A600 has been loaded in each well. Samples were first run on a 15% (cell) or 8% (supernatant) SDS-PAGE, the membrane was revealed by anti-HxcT (left) or anti-LapA (right) antibodies. An arrow indicates the position of the proteins.
The presence of P. aeruginosa HxcT close homologs in six Pseudomonas species indicates that the transfer might be ancient in this genus or that secondary horizontal gene transfers occurred among those Pseudomonas species after the initial acquisition of an HxcT gene by one of them. The high similarity among those seven Pseudomonas sequences (in particular the conservation of the nine residues) (Fig. 7B) together with the conservation of the singular genetic organization found in P. aeruginosa PAO1 Hxc T2SS (Fig. 7C) underline the similarity of the T2SS systems in those species. The functional redundancy among the seven identified HxcT homologs is confirmed by the complementation of a P. aeruginosa PAO1 hxcT mutant by the hxcT allele of P. fluorescens (Fig. 7D).
DISCUSSION
Among the different protein secretion mechanisms found in Gram-negative bacteria, the type II secretion process remains one of the less understood. In analogy with the type IV piliation system, several reports are proposing the involvement of a pilus-like structure, called pseudopilus, in the type II secretion process. Since the discovery in P. aeruginosa of a second T2SS secreting its own substrate (16), we wondered why the bacteria acquired a complete second system for the secretion of one more T2SS-dependent protein instead of adding LapA to the dozen of others exoproteins traveling through the Xcp system. Our phylogenetic analyses provide an explanation: HxcT and likely the whole Hxc system, including LapA, was acquired by P. aeruginosa PAO1 and a few other Pseudomonas species through horizontal gene transfers from a betaproteobacterium and is not the result of an evolutionary process that occurred after a duplication event in the Pseudomonas lineage. The conservation of both Hxc and Xcp systems in P. aeruginosa PAO1 suggests that they may represent two distinct subfamilies of T2SSs.
We are proposing that the secretion process of the Hxc T2SS of P. aeruginosa is animated by a pseudopilus whose structure and stability may differ from the one commonly found in Xcp and other known T2SSs. Our study shows that modifying HxcT for nine selected residues conserved in XcpT-like protein is sufficient to convert HxcT into a classical compatible major pseudopilin. We gradually substituted the nine residues and observed after introducing the fifth mutation that we were able to restore HPP formation and Xcp-dependent secretion. In our view, the first five mutations are of the most importance because from the HxcTmut5 mutant the phenotypic conversion is obtained, and further substitutions in this mutant form only improved HPP formation and Xcp-dependent secretion. These five mutations are K23Q (n1), E84K (n2), S77K (n3), N123D (n4), and K48D (n5). We hypothesized that restoring those substitutions would restore favorable interactions that would allow the HPP formation. A recent report (28) presents a detailed assembly model of the K. oxytoca Pul-dependent type II secretion pilus. Because XcpT is able to substitute PulG for HPP formation in the Pul system (15), the pseudopilus model proposed in this study could be applicable to the XcpT pseudopilus. Strikingly, our structure-based mutagenesis that turned HxcT into an XcpT-compatible pseudopilin indeed highlighted amino acids known to be essential for the PulG/XcpT type of pseudopilus (28). By mapping our mutations onto the PulG pseudopilus model, we are able to describe the effect of each mutation and therefore their potential impact on pilus formation. Taken together, the mutations n2 and n5 reestablish an intermolecular salt bridge between Lys84 and Asp48. This salt bridge has been demonstrated to be essential for PulG/XcpT pseudopilus formation (28). The mutations n3 and n4 are also correlated because substituting these two residues (S77K and N123D) has two consequences. Indeed, the residue Lys77 in XcpT (Arg78 in PulG) is involved in (i) the establishment of a salt bridge with the residue Asp32 of another protomer and (ii) the restoration of a positive patch, composed of residues Arg51, Arg56, and Lys77, which interacts with a negative patch of another protomer composed of acidic residues Asp114 and Asp123. Interestingly Campos et al. (28) proceeded to the charge inversion of Arg87 and Asp48 into Glu87 and Lys48, which exactly correspond to the residues present in HxcT. These residues Lys48 (Asp48 of PulG) and Glu84 (Arg87 in PulG) are also able to form a salt bridge, but this potential salt bridge is not important for Hxc pseudopilus because the HxcTmut2 variant (Lys84/Lys48) cannot form a salt bridge but is still functional in Hxc-dependent secretion (Fig. 5). The mutation n2 disrupts this interaction by replacing Glu84 by Lys84 (the couple Lys48/Lys84 precludes the formation of a salt bridge). We confirm the absolute requirement of this Lys84/Asp48 salt bridge for Xcp secretion because its absence in the first six substitutions fully abolished the phenotypic restoration, as demonstrated by the mutant HxcTmut6Δn5 (Fig. 4).
Interestingly, our study reveals a clear coupling between pseudopilus formation and protein secretion in classical T2SS. We indeed observed that during the process of the mutagenesis both protein secretion and HPP formation phenotypes were concomitantly reverted, thus suggesting that a high degree of major pseudopilin multimerization is not only required but essential for protein secretion in classical T2SSs such as Xcp. We are showing that HxcT is unable to reach such a level of multimerization, indicating that the multimerization capacity associated with the nine conserved XcpT residues is dispensable for Hxc secretion, strengthening our idea that different core assembly might exist between Hxc-like and Xcp-like pseudopili.
Major pseudopilins are not directly involved in substrate recognition because various GspGs are able to replace PulG in Pullulanase secretion by the Pul secreton (15). Therefore, we do not think that XcpT and HxcT are involved in the direct recognition of the substrate. However, we do think that the degree of pseudopilus polymerization does indirectly matter in substrate recognition through another secreton component. The fact that Xcp-compatible HxcT variants are still able to support secretion of the Hxc substrate indicates that other motifs are involved in substrate recognition.
In conclusion, we clearly established that two subfamilies of T2SS which differ by the assembly characteristic of their pseudopili could now be identified. We therefore propose to name them T2aSS and T2bSS for Xcp- and Hxc-type systems, respectively. The next challenge will be to understand why specific residues allowing HPP formation in T2aSS are not required in T2bSS secretion. Such evolution of a secretory machine was previously reported for type V secretion and revealed in one case the alternative widespread T5dSS (29) and in the other case the singular P-Usher transporter (30). Those findings together with the present T2bSS demonstrate how similar bacterial machineries evolve toward distinct mechanisms that may contribute specific functions.
Supplementary Material
Acknowledgment
We are grateful to Mariella Tegoni for constant interest in this work.
This work was supported by 3D-Pilus Young Researcher Grant ANR-JC 07-183230 (to E. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3, Tables S1–S4, and additional references.
- T2SS
- type II secretion system
- HPP
- hyper-pseudopilus
- IMF
- immunofluorescence
- ML
- maximum likelihood.
REFERENCES
- 1. Economou A., Christie P. J., Fernandez R. C., Palmer T., Plano G. V., Pugsley A. P. (2006) Mol. Microbiol. 62, 308–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Voulhoux R., Ball G., Ize B., Vasil M. L., Lazdunski A., Wu L. F., Filloux A. (2001) EMBO J. 20, 6735–6741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Michel G. P., Voulhoux R. (2009) in Bacterial Secreted Proteins (Wooldridge K. ed) Vol. 1, pp. 67–92, Caister Academic Press, Norfolk, UK [Google Scholar]
- 4. Filloux A. (2004) Biochim. Biophys. Acta 1694, 163–179 [DOI] [PubMed] [Google Scholar]
- 5. Bally M., Filloux A., Akrim M., Ball G., Lazdunski A., Tommassen J. (1992) Mol. Microbiol. 6, 1121–1131 [DOI] [PubMed] [Google Scholar]
- 6. Bleves S., Voulhoux R., Michel G., Lazdunski A., Tommassen J., Filloux A. (1998) Mol. Microbiol. 27, 31–40 [DOI] [PubMed] [Google Scholar]
- 7. Alphonse S., Durand E., Douzi B., Waegele B., Darbon H., Filloux A., Voulhoux R., Bernard C. (2010) J. Struct. Biol. 169, 75–80 [DOI] [PubMed] [Google Scholar]
- 8. Korotkov K. V., Hol W. G. (2008) Nat. Struct. Mol. Biol. 15, 462–468 [DOI] [PubMed] [Google Scholar]
- 9. Lam A. Y., Pardon E., Korotkov K. V., Hol W. G., Steyaert J. (2009) J. Struct. Biol. 166, 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yanez M. E., Korotkov K. V., Abendroth J., Hol W. G. (2008) J. Mol. Biol. 377, 91–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yanez M. E., Korotkov K. V., Abendroth J., Hol W. G. (2008) J. Mol. Biol. 375, 471–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nunn D. N., Lory S. (1993) J. Bacteriol. 175, 4375–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Durand E., Bernadac A., Ball G., Lazdunski A., Sturgis J. N., Filloux A. (2003) J. Bacteriol. 185, 2749–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu N. T., Leu W. M., Lee M. S., Chen A., Chen S. C., Song Y. L., Chen L. Y. (2002) Biochem. J. 365, 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vignon G., Köhler R., Larquet E., Giroux S., Prévost M. C., Roux P., Pugsley A. P. (2003) J. Bacteriol. 185, 3416–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ball G., Durand E., Lazdunski A., Filloux A. (2002) Mol. Microbiol. 43, 475–485 [DOI] [PubMed] [Google Scholar]
- 17. Viarre V., Cascales E., Ball G., Michel G. P., Filloux A., Voulhoux R. (2009) J. Biol. Chem. 284, 33815–33823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Durand E., Michel G., Voulhoux R., Kürner J., Bernadac A., Filloux A. (2005) J. Biol. Chem. 280, 31378–31389 [DOI] [PubMed] [Google Scholar]
- 19. Kagami Y., Ratliff M., Surber M., Martinez A., Nunn D. N. (1998) Mol. Microbiol. 27, 221–233 [DOI] [PubMed] [Google Scholar]
- 20. Douzi B., Durand E., Bernard C., Alphonse S., Cambillau C., Filloux A., Tegoni M., Voulhoux R. (2009) J. Biol. Chem. 284, 34580–34589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Katoh K., Toh H. (2008) Brief Bioinform. 9, 286–298 [DOI] [PubMed] [Google Scholar]
- 23. Philippe H. (1993) Nucleic Acids Res. 21, 5264–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de Groot A., Koster M., Gérard-Vincent M., Gerritse G., Lazdunski A., Tommassen J., Filloux A. (2001) J. Bacteriol. 183, 959–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. de Groot A., Krijger J. J., Filloux A., Tommassen J. (1996) Mol. Gen. Genet. 250, 491–504 [DOI] [PubMed] [Google Scholar]
- 26. Craig L., Volkmann N., Arvai A. S., Pique M. E., Yeager M., Egelman E. H., Tainer J. A. (2006) Mol. Cell 23, 651–662 [DOI] [PubMed] [Google Scholar]
- 27. Korotkov K. V., Gray M. D., Kreger A., Turley S., Sandkvist M., Hol W. G. (2009) J. Biol. Chem. 284, 25466–25470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Campos M., Nilges M., Cisneros D. A., Francetic O. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 13081–13086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Salacha R., Kovaciæ F., Brochier-Armanet C., Wilhelm S., Tommassen J., Filloux A., Voulhoux R., Bleves S. (2010) Environ. Microbiol. 12, 1498–1512 [DOI] [PubMed] [Google Scholar]
- 30. Ruer S., Ball G., Filloux A., de Bentzmann S. (2008) EMBO J. 27, 2669–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.