Abstract
Inositol 1,4,5-trisphosphate (IP3) receptors are endoplasmic reticulum membrane calcium channels that, upon activation, are degraded via the ubiquitin-proteasome pathway. While searching for novel mediators of IP3 receptor processing, we discovered that RNF170, an uncharacterized RING domain-containing protein, associates rapidly with activated IP3 receptors. RNF170 is predicted to have three membrane-spanning helices, is localized to the ER membrane, and possesses ubiquitin ligase activity. Depletion of endogenous RNF170 by RNA interference inhibited stimulus-induced IP3 receptor ubiquitination, and degradation and overexpression of a catalytically inactive RNF170 mutant suppressed stimulus-induced IP3 receptor processing. A substantial proportion of RNF170 is constitutively associated with the erlin1/2 (SPFH1/2) complex, which has been shown previously to bind to IP3 receptors immediately after their activation. Depletion of RNF170 did not affect the binding of the erlin1/2 complex to stimulated IP3 receptors, whereas erlin1/2 complex depletion inhibited RNF170 binding. These results suggest a model in which the erlin1/2 complex recruits RNF170 to activated IP3 receptors where it mediates IP3 receptor ubiquitination. Thus, RNF170 plays an essential role in IP3 receptor processing via the ubiquitin-proteasome pathway.
Keywords: E3 Ubiquitin Ligase, Endoplasmic Reticulum (ER), Proteasome, Protein Degradation, Ubiquitination, Inositol Trisphosphate Receptor
Introduction
Inositol 1,4,5-trisphosphate (IP3)3 receptors form tetrameric, IP3- and Ca2+-gated Ca2+ channels in endoplasmic reticulum (ER) membranes of mammalian cells and play a key role in cell signaling (1). Stimulation of certain cell surface receptors triggers IP3 formation at the plasma membrane, which then diffuses through the cytosol and binds to IP3 receptors (1). This, in concert with Ca2+ binding, induces yet to be defined conformational changes in the tetrameric channel that permit Ca2+ to flow from stores within the ER lumen into the cytosol (1). There are three IP3 receptor types in mammals, IP3R1, IP3R2, and IP3R3, and although they differ considerably in their tissue distribution, they have broadly similar properties and are often coexpressed (1).
In recent years it has become clear that G-protein-coupled receptor-induced activation of endogenous IP3 receptors can lead to their rapid degradation via the ubiquitin-proteasome pathway (UPP) (2). This IP3 receptor “down-regulation” has been demonstrated in many mammalian cell types, including gonadotropin-releasing hormone (GnRH)-stimulated αT3-1 mouse pituitary gonadotropes (3), endothelin 1 (ET1)-stimulated Rat1 fibroblasts (4), and muscarinic agonist-stimulated SH-SY5Y human neuroblastoma cells (5) and mHeLa cells (6).
The generally accepted summary of the UPP is that substrates are first polyubiquitinated and then processed by the proteasome, a multi-subunit protease that can recognize and degrade polyubiquitinated proteins (7, 8). Ubiquitination, the key step in targeting a protein for proteasomal degradation, is achieved through the hierarchical action of three enzymes, termed ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (Ubc or E2), and ubiquitin ligase (E3) (7, 8). Although there are only two E1s, there are dozens of E2s, and, in mammalian cells, hundreds of E3s (7–10). In summary, E1-activated ubiquitin is transferred to an E2, and with the guidance of an E3, the ubiquitin moiety is coupled to the ϵ-amino group of a lysine residue in the substrate through an isopeptide bond. A polyubiquitin chain can be formed through multiple rounds of ubiquitination. The C terminus of incoming ubiquitin moieties are isopeptide-bonded to lysine residues in the already attached ubiquitin. The selection of substrates for ubiquitination appears to rest primarily with E3s, and two major families are known: those that contain RING domains (9), and those that contain HECT domains (10). The RING domain is a 40–100 amino acid motif that coordinates zinc and whose presence in RING E3s is essential for ubiquitin ligase activity. Remarkably, there are over 600 RING domain-containing proteins encoded by the human genome, allowing for the recognition of a diverse array of substrates (9).
The ERAD pathway is a facet of the UPP responsible for the degradation of aberrant proteins in the ER (11), and a range of evidence indicates that the ERAD pathway also accounts for the ubiquitination and proteasomal degradation of certain metabolically regulated native ER membrane proteins, including IP3 receptors (2) and HMG-CoA reductase (12). A good deal is known about the enzymes, particularly the E3s, that catalyze ERAD substrate ubiquitination (11–13), and it has also been established that a cytosolic complex composed of the ATPase p97 and its cofactors Ufd1 and Npl4 mediates the transfer of ERAD substrates to the proteasome (11). This p97-Ufd1-Npl4 complex participates in the delivery of polyubiquitinated IP3 receptors to the proteasome (4). Additionally, we have recently found that the ER membrane-located erlin1/2 (SPFH1/2) complex associates rapidly with activated IP3 receptors and appears to be the element that selects them for ERAD (14, 15). However, the E3 responsible for IP3 receptor ubiquitination has remained elusive (2). Here we report that RNF170, a previously uncharacterized RING domain-containing protein, resides in the ER membrane, exhibits ubiquitin ligase activity, associates rapidly with activated IP3 receptors, and mediates IP3 receptor ubiquitination and processing by the ERAD pathway.
EXPERIMENTAL PROCEDURES
Materials
αT3-1 cells, HeLa cells, Rat1 cells, SH-SY5Y cells, and muscarinic HeLa (mHeLa) cells, which stably express m3-muscarinic receptors (6), were cultured as described (14, 5, 6). Already available antibodies used were rabbit polyclonal anti-IP3R1 (16), anti-erlin2 (14), anti-FLAG epitope (15), anti-α-transaldolase (a kind gift from Dr. A. Perl, State University of New York Upstate Medical University, Syracuse, NY), mouse monoclonal anti-p97 (Research Diagnostics, Inc.), anti-ubiquitin clone FK2 (BioMol International), anti-FLAG epitope clone M2 or M5 (Sigma), anti-HA epitope clone HA11 (Covance), and horseradish peroxidase- and fluorophore-conjugated secondary antibodies raised in goat (Sigma). Rabbit polyclonal anti-RNF170 and anti-HA epitope were raised against peptides NIHPENQELVRVLREQ and YPYDVPDYA, respectively, as described (16). GnRH, carbachol, N-ethylmaleimide, protease inhibitors, Triton X-100, CHAPS, puromycin, and Na2CO3 were purchased from Sigma. ET1 was from Calbiochem. DTT, Precision PlusTM protein standards and SDS-PAGE reagents were from Bio-Rad. Protein A-Sepharose CL-4B was from Amersham Biosciences, and LipofectamineTM 2000 (LF) and the Neon electroporation system were from Invitrogen.
Cell Lysis, Immunoprecipitation, SDS-PAGE, and Mass Spectrometry
For immunoprecipitation experiments, cells were harvested with lysis buffer (150 mm NaCl, 50 mm Tris-HCl, 1 mm EDTA, 1% CHAPS or 1% Triton X-100, 10 μm pepstatin, 0.2 mm PMSF, and 0.2 μm soybean trypsin inhibitor (pH 8.0)) usually supplemented with 1 mm DTT. When IP3 receptor polyubiquitination was to be measured, cells were harvested with DTT-free lysis buffer, and then 2.5 mm N-ethylmaleimide was added to the lysates for 1 min, followed by 5 mm DTT. For IP3 receptor down-regulation experiments, cells were harvested with 1% Triton X-100 lysis buffer plus 1 mm DTT. Lysates were incubated on ice for 30 min and clarified by centrifugation at 16,000 × g for 10 min at 4 °C. To immunoprecipitate specific proteins, clarified lysates were incubated with antisera and Protein A-Sepharose CL-4B for 4–16 h at 4 °C, washed thoroughly with lysis buffer, resuspended in gel-loading buffer, incubated at 100 °C for 3 min or 37 °C for 30 min, subjected to SDS-PAGE, and either transferred to nitrocellulose for immunoblotting or silver-stained with the Proteosilver Plus silver stain kit (Sigma). Immunoreactivity was detected using Pierce ECL reagents and a Genegnome imager (Syngene Bio Imaging) or x-ray film. Silver-stained protein bands were cut from SDS-PAGE gels, destained, and analyzed by mass spectrometry at the Taplin Biological Mass Spectrometry Facility, Harvard Medical School.
RT-PCR of the RNF170 Coding Sequence and Generation of Expression Constructs
Using Qiagen reagents, 1 μg of purified αT3-1 cell mRNA was first reverse-transcribed at 50 °C for 35 min in 50 μl of reaction volume containing 2.0 units of RT enzyme mix followed by 15 min at 94 °C for PCR amplification to generate RNF170 cDNA. For protein expression, this cDNA was subjected to PCR to amplify specifically the sequence encoding the predicted 257 amino acid mouse RNF170 protein and then ligated into the pcDNA3 expression vector. To generate RNF170 with a C-terminal triple FLAG tag (RNF170FLAG), the RNF170-encoding sequence in pcDNA3 was PCR-amplified and ligated into the pCMV14–3xFLAG expression vector. *RNF170FLAG, in which Cys-101 and His-103 are mutated to Ser and Ala, respectively, was made using RNF170FLAG as a template and mutagenic primers.
Ubiquitin Ligase Activity
Assays were performed essentially as described (17) using UBE1, UbcH5b, and HA-ubiquitin from Boston Biochem. HeLa cells transiently transfected using LF to express either RNF170FLAG or *RNF170FLAG were harvested with 1% Triton X-100-containing lysis buffer plus 1 mm DDT, and the tagged proteins were immunopurified with rabbit polyclonal anti-FLAG. Immune complexes containing ∼0.5 ng of tagged protein were then incubated with 0.1 μg of UBE1, 0.2 μg of UbcH5b, 2.5 μg of HA-ubiquitin, 2 mm ATP, 50 mm Tris-HCl (pH 7.5), 2.5 mm MgCl2, and 0.5 mm DTT (30 μl of reaction volume). Following addition of gel-loading buffer, SDS-PAGE, and immunoblotting, HA and FLAG immunoreactivity were detected using rabbit polyclonal anti-HA and anti-FLAG.
Immunofluorescence Microscopy and Subcellular Fractionation
HeLa cells on coverslips were transiently cotransfected with pDsRed2-ER (Clontech) and RNF170FLAG cDNAs using LF. 24 h post-transfection, the cells were fixed with 3.7% paraformaldehyde for 10 min at 25 °C, washed with PBS, incubated in blocking solution (10% goat serum, 0.1% BSA in PBS) for 45 min at 25 °C, incubated with rabbit polyclonal anti-FLAG for 16 h at 4 °C, washed three times with PBS, incubated with FITC-conjugated anti-rabbit for 1 h at 25 °C, washed with PBS, mounted on glass slides using Vectashield mounting medium (Vector Laboratories), and then images were acquired on a Zeiss AxioPlan 2 microscope equipped with a 63× oil immersion objective.
Subcellular fractionation was performed as described (14) using 50,000 × g for 20 min at 4 °C for both centrifugation steps.
Stable Expression of RNF170FLAG and *RNF170FLAG in Rat1 Cells
Rat1 cells were transfected with pCMV14–3xFLAG vectors (2 μg of cDNA plus 5 μl of LF per 1.6 × 105 cells in wells of a 12-well plate), subcultured 24 h later, and diluted into 96-well plates. Cells were then incubated with culture medium supplemented with 520 μg/ml G418 for ∼2 weeks. Viable clones were screened for FLAG epitope expression in immunoblots with mouse monoclonal anti-FLAG clone M2 and were maintained in 250 μg/ml G418. Expression levels of RNF170FLAG were consistently higher than those of *RNF170FLAG, and *RNF170FLAG-expressing cells grew relatively slowly, suggesting that *RNF170FLAG may be detrimental to cell health. For analysis of IP3 down-regulation, cells in 6-well plates were serum-starved for 16 h prior to incubation with ET1 and harvested with 1% Triton X-100 lysis buffer.
RNA Interference
A siRNA sequence designed against mouse RNF170 mRNA, encoded by tagacaaacggtaactctt was expressed from the 6OH1O-pSUPER.retro.puro vector (14). This vector and a vector encoding a non-targeting (Random) siRNA (14) were introduced into αT3-1 cells using the Neon electroporation system (∼2.5 × 106 cells plus 15 μg of DNA per 100 μl exposed to a single 1500-V, 20-ms pulse). Electroporated cells were transferred to DMEM plus 5% FBS. Puromycin (0.5 μg/ml) was added 24 h later, and cells were stimulated and harvested with 1% Triton X-100 lysis buffer 72–96 h after transfection. ON-TARGETplus siRNAs (Random non-targeting pool and human RNF170 SMARTpool) were purchased from Dharmacon and were transiently transfected into mHeLa cells; for ubiquitination experiments, 12 × 106 cells per 10-cm diameter dish plus 10 ml of medium with 34 μl of LF/50 nm siRNA, and for down-regulation experiments, 5 × 105 cells per well of a 6-well plate plus 2 ml of medium with 7 μl of LF/50 nm siRNA. Erlin2 was depleted using 12 × 106 cells per 10-cm diameter dish plus 10 ml of medium with 34 μl of LF/14 μg siRNA-encoding vector as described (6). Cells were harvested 72–96 h after transfection with 1% CHAPS lysis buffer for ubiquitination experiments or with 1% Triton X-100 lysis buffer for down-regulation experiments.
Data Analysis
All experiments were repeated at least once, and representative images of gels or micrographs are shown. Quantitated data are graphed as mean ± S.E. of n independent experiments, with unpaired Student's t test used to obtain p values.
RESULTS
Identification of RNF170 as an IP3R1-associating Protein
To identify novel mediators of IP3 receptor ERAD, we examined IP3R1 immunoprecipitates from control and GnRH-stimulated αT3-1 cells by SDS-PAGE and silver staining (Fig. 1A). In addition to erlin1 (41 kDa) and erlin2 (43 kDa), which we characterized previously (14, 15), one of the most abundant proteins that associated with IP3R1 in a stimulus-dependent manner migrated as a band at 21.5 kDa (lane 2), and excision of this band followed by mass spectral analysis revealed it to be an uncharacterized protein termed RING finger protein 170 (RNF170). Database searching and sequence analysis (Fig. 1B) indicated that RNF170 is ∼257 amino acids in length, is highly conserved in vertebrates, has a predicted molecular mass of ∼30 kDa, contains a canonical RING-HC domain, and has three putative transmembrane domains. Topological predictions indicate that RNF170 resides in the ER membrane with its N terminus in the ER lumen and with the RING domain and the C terminus in the cytosol (Fig. 1C).
FIGURE 1.
RNF170 is a RING domain-containing protein that associates with activated IP3 receptors. A, αT3-1 cells were incubated without or with 100 nm GnRH for 7 min and harvested with 1% CHAPS lysis buffer, and then anti-IP3R1 immunoprecipitates were subjected to SDS-PAGE and silver staining. The 20- to 25-kDa region of a representative gel is shown, and the 21.5-kDa band marked with an arrow was identified as RNF170. B, a multiple-sequence alignment of RNF170 homologs from selected vertebrates using ClustalW. Amino acid identity among all species shown is indicated with asterisks, whereas identity between at least rodents and humans is indicated with dots. Predicted are three putative transmembrane domains (yellow, TMHMM Server v 2.0), a RING-HC domain (pink, with zinc-coordinating residues in orange (9)), ER membrane localization (PSORTb v 3.0), and a molecular weight of ∼30 kDa (ExPASy Compute pI/MW). The peptide sequence boxed was used to raise anti-RNF170, and the individual residues marked with arrowheads were mutated in *RNF170FLAG. C, topological model of RNF170 based on predictions of three transmembrane domains and the RING domain facing the cytosol. D, HeLa cells were transfected with either vector (lane 1) or with a construct containing the RNF170 encoding sequence isolated from αT3-1 cells (lane 2), and RNF170 expression was assessed in immunoblots (IB) with anti-RNF170. E, αT3-1 cells were treated for the indicated times with 100 nm GnRH and harvested with 1% CHAPS lysis buffer, and then anti-IP3R1 immunoprecipitates were probed for ubiquitin, IP3R1, p97, erlin2, and RNF170. F, the kinetics of GnRH-induced IP3R1 ubiquitination and coimmunoprecipitation of p97, erlin2, and RNF170 were quantitated and graphed. G, SH-SY5Y cells were treated for the indicated times with 1 mm carbachol and harvested with 1% CHAPS lysis buffer, and then anti-IP3R1 immunoprecipitates were probed for ubiquitin, IP3R1, erlin2, and RNF170. In E and G, polyubiquitinated IP3R1, unmodified IP3R1, p97, erlin2, and RNF170 migrated at 275–380 kDa, 260 kDa, 97 kDa, 43 kDa, and 21.5 kDa, respectively.
In view of the discrepancy between the apparent and predicted sizes of RNF170 (21.5 kDa versus 30 kDa, respectively) we used RT-PCR to isolate the RNF170-encoding sequence from αT3-1 cells. This identified an mRNA with an open reading frame corresponding to the 257-amino acid sequence shown in Fig. 1B, and transfection of cells with a construct containing this open reading frame (D) generated a 21.5-kDa anti-RNF170 immunoreactive band (lane 2) that was much stronger than the 21.5-kDa band seen in vector-transfected cells (lane 1), which likely represents endogenous RNF170. Thus, the 257-amino acid sequence depicted in Fig. 1B migrates at 21.5 kDa and is equivalent to endogenous RNF170. Why RNF170 migrates more rapidly than predicted is presently unclear but may reflect a posttranslational modification or the maintenance of protein folding during SDS-PAGE.
Examination of the association kinetics of RNF170 with IP3R1 in activated αT3-1 cells revealed that it bound rapidly and similarly to the rate at which erlin2 binds (Fig. 1, E and F). The binding of erlin2 (which provides a measure of erlin1/2 complex binding (15)) and RNF170 was more rapid than both the accumulation of polyubiquitinated IP3R1 and the binding of p97 (Fig. 1, E and F), which associates with IP3 receptors after they have been polyubiquitinated (4, 14, 15). To assess the universality of RNF170 binding, we also examined SH-SY5Y cells in which carbachol has been shown previously to induce IP3 receptor ubiquitination and down-regulation (5). As in αT3-1 cells, RNF170 associated rapidly with IP3 receptors after cell stimulation, and the association paralleled that of erlin2 (Fig. 1G). Overall, these data indicate that RNF170 binds to activated IP3 receptors prior to their polyubiquitination, possibly in conjunction with the erlin1/2 complex.
RNF170 Is an E3 Localized to the ER Membrane
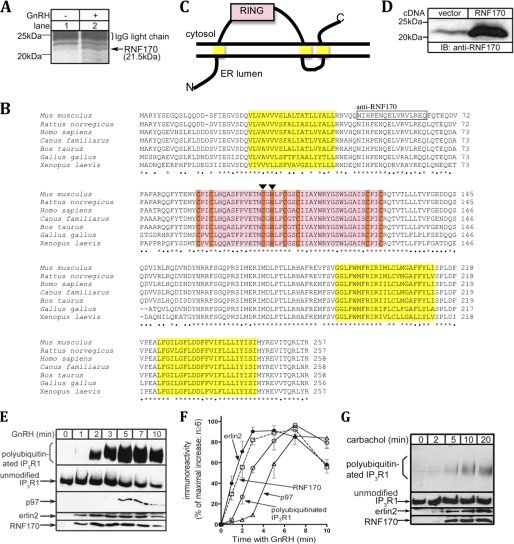
To facilitate the identification and purification of RNF170, a triple FLAG tag was appended to the C terminus, generating RNF170FLAG that migrated at 26 kDa (Fig. 2, lower panel, lanes 1–4). To examine whether RNF170 is an E3, immunopurified RNF170FLAG was incubated with E1, E2, and HA-ubiquitin. This generated an anti-HA immunoreactive smear from 40–250 kDa (Fig. 2, upper panel, lane 1), indicating that RNF170 possesses ubiquitin ligase activity. The fidelity of this activity was demonstrable by the observations that omission of E1, E2, or HA-ubiquitin blocked the formation of the high molecular mass species (lanes 2–4) and that *RNF170FLAG (in which the zinc-coordinating residues Cys-101 and His-103 are mutated to Ser and Ala, respectively) did not exhibit ubiquitin ligase activity (lane 5). The in vitro ubiquitin ligase activity of wild-type protein and the inhibitory effect of mutation of residues cognate with Cys-101 and His-103 has been seen with previously authenticated RING domain-containing E3s (9, 18), and shows that RNF170 is an E3.
FIGURE 2.
RNF170 possesses ubiquitin ligase activity. RNF170FLAG (lanes 1–4) and *RNF170FLAG (lane 5), immunopurified from transfected HeLa cells, were incubated with E1 (UBE1), E2 (UbcH5b) and HA-ubiquitin as indicated for 30 min at 30 °C. Samples were then subjected to SDS-PAGE and were probed in immunoblots (IB) with anti-HA epitope to assess ubiquitination (upper panel) or anti-FLAG epitope to assess RNF170FLAG/*RNF170FLAG levels (lower panel).
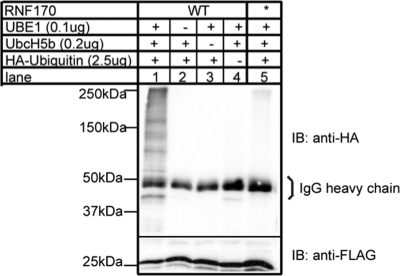
Examination of the subcellular distribution of RNF170FLAG by immunofluorescence microscopy (Fig. 3A) revealed an ER-like staining pattern similar to that of the ER marker DsRed2-ER and that of erlin1 and erlin2 (14, 15). Centrifugation of hypotonically lysed cells showed that RNF170FLAG localizes to the pellet together with the integral ER membrane E3 Hrd1 (19) and not to the supernatant with the cytosolic marker α-transaldolase (14, 15) (Fig. 3B, lanes 1 and 2). SDS, but not Na2CO3, released RNF170FLAG from the pellet (Fig. 3B, lanes 3–8). Together, these data indicate that RNF170 is an integral ER membrane protein.
FIGURE 3.
RNF170 is an integral ER membrane protein. A, HeLa cells were cotransfected with cDNAs encoding the ER marker DsRed2-ER and wild-type RNF170FLAG and were examined by confocal microscopy. B, HeLa cells transfected with cDNA encoding RNF170FLAG were harvested in hypotonic buffer, sonicated, and fractionated by centrifugation (lanes 1 and 2). Pellets were then resuspended in hypotonic buffer, 0.1% SDS, or 0.1 m Na2CO3, and were recentrifuged (lanes 3–8). The supernatants (S) and pellets (P) from each centrifugation were then probed in immunoblots for the indicated proteins.
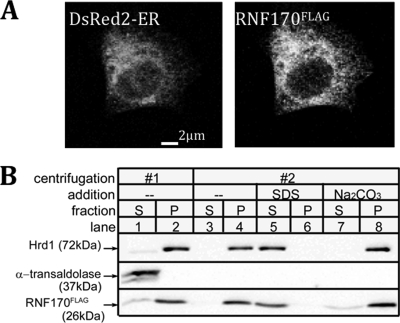
Interaction of RNF170 with the erlin1/2 Complex
To determine how RNF170 might interact with activated IP3 receptors, we first examined whether it interacts with the erlin1/2 complex. Silver staining of erlin1/2 complex immunopurified from unstimulated αT3-1 cells showed that the most abundant specifically coimmunoprecipitating protein migrated at 21.5 kDa (Fig. 4A, lane 3), and mass spectral analysis identified this band as RNF170. This association was confirmed by probing immunoblots with anti-RNF170 (Fig. 4B, lane 3) and in the reciprocal immunoprecipitation of RNF170 followed by probing with anti-erlin2 (lane 6). Interestingly, a large proportion of cellular RNF170 was associated with the erlin1/2 complex because immunodepletion of erlin2 from αT3-1 cell lysates also depleted an equivalent amount of RNF170 (Fig. 4C, lane 2). The interaction of RNF170 with the erlin1/2 complex was confirmed using exogenous proteins. Coimmunoprecipitation was seen between RNF170FLAG and erlin1HA/erlin2HA when these proteins were expressed in HeLa cells and immunoprecipitated with either anti-FLAG or anti-HA (Fig. 4D, lanes 8 and 9). Overall, these data indicate that the majority of cellular RNF170 is constitutively associated with the erlin1/2 complex.
FIGURE 4.
Most RNF170 is constitutively associated with the erlin1/2 complex. Cells were harvested with 1% CHAPS lysis buffer. A, anti-erlin2 immunoprecipitates from αT3-1 cells (lane 3) plus controls (lanes 1 and 2) were subjected to SDS-PAGE and silver staining. The 15- to 50-kDa region of a representative gel is shown, and the 21.5-kDa band marked with an arrow was identified by mass spectrometry as RNF170. B, anti-erlin2 or anti-RNF170 immunoprecipitates from αT3-1 cells (lanes 3 and 6) plus controls (lanes 1, 2, 4 and 5) were subjected to SDS-PAGE and probed in immunoblots (IB) with anti-RNF170 or anti-erlin2 as indicated. The 15- to 50-kDa is shown, and the migration positions of RNF170 and erlin2 are indicated with arrows. C, αT3-1 cell lysates were incubated without antibody (lane 1) or with anti-erlin2 (lane 2), and the immunodepleted lysates were subjected to SDS-PAGE and probed in immunoblots with anti-erlin2 or with anti-RNF170. D, HeLa cells were transfected with vectors encoding RNF170FLAG and/or erlin1HA/erlin2HA; cell lysates were incubated without antibody (lanes 1, 3, and 6), with anti-FLAG (F, lanes 2, 5, and 8), or with anti-HA (H, lanes 3, 6, and 9), and immunoprecipitates were probed in immunoblots with anti-HA or anti-FLAG.
Depletion of Endogenous RNF170 or Overexpression of *RNF170FLAG Inhibits IP3 Receptor Ubiquitination and Degradation
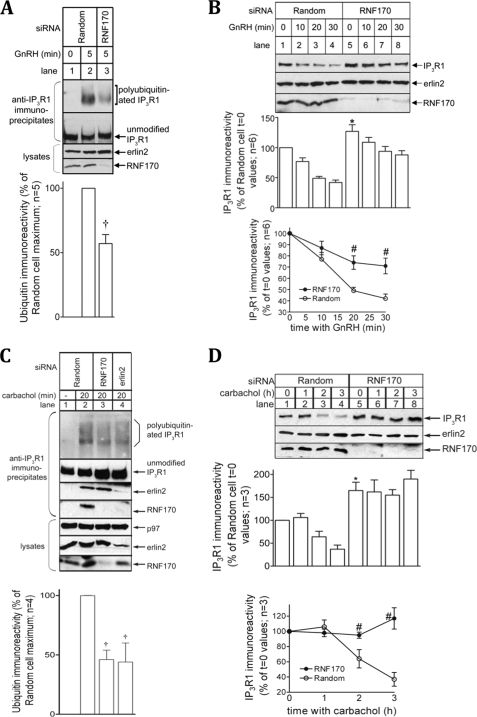
To assess the role of RNF170 in IP3 receptor processing, we used RNA interference to deplete endogenous RNF170. In αT3-1 cells, introduction by electroporation of a vector encoding a siRNA targeting RNF170 resulted in specific depletion of RNF170. RNF170 immunoreactivity was markedly reduced without a change in the levels of erlin2, which served as a loading control (Fig. 5A and B, lower panels). This RNF170 depletion inhibited GnRH-induced IP3R1 polyubiquitination (Fig. 5A, lane 3, uppermost panel) and down-regulation (Fig. 5B, lanes 5–8, uppermost panel). Quantitation revealed that RNF170 depletion inhibited IP3R1 polyubiquitination significantly to 57 ± 7% of control values (Fig. 5A, histogram), inhibited IP3R1 down-regulation significantly by ∼ 50% (Fig. 5B, line graph), and increased basal IP3R1 levels significantly by 27 ± 11% (Fig. 5B, compare lanes 1 and 5, and histogram). To authenticate these results we also examined mHeLa cells, in which we have previously shown that carbachol, a muscarinic receptor agonist, induces IP3 receptor ubiquitination and down-regulation (6). Consistent with the data from αT3-1 cells (Fig. 1E), RNF170 (and erlin2) coimmunoprecipitated with IP3 receptors in stimulated mHeLa cells (Fig. 5C, lane 2). Depletion of RNF170 from mHeLa cells using a transiently transfected oligonucleotide siRNA pool significantly inhibited IP3 receptor ubiquitination (Fig. 5C, lane 3 and histogram) and down-regulation (Fig. 5D, lanes 5–8 and line graph), and significantly elevated basal IP3R1 levels (Fig. 5D, compare lanes 1 and 5, and histogram). Thus, depletion RNF170 in two different cell lines, using different methods for siRNA delivery, inhibits stimulus-induced IP3 receptor ubiquitination and degradation, showing that RNF170 is critical to IP3 receptor processing by the ERAD pathway. Further, RNF170 depletion also increased basal IP3R1 levels, indicating that RNF170 also plays a role in IP3R1 turnover in resting cells.
FIGURE 5.
Depletion of endogenous RNF170 inhibits IP3 receptor processing. A and B, αT3-1 cells were electroporated to express vectors encoding either Random or RNF170 siRNAs, and effects on 100 nm GnRH-induced IP3 receptor polyubiquitination in anti-IP3R1 immunoprecipitates (A) or down-regulation in cell lysates (B) were assessed. Erlin2 served as a loading control. The histograms and graph depict combined quantitated data. C and D, mHeLa cells were transfected to express either Random or RNF170 oligonucleotide siRNAs or vectors encoding erlin2 siRNA (6), and effects on 10 μm carbachol-induced IP3 receptor ubiquitination in immunoprecipitates (C) or down-regulation in cell lysates (D) were assessed. Erlin2 and p97 served as loading controls. The histograms and graph depict combined quantitated data. †, p < 0.05 comparing polyubiquitination in Random versus RNF170 or erlin2 siRNA-expressing cells; *, p < 0.05 comparing IP3R1 immunoreactivity in unstimulated Random versus RNF170 siRNA expressing cells; #, p < 0.05 comparing IP3R1 down-regulation at each time point in Random versus RNF170 siRNA expressing cells. Polyubiquitinated IP3R1, unmodified IP3R1, p97, erlin2, and RNF170 migrated at 275–380 kDa, 260 kDa, 97 kDa, 43 kDa, and 21.5 kDa, respectively.
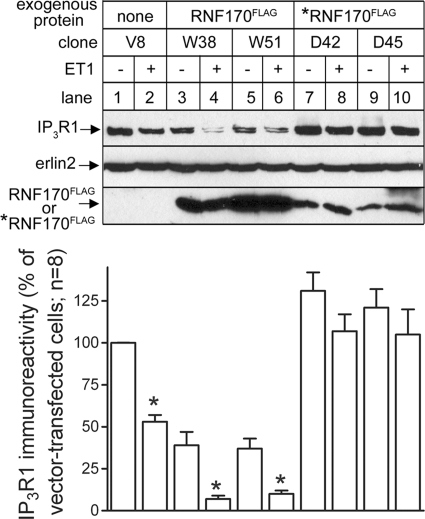
We also reasoned that, in view of the inability of *RNF170FLAG to catalyze ubiquitination, this mutant might exhibit dominant-negative activity, parallel to that seen with other E3s mutated in their RING domains (18, 19). This turned out to be the case, as stable expression of *RNF170FLAG in Rat1 cells inhibited ET1-induced IP3R1 down-regulation (Fig. 6, lanes 7–10), whereas significant and enhanced down-regulation was seen in RNF170FLAG-expressing cells (lanes 3–6). Interestingly, *RNF170FLAG expression raised basal IP3 receptor levels, whereas RNF170FLAG expression suppressed basal IP3 receptor levels (Fig. 6, lane 3-10). Again, these data support the view that RNF170 is critical to the processing of IP3 receptors under both basal and stimulated conditions.
FIGURE 6.
Overexpression of RNF170FLAG or *RNF170FLAG modulates IP3 receptor down-regulation. Rat1 cell clones stably expressing either RNF170 FLAG, *RNF170FLAG, or empty vector were incubated without or with 10 nm ET1 for 1 h, and immunoreactivity of IP3R1, erlin2 (loading control), or FLAG epitope (to monitor RNF170 FLAG or *RNF170FLAG expression) was assessed. Data shown are representative of at least six vector-, RNF170 FLAG-, or *RNF170FLAG-expressing clones. *, p < 0.05 comparing IP3R1 immunoreactivity without or with ET1 for each clone.
Priority of Binding to Activated IP3 Receptors
As RNF170 and the erlin1/2 complex appear to interact constitutively (Fig. 4) and bind to IP3 receptors with the same kinetics (Fig. 1, E–G), we sought to determine whether one mediates the binding of the other to activated IP3 receptors. siRNA-mediated depletion of RNF170 did not inhibit the association of erlin2 and, by extrapolation, the erlin1/2 complex (15) with activated IP3 receptors (Fig. 5C, lane 3), whereas depletion of erlin2 strongly inhibited association of RNF170 with activated IP3 receptors (Fig. 5C, lane 4). Thus, the erlin1/2 complex appears to recruit RNF170 to activated IP3 receptors.
DISCUSSION
That IP3 receptors are ubiquitinated and degraded in response to cell stimulation has been known for over a decade, yet the E3 that mediates this process has remained elusive (2). Here we provide evidence that RNF170 is involved because it associates with activated IP3 receptors, possesses E3 ligase activity, depletion of endogenous RNF170 inhibits IP3 receptor ubiquitination and degradation, and overexpression of a catalytically inactive RNF170 mutant is also inhibitory. In view of the findings that RNF170 interacts constitutively with the erlin1/2 complex and that depletion of the erlin1/2 complex inhibits RNF170 association with activated IP3 receptors but not vice-versa, it appears that RNF170 is recruited to activated IP3 receptors by the erlin1/2 complex.
Because IP3 receptors (1) and the erlin1/2 complex (15) are located in the ER membrane, the finding that RNF170 associates with these proteins and is predicted to contain three transmembrane domains immediately suggests that it is located in the ER membrane. This was confirmed by experiments showing that RNF170 is an integral membrane protein and that it has an ER-like distribution pattern. RNF170 can thus be added to the list of known mammalian ER membrane E3 ligases (13, 20). These include Hrd1 and gp78 (the mammalian homologs of yeast Hrd1p) (11, 13), TEB4 (the mammalian homolog of yeast Doa10) (11, 13), RNF5 (21), Kf-1 (22), TRC8 (23), RNF121 (17), RNF122 (24), and RNF180 (25). The substrates for some of these mammalian ligases are known (for example, TRC8 ubiquitinates MHC class I (23)), and it seems likely that each may target a fairly restricted group of substrates. This contrasts with the situation in yeast, where the ER contains only two E3 ligases, Hrd1p and Doa10, which, via adapters, mediate the processing of a broad range of substrates (11–13). It is noteworthy that we previously investigated several of these mammalian E3 ligases (Hrd1, gp78, TEB4, and RNF5) for a role in IP3 receptor processing using RNA interference, but depletion of none had a significant effect on IP3 receptor ubiquitination or degradation4. To date, RNF170 is the only ubiquitin ligase shown to exhibit activity toward IP3 receptors. Whether RNF170 is the only E3 for IP3 receptors is presently unclear, as depletion of endogenous RNF170 did not completely block IP3 receptor ubiquitination. This lack of complete inhibition could be due to the action of residual RNF170 after RNA interference or from the activity of another ligase or other ligases. Interestingly, whereas stimulus-induced IP3 receptor degradation was only partially blocked by RNF170 depletion in αT3-1 cells, it was completely blocked in mHeLa cells. This suggests that, in mHeLa cells, in addition to catalyzing IP3 receptor ubiquitination, RNF170 may govern the activity of an additional protein, perhaps an ERAD pathway component, that is required for IP3 receptor degradation.
In addition to inhibiting IP3 receptor processing by the UPP in stimulated cells, RNF170 depletion also raised basal IP3 receptor levels in resting cells. This suggests that RNF170 also plays a role in basal IP3 receptor turnover, which thus far has appeared to be independent of the UPP and which available data suggest is mediated by transfer to lysosomes (26). Consistent with the effect of RNF170 depletion, erlin2 depletion also raises basal IP3 receptor levels (6). Overall, these results point toward a hitherto unrecognized role for RNF170 and the UPP in basal IP3 receptor turnover.
A very recent report presented evidence that an Arg-to-Cys point mutation in human RNF170 is the cause of autosomal dominant sensory ataxia, a rare, progressive ataxia caused by degeneration of the posterior columns of the spinal cord (27). The effects of this point mutation on RNF170 activity and the substrates for RNF170 in neuronal cells have yet to be examined, but our studies show that in cultured SH-SY5Y neuroblastoma cells, RNF170 interacts with IP3 receptors as it does in other cell types. Thus, the potential is there for misregulation of IP3 receptors in individuals expressing mutant RNF170, which could be pathogenic, as deranged IP3 receptor function and Ca2+ signaling contributes to various neurodegenerative disorders (28).
In summary, here we present evidence that RNF170 is an E3 ligase that mediates IP3 receptor ubiquitination and processing by the UPP and that it is recruited to activated IP3 receptors by the erlin1/2 complex to which it is constitutively bound. Interestingly, erlin1 and erlin2 are anchored to the ER membrane via N-terminal transmembrane domains, and the vast majority (>90%) of the polypeptides, and the erlin1/2 complexes they form, lie within the ER lumen (15). It is thought that the intralumenal regions of the erlin1/2 complex interact with intralumenal regions of activated IP3 receptors and that the erlin1/2 complex acts as a “recognition factor” that selects activated IP3 receptors for degradation via the ERAD pathway (2, 15). IP3 receptors are ubiquitinated at specific, exposed lysine residues in their cytosolic domains (29, 30), and although it has yet to be defined how the erlin1/2 complex and RNF170 interact, it is reasonable to think that this interaction juxtaposes the cytosolic RING domain of RNF170 with the cytosolic regions of activated IP3 receptors that become ubiquitinated.
Acknowledgments
We thank Jennifer Vella for assisting with the initial identification of RNF170, Forrest Wright for help with SH-SY5Y cell culture, Tanya Fedotova for helpful suggestions, and Randy Hampton for advice regarding ligase assays.
This work was supported, in whole or in part, by National Institutes of Health Grant DK049194.
M. M. P. Pearce and R. J. H. Wojcikiewicz, unpublished data.
- IP3
- inositol 1,4,5-trisphosphate
- ER
- endoplasmic reticulum
- IP3R1
- type I inositol 1,4,5-trisphosphate receptor
- UPP
- ubiquitin-proteasome pathway
- GnRH
- gonadotropin-releasing hormone
- ERAD
- endoplasmic reticulum-associated degradation
- ET1
- endothelin-1
- LF
- LipofectamineTM 2000.
REFERENCES
- 1. Foskett J. K., White C., Cheung K. H., Mak D. O. (2007) Physiol. Rev. 87, 593–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wojcikiewicz R. J., Pearce M. M., Sliter D. A., Wang Y. (2009) Cell Calcium 46, 147–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wojcikiewicz R. J., Xu Q., Webster J. M., Alzayady K., Gao C. (2003) J. Biol. Chem. 278, 940–947 [DOI] [PubMed] [Google Scholar]
- 4. Alzayady K. J., Panning M. M., Kelley G. G., Wojcikiewicz R. J. H. (2005) J. Biol. Chem. 280, 34530–34537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhu C. C., Wojcikiewicz R. J. (2000) Biochem. J. 348, 551–556 [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y., Pearce M. M., Sliter D. A., Olzmann J. A., Christianson J. C., Kopito R. R., Boeckmann S., Gagen C., Leichner G. S., Roitelman J., Wojcikiewicz R. J. (2009) Biochim. Biophys. Acta. 1793, 1710–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Finley D. (2009) Annu. Rev. Biochem. 78, 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Komander D. (2009) Biochem. Soc. Trans. 37, 937–953 [DOI] [PubMed] [Google Scholar]
- 9. Deshaies R. J., Joazeiro C. A. P. (2009) Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 10. Bernassola F., Karin M., Ciechanover A., Melino G. (2008) Cancer Cell 14, 10–21 [DOI] [PubMed] [Google Scholar]
- 11. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hampton R. Y., Garza R. M. (2009) Chem. Rev. 109, 1561–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mehnert M., Sommer T., Jarosch E. (2010) BioEssays 32, 905–913 [DOI] [PubMed] [Google Scholar]
- 14. Pearce M. M., Wang Y., Kelley G. G., Wojcikiewicz R. J. (2007) J. Biol. Chem. 282, 20104–20115 [DOI] [PubMed] [Google Scholar]
- 15. Pearce M. M., Wormer D. B., Wilkens S., Wojcikiewicz R. J. (2009) J. Biol. Chem. 284, 10433–10445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wojcikiewicz R. J. H. (1995) J. Biol. Chem. 270, 11678–11683 [DOI] [PubMed] [Google Scholar]
- 17. Darom A., Bening-Abu-Shach U., Broday L. (2010) Mol. Biol. Cell 21, 1788–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fang S., Jensen J. P., Ludwig R. L., Vousden K. H., Weissman A. M. (2000) J. Biol. Chem. 275, 8945–8951 [DOI] [PubMed] [Google Scholar]
- 19. Kikkert M., Doolman R., Dai M., Avner R., Hassink G., van Voorden S., Thanedar S., Roitelman J., Chau V., Wiertz E. (2004) J. Biol. Chem. 279, 3525–3534 [DOI] [PubMed] [Google Scholar]
- 20. Neutzner A., Neutzner M., Benischke A. S., Ryu S. W., Frank S., Youle R. J., Karbowski M. (2011) J. Biol. Chem. 286, 8633–8643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., Cyr D. M. (2006) Cell 126, 571–582 [DOI] [PubMed] [Google Scholar]
- 22. Maruyama Y., Yamada. M., Takahashi. K., Yamada M. (2008) Biochem. Biophys. Res. Commun. 374, 737–741 [DOI] [PubMed] [Google Scholar]
- 23. Stagg H. R., Thomas M., van den Boomen D., Wiertz E. J., Drabkin H. A., Gemmill R. M., Lehner P. J. (2009) J. Cell Biol. 186, 685–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peng Z., Shi T., Ma D. (2010) BMC Cell Biol. 11, 41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ogawa M., Mizugishi K., Ishiguro A., Koyabu Y., Imai Y., Takahashi R., Mikoshiba K., Aruga J. (2008) Genes Cells 13, 397–409 [DOI] [PubMed] [Google Scholar]
- 26. Kahn M. T., Joseph S. K. (2003) Biochem. J. 375, 603–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Valdmanis P. N., Dupré N., Lachance M., Stochmanski S. J., Belzil V. V., Dion P. A., Thiffault I., Brais B., Weston L., Saint-Amant L., Samuels M. E., Rouleau G. A. (2011) Brain. 134, 602–607 [DOI] [PubMed] [Google Scholar]
- 28. Bezprozvanny I. (2011) Neurochem. Res. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sliter D. A., Kubota K., Kirkpatrick D. S., Alzayady K. J., Gygi S. P., Wojcikiewicz R. J. (2008) J. Biol. Chem. 283, 35319–35328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sliter D. A., Aguiar M., Gygi S. P., Wojcikiewicz R. J. H. (2011) J. Biol. Chem. 286, 1074–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]