Abstract
Human bone marrow-derived mesenchymal stromal cells (hMSCs) have the capacity to differentiate into several cell types including osteoblasts and are therefore an important cell source for bone tissue regeneration. A crucial issue is to identify mechanisms that trigger hMSC osteoblast differentiation to promote osteogenic potential. Casitas B lineage lymphoma (Cbl) is an E3 ubiquitin ligase that ubiquitinates and targets several molecules for degradation. We hypothesized that attenuation of Cbl-mediated degradation of receptor tyrosine kinases (RTKs) may promote osteogenic differentiation in hMSCs. We show here that specific inhibition of Cbl interaction with RTKs using a Cbl mutant (G306E) promotes expression of osteoblast markers (Runx2, alkaline phosphatase, type 1 collagen, osteocalcin) and increases osteogenic differentiation in clonal bone marrow-derived hMSCs and primary hMSCs. Analysis of molecular mechanisms revealed that the Cbl mutant increased PDGF receptor α and FGF receptor 2 but not EGF receptor expression in hMSCs, resulting in increased ERK1/2 and PI3K signaling. Pharmacological inhibition of FGFR or PDGFR abrogated in vitro osteogenesis induced by the Cbl mutant. The data reveal that specific inhibition of Cbl interaction with RTKs promotes the osteogenic differentiation program in hMSCs in part by decreased Cbl-mediated PDGFRα and FGFR2 ubiquitination, providing a novel mechanistic approach targeting Cbl to promote the osteogenic capacity of hMSCs.
Keywords: ERK, PI 3-Kinase, Proteasome, Receptor Tyrosine Kinase, Ubiquitin Ligase, Cbl
Introduction
Human bone marrow-derived mesenchymal stromal cells (MSCs)3 are adherent cells that can differentiate into multiple lineages including chondroblasts, osteoblasts, and adipocytes in a specific environment (1–3). Adult human MSCs (hMSCs) are an important cell source for tissue repair and therapy in regenerative medicine (4, 5). One important limitation to using hMSCs is their limited potential to differentiate into functional bone-forming cells. An important challenge is therefore to develop strategies that can promote the ex vivo osteogenic potential of hMSCs for bone tissue regeneration (6, 7). This requires a better understanding of the signaling molecules that trigger MSC osteogenic differentiation.
The osteogenic differentiation of MSCs is characterized by the expression of timely expressed genes such as Runx2, alkaline phosphatase (ALP), and type I collagen (Col1A1) followed by extracellular matrix mineralization (8, 9). The osteogenic potential of hMSCs can be promoted by proteins such as bone morphogenetic proteins that increase Runx2 expression and downstream osteoblast marker genes (10). Very recent studies indicate that activation of receptor tyrosine kinases (RTKs) such as platelet-derived growth factor (PDGFR) (11) or fibroblast growth factor receptor 2 (FGFR2) (12) promotes the osteogenic differentiation of human or murine MSCs. This effect results in part from activation of signaling pathways such as extracellular signal-regulated protein kinase (ERK1/2) and phosphatidylinositol kinase (PI3K), leading to activation of osteoblast marker gene expression in MSCs and osteogenic differentiation (11, 12). This suggests that finding approaches to activate these receptors may potentially promote the osteogenic capacity of hMSCs.
Ubiquitin ligases are important proteins that act as regulators of signal transduction pathways. These proteins ubiquitinate and target several signaling molecules for degradation (13–16). The E3 ubiquitin ligase Cbl (Casitas B-lineage lymphoma) is a 120-kDa cytoplasmic polypeptide that is responsible for the down-regulation of RTKs and nonreceptor tyrosine kinases that undergo proteasome-mediated degradation after being ubiquitinated by Cbl (17, 18). A number of studies indicate that Cbl plays a potential role as a negative regulator of RTKs, including epidermal growth factor (EGFR), PDGFR, and FGFR (19–23). This negative regulatory function is mediated by two Cbl domains, the phosphotyrosine kinase-binding (PTB) domain and the RING finger domain (24). The PTB domain allows interaction of the Cbl protein with activated RTK, and the RING finger domain allows recruitment of ubiquitin-conjugated enzymes (E2), resulting in ubiquitination of RTKs and their proteasome degradation (25). Alteration of Cbl activity using the Cbl G306E mutant which inactivates the PTB domain (26, 27) results in loss of function, indicating that the negative regulation of RTK signaling pathways by Cbl requires direct interaction of RTK with intact PTB domain (28).
Although the role of Cbl as a negative regulator of RTKs is well documented, the potential impact of Cbl-mediated regulatory mechanisms in the physiological control of cell proliferation, differentiation, and survival remains to be determined. In previous studies, we showed that Cbl plays a role in the control of osteoblasts by regulating the degradation of FGFR2, Src proteins, PI3K, and α5 integrin subunit, indicating that Cbl-mediated attenuation of signaling pathways is an important physiological mechanism controlling osteoblastogenesis (20, 29–31). Here, we hypothesized that attenuation of Cbl interaction with RTK in hMSC may promote MSC osteoblast differentiation via increased RTK signaling, which could be used as a therapeutic tool for promoting osteogenic differentiation. In this study, we report that specific attenuation of Cbl-mediated degradation of some RTKs using a Cbl-inactive mutant promotes osteogenic differentiation in hMSCs without adversely affecting cell proliferation or survival. We show that the osteogenic differentiation program induced by the Cbl mutant is mediated in part by enhanced FGFR2 and PDGFRα expression and signaling. These results indicate that specific attenuation of Cbl interaction with RTK can be an efficient strategy for promoting osteogenic differentiation of hMSCs.
EXPERIMENTAL PROCEDURES
Cells and Treatments
Cultured immortalized clonal human bone marrow stroma-derived Stro-1-positive cells (F/Stro-1+ hMSCs) (32) were obtained as previously described (33). These cells display osteogenic differentiation potential (33, 34). Primary hMSCs were purchased from PromoCell (Heidelberg, Germany). HEK293T cells were purchased from American Type Culture Collection. Cells were routinely cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% heat-inactivated fetal calf serum (FCS), 1% l-glutamine, and penicillin/streptomycin (10,000 units/ml and 10,000 μg/ml, respectively) at 37 °C in humidified atmosphere containing 5% CO2 in air. Culture media were changed three times a week. In some experiments, cells were treated with the PDGFR inhibitor Imatinib® (Novartis, Basel, Switzerland) used at 0.5 μm, or the FGFR inhibitor PD173074 (Sigma) used at 5 nm.
Lentiviral Particles Production and Transduction
The pBK plasmid containing the human G306E Cbl complete sequence that abolishes the binding ability of Cbl PTB domain (19) was amplified by PCR using forward primer 5′-CCCTCGAGCGGCCACCATGGATTACAAGGATGACGACGATAAGTGAGCCGGCAACGTGAAGAAGAGCTCT-3′ and reverse primer 5′-CGGGGTACCCTAGGTAGCTACATGGGCAGGAGA-3′, then cloned into the lentiviral TRIP vector previously described (35) between the Xho1-Kpn1 sites. Lentiviral production was performed using HEK293T cells plated at 8 · 104 cells/cm2 and grown overnight at 37 °C in DMEM supplemented with 10% heat-inactivated FCS and penicillin/streptomycin (10,000 units/ml and 10,000 μg/ml, respectively). Virion particles containing the TRIP-G306E Cbl or the control empty vector TRIP-EV were produced by transient calcium phosphate cotransfection of HEK293T cells with 25 μg of the vector plasmid, 25 μg of the encapsidation plasmid (pCMVR8.91) and 5 μg of the vesicular stomatitis virus G protein envelope expression plasmid (pHCMV-G). F/Stro-1+ or primary hMSCs were transduced at 50% confluence with lentiviral particles in the presence of Polybrene (10 μg/ml or 5 μg/ml, respectively) for 48 h. Lentiviral transduction efficacy was evaluated by GFP level under fluorescence microscopy (36). Human shCbl particles were purchased from CliniSciences (Montrouge, France).
Proliferation and Apoptosis Assays
Cells were seeded at 2.5 · 104 cells/cm2, and cell proliferation was evaluated by cell counting or BrdU incorporation according to the manufacturer's recommendations (GE Healthcare). For the apoptosis assay, 2 · 104 cells/cm2 were cultured in the presence of 0 or 10% FCS, and DNA fragmentation was detected using TUNEL staining (37). Cell viability was evaluated by the colorimetric MTT microassay as described (38).
Differentiation Assays
Alkaline phosphatase activity was determined using a FAST kit according to the manufacturer's recommendations (Sigma). For the in vitro osteogenic assay, cell culture medium was supplemented with 50 μg/ml ascorbic acid and 3 mm inorganic phosphate (NaH2PO4) to allow matrix synthesis and mineralization. At the indicated time point, cells were fixed in 70% ethanol at 4 °C. Matrix mineralization was evaluated by alizarin red staining and microphotographed using an Olympus microscope (12).
Quantitative RT-PCR Analysis
Total RNAs were isolated using TRIzol reagent (Invitrogen) according to the manufacturer's recommendation and stored at −80 °C in RNAsecure Reagent-treated H2O. Three μg of total RNA from each sample were denatured for 10 min at 70 °C and then reversed transcribed at 37 °C for 90 min using 300 units Moloney murine leukemia virus reverse transcriptase, 1.5 μg of oligo(dT) primers, 1 mm dNTP in 20 μl of total and finally inactivated at 85 °C for 5 min. Relative mRNA levels were evaluated by quantitative PCR (LightCycler; Roche Applied Science) using a SYBR Green PCR kit (ABGen, Courtabœuf, France) and specific primers (12). Thermal conditions were: activation for 15 min at 95 °C then 45 cycles of denaturation at 95 °C for 20 s, 60 °C annealing for 15 s, and 72 °C extension for 15 s. Signals were normalized to GAPDH as internal control. The relative amount of RNA was calculated by the 2-ΔΔCt method.
Western Blot and Immunoprecipitation Analyses
Total cell lysates were prepared as described (12). Proteins (30 μg) were resolved on 4–12% SDS-PAGE and transferred onto PVDF nitrocellulose membranes (Millipore). Filters were incubated for 2 h in 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 0.1% (v/v) Tween 20, 0.5% (w/v) bovine serum albumin (TBST/BSA), then overnight at 4 °C on a shaker with the following primary antibodies (1/500–1/1000 in TBST/BSA): anti-c-Cbl, anti-FGFR2, anti-phosphotyrosine (Tyr(P)), anti-Cbl-b, anti-EGFR and anti-PDGFRα, anti-ERK1/2 and anti-p-ERK1/2, anti-ubiquitin (Santa Cruz Biotechnology, Santa Cruz, CA), anti-AKT, anti-p-AKT, anti-PI3K p85, anti-p-PI3K p85, anti-p-FRS2α (Cell Signaling, Danvers, MA), anti-β-actin (Sigma), anti-Grb2 (BD Biosciences), or anti-phosphotyrosine Sprouty2 (Spy2; BD Biosciences). Membranes were washed twice with TBST and incubated for 2 h with the appropriate HRP-conjugated secondary antibody (1/10,000–1/20,000 in TBST/BSA). After final washes, the signals were visualized with enhanced chemiluminescence Western blotting detection reagent (ECL, Amersham Biosciences) and autoradiographic film (X-Omat-AR, Eastman Kodak). Densitometric analysis using ImageQuant software was performed following digital scanning (Agfa). Representative images of immunoblots are shown. For immunoprecipitation analysis, cell lysates were prepared as for Western blot analysis, and aliquots of total protein (250 μg) were incubated overnight under weak agitation at 4 °C with 2 μg of specific antibody and 20 μl of Dynabeads protein G (Invitrogen). Components of the bound immune complex (both antigen and antibody) were eluted from the Dynabeads and analyzed by SDS-PAGE according to the manufacturer's recommendations.
Statistical Analysis
The data are the mean ± S.D. of an average of six samples and are representative of at least three distinct experiments. The data were analyzed by Student's test, and a minimal level of p < 0.05 was considered significant.
RESULTS
The G306E Cbl Mutant Moderately Modulates hMSC Proliferation and Survival
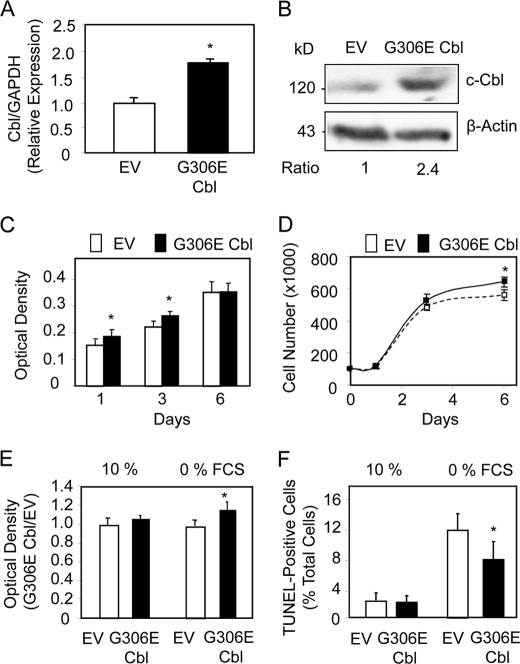
We have used the G306E Cbl mutant specifically to target the link between Cbl and RTK because knocking down Cbl may have undesirable effects on several proteins in addition to RTKs. We first determined the expression levels of Cbl in hMSCs transduced with the G306E Cbl mutant. Quantitative PCR analysis showed that the mutant Cbl induced a 2-fold increase in Cbl mRNA levels in F/Stro-1+ MSCs (Fig. 1A). Western blot analysis confirmed that total Cbl (c-Cbl and G306E Cbl) was increased about 2-fold in transduced hMSCs, thus validating the assay (Fig. 1B). A similar effect was obtained in primary hMSCs (supplemental Fig. 1A). To determine the cellular effect of the mutant Cbl in hMSCs, we first investigated the changes in cell proliferation in F/Stro-1+ MSCs transduced with G306E Cbl. Determination of DNA replication using the BrdU assay showed that the mutant Cbl induced only a slight (<15%) increase in DNA replication at early time points (Fig. 1C). Analysis of cell number confirmed that the G306E Cbl mutant only slightly (<10%) increased cell number at 6 days of culture (Fig. 1D). Similar effects of the Cbl mutant were obtained in primary hMSCs (supplemental Fig. 1B). We then determined the cellular effect of the mutant Cbl on mesenchymal cell survival. Analysis of cell apoptosis showed that the G306E Cbl mutant had no effect on F/Stro-1+ MSC survival cultured in basal conditions (10% FCS) and only slightly increased cell apoptosis in a serum-deprived (0% FCS) condition, as determined by both MTT test (Fig. 1E) and TUNEL staining (Fig. 1F). A similar effect was observed in primary hMSCs using the MTT test (supplemental Fig. 1C). These results show that specific attenuation of Cbl interaction with RTKs using the G306E Cbl mutant has only a marginal effect on cell proliferation and survival in hMSCs.
FIGURE 1.
The G3036E Cbl mutant marginally modulates hMSC proliferation and survival. A and B, quantitative PCR analysis (A) and Western blot analysis (B) showing increased Cbl mRNA and protein levels, respectively, in clonal hMSCs transduced with the G306E Cbl mutant compared with the empty vector (EV). C and D, effect of the G3036E Cbl mutant on hMSC cell replication as shown by BrdU assay (C) and cell number (D). E and F, change in cell survival induced by the G3036E Cbl mutant in clonal hMSCs as shown by MTT assay (E) and TUNEL staining in serum-deprived conditions (F). *, significant difference with EV-transduced cells (p < 0.05). Error bars, S.D.
The G306E Cbl Mutant Promotes hMSC Osteogenic Differentiation
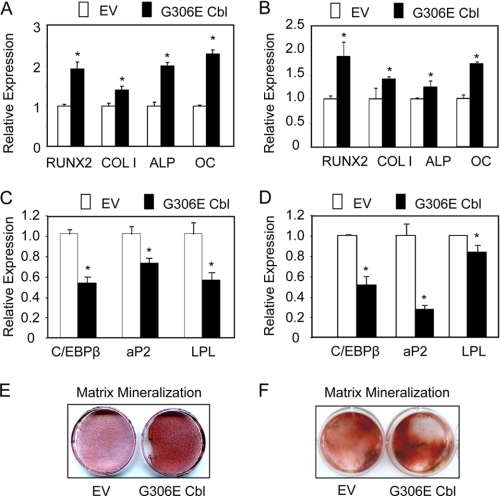
We then investigated the effects of the Cbl mutant on osteogenic differentiation of F/Stro-1+ and primary hMSCs. Quantitative PCR analysis showed that the Cbl mutant induced a 2-fold increase in mRNA expression for Runx2, a specific osteoblast transcription factor. Consistently, the Cbl mutant increased the expression of alkaline phosphatase (ALP), an early osteoblast marker, type 1 collagen (Col1A1), a functional osteoblast marker, as well as osteocalcin (OC), a late osteoblast marker (Fig. 2A). Similar effects of the G306E Cbl mutant on osteoblast differentiation markers were obtained in primary hMSCs (Fig. 2B), confirming that the mutant Cbl promoted osteoblast differentiation in hMSCs. We also showed that the G306E Cbl mutant decreased the expression of adipocyte genes such as C/EBPα, aP2, and LPL in clonal hMSCs (Fig. 2C) and primary hMSCs (Fig. 2D), indicating decreased adipogenic differentiation. We also determined the effects of the mutant Cbl on in vitro matrix mineralization. As shown in Fig. 2, the mutant Cbl increased matrix mineralization in clonal hMSC (Fig. 2E) and primary hMSC cultures (Fig. 2F). In marked contrast, we found that a shCbl that effectively decreased Cbl level, as revealed by qPCR and Western blot analyses (supplemental Fig. 2, A and B) reduced the expression of most osteoblast differentiation marker genes (supplemental Fig. 2C). This effect of Cbl knocking down presumably results from undesirable effects on several proteins besides RTKs. Thus, in contrast to general Cbl knocking down, specific attenuation of Cbl interaction with RTKs using the G306E Cbl mutant increases osteoblast marker expression and osteogenic potential and decreases adipogenic differentiation of hMSCs.
FIGURE 2.
The G3036E Cbl mutant promotes hMSC osteogenic differentiation. A–D, quantitative PCR analysis showing that the G306E Cbl mutant increased the expression of osteoblast markers and decreased adipocyte gene expression in clonal hMSCs (A and C) and primary hMSCs (B and D) compared with empty vector (EV) transduced cells. E and F, G306E Cbl mutant-increased matrix mineralization as shown by alizarin red staining at 14 days of culture in clonal hMSCs (E) and primary hMSCs (F). *, significant difference with EV-transduced cells (p < 0.05). Error bars, S.D.
The G306E Cbl Mutant Affects FGFR2 and PDGFRα Signaling
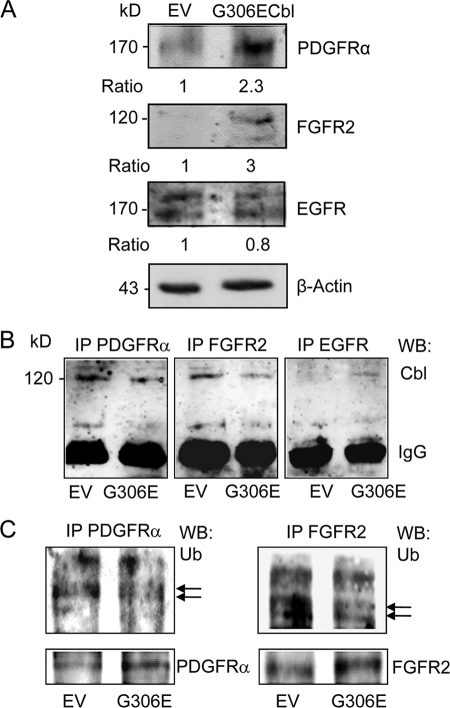
The ubiquitin ligase Cbl acts by recruiting proteins such as RTKs for ubiquitination and proteasome degradation. We therefore sought to determine the effect of the G306E Cbl mutant that abolishes the binding ability of Cbl PTB domain (19) on RTK levels. Western blot analysis showed that transduction of F/Stro-1+ MSCs with G306E Cbl resulted in increased PDGFRα and FGFR2 protein levels, whereas EGFR level was unchanged (Fig. 3A). Similar effects were observed in primary hMSCs (supplemental Fig. 3A). Further immunoprecipitation analyses showed that the mutant Cbl decreased the amount of Cbl associated with PDGFRα and FGFR2 but not EGFR (Fig. 3B). The recruitment of Cbl to RTKs leads to ubiquitination and degradation. To investigate the resulting effect of the mutant Cbl on RTKs, we performed immunoprecipitation analyses in hMSCs. As shown in Fig. 3C, the mutant Cbl decreased ubiquitin levels associated with PDGFRα and FGFR2, resulting in increased PDGFRα and FGFR2 protein levels. Overall, these results show that the G306E Cbl mutant decreases ubiquitination of at least two receptors (PDGFRα and FGFR2), resulting in increased expression of these receptors in hMSCs.
FIGURE 3.
Effects of the G3036E Cbl mutant on FGFR2, PDGFRα, and EGFR signaling. A, Western blot analysis showing that the G306E Cbl mutant increased PDGFRα and FGFR2, but not EGFR levels in clonal hMSCs. B, immunoprecipitation (IP) analysis showing decreased PDGFRα and FGFR2 associated with Cbl in clonal hMSCs expressing the G306E Cbl mutant. IgG served as internal control. WB, Western blotting. C, immunoprecipitation analysis showing decreased PDGFRα and FGFR2 associated with ubiquitin (Ub) in clonal hMSCs transduced with the G306E Cbl mutant. The same protein lysates showing increased PDGFRα and FGFR2 in G306E Cbl transduced cells were used as internal controls.
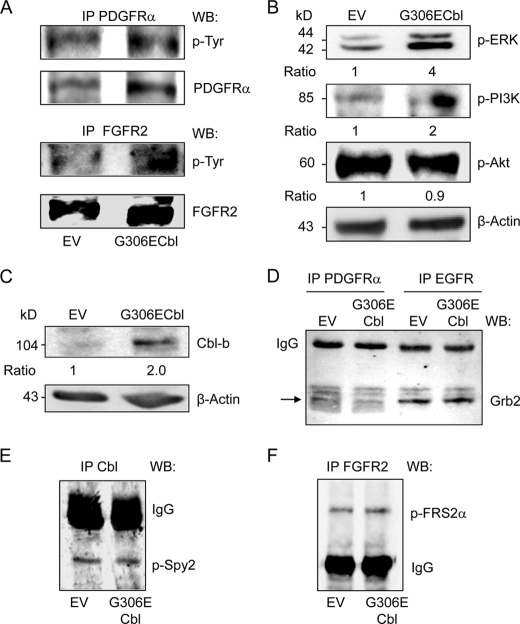
Having shown that the expression of the mutant Cbl results in increased PDGFRα and FGFR2 protein levels in hMSCs, we sought to determine whether this effect is associated with increased RTK signaling. As shown in Fig. 4A, the increased PDGFRα induced by the mutant Cbl was associated with an increased phosphorylated PDGFRα level. Similarly, the Cbl mutant increased both FGFR2 and phosphorylated FGFR2 levels (Fig. 4A), indicating that the up-regulation of PDGFRα and FGFR2 translated into activation of these receptors in hMSCs. We next determined whether receptor activation induced by the mutant Cbl triggered signaling pathways known to be activated by PDGFR and FGFR2 in osteogenic cells. As shown in Fig. 4B, activation of PDGFRα and FGFR2 induced by the Cbl mutant increased ERK1/2 and PI3K phosphorylation whereas Akt phosphorylation was unchanged. Again, all effects of the mutant Cbl on cell signaling were confirmed in primary hMSCs, although the effects were lower than in clonal hMSCs (supplemental Fig. 3B). These results show that specific attenuation of Cbl interaction with RTKs in hMSCs increased the expression and signaling of at least PDGFRα and FGFR2, resulting in activation of two signaling pathways, namely ERK1/2 and PI3K.
FIGURE 4.
Effect of the G3036E Cbl mutant on RTK signaling and ancillary proteins. A, immunoprecipitation (IP) analysis showing that the G306E Cbl mutant increased PDGFRα and FGFR2 phosphorylation in clonal hMSCs. B, Western blot analysis showing that the G306E Cbl mutant increased ERK1/2 and PI3K, but not Akt phosphorylation in clonal hMSCs. C–F, Western blot (WB) analysis showing increased Cbl-b (C) but unchanged Grb2 (D), phospho-Spy2 (E) and phospho-FRS2α (F) in clonal hMSCs expressing the G306E Cbl mutant. IgG served as internal control.
Lack of Effect of the G306E Cbl Mutant on RTK-associated Ancillary Proteins
The degradation of activated tyrosine kinase receptors is regulated by several mechanisms involving multiple proteins. We therefore investigated whether the mutant Cbl altered RTK interactions with regulatory proteins. We found that transduction of hMSCs with the G306E Cbl mutant increased protein level of Cbl-b, another Cbl ubiquitin ligase that is involved in protein degradation (Fig. 4C), suggesting that attenuation of Cbl function by the mutant may be, in part, compensated by increased Cbl-b level. We also determined whether the mutant Cbl caused changes in ancillary proteins that are recruited by activated RTKs and modulate RTK activity (39). Because Cbl can form a complex with PDGFR or EGFR indirectly by means of Grb2 (40), we investigated whether Grb2-mediated binding of Cbl to PDGFR was altered by the mutant Cbl. We found that the Grb2 level associated with PDGFRα or EGFR was not significantly changed by the mutant Cbl in hMSCs (Fig. 4D), indicating normal Cbl-Grb2 binding of this complex. Spy2 is another signaling attenuator controlling RTK signaling (41). Upon stimulation by RTKs, phosphorylation of Spy2 on a conserved tyrosine residue 55 leads to binding to Cbl, resulting in reduced Cbl interaction with RTKs and reduced RTK ubiquitination and degradation (42). We found that the level of p-Spy2 associated with Cbl was not affected in hMSCs transduced with the mutant Cbl (Fig. 4E). Finally, we investigated whether the mutant Cbl affected the docking protein FGFR substrate 2 (FRS2), which is required for Spy2 phosphorylation upon FGFR activation (40), resulting in interaction with Cbl and receptor ubiquitination (43). As shown in Fig. 4F, p-FRS2α associated with FGFR2 was unchanged by the mutant Cbl in hMSCs. Overall, these results indicate that the G306E mutant does not appear to alter Spy2-FRS2α-FGFR2 interactions in hMSCs.
FGFR2 and PDGFR Signaling Mediates hMSC Osteogenic Differentiation Induced by the G306E Cbl Mutant
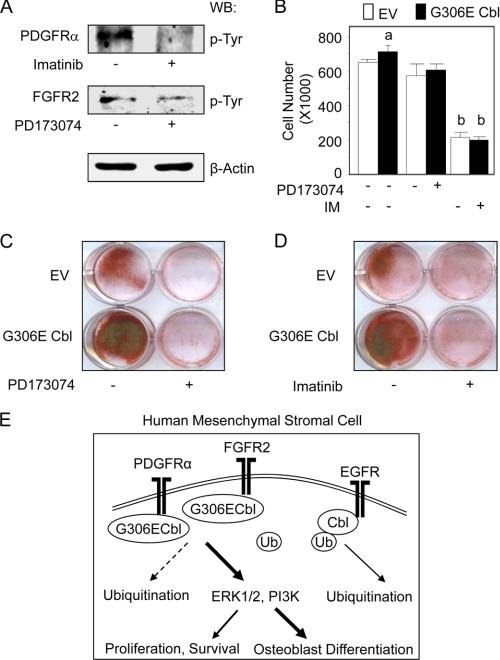
The above results indicate that specific attenuation of Cbl interaction with RTKs in hMSCs triggers up-regulation of PDGFRα and FGFR2, leading to increased ERK1/2 and PI3K signaling. To determine whether these signaling pathways are functionally involved in the cellular phenotype induced by the Cbl mutant in hMSCs, we inhibited PDGFR and FGFR in hMSCs expressing the Cbl mutant. As shown in Fig. 5A, the pharmacological inhibitors abolished PDGFRα and FGFR2 phosphorylation, thus validating the methodological approach. We found that both PDGFR and FGFR inhibitors abrogated the slight increase in osteoblast proliferation induced by the Cbl mutant (Fig. 5B). Additionally, long term pharmacological inhibition of PDGFR (Fig. 5C) or FGFR (Fig. 5D) reduced the basal in vitro osteogenesis induced by MSCs, which is consistent with the role of PDGFR and FGFR2 signal transduction in MSC osteogenesis (11, 12). Furthermore, pharmacological blockade of PDGFR and FGFR2 phosphorylation blunted in vitro osteogenesis induced by the G3060E Cbl mutant in clonal hMSCs (Fig. 5, C and D). These results indicate that PDGFR and FGFR mediate, at least in part, the osteogenic differentiation program induced by the Cbl G306E mutant in hMSCs. Overall, the results indicate that reduction of PDGFRα and FGFR2 ubiquitination induced by the G306E Cbl mutant leads to enhanced ERK1/2 and PI3K signaling and increased osteoblast differentiation and osteogenesis in hMSCs (Fig. 5E).
FIGURE 5.
hMSC osteogenic differentiation induced by the G3036E Cbl mutant involves FGFR and PDGFR signaling. A, Western blot (WB) analysis showing that the PDGFR inhibitor Imatinib® (IM, 0.5 μm) and the FGFR inhibitor PD173074 (5 nm) reduced PDGFRα and FGFR2 phosphorylation in clonal hMSCs. B, cell proliferation assay showing that the two inhibitors abrogated the increased cell number induced by the Cbl mutant in clonal hMSCs at 6 days of culture. C and D, both PDGFR (C) and FGFR (D) inhibitors blunted in vitro matrix mineralization induced by the G306E Cbl mutant in clonal hMSCs as shown by alizarin red staining at 14 days of culture. EV, empty vector. E, proposed signaling mechanism underlying the osteogenic differentiation induced by the G3036E Cbl mutant in hMSCs. Specific attenuation of Cbl interaction with RTKs induced by the G306E Cbl mutant decreases PDGFRα and FGFR2 ubiquitination (dotted line), causing increased PDGFRα and FGFR2 levels and signaling and resulting in increased osteoblast marker expression and osteogenic differentiation.
DISCUSSION
Efficient cell-based therapy for bone tissue engineering and therapeutic applications requires to use approaches that promote differentiation of hMSCs without adverse effects on cell proliferation or survival. Here, we demonstrate that targeting the ubiquitin ligase Cbl-RTK interaction attenuates RTK ubiquitination and increases RTK signaling and thereby promotes the osteogenic differentiation program in hMSCs. We first found that targeting Cbl function using the Cbl mutant G306E that abrogates binding to RTK via the PTB domain promoted osteoblast marker gene expression and in vitro osteogenesis in bone marrow-derived hMSCs. Interestingly, this effect was not associated with marked effects on cell proliferation or survival. These results provide the first evidence that attenuation of a specific interaction of a single E3 ubiquitin ligase, namely Cbl, with RTK is sufficient to promote the osteogenic program in hMSCs without adversely affecting cell proliferation or survival. Our finding that the Cbl-RTK interaction plays a role in hMSC osteoblast differentiation indicates that this ubiquitin ligase is involved in the control of early stages of osteoblast differentiation, in addition to its implication in later stages of osteoblastogenesis (20, 29–31). Recent studies showed that proteasome inhibitors increase osteoblast differentiation (44, 45), although specific targets have not been established. A more recent study indicates that targeting the proteasome results in increased Runx2 expression and enhanced MSC osteoblast differentiation (46). The present data indicate that targeting specifically the ubiquitin ligase Cbl for decreasing RTK ubiquitination results in increased Runx2 expression and osteogenic differentiation in hMSCs. We also found that attenuation of Cbl interaction with RTKs reduced adipogenic differentiation in hMSCs. Previous data indicated that deletion of Cbl in mice results in reduction in adipose tissue which can be explained by increased whole body energy expenditure and increased insulin action (47). The present results suggest that inhibition of Cbl interaction with RTKs can also reduce adiposity through activation of hMSC differentiation toward osteoblasts rather than toward adipocytes.
We next identified the mechanisms responsible for the positive effect of the Cbl mutant on hMSC osteogenic differentiation. Our data indicate that the Cbl mutant attenuated RTK ubiquitination and thereby increased expression and signalization of specific RTKs. This effect resulted directly from decreased Cbl interaction with some RTKs rather than alteration of its interaction with ancillary proteins such as Spy2, FRS2α, or Grb2. Interestingly, we did not find that EGFR protein expression was altered by the Cbl mutant, although EGFR is known to be down-regulated by Cbl after activation of the receptor (48, 49). Possible explanations for the absence of EGFR up-regulation by the mutant Cbl include increased binding to native Cbl or to Cbl-b which may have compensated for the defective mutant Cbl (Fig. 4C). In contrast to EGFR, our data revealed that FGFR2, one important signaling pathway involved in osteoblastogenesis (50), was up-regulated by the mutant Cbl in hMSCs and thereby triggered osteoblast differentiation in hMSCs. This is consistent with our recent finding that activation of FGFR2 promotes osteoblast differentiation in murine MSCs (12). Besides FGFR2, we found that the mutant Cbl up-regulated PDGFRα signaling, resulting in increased hMSC osteoblast differentiation. This is another important finding because divergent effects of PDGFR have been reported in MSC osteogenic differentiation. Although PDGF was reported to have positive or negative effects on bone marrow-derived mesenchymal progenitor cell differentiation in vitro (51, 52), PDGFR signaling was shown to promote bone formation in vivo (53–55). Consistently, we found that inhibition of PDGFR and FGFR abrogated hMSC osteogenic differentiation induced by the mutant Cbl, which strongly supports a role for these receptors in hMSC osteogenic differentiation. Besides PDGFRα and FGFR2, other RTKs, such as VEGFR and insulin-like growth factor receptor, may possibly interact with Cbl in MSCs. However, VEGFR seems to be indirectly regulated by Cbl (56, 57), and insulin-like growth factor receptor down-regulation was found to involve Cbl-b in osteoblasts (58). Whether these RTKs are regulated by Cbl in MSC and may contribute to MSC osteogenic differentiation remains to be determined.
Our data allowed to identify the signaling molecules that trigger MSC osteogenic differentiation induced by attenuation of Cbl interaction with RTKs. We found that the G306E mutant Cbl increased ERK1/2 and PI3K signaling, indicating that its positive effect on MSC osteogenic differentiation is dependent, at least in part, on FGFR2- and PDGFRα-induced activation of ERK and PI3K signaling pathways. These results are consistent with the recent finding that ERK1/2 and PI3K play important roles in MSC osteoblast differentiation (12, 59, 60). Although we cannot rule out that other signaling pathways may be induced by the mutant Cbl, our data identified the main signaling mechanisms that are induced by inhibition of Cbl interaction with RTKs in hMSCs. It has been recently proposed that cross-talks between PDGFR and FGFR may converge to induce ERK1/2 and PI3K signaling pathways (39). In support of this concept, we recently showed that constitutive FGFR2 activation results in increased PDGFRα expression and ERK1/2 and PI3K signaling in human osteoblasts (55, 61). It is thus possible that the positive effect on hMSC osteoblast differentiation induced by the G306E Cbl mutant may result from convergent signaling pathways induced by PDGFR-FGFR2 cross-talks, in addition to the effects on individual receptors.
In summary, the present data reveal that the G306E Cbl mutant triggers hMSC differentiation in part by up-regulating PDGFRα and FGFR2 and downstream ERK and PI3K signaling (Fig. 5E). This reveals that Cbl, via the control of these RTKs, plays a positive regulatory role in the early stages of hMSC osteoblast differentiation. We suggest that Cbl-RTK interaction could be targeted for promoting osteoblast differentiation of hMSCs, which may be an effective approach for increasing the osteogenic potential of hMSCs for cellular transplantation in therapeutic approaches.
Supplementary Material
Acknowledgment
We thank Novartis (Basle, Switzerland) for the Imatinib®.
This work was supported in part by Institut National de la Santé et de la Recherche Médicale.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- MSC
- mesenchymal stromal cell
- Cbl
- Casitas B lineage lymphoma
- FRS2
- FGFR substrate 2
- hMSC
- human MSC
- PTB
- phosphotyrosine kinase-binding
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- R
- receptor
- Spy2
- Sprouty2.
REFERENCES
- 1. Bianco P., Riminucci M., Gronthos S., Robey P. G. (2001) Stem Cells 19, 180–192 [DOI] [PubMed] [Google Scholar]
- 2. Kassem M. (2004) Cloning Stem Cells 6, 369–374 [DOI] [PubMed] [Google Scholar]
- 3. Pittenger M. F., Martin B. J. (2004) Circ. Res. 95, 9–20 [DOI] [PubMed] [Google Scholar]
- 4. Prockop D. J., Gregory C. A., Spees J. L. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11917–11923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Charbord P., Livne E., Gross G., Häupl T., Neves N. M., Marie P., Bianco P., Jorgensen C. (2011) Stem Cell Rev. 7, 32–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lian J. B., Javed A., Zaidi S. K., Lengner C., Montecino M., van Wijnen A. J., Stein J. L., Stein G. S. (2004) Crit. Rev. Eukaryot. Gene Expr. 14, 1–41 [PubMed] [Google Scholar]
- 7. Marie P. J., Fromigué O. (2006) Regen. Med. 1, 539–548 [DOI] [PubMed] [Google Scholar]
- 8. Cheng S. L., Yang J. W., Rifas L., Zhang S. F., Avioli L. V. (1994) Endocrinology 134, 277–286 [DOI] [PubMed] [Google Scholar]
- 9. Franceschi R. T. (1999) Crit. Rev. Oral Biol. Med. 10, 40–57 [DOI] [PubMed] [Google Scholar]
- 10. Phimphilai M., Zhao Z., Boules H., Roca H., Franceschi R. T. (2006) J. Bone Miner. Res. 21, 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kratchmarova I., Blagoev B., Haack-Sorensen M., Kassem M., Mann M. (2005) Science 308, 1472–1477 [DOI] [PubMed] [Google Scholar]
- 12. Miraoui H., Oudina K., Petite H., Tanimoto Y., Moriyama K., Marie P. J. (2009) J. Biol. Chem. 284, 4897–4904 [DOI] [PubMed] [Google Scholar]
- 13. Ryan P. E., Davies G. C., Nau M. M., Lipkowitz S. (2006) Trends Biochem. Sci. 31, 79–88 [DOI] [PubMed] [Google Scholar]
- 14. Sanjay A., Horne W. C., Baron R. (2001) Sci. STKE 2001, pe40. [DOI] [PubMed] [Google Scholar]
- 15. Schmidt M. H., Dikic I. (2005) Nat. Rev. Mol. Cell Biol. 6, 907–918 [DOI] [PubMed] [Google Scholar]
- 16. Thien C. B., Langdon W. Y. (2001) Nat. Rev. Mol. Cell Biol. 2, 294–307 [DOI] [PubMed] [Google Scholar]
- 17. Miyake S., Lupher M. L., Jr., Andoniou C. E., Lill N. L., Ota S., Douillard P., Rao N., Band H. (1997) Crit. Rev. Oncog. 8, 189–218 [DOI] [PubMed] [Google Scholar]
- 18. Thien C. B., Langdon W. Y. (1997) Oncogene 14, 2239–2249 [DOI] [PubMed] [Google Scholar]
- 19. Bonita D. P., Miyake S., Lupher M. L., Jr., Langdon W. Y., Band H. (1997) Mol. Cell. Biol. 17, 4597–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kaabeche K., Lemonnier J., Le Mée S., Caverzasio J., Marie P. J. (2004) J. Biol. Chem. 279, 36259–36267 [DOI] [PubMed] [Google Scholar]
- 21. Miyake S., Lupher M. L., Jr., Druker B., Band H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7927–7932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Miyake S., Mullane-Robinson K. P., Lill N. L., Douillard P., Band H. (1999) J. Biol. Chem. 274, 16619–16628 [DOI] [PubMed] [Google Scholar]
- 23. Ueno H., Sasaki K., Miyagawa K., Honda H., Mitani K., Yazaki Y., Hirai H. (1997) J. Biol. Chem. 272, 8739–8743 [DOI] [PubMed] [Google Scholar]
- 24. Swaminathan G., Tsygankov A. Y. (2006) J. Cell. Physiol. 209, 21–43 [DOI] [PubMed] [Google Scholar]
- 25. Kassenbrock C. K., Anderson S. M. (2004) J. Biol. Chem. 279, 28017–28027 [DOI] [PubMed] [Google Scholar]
- 26. Lupher M. L., Jr., Reedquist K. A., Miyake S., Langdon W. Y., Band H. (1996) J. Biol. Chem. 271, 24063–24068 [DOI] [PubMed] [Google Scholar]
- 27. Lupher M. L., Jr., Songyang Z., Shoelson S. E., Cantley L. C., Band H. (1997) J. Biol. Chem. 272, 33140–33144 [DOI] [PubMed] [Google Scholar]
- 28. Langdon W. Y., Blake T. J. (1990) Curr. Top. Microbiol. Immunol. 166, 159–164 [DOI] [PubMed] [Google Scholar]
- 29. Dufour C., Guenou H., Kaabeche K., Bouvard D., Sanjay A., Marie P. J. (2008) Bone 42, 1032–1039 [DOI] [PubMed] [Google Scholar]
- 30. Guenou H., Kaabeche K., Dufour C., Miraoui H., Marie P. J. (2006) Am. J. Pathol. 169, 1303–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kaabeche K., Guenou H., Bouvard D., Didelot N., Listrat A., Marie P. J. (2005) J. Cell Sci. 118, 1223–1232 [DOI] [PubMed] [Google Scholar]
- 32. Gronthos S., Fitter S., Diamond P., Simmons P. J., Itescu S., Zannettino A. C. (2007) Stem Cells Dev. 16, 953–963 [DOI] [PubMed] [Google Scholar]
- 33. Oyajobi B. O., Lomri A., Hott M., Marie P. J. (1999) J. Bone Miner. Res. 14, 351–361 [DOI] [PubMed] [Google Scholar]
- 34. Ahdjoudj S., Lasmoles F., Oyajobi B. O., Lomri A., Delannoy P., Marie P. J. (2001) J. Cell. Biochem. 81, 23–38 [DOI] [PubMed] [Google Scholar]
- 35. Dardalhon V., Herpers B., Noraz N., Pflumio F., Guetard D., Leveau C., Dubart-Kupperschmitt A., Charneau P., Taylor N. (2001) Gene Ther. 8, 190–198 [DOI] [PubMed] [Google Scholar]
- 36. Orosco A., Fromigué O., Bazille C., Entz-Werle N., Levillain P., Marie P. J., Modrowski D. (2007) Cancer Res. 67, 3708–3715 [DOI] [PubMed] [Google Scholar]
- 37. Fromigué O., Haÿ E., Modrowski D., Bouvet S., Jacquel A., Auberger P., Marie P. J. (2006) Cell Death Differ. 13, 1845–1856 [DOI] [PubMed] [Google Scholar]
- 38. Fromigué O., Kheddoumi N., Lomri A., Marie P. J., Body J. J. (2001) J. Bone Miner. Res. 16, 1600–1610 [DOI] [PubMed] [Google Scholar]
- 39. Schlessinger J. (2004) Science 306, 1506–1507 [DOI] [PubMed] [Google Scholar]
- 40. Wong A., Lamothe B., Lee A., Schlessinger J., Lax I., Li A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6684–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mason J. M., Morrison D. J., Basson M. A., Licht J. D. (2006) Trends Cell Biol. 16, 45–54 [DOI] [PubMed] [Google Scholar]
- 42. Fong C. W., Leong H. F., Wong E. S., Lim J., Yusoff P., Guy G. R. (2003) J. Biol. Chem. 278, 33456–33464 [DOI] [PubMed] [Google Scholar]
- 43. Wong E. S., Fong C. W., Lim J., Yusoff P., Low B. C., Langdon W. Y., Guy G. R. (2002) EMBO J. 21, 4796–4808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Garrett I. R., Chen D., Gutierrez G., Zhao M., Escobedo A., Rossini G., Harris S. E., Gallwitz W., Kim K. B., Hu S., Crews C. M., Mundy G. R. (2003) J. Clin. Invest. 111, 1771–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Giuliani N., Morandi F., Tagliaferri S., Lazzaretti M., Bonomini S., Crugnola M., Mancini C., Martella E., Ferrari L., Tabilio A., Rizzoli V. (2007) Blood 110, 334–338 [DOI] [PubMed] [Google Scholar]
- 46. Mukherjee S., Raje N., Schoonmaker J. A., Liu J. C., Hideshima T., Wein M. N., Jones D. C., Vallet S., Bouxsein M. L., Pozzi S., Chhetri S., Seo Y. D., Aronson J. P., Patel C., Fulciniti M., Purton L. E., Glimcher L. H., Lian J. B., Stein G., Anderson K. C., Scadden D. T. (2008) J. Clin. Invest. 118, 491–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Molero J. C., Jensen T. E., Withers P. C., Couzens M., Herzog H., Thien C. B., Langdon W. Y., Walder K., Murphy M. A., Bowtell D. D., James D. E., Cooney G. J. (2004) J. Clin. Invest. 114, 1326–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Visser Smit G. D., Place T. L., Cole S. L., Clausen K. A., Vemuganti S., Zhang G., Koland J. G., Lill N. L. (2009) Sci. Signal. 2, ra86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Waterman H., Levkowitz G., Alroy I., Yarden Y. (1999) J. Biol. Chem. 274, 22151–22154 [DOI] [PubMed] [Google Scholar]
- 50. Marie P. J. (2003) Gene 316, 23–32 [DOI] [PubMed] [Google Scholar]
- 51. Gruber R., Karreth F., Kandler B., Fuerst G., Rot A., Fischer M. B., Watzek G. (2004) Platelets 15, 29–35 [DOI] [PubMed] [Google Scholar]
- 52. Cárcamo-Orive I., Gaztelumendi A., Delgado J., Tejados N., Dorronsoro A., Fernández-Rueda J., Pennington D. J., Trigueros C. (2010) J. Bone Miner. Res. 25, 2115–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mitlak B. H., Finkelman R. D., Hill E. L., Li J., Martin B., Smith T., D'Andrea M., Antoniades H. N., Lynch S. E. (1996) J. Bone Miner. Res. 11, 238–247 [DOI] [PubMed] [Google Scholar]
- 54. Moenning A., Jäger R., Egert A., Kress W., Wardelmann E., Schorle H. (2009) Mol. Cell. Biol. 29, 881–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miraoui H., Ringe J., Häupl T., Marie P. J. (2010) Hum. Mol. Genet. 19, 1678–1689 [DOI] [PubMed] [Google Scholar]
- 56. Duval M., Bédard-Goulet S., Delisle C., Gratton J. P. (2003) J. Biol. Chem. 278, 20091–20097 [DOI] [PubMed] [Google Scholar]
- 57. Rahimi N. (2009) Biochem. Soc. Trans. 37, 1189–1192 [DOI] [PubMed] [Google Scholar]
- 58. Suzue N., Nikawa T., Onishi Y., Yamada C., Hirasaka K., Ogawa T., Furochi H., Kosaka H., Ishidoh K., Gu H., Takeda S., Ishimaru N., Hayashi Y., Yamamoto H., Kishi K., Yasui N. (2006) J. Bone Miner. Res. 21, 722–734 [DOI] [PubMed] [Google Scholar]
- 59. Franceschi R. T., Xiao G. (2003) J. Cell. Biochem. 88, 446–454 [DOI] [PubMed] [Google Scholar]
- 60. Jaiswal R. K., Jaiswal N., Bruder S. P., Mbalaviele G., Marshak D. R., Pittenger M. F. (2000) J. Biol. Chem. 275, 9645–9652 [DOI] [PubMed] [Google Scholar]
- 61. Miraoui H., Marie P. J. (2010) Sci. Signal. 3, re9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.