Abstract
Regulation of RANKL (receptor activator of nuclear factor κB ligand)-induced osteoclast differentiation is of current interest in the development of antiresorptive agents. Osteoclasts are multinucleated cells that play a crucial role in bone resorption. In this study, we investigated the effects of N-methylpyrrolidone (NMP) on the regulation of RANKL-induced osteoclastogenesis. NMP inhibited RANKL-induced tartrate-resistant acid phosphatase activity and the formation of tartrate-resistant acid phosphatase-positive multinucleated cells. The RANKL-induced expression of NFATc1 (nuclear factor of activated T cells, cytoplasmic 1) and c-Fos, which are key transcription factors for osteoclastogenesis, was also reduced by treatment with NMP. Furthermore, NMP induced disruption of the actin rings and decreased the mRNAs of cathepsin K and MMP-9 (matrix metalloproteinase-9), both involved in bone resorption. Taken together, these results suggest that NMP inhibits osteoclast differentiation and attenuates bone resorption. Therefore, NMP could prove useful for the treatment of osteoporosis or other bone diseases associated with excessive bone resorption.
Keywords: Bone, Fos, Jun Transcription Factor, NF-κB Transcription Factor, NFAT Transcription Factor, Osteoclast Acidification, p38 MAPK, Tumor Necrosis Factor (TNF), Osteoclast Maturation, Osteoporosis
Introduction
Bone remodeling is a physiological process that involves the resorption and synthesis of bone by osteoclasts and osteoblasts, respectively (1, 2). Osteoclasts are known to be formed by the fusion of hematopoietic cells of the monocyte-macrophage lineage during the early stage of the differentiation process (3). This process consists of multiple steps, including differentiation of osteoclast precursors into mononuclear osteoclasts, fusion of mononuclear preosteoclasts into mature multinucleated osteoclasts, and activation of osteoclasts to resorb bone (4–7). The terminal differentiation in this lineage is characterized by acquisition of mature phenotypic markers such as expression of tartrate-resistant acid phosphatase (TRAP),2 the calcitonin receptor, MMP-9, and cathepsin K, as well as morphological conversion into large multinucleated cells and the ability to form resorption lacunae on bone (8–10). The essential signaling molecules for osteoclast differentiation include RANKL and M-CSF (macrophage colony-stimulating factor) in bone marrow-derived macrophage precursor cells (11, 12). RANKL is a member of the TNF superfamily that is expressed in osteoblasts. It interacts with the osteoclast cell surface receptor RANK, which in turn recruits TNF receptor-associated factors and plays a crucial role in the osteoclast differentiation axis (13, 14). The downstream intracellular signaling mediated by RANK in osteoclast progenitor cells includes TRAF6 (TNF receptor-associated factor 6)-dependent activation of NF-κB via the IκB kinase complex and MAPKs such as ERK, p38 MAPK, and JNK (7, 11, 15). In addition, RANKL induces the key transcription factors for osteoclastogenesis, NFATc1 and c-Fos (9, 16, 17). Therefore, chemical or natural compounds that specifically inhibit these steps could be developed as antiresorptive drugs for the treatment of metabolic bone disorders characterized by excessive osteoclastic bone resorption.
Recently, we found that N-methylpyrrolidone (NMP) is a bioactive drug because it enhances bone regeneration in vivo in a rabbit calvarial defect model (18). At the cellular level, the pharmaceutical effect of NMP was confirmed because NMP increased early and late markers for maturation of preosteoblasts and human bone marrow-derived stem cells in vitro (18).
In this study, we investigated the effects of NMP on RANKL-induced osteoclast differentiation in vitro and characterized the role of NMP in osteoclast differentiation. We provide the first evidence that NMP inhibits RANKL-stimulated osteoclastogenesis by suppressing NFATc1 expression and RANKL-induced osteoclast function by disturbing actin ring formation and decreasing MMP-9 activity.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Recombinant RANKL was purchased from Invitrogen. Anti-NFATc1 (H-110) and anti-c-Fos (H-125) polyclonal antibodies were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Anti-ERK1/2 and anti-phospho-ERK1/2 polyclonal antibodies were obtained from Cell Signaling Technology. The TransAM AP-1 (activator protein 1)/c-Fos transcription factor kit was from Active Motif (Rixensart, Belgium). The RNA extraction kit (RNeasy kit) was from Qiagen. The BCA kit for protein determination was from Pierce. TRAP solution was from Sigma. All other chemicals were obtained from Sigma.
Cell Cultures
RAW264.7 cells were cultured in DMEM supplemented with 10% FBS and antibiotics (100 units/ml penicillin G and 100 mg/ml streptomycin). The cultures were never allowed to become confluent. Incubations were performed at 37 °C in 5% CO2 in humidified air. Bone marrow-derived macrophages (BMMs) were isolated from the long bones of 6-week-old mice and were maintained in α-minimal essential medium containing 10% heat-inactivated FBS in the presence of M-CSF (100 ng/ml) as described previously (19). To generate osteoclasts from BMMs, cells were plated in 24-well tissue culture plates and cultured in the presence of 25 ng/ml RANKL and 25 ng/ml M-CSF.
Cell Viability and Proliferation Assay
The effect of different concentrations of NMP on RAW264.7 cell proliferation/viability was analyzed using a nonradioactive WST-1 cell proliferation assay kit (Roche Diagnostics) according to the manufacturer's instruction.
TRAP Activity and TRAP Staining
RAW264.7 cells were plated in a 12-well culture dish (Corning) with different concentrations of NMP in the presence of 25 ng/ml RANKL. The medium and factors were replaced every 2 days. After 6 days of culture, the medium was removed, and the cell monolayer was gently washed twice with PBS. The cells were then lysed with 200 μl of 0.1% Triton X-100. TRAP activity in the cell lysate was determined using TRAP solution (0.1 m sodium acetate (pH 5.8), 1 mm ascorbic acid, 0.15 m KCl, 10 mm disodium tartrate, and 10 mm p-nitrophenyl phosphate). An aliquot of the cell lysate was added to TRAP solution and subsequently incubated for 30 min at 37 °C. The reaction was stopped with 0.3 n NaOH, and the absorbance was measured at 405 nm using a Synergy HT microplate reader (BioTek). Results (normalized to protein content) are expressed as a percentage of the activity obtained in RANKL-stimulated cells.
TRAP histochemical staining of the cells was performed using a leukocyte acid phosphatase kit (Sigma). Cultured cells were fixed with formaldehyde for 5 min at room temperature, washed with PBS, and air-dried. After TRAP staining, TRAP-positive multinucleated cells (more than three nuclei) were counted under a phase-contrast microscope.
Pit Formation Assay
RAW264.7 cells were placed on bone slices in 24-well plates. After preincubation for 6 h, the bone slices were transferred to 12-well plates (one bone slice/well) and further cultured in the presence or absence of 25 ng/ml RANKL and 5 mm NMP. The medium was replaced with fresh medium every 2 days. After 9 days of culture, cells were removed, and the pit area was visualized by toluidine blue staining and quantified using an image analysis system. Resorption pits of representative cultures are shown.
Actin Ring Formation Assay
Actin rings of osteoclasts were detected by staining actin filaments with rhodamine-conjugated phalloidin. Osteoclasts were formed from RAW264.7 cell cultures in the presence of RANKL (25 ng/ml) and NMP (1, 5, and 10 mm). At the end of incubation, osteoclasts were stained with rhodamine-conjugated phalloidin for actin and with DAPI for the nucleus. The distribution of actin rings was visualized and detected under a fluorescence microscope.
Quantitative Real-time Reverse Transcription-PCR
RNA from RAW264.7 cells was extracted using the RNeasy kit. The mRNA was reverse-transcribed into cDNA. The resultant cDNA was subjected to real-time PCR with gene-specific primers using iQ SYBR Green Supermix and an iCycler real-time PCR machine (both from Bio-Rad) according to the manufacturer's instructions. The primer sequences are presented in Table 1.
TABLE 1.
Primer sequences used in real-time PCR
| Gene | Sequence (5′ → 3′) | Amplicon size |
|---|---|---|
| bp | ||
| GAPDH | ||
| Forward | AGGTCGGTGTGAACGGATTTG | 123 |
| Reverse | TGTAGACCATGTAGTTGAGGTCA | |
| NFATc1 | ||
| Forward | GGAGCGGAGAAACTTTGCG | 94 |
| Reverse | GTGACACTAGGGGACACATAACT | |
| c-Fos | ||
| Forward | CGGGTTTCAACGCCGACTA | 166 |
| Reverse | TTGGCACTAGAGACGGACAGA | |
| MMP-9 | ||
| Forward | CTGGACAGCCAGACACTAAAG | 145 |
| Reverse | CTCGCGGCAAGTCTTCAGAG | |
| Cathepsin K | ||
| Forward | GAAGAAGACTCACCAGAAGCAG | 102 |
| Reverse | TCCAGGTTATGGGCAGAGATT |
Protein Preparation and Western Blot Analysis
RAW264.7 cells treated with the different compounds were rapidly frozen in liquid nitrogen and stored at −80 °C for further analysis. Cells were lysed as described previously (20). Proteins were fractionated onto a 12% SDS-polyacrylamide gel, transferred to Immobilon P membranes (Millipore), and immunoblotted with specific antibodies. Detection was performed using peroxidase-coupled secondary antibody, an enhanced chemiluminescence reaction (Amersham Biosciences ECL Western blotting detection reagents, GE Healthcare Europe GmbH, Otelfingen, Switzerland), and visualization by autoradiography. Membranes that were reprobed had been stripped in stripping buffer (62.5 mm Tris-HCl (pH 6.8), 2% (w/v) SDS, and 100 mm β-mercaptoethanol) according to the manufacturer's protocol (Millipore).
Zymography
Equal volumes of conditioned medium were loaded onto a 10% SDS-polyacrylamide gel containing 0.1% porcine gelatin (Sigma). After electrophoresis, the gels were washed twice for 15 min each with 2.5% Triton X-100 and then incubated overnight at 37 °C in substrate buffer (50 mm Tris-HCl, 0.2 m NaCl, 5 mm CaCl2, and 0.02% Brij 35). The gels were stained with Coomassie Blue R-250 (Sigma) for 1 h. The gels were destained briefly with 50% methanol and 10% acetic acid. Areas of gelatinolytic activity appeared to be clear against the blue background of the blue-stained undigested gelatin.
AP-1 Activation Analysis
To detect AP-1 activation in RAW264.7 cells, we used the ELISA-based TransAM AP-1 transcription factor kit (Active Motif). Cells were plated in Petri dishes for 48 h. Cells were preincubated for 4 h in serum-free fresh medium and stimulated for 30 min as indicated in the figures. Preparation of nuclear cell extract was done according to the manufacturer's instructions. The active form of c-Fos in nuclear extracts can be detected using an antibody specific for an epitope that is accessible only when the nuclear factor is activated and bound to its target DNA. By using an antibody directed against c-Fos, the AP-1 dimer bound to the oligonucleotide is detected. A secondary antibody conjugated to horseradish peroxidase provides colorimetric visualization. Absorbance was determined with a Synergy HT microplate reader.
Statistical Analysis
Experiments were carried out independently at least three times. Results are expressed as the mean ± S.D. and were compared by Student's t test. Results were considered significantly different for p < 0.05.
RESULTS
NMP Suppresses RANKL-induced Osteoclastogenesis
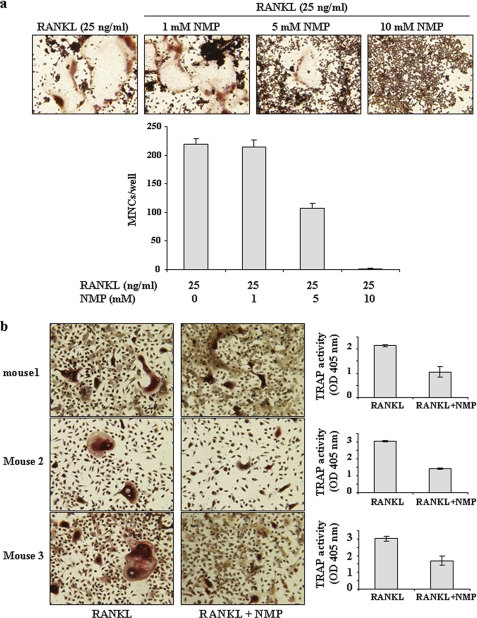
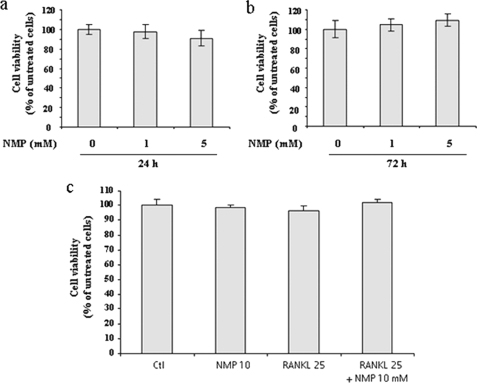
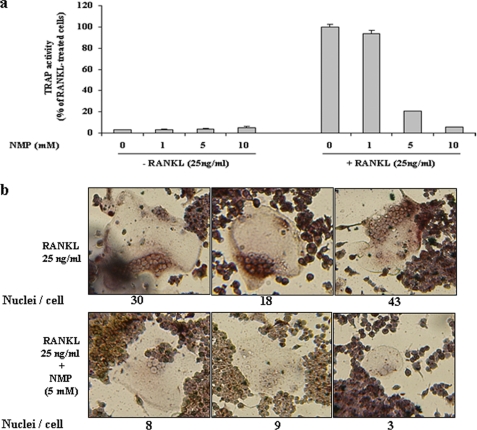
To clarify the effects of NMP on osteoclastogenesis, we used RAW264.7 cells and BMMs (Fig. 1). Cells were incubated with NMP in the presence of RANKL for RAW264.7 cells (Fig. 1a) or with NMP in the presence of RANKL and M-CSF for BMM cells (Fig. 1b) as indicated. In the absence of NMP, RAW264.7 cells differentiated into mature TRAP-positive multinucleated cells (MNCs), whereas NMP (5 and 10 mm) reduced the formation and numbers of TRAP-positive MNCs in a concentration-dependent manner (Fig. 1a). The number of MNCs was quantified microscopically. In the presence of NMP (5 mm), the MNC number induced by RANKL was reduced by ∼50%, whereas 10 mm NMP completely abolished the formation of MNCs. In BMM cells, NMP appeared to have the same effect (Fig. 1b). Indeed, treatment of BMMs with RANKL and M-CSF induced the formation of TRAP-positive MNCs and increased TRAP activity. NMP treatment completely blocked this effect. To exclude the possibility that the inhibition was due to cytotoxicity of NMP, cell cytotoxicity/viability was analyzed using a nonradioactive WST-1 assay (Fig. 2). NMP demonstrated no cytotoxic effects after 24 and 72 h of treatment at the concentration that effectively inhibited osteoclast formation by 50% (5 mm). We next examined in more detail the effect of NMP on TRAP activity induced by RANKL in RAW264.7 cells (Fig. 3). We found that TRAP activity was significantly reduced in the cells treated with RANKL and NMP compared with the cells treated with RANKL alone (Fig. 3a). Furthermore, NMP dramatically reduced the number of nuclei per osteoclast (Fig. 3b), suggesting that NMP could modulate the fusion process.
FIGURE 1.
Effects of NMP on osteoclastogenesis. a, RAW264.7 cells were seeded in 12-well culture plates and treated for 6 days with NMP, RANKL, or both as indicated. For visualization of TRAP-positive MNCs, the cells were fixed and stained for TRAP activity as described under “Experimental Procedures.” Stained cells were photographed, and TRAP-positive MNCs containing three or more nuclei were counted as osteoclasts. Data are expressed as the mean ± S.D. (n = 3). b, BMMs from three different mice were treated with NMP (5 mm) in the presence of M-CSF (25 ng/ml) and RANKL (25 ng/ml) for 4 days. After culturing, the cells were fixed and stained for TRAP. In parallel, TRAP activity was determined as described under “Experimental Procedures.” Data are expressed as the mean ± S.D. (n = 4) from a representative experiment.
FIGURE 2.
NMP had no cytotoxic effect on RAW264.7 cells. Cells were seeded on a 96-well plate and treated for either 24 h (a) or 72 h (b) with different concentrations of NMP or with RANKL and NMP (c) as indicated. Cell viability/cytotoxicity was measured using WST-1 reagent as described under “Experimental Procedures.” The results were normalized to cells grown in DMEM alone (control (Ctl)). Data are expressed as the mean ± S.D. (n = 4) from a representative experiment.
FIGURE 3.
NMP suppresses osteoclast differentiation and decreases the fusion process. a, TRAP activity. b, microscopic view of MNCs after TRAP staining. RAW264.7 cells were seeded on a 24-well culture plate and treated with RANKL alone or with different concentrations of NMP as indicated. After 6 days of incubation, TRAP activity was measured as described under “Experimental Procedures.” Data are expressed as the mean ± S.D. (n = 3). In parallel, cells were stained for TRAP after differentiation into mature osteoclasts.
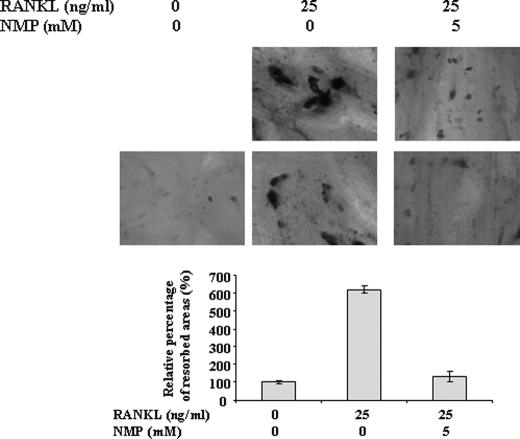
NMP Inhibits Bone Resorption and Actin Ring Formation
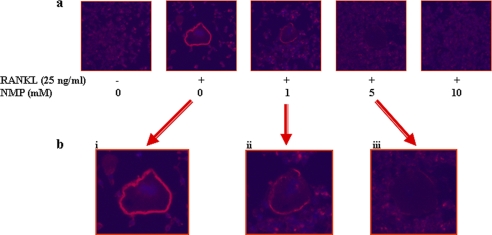
We further examined if NMP has an effect on the ability of mature osteoclasts to resorb bone. RAW264.7 cells were plated on bone slices and stimulated with RANKL in the presence or absence of NMP. RANKL-stimulated cells formed a number of pits (Fig. 4), suggesting that the bone resorption activity of RANKL-treated cells made them into functionally active state-resembling osteoclasts. Treatment with 5 mm NMP significantly reduced the formation of resorption pits in number and in overall area compared with treatment with RANKL alone. Bone resorption occurs within the sealing zone, which is formed by an actin ring structure. To investigate the effect of NMP on actin ring formation, immunofluorescence analysis was performed (Fig. 5). The majority of RANKL-treated cells revealed well formed actin rings (Fig. 5a). Cells treated with RANKL in the presence of 5 mm NMP displayed mainly disrupted actin rings (Fig. 5b). As expected, cells treated with RANKL in the presence of 10 mm NMP showed no actin rings compared with cells treated with RANKL alone. Wilson et al. (21) reported that osteoclasts displaying a full actin ring or disrupted actin rings with >50% intact were identified as active. Our results are in line with this observation because with 1 mm NMP, the concentration that ineffectively inhibited osteoclast differentiation, the actin ring was not completely disrupted (Fig. 5b).
FIGURE 4.
NMP inhibits RANKL-induced bone resorption. RAW264.7 cells were cultured on bone slices and treated as indicated. After 9 days of culture, the cells were completely removed, and the bone slices were stained with toluidine blue, followed by photography (upper). Histograms represent the percentage of the resorbed area (lower). Data are expressed as the mean ± S.D. (n = 3) from a representative experiment.
FIGURE 5.
NMP disrupts RANKL-induced actin ring formation. RAW264.7 cells were differentiated into osteoclasts in the presence of RANKL alone or with 1, 5, or 10 mm NMP for 6 days. Cells were fixed and stained for actin rings as described under “Experimental Procedures.”
NMP Suppresses RANKL-induced MMP-9 and Cathepsin K
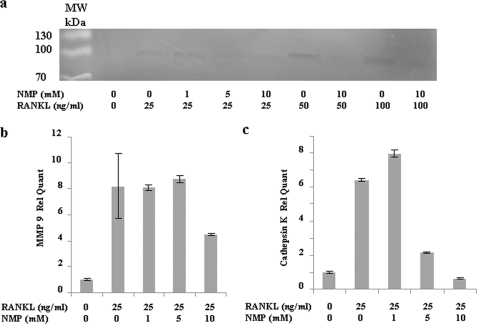
The bone resorption-related enzymes MMP-9 and cathepsin K are highly expressed in osteoclastic cells and play an important role in skeletal remodeling (22, 23). Therefore, we investigated the effect of NMP on the expression of MMP-9 and cathepsin K expression (Fig. 6) in detail. First, we evaluated MMP-9 activity by gelatin zymography (Fig. 6a). RAW264.7 cells treated with RANKL showed a concentration-dependent increase in MMP-9 gelatinolytic activity. This gelatinolytic activity was significantly decreased by NMP treatment and correlated with the suppression of osteoclast differentiation visualized when the same cultures were stained for TRAP (data not shown). Fig. 6b demonstrates that RANKL induced an increase in MMP-9 mRNA and that only treatment with 10 mm NMP was able to decrease the RANKL-induced MMP-9 mRNA. Cathepsin K mRNA expression was also increased by RANKL treatment. In the presence of NMP (5 and 10 mm), RANKL-induced cathepsin K mRNA was significantly suppressed (Fig. 6b) already at 5 mm NMP.
FIGURE 6.
NMP inhibits RANKL-induced MMP-9 and cathepsin K expression. a, representative gelatin zymography assay of MMP-9 activity in RAW264.7 cells. RAW264.7 cells were cultured in serum-free DMEM supplemented with SITE liquid medium (Sigma) as indicated. After 48 h of stimulation, aliquots from supernatants were used in zymography assays as described under “Experimental Procedures.” b and c, MMP-9 and cathepsin K mRNAs were determined by real-time PCR after 48 h of stimulation with RANKL alone or in the presence of NMP (as indicated). Data are expressed as the mean ± S.D. (n = 3) from a representative experiment. Rel Quant, relative quantity.
NMP Inhibits NFATc1 and c-Fos Expression and Decreases AP-1 Activation
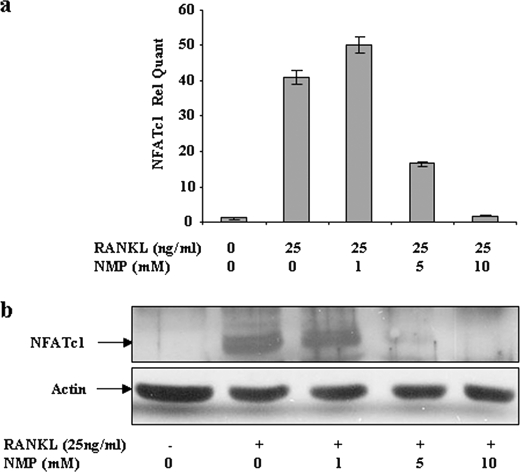
NFATc1, a member of the NFAT family of transcription factor, has been shown to be up-regulated after RANKL stimulation and is important for osteoclast differentiation (10, 16). Therefore, we examined the effect of NMP on the expression of NFATc1 (Fig. 7). Stimulation of RAW264.7 cells with RANKL induced a high level of expression of NFATc1 mRNA (Fig. 7a). Treatment with NMP (5 mm) significantly decreased RANKL-induced NFATc1 mRNA. RANKL-induced NFATc1 mRNA was completely blocked by 10 mm NMP treatment. As shown in Fig. 7b, cellular protein expression correlated with mRNA expression. Indeed, RANKL-induced NFATc1 expression was significantly and concentration-dependently attenuated by NMP treatment. With 10 mm NMP, the expression of NFATc1 induced by RANKL was similar to that in untreated cells.
FIGURE 7.
NMP suppresses RANKL-induced NFATc1 expression. RAW264.7 cells were stimulated with RANKL alone or with different concentrations of NMP as indicated (48 h for mRNA and 72 h for proteins). a, total RNA was extracted, and NFATc1 mRNA was determined by real-time PCR as described under “Experimental Procedures.” Data are expressed as the mean ± S.D. (n = 3) from a representative experiment. b, whole cells were subjected to Western blot analysis with antibody against NFATc1. Anti-actin antibody was used to visualize the loading control. Rel Quant, relative quantity.
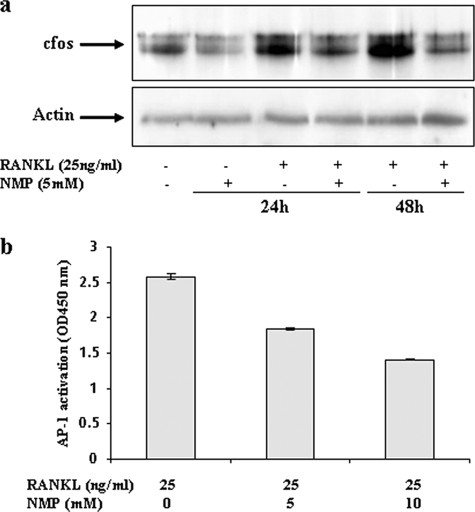
It is well known that c-Fos, a transcription factor for the AP-1 complex, is required for osteoclast differentiation. This factor has been shown to bind to the NFATc1 promoter and to be important for its activation (17, 24). As shown in Fig. 8a, c-Fos expression was increased by RANKL. NMP treatment (5 mm) significantly decreased c-Fos expression induced by RANKL. Next, we examined whether NMP could suppress AP-1 activity by TransAM AP-1 assays (Fig. 8b). The results show that AP-1 transcriptional activity was increased when cells were exposed to RANKL, whereas NMP suppressed AP-1 activity, suggesting that NMP can inhibit RANKL-induced AP-1 activation. The specificity of the assay was confirmed by using positive control, wild-type, and mutant oligonucleotides (data not shown).
FIGURE 8.
NMP inhibits RANKL-mediated c-Fos expression and AP-1 activation. RAW264.7 cells were treated with RANKL alone or with NMP as indicated. a, the c-Fos protein level was analyzed by Western blotting using anti-c-Fos and anti-actin antibodies as described under “Experimental Procedures.” b, nuclear extracts from RAW264.7 cells were used to detect AP-1 activation with an ELISA-based method (TransAM AP-1) as described under “Experimental Procedures.” The specificity of the assay was monitored by using the K-562 (12-O-tetradecanoylphorbol-13-acetate-stimulated) nuclear extract and free wild-type or mutant oligonucleotides according to the manufacturer's instructions. Data are expressed as the mean ± S.D. (n = 4) from two different experiments.
NMP Inhibits RANKL-induced ERK Pathway Activation
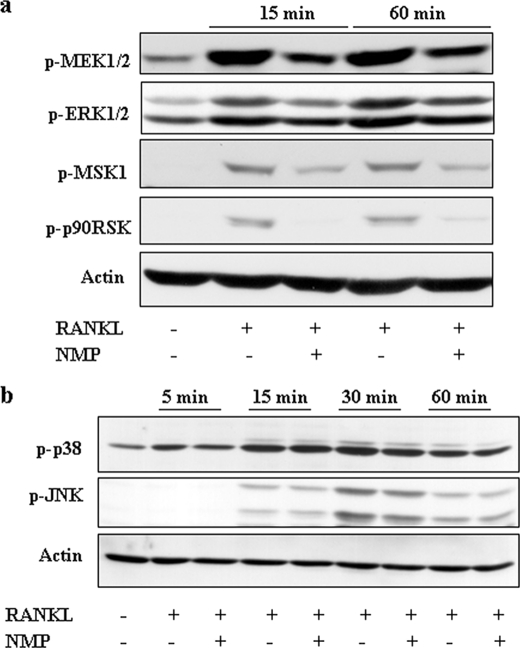
Growth factors that stimulate ERK may ultimately control the activity of c-Fos and AP-1-dependent transcription (25). Recently, it was reported that MIP1α (macrophage inflammatory protein 1α) induces osteoclast formation by activating the MEK/ERK/c-Fos pathway (26) and that vitamin E inhibits RANKL-induced osteoclast differentiation from precursors by suppression of c-Fos expression, possibly through inhibiting ERK (27). To investigate the role of the ERK, JNK, and p38 pathways in the inhibitory effect of NMP, RAW264.7 cells were treated as indicated in Fig. 9a, and whole cell extract was subjected to Western blotting using anti-phospho-ERK pathway antibodies. RANKL treatment induced ERK1/2 phosphorylation, and the presence of NMP resulted in a decrease in ERK1/2 phosphorylation. We also examined the signaling steps upstream and downstream of ERK1/2. As shown in Fig. 9a, NMP suppressed the RANKL-induced phosphorylation of MEK1/2 as well as that of two downstream targets of ERK1/2, p90RSK and MSK1. NMP reduced the RANKL-induced activation of ERK but not that of p38 and JNK (Fig. 9b), suggesting that the effect of NMP on the ERK pathway is specific.
FIGURE 9.
NMP inhibits RANKL-induced ERK activation but not p38 and JNK activation. a, RAW264.7 cells were stimulated as indicated, and whole cell extracts were separated by SDS-PAGE, transferred to PVDF membranes, and probed sequentially with anti-phospho-ERK1/2 pathway antibodies. b, RAW264.7 cells were stimulated with RANKL alone or with 10 mm NMP for the indicated time points, and the activities of p38 and JNK were examined by Western blot analysis using phospho-specific (p) antibodies. Anti-actin antibody was used as a loading control.
DISCUSSION
The major modalities currently used in osteoporosis treatment include primarily estrogen replacement therapy along with bisphosphonates, selective estrogen receptor modulators, and calcitonin. However, such therapies are associated with adverse effects, including breast cancer, hypercalcemia, and hypertension (5, 28–32).
In medicine, NMP has a long track record as a constituent in medical devices approved by the Food and Drug Administration and thus can be considered as a safe and biologically inactive small chemical. Recently, we revealed that NMP enhances bone morphogenetic protein activity by increasing the kinase activity of the bone morphogenetic protein receptor complex for Smad1, Smad5, Smad8, and p38 and could be employed as a potent drug for bone tissue regeneration and engineering (18). Bone remodeling is tightly regulated by two processes: bone formation and bone resorption. The balance between both processes is key for maintaining bone density. In this study, we have reported that, in addition to its role in bone formation, NMP is also able to inhibit osteoclast differentiation in vitro because NMP attenuates the formation of TRAP-positive MNCs from precursor cells stimulated with RANKL. Our results suggest that NMP inhibits MNC formation, including fusion of the mononuclear precursor cells, because it decreases the number of nuclei per MNC. The number of nuclei per MNC should reflect the relative rate of fusion of mononuclear precursors to form MNCs.
NFATc1 and c-Fos are crucial transcription factors in RANKL-induced osteoclastogenesis. The role of the c-Fos transcription factor in osteoclastogenesis has been revealed by knock-out experiments (33). c-Fos knock-out mice exhibit a serious osteopetrotic phenotype due to a failure to form osteoclasts. In addition, previous reports demonstrated that NFATc1 is not induced by RANKL stimulation in osteoclasts lacking c-Fos. The importance of NFATc1 in osteoclastogenesis was supported by in vitro experiments in which NFATc1 knock-out mouse-derived stem cells failed to differentiate into osteoclasts and BMM cells, when forced to express NFATc1, differentiated to osteoclasts even in the absence of RANKL (24, 33). In our study, NMP significantly suppressed the NFATc1 up-regulation normally seen after RANKL treatment. Previous studies have shown that NFATc1, the master regulator of osteoclastogenesis, is regulated by the AP-1 complex. AP-1 is a dimeric transcription factor composed of members of the Jun and Fos protein family. AP-1 converts extracellular signals in bone and immune cells into changes in the expression of specific target genes that harbor an AP-1-binding site(s) in their promoter or enhancer regions (34).
Because c-Fos up-regulation is needed for NFATc1 induction, it is possible that the suppression of NFATc1 expression by NMP is the consequence of the down-regulation of c-Fos, with the subsequent down-regulation of AP-1 activity and inhibition of osteoclast-specific gene expression normally required for efficient osteoclast differentiation and bone resorption. This finding is important because the AP-1 transcription factor links inflammatory processes to the activation of signaling pathways in osteoclasts and therefore can be considered an important contributor to inflammatory bone diseases.
Active osteoclasts display characteristic membranes, including the ruffled border, attachment zone, and basolateral secretory membrane. After attachment to bone, the ruffled border secretes enzymes and protons enabling the solubilization and digestion of the bone matrix. Rapid cytoskeletal reorganization is essential for osteoclast function and formation of the specialized membranes (21, 35). NMP treatment disrupted actin ring formation in osteoclasts. More importantly, osteoclastic bone resorption was strongly inhibited by NMP. Moreover, NMP inhibited the RANKL-induced up-regulation of MMP-9 and cathepsin K, both highly expressed in osteoclastic cells and playing an important role in skeletal remodeling. Recently, Wilson et al. (21) reported that cathepsin K activity is required for the initial formation of actin rings and thus for the activation of osteoclasts. Our results are in line with this observation because at 5 and 10 mm NMP, concentrations that effectively suppressed RANKL-induced cathepsin K mRNA production and inhibited osteoclast differentiation, the actin rings were completely disrupted. Together, our results indicate that NMP plays a critical role in osteoclastic bone resorption by regulating actin ring formation and decreasing bone resorption-related enzymes.
MAPK signaling cascades regulate transcription by multiple mechanisms, including the phosphorylation and activation of transcription factors, coactivators, corepressors, and the basal transcriptional machinery. Activated ERK1/2 and/or p90RSK has been shown to target c-Fos (36–38) as well as to participate in the activation of NF-κB (39). In this study, we have reported that NMP decreased RANKL-induced activation of the ERK pathway, and this inhibition might significantly reduce the induction of the c-Fos transcription factor and subsequently AP-1 activation by RANKL.
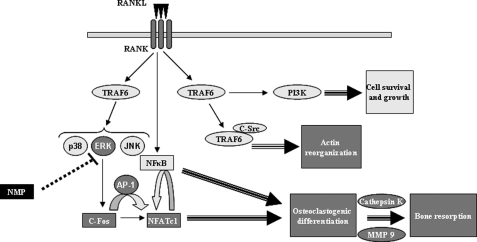
In this study, we have demonstrated for the first time the inhibitory effect of NMP on osteoclast differentiation and function. NMP inhibits RANKL-induced osteoclast differentiation and function. Accordingly, we propose a model to explain the bioactivity of NMP (Fig. 10). NMP might act as an inhibitor of several key events. In particular, NMP inhibits RANKL-induced ERK p42/44 phosphorylation, which is important for c-Fos expression and AP-1 activation. Thus, the expression of NFATc1, which is regulated by AP-1, is decreased by NMP treatment. NMP is also able to inhibit osteoclast function by disturbing actin ring formation and reducing the expression of MMP-9 and cathepsin K, both involved in bone digestion. Therefore, NMP inhibits osteoclast differentiation by modulating the AP-1 and NFATc1 transcription factors and inhibits osteoclast function by reducing MMP-9 and cathepsin K expression and disrupting actin ring formation.
FIGURE 10.
Schematic model for the osteoclastogenesis inhibitory function of NMP. RANKL induces activation of the ERK pathway. Inhibition of this pathway by NMP significantly reduces induction of the c-Fos transcription factor and subsequently AP-1 activation by RANKL. Blockade of the AP-1 transcription factor considerably decreases NFATc1 transcription factor expression, leading to suppressed osteoclastogenic gene expression and subsequently osteoclast formation and function.
Taken together, our results demonstrate that NMP has an inhibitory activity on both osteoclast differentiation and function through mechanisms involving inhibition of RANKL-induced AP-1 activation. Additional work is needed to further pinpoint the exact site(s) where NMP exerts its suppressive activity on osteoclasts.
Molecules such as NMP that enhance bone regeneration on the one hand and suppress osteoclast differentiation on the other may have great therapeutic value in treating osteoporosis and other bone-erosive diseases such as rheumatoid arthritis and metastasis associated with bone loss. Our findings suggest that NMP may potentially be useful for the treatment of bone diseases associated with excessive bone resorption.
Acknowledgments
We thank Dr. M. Ehrbar (UniversitätsSpital Zürich) for providing expert technical assistance with fluorescence microscopy. We also thank Yvonne Bloemhard and Alexandre Tchouboukov for excellent technical assistance.
This work was supported in part by Swiss National Science Foundation Grant 310000-116240.
- TRAP
- tartrate-resistant acid phosphatase
- M-CSF
- macrophage colony-stimulating factor
- NMP
- N-methylpyrrolidone
- BMM
- bone marrow-derived macrophage
- MNC
- multinucleated cell.
REFERENCES
- 1. Corral D. A., Amling M., Priemel M., Loyer E., Fuchs S., Ducy P., Baron R., Karsenty G. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13835–13840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Karsenty G., Wagner E. F. (2002) Dev. Cell 2, 389–406 [DOI] [PubMed] [Google Scholar]
- 3. Boyle W. J., Simonet W. S., Lacey D. L. (2003) Nature 423, 337–342 [DOI] [PubMed] [Google Scholar]
- 4. Chambers T. J. (2000) J. Pathol. 192, 4–13 [DOI] [PubMed] [Google Scholar]
- 5. Rodan G. A., Martin T. J. (2000) Science 289, 1508–1514 [DOI] [PubMed] [Google Scholar]
- 6. Teitelbaum S. L. (2000) Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 7. Teitelbaum S. L., Ross F. P. (2003) Nat. Rev. Genet. 4, 638–649 [DOI] [PubMed] [Google Scholar]
- 8. Motyckova G., Weilbaecher K. N., Horstmann M., Rieman D. J., Fisher D. Z., Fisher D. E. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5798–5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takayanagi H. (2007) Ann. N.Y. Acad. Sci. 1116, 227–237 [DOI] [PubMed] [Google Scholar]
- 10. Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 11. Darnay B. G., Haridas V., Ni J., Moore P. A., Aggarwal B. B. (1998) J. Biol. Chem. 273, 20551–20555 [DOI] [PubMed] [Google Scholar]
- 12. Wada T., Nakashima T., Hiroshi N., Penninger J. M. (2006) Trends Mol. Med. 12, 17–25 [DOI] [PubMed] [Google Scholar]
- 13. Lerner U. H. (2004) Crit. Rev. Oral Biol. Med. 15, 64–81 [DOI] [PubMed] [Google Scholar]
- 14. Wong B. R., Josien R., Lee S. Y., Vologodskaia M., Steinman R. M., Choi Y. (1998) J. Biol. Chem. 273, 28355–28359 [DOI] [PubMed] [Google Scholar]
- 15. Yamashita T., Yao Z., Li F., Zhang Q., Badell I. R., Schwarz E. M., Takeshita S., Wagner E. F., Noda M., Matsuo K., Xing L., Boyce B. F. (2007) J. Biol. Chem. 282, 18245–18253 [DOI] [PubMed] [Google Scholar]
- 16. Ishida N., Hayashi K., Hoshijima M., Ogawa T., Koga S., Miyatake Y., Kumegawa M., Kimura T., Takeya T. (2002) J. Biol. Chem. 277, 41147–41156 [DOI] [PubMed] [Google Scholar]
- 17. Mohamed S. G., Sugiyama E., Shinoda K., Taki H., Hounoki H., Abdel-Aziz H. O., Maruyama M., Kobayashi M., Ogawa H., Miyahara T. (2007) Bone 41, 592–602 [DOI] [PubMed] [Google Scholar]
- 18. Miguel B. S., Ghayor C., Ehrbar M., Jung R. E., Zwahlen R. A., Hortschansky P., Schmoekel H. G., Weber F. E. (2009) Tissue Eng. Part A 15, 2955–2963 [DOI] [PubMed] [Google Scholar]
- 19. Takahashi N., Udagawa N., Kobayashi Y., Suda T. (2007) Methods Mol. Med. 135, 285–301 [DOI] [PubMed] [Google Scholar]
- 20. Ghayor C., Ehrbar M., San Miguel B., Grätz K. W., Weber F. E. (2009) Biochem. Biophys. Res. Commun. 381, 247–252 [DOI] [PubMed] [Google Scholar]
- 21. Wilson S. R., Peters C., Saftig P., Brömme D. (2009) J. Biol. Chem. 284, 2584–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reponen P., Sahlberg C., Munaut C., Thesleff I., Tryggvason K. (1994) Ann. N.Y. Acad. Sci. 732, 472–475 [DOI] [PubMed] [Google Scholar]
- 23. Sundaram K., Nishimura R., Senn J., Youssef R. F., London S. D., Reddy S. V. (2007) Exp. Cell Res. 313, 168–178 [DOI] [PubMed] [Google Scholar]
- 24. Matsumoto M., Sudo T., Saito T., Osada H., Tsujimoto M. (2000) J. Biol. Chem. 275, 31155–31161 [DOI] [PubMed] [Google Scholar]
- 25. Monje P., Hernández-Losa J., Lyons R. J., Castellone M. D., Gutkind J. S. (2005) J. Biol. Chem. 280, 35081–35084 [DOI] [PubMed] [Google Scholar]
- 26. Tsubaki M., Kato C., Isono A., Kaneko J., Isozaki M., Satou T., Itoh T., Kidera Y., Tanimori Y., Yanae M., Nishida S. (2010) J. Cell. Biochem. 111, 1661–1672 [DOI] [PubMed] [Google Scholar]
- 27. Ha H., Lee J. H., Kim H. N., Lee Z. H. (2011) Biochem. Biophys. Res. Commun. 406, 546–551 [DOI] [PubMed] [Google Scholar]
- 28. Body J. J. (2002) Bone 30, 75S–79S [DOI] [PubMed] [Google Scholar]
- 29. Goldhahn J., Little D., Mitchell P., Fazzalari N. L., Reid I. R., Aspenberg P., Marsh D. (2010) Bone 46, 267–271 [DOI] [PubMed] [Google Scholar]
- 30. Howard P. A., Barnes B. J., Vacek J. L., Chen W., Lai S. M. (2010) Am. J. Cardiovasc. Drugs 10, 359–367 [DOI] [PubMed] [Google Scholar]
- 31. Odvina C. V. (2006) J. Investig. Med. 54, 114–122 [DOI] [PubMed] [Google Scholar]
- 32. O'Regan R. M., Gradishar W. J. (2001) Oncology 15, 1177–1185, 1189–1190; discussion 1190–1194 [PubMed] [Google Scholar]
- 33. Matsuo K., Galson D. L., Zhao C., Peng L., Laplace C., Wang K. Z., Bachler M. A., Amano H., Aburatani H., Ishikawa H., Wagner E. F. (2004) J. Biol. Chem. 279, 26475–26480 [DOI] [PubMed] [Google Scholar]
- 34. Wagner E. F., Eferl R. (2005) Immunol. Rev. 208, 126–140 [DOI] [PubMed] [Google Scholar]
- 35. Mulari M. T., Zhao H., Lakkakorpi P. T., Väänänen H. K. (2003) Traffic 4, 113–125 [DOI] [PubMed] [Google Scholar]
- 36. Bjørbaek C., Zhao Y., Moller D. E. (1995) J. Biol. Chem. 270, 18848–18852 [DOI] [PubMed] [Google Scholar]
- 37. Chen R. H., Chung J., Blenis J. (1991) Mol. Cell. Biol. 11, 1861–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frödin M., Gammeltoft S. (1999) Mol. Cell. Endocrinol. 151, 65–77 [DOI] [PubMed] [Google Scholar]
- 39. Ghoda L., Lin X., Greene W. C. (1997) J. Biol. Chem. 272, 21281–21288 [DOI] [PubMed] [Google Scholar]